Abstract
Purpose
Acute myeloid leukemia (AML) is associated with a poor overall prognosis. PIM family genes, including PIM1, PIM2, and PIM3, are proto-oncogenes that are aberrantly overexpressed in different types of human cancers. In this study, we aimed to explore and clarify the function of PIM3 in AML.
Patients and Methods
The expression of the three PIM genes in AML was detected using the Gene Expression Omnibus. The expression of PIM3 and PIM3 in patient samples and AML cell lines was measured using quantitative real-time polymerase chain reaction or Western blot analyses. The cellular behaviors of PIM3-overexpressing AML cell lines were detected using a CCK-8 assay, flow cytometry, Western blotting, immunofluorescence staining, and a cell migration assay. The interactions between PIM3 and phosphorylated CXCR4 (pCXCR4) were explored via immunoprecipitation.
Results
Higher PIM3 expression was detected in primary AML cells than in healthy donor cells. Second, PIM3 overexpression promoted AML cell proliferation and protected against spontaneous apoptosis by phosphorylating BAD (pBAD) at Ser112. Furthermore, PIM3 overexpression might promote the migration of AML cells via CXCR4. PIM3-overexpressing AML cell lines exhibited increased CXCR4 phosphorylation at Ser339, and pCXCR4 interacted with PIM3.
Conclusion
Our findings suggest that PIM3 regulates the proliferation, survival, and chemotaxis of AML cell lines. Moreover, pCXCR4 might mediate the regulation of PIM3-induced chemotaxis. Therefore, the inhibition of PIM3 expression may be a promising therapeutic target in AML.
Keywords: pBAD, pCXCR4, PIM1, PIM2, PIM3
Introduction
Acute myeloid leukemia (AML), a malignancy with a poor overall prognosis, is characterized by the expansion of malignant myeloid precursor cells in the peripheral blood and bone marrow.1 Provirus integrating site moloney murine leukemia virus (PIM) family genes, including PIM1, PIM2, and PIM3, encode serine/threonine kinases that integrate into cell signaling networks and regulate various cell behaviors, such as proliferation, survival, and migration.2–5 Previous research findings suggest that PIM family genes are proto-oncogenes and are expressed aberrantly in different human cancers.
Of the three known PIM genes, Pim1 has been intensively investigated. Overexpression of this gene has been reported in prostate cancer,6 pancreatic cancer,7 squamous cell carcinoma,8 gastric cancer,9 diffuse large B-cell lymphoma,10 acute myeloid leukemia (AML),11 multiple myeloma, and other malignancies. High PIM1 expression has been linked to cancer progression and poor treatment outcomes.12 PIM2 expression is strongly elevated in AML cells and multiple B-cell tumors, including chronic lymphocytic leukemia and nested cell lymphoma.13–15 In multiple myeloma, PIM2 is highly expressed and regulates mTOR-C1 by phosphorylating TSC2 to promote the proliferation of MM cells.16,17 Therefore, PIM kinases, particularly PIM1, are considered promising anti-cancer therapeutic targets.
PIM3 shares a high level of amino acid sequence similarity with PIM1 and PIM2.3,18,19 Forced PIM3 overexpression also promotes cell growth, and aberrant expression of this kinase has been observed in many human cancers. However, PIM3 expression appears to exhibit a different pattern of tissue specificity; for example, PIM1, but not PIM3, is expressed in human colon, pancreas, liver, and small intestine.19,20 This expression pattern suggests that PIM3 has unique functions under physiological conditions. However, the expression and function of PIM3 in leukemia are relatively less understood. Previously, Ishikawa et al observed that PIM3 knockdown suppressed cell growth in T cell leukemia cell lines,21 whereas Zhou et al compared PIM3 expression in AML patients before and after chemotherapy and observed that changes in this parameter during the treatment correlated with the patient’s remission status.22 In this study, we performed both clinical data analyses and cell line studies to further elucidate the function of PIM3 in AML, and observed regulatory effects on proliferation, survival and chemotaxis.
Patients and Methods
Patient Samples
Bone marrow samples were collected from 40 patients with AML who were hospitalized at West China Hospital, Sichuan University, China, as well as from 26 healthy volunteers. All the patients with AML had bone marrow blast frequencies >50%. Mononuclear cells were isolated from the samples by Ficoll density gradient centrifugation. These specimens were collected in the early time. Informed consent was obtained and the study protocol was approved by the Ethical Committee of West China Hospital of Sichuan University and conformed to the Declaration of Helsinki.
Cell Lines
The K562, U937, and THP-1 human leukemia cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were maintained in RPMI-1640 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37ºC and 5% CO2.
Cell Treatments
The pCDNA4 plasmid backbone ligated with PIM3 cDNA was kindly provided by Kanazawa University, Japan. PIM3 cDNA, including the open reading frame, was subcloned into the pIRES2-EGFP vector. The resulting construct was transfected into K562 cells to induce PIM3 overexpression (Pim3-OE) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. K562 cells were also transfected with a vector overexpressing GFP only as a control (CTR-OE). The cells were incubated with normal growth medium for another 24 hours prior to Western blotting, immunoprecipitation, immunofluorescence staining, cell proliferation, and apoptosis assays. The lentiviral vector LV-Pim3 (engineered to overexpress PIM3) and a negative control vector (LV-NC) were purchased from Western (Chongqing, China). U937 cells were infected with LV-Pim3 or LV-NC using polybrene (Sigma, St. Louis, MO, USA) to establish U937-CTR and U937-Pim3 cell lines, as previously described.23
Real-Time Quantitative PCR
Total RNA was isolated from the cells using TRIzol agent (Life Technologies, Carlsbad, CA, USA) According to the manufacturer’s protocol. Next, cDNA was synthesized using the PrimerScript RT Kit (Takara, Shiga, Japan). For each real-time quantitative PCR (RT-PCR), 50 ng cDNA were mixed with SYBR Green PCR MasterMix (Takara). The reactions were run in a CFX96 real-time PCR thermocycler (Bio-Rad, US). The following oligonucleotides were used as RT-PCR primers:
Pim3 forward, 5´-GCACACACAATGCAAGTCCT-3´
Pim3 reverse, 5´-AGAGGCAGACTGCTCAGAGG-3´
GAPDH forward, 5´-ACCACAGTCCATGCCATCAC-3´
GAPDH reverse, 5´-TCCACCACCCTGTTGCTGTA-3´
Western Blotting and Immunoprecipitation
The cells were harvested, washed with ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (Beyotime, Shanghai, China). Target proteins were analyzed by Western blotting as previously described.10 Antibodies specific for phosphorylated BAD (pBAD)(Ser112) and total BAD were purchased from Cell Signaling, Inc. (Danvers, MA, USA). Antibodies specific for pCXCR4 (Ser339) and CXCR4 were purchased from Abcam, Inc. (Cambridge, UK). A Pim3-specific antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA).
For immunoprecipitation, the cells were lysed as described above.20 The lysates were pre-cleaned by incubation with protein A/G agarose beads for 3 hours at 4ºC and then incubated with a pCXCR4 (Ser339) antibody overnight at 4ºC. The protein A/G beads were added to the mixture, which was incubated at 4ºC for 3 hours. The beads were washed 5 times with lysis buffer, resuspended in 2×loading buffer, and boiled for 10 min before Western blotting analyses.
Immunofluorescence Staining and Confocal Microscopy
The cells were adhered to a glass slide, fixed with 4% formaldehyde for 5 min on ice, and washed 3 times with ice-cold PBS. Next, the cells were permeabilized with 0.1% Triton X-100 for 5 min and blocked with 5% bovine serum albumin in PBS at room temperature. After a washing step, the cells were incubated with anti-pBAD (Ser112), and anti-PIM3 antibodies overnight at 4ºC. After washing, the cells were incubated with fluorescently labeled secondary antibodies (Invitrogen). The cell nuclei were stained with DAPI. Fluorescence staining was visualized using a confocal microscope (Carl Zeiss, Oberkochen, Germany).
Flow Cytometry Analysis
Flow cytometry was used to examine the expression of cellular proteins, as previously described.24 Antibody specific for pCXCR4 (Ser339) was purchased from Abcam, Inc. (Cambridge, UK). The labeled cells were analyzed using a Navios flow cytometer (Beckman Coulter, Brea, CA, USA).
Cell Proliferation and Apoptosis Assays
The cells were seeded in 96-well plates at a density of 4000 cells in 100 µL medium/well, and were cultured for 24 hours, 48 hours, 72 hours, 96 hours and 120 hours. To examine cell proliferation, 10 µL of CCK-8 solution were added to each well, and incubated for 2 hours. The absorbance in each well at 450 nm was examined using a microplate reader. To evaluate cell apoptosis, the cells were harvested and stained using an Annexin-V Alexa Fluor 647/PI kit according to the manufacturer’s instructions (4A Biotech, Beijing, China). The cells were analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA).
Cell Migration Assay
To examine cell migration, 2 × 104 cells were loaded into the upper chamber of a two-chamber Transwell with a pore size of 8.0 µm (Costar, Corning, NY, USA) and allowed to migrate toward a SDF1a (10 nM) gradient for 4 hours as previously described.25 The cells that migrated to the lower layer were then subjected to a CCK-8 viability analysis.
Bioinformatics and Statistics
The AML GEP dataset GSE13159 was downloaded from the NCBI Gene Expression Omnibus database.26 Clinical information about the patients in the dataset was downloaded from Oncomine (www.oncomine.org). All expression values were reported as means ± standard errors of the means of multiple experiments or as means ± standard deviations from representative experiments. Student’s t test was used to compare two experimental groups, and a p value of <0.05 was considered statistically significant.
Results
PIM3 Expression in Adult AML
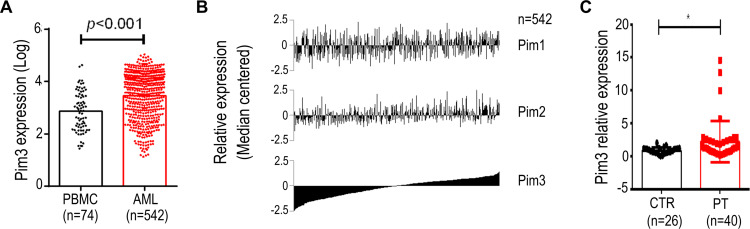
We initially performed a microarray-based analysis using the GSE13159 dataset to examine the expression of PIM3 in adult AML cells. Notably, stronger PIM3 expression was observed in these cells than in peripheral blood mononuclear cells (PBMCs) collected from healthy donors (Figure 1A, p < 0.001). Next, we examined the expression of PIM1 and PIM2 using the same dataset and sorted the results by PIM3 expression. As shown in Figure 1B, we observed no correlation between the expression of PIM3 and that of PIM1 or PIM2, suggesting that the gene expression each PIM family member might be regulated differently. Finally, we validated our microarray-based analysis findings using AML patient samples from West China Hospital. As shown in Figure 1C, stronger PIM3 expression was observed in patient samples than in bone marrow mononuclear cells from healthy volunteers (The fold-changes: 2.227 ± 0.4998 versus 0.8667 ± 0.09480; p < 0.05). Overall, our results demonstrate increased PIM3 expression in AML.
Figure 1.
PIM3 expression in adult acute myeloid leukemia (AML). Microarray-based analysis of PIM family gene expression in AML patient samples. (A) PIM3 expression in peripheral blood mononuclear cells from healthy donors versus AML cells. (B) Expression of PIM1, PIM2, and PIM3 in AML patient samples. (C) Expression of PIM3 mRNA in AML cells from patients (PT) and normal bone marrow mononuclear cells (CTR) from healthy donors via qRT-PCR analysis. *p < 0.05.
Forced PIM3 Overexpression in AML Cell Lines
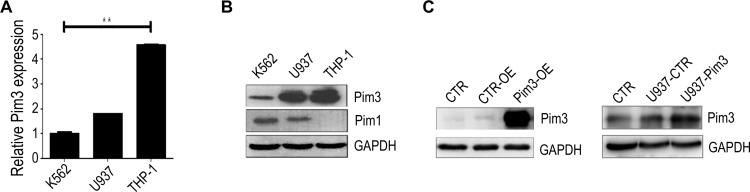
Next, we examined PIM3 expression in human leukemia cell lines to investigate the potential role of this protein in AML. Quantitative real-time PCR (qRT-PCR) and Western blotting analyses revealed that the K562, U937 and THP-1 leukemia lines all expressed PIM3, although K562 cells expressed a relatively lower level (Figure 2A and B). In addition, the patterns of PIM1 and PIM3 expression differed among the tested cell lines (Figure 2B). Next, we established transient control-overexpressing (CTR-OE, overexpressing GFP only) and PIM3-overexpressing (Pim3-OE) K562 cells (Figure 2C, left panel) and compared these to normal K562 cells (blank CTR). The construction and transfection of vectors are described in the Material and Methods section. Meanwhile, U937 cells were infected with lentiviral to generate U937-Pim3 cells with consistent overexpressing PIM3 (Figure 2C, right panel).
Figure 2.
PIM3 expression in human acute myeloid leukemia (AML) cell lines. (A) Expression of PIM3 mRNA in K562, U937, and THP-1 cells analyzed by qRT-PCR. (B) Western blotting of PIM3 and PIM1 expression in different human AML cell lines. (C) Left panel: Western blotting of PIM3 in control (CTR-OE) versus PIM3-overexpressing (Pim3-OE) K562 AML cells. Blank CTR: normal K562 cells. Right panel: Western blotting of PIM3 in control (U937-CTR) versus PIM3-overexpressing (U937-Pim3) U937 AML cells. Blank CTR: normal U937 cells. **p < 0.01.
PIM3 Overexpression Promotes AML Cell Proliferation
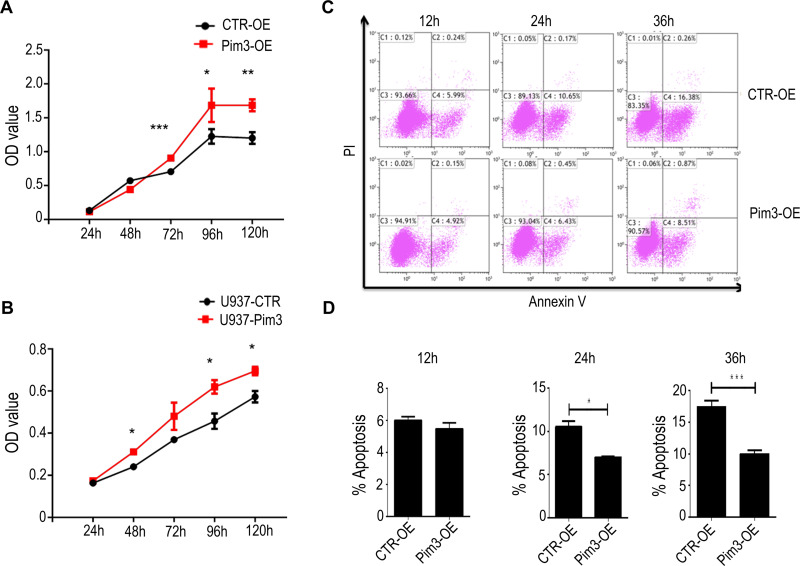
Next, we examined cell proliferation in PIM3-overexpressing cells using the CCK8 method. Notably, PIM3 overexpression promoted the proliferation of K562 cells in a time-dependent manner, as demonstrated by a higher proliferation rate relative to the control group after 72 hours of culture (Figure 3A). Similar results were observed in U937 cell line (Figure 3B). Next, we examined the effect of PIM3 on the spontaneous apoptosis of K562 cells. As shown in Figure 3C, leukemia cells exhibited increased apoptosis during culture. However, Pim3-OE cells exhibited a significant lower rate of cell apoptosis than CTR-OE cells, as presented quantitatively in Figure 3D (p < 0.05).
Figure 3.
PIM3 overexpression promotes acute myeloid leukemia (AML) cell proliferation and survival. (A) Growth of control (CTR-OE) and PIM3-overexpressing (Pim3-OE) K562 AML cells. (B) Growth of control (U937-CTR) and PIM3-overexpressing (U937-Pim3) U937 AML cells. (C) The apoptotic cells were detected by staining with annexin V and PI and flow cytometry. The results are quantified in (D). *p < 0.05, **p < 0.01, ***p < 0.001.
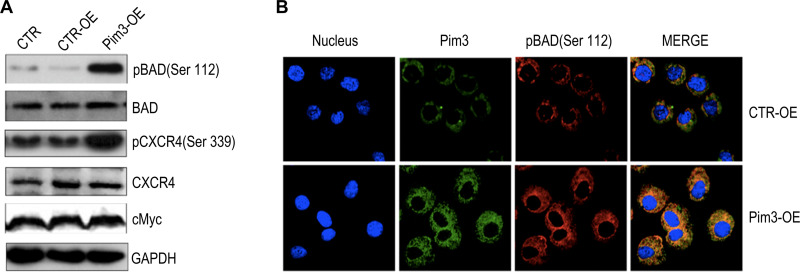
Data from previous studies of other cells suggest that PIM kinases promote cell survival by regulating the activity of BAD,27–29 a known pro-apoptotic factor that interacts with and attenuates the anti-apoptotic activities of BCL-2 family members. BAD can be phosphorylated at different residues, which blocks BCL-2 binding activity and thus promotes cell survival.4,29 However, it remained unclear whether PIM3 promoted BAD phosphorylation in AML. We observed that in AML cells, PIM3-overexpression was associated with an increased level of pBAD (Ser112) (Figure 4A), but did not affect the levels of total Bad and cMyc. Interestingly, PIM3 overexpression also increased the level of pCXCR4 (Ser339) but did not affect the level of total CXCR4. We further examined the level of pBAD using immunofluorescent staining. As shown in Figure 4B, PIM3-overexpressing cells produced stronger PIM3 and pBAD (Ser112) signals, and these proteins were found to colocalize. In summary, we observed that the overexpression of PIM3 promoted the proliferation and survival of AML cells, possibly by inducing the phosphorylation of BAD.
Figure 4.
PIM3 promotes BAD phosphorylation in acute myeloid leukemia (AML) cells. (A) Phosphorylated BAD (pBAD) (Ser112), BAD, pCXCR4 (Ser339), CXCR4 and cMyc were detected in control (CTR-OE) versus PIM3-overexpressing (Pim3-OE) K562 AML cells by Western blotting. Blank CTR: normal K562 cells. (B) Confocal fluorescence microscopy analysis of immunofluorescently stained cells. Red, green, and blue correspond to pBAD (Ser112), PIM3 and DAPI (nuclear) staining.
PIM3 Overexpression Promotes AML Cell Migration
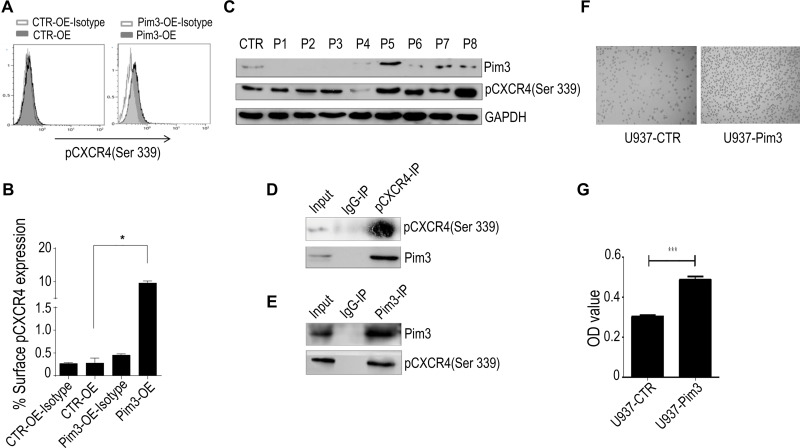
As shown in Figure 4A, PIM3-overexpressing K562 cells contained an increased level of pCXCR4 (Ser339). Similar results were observed in U937 cells (Supplementary Figure 1A). We additionally performed flow cytometry to detect the expression of pCXCR4 (Ser339) on the cell surface. Our results demonstrated a significantly higher level of pCXCR4 (Ser339) in Pim3-OE cells (Figure 5A; quantitative results in Figure 5B). Next, we used Western blotting to detect the expression of PIM3 and pCXCR4 (Ser339) and further verify the relationship between these molecules in AML patients. We discovered significantly higher levels of pCXCR4 (Ser339) in Patient 5 and Patient 8 (PIM3-overexpressing) than in healthy volunteers (CTR) and other AML patients (Figure 5C), consistent with the results obtained in vitro. Furthermore, the results of an immunoprecipitation experiment suggested that Pim3 and pCXCR4 (Ser339) formed a complex in K562 cells and U937 cells (Figure 5D, E and Supplementary Figure 1B, 1C).
Figure 5.
PIM3 promotes the chemotaxis of acute myeloid leukemia (AML) cells via CXCR4. (A) Flow cytometry analysis of surface pCXCR4 (Ser339) expression on CTR-OE and Pim3-OE K562 AML cells. (B) Quantitative analysis of results from (A and C) Western blotting of PIM3 and pCXCR4 (Ser339) in samples from AML patients and healthy volunteers. (D) Immunoprecipitation of PIM3 and pCXCR4 (Ser339) and Western blotting of pCXCR4 (Ser339) and PIM3 (upper and lower panels, respectively). (E) Immunoprecipitation of PIM3 and pCXCR4 (Ser339) and Western blotting of PIM3 and pCXCR4 (Ser339) (upper and lower panels, respectively). (F) Imaging of control (U937-CTR, left panel) and PIM3-overexpressing (U937-Pim3, right panel) U937 cells subjected to a migration assay. (G) The cells in the lower migration chambers were collected and subjected to a quantitative CCK8 assay. *p< 0.05, ***p < 0.001.
CXCR4 is the receptor that binds SDF-1a. Based on the above results, we hypothesized that the regulatory effect of PIM3 on AML cell migration might be mediated by CXCR4. To investigate the exact biological role of PIM3 in AML, we also examined the migration of PIM3-overexpressing AML cells in response to a gradient of the chemotactic factor SDF-1a, as described in the Material and Methods section. However, U937 cells were used because K562 exhibited a poor chemotactic response. Representative photographs of the migrated cells are shown in Figure 5F. A CCK8 assay of cells in the lower chamber revealed that PIM3-overexpressing U937 cells exhibited increased migration (Figure 5G).
Discussion
Very few previous studies have evaluated the expression and function of PIM3 in AML.21,22 Here, we revealed a high level of PIM3 expression in AML cells relative to healthy donor PBMCs. Moreover, we observed distinctive expression patterns of PIM1, PIM2, and PIM3 in AML,10,11,13,22 and our results suggest that PIM3 expression is subject to regulation via a unique upstream process. A previous study of the PIM3 promoter revealed a canonical TATA box and binding sites for the transcription factors Stat3, Sp1, Ets1 and NF-&B.30 In contrast, PIM1 promoter activity is regulated by NF1,31 HOXA9,32 ERG,33 and some other downstream factors in the Akt pathway. These differences in promotor regulatory elements might explain the differential regulation of these 2 PIM kinases in AML cells. Furthermore, the differential expression of PIM3 among different patients with AML indicated a transcription-level regulation of this gene. In summary, multiple transcription factors appear to determine the expression of PIM3. In this study, we focused mainly on the function of PIM3 in the regulation of AML cell proliferation and survival. Notably, forced overexpression of PIM3 promoted the proliferation and survival of PIM3L°w K562 AML cells. BAD is a member of the BCL-2 family of proteins, which play crucial roles in apoptosis regulation. Some proteins within this family have proapoptotic effects (eg, BIM, BAD, BAX, BAK), whereas others play antiapoptotic roles (eg, BCL-2, BCL-xL, MCL-1, A1, BAG-1).28 Previous studies revealed that PIM kinases can inactive BAD via phosphorylation, retain the expression of BCL-xL and ultimately inhibit the apoptosis of cancer cells. Furthermore, PIM kinases phosphorylate BAD at multiple sites, including the Ser112 gatekeeper site, Ser136, and Ser155.27 Like other PIM kinases, PIM3high promoted cell survival in a process mediated by BAD phosphorylation. In this study, we demonstrated for the first time that PIM3 overexpression stimulates BAD phosphorylation in AML, consistent with similar findings in many solid tumors, such as hepatocellular carcinoma,34 pancreatic cancer,35 colon cancer and gastric adenoma.20 We further observed that PIM3 overexpression promoted AML cell migration. Previous studies demonstrated that PIM1 stimulated the surface expression of CXCR4 and promoted the migration and homing of AML cells to the bone marrow.36,37 The SDF1a-CXCR4 axis plays a critical role in AML cell migration,38 and our findings indicate that PIM3 interacts with pCXCR4 in AML cells. Notably, the overexpression of PIM3 upregulated the level of pCXCR4 (Ser339) without affecting the level of total CXCR4. CXCR4 contains many potential phosphorylation sites, each of which is responsible for different molecular functions. A previous study suggested that the phosphorylation of CXCR4 at Ser339 was catalyzed primarily by the kinase GRK6.5 Our observations that PIM3 overexpression increased pCXCR4 (Ser339) in AML cells and that these two proteins were co-immunoprecipitated strongly suggest a functional crosstalk between PIM3 and pCXCR4 (Ser339). We are conducting further investigations to demonstrate the mechanism by which PIM3 functionally interacts with and regulates CXCR4.
PIM family kinases are considered therapeutic targets in many human cancers.3,5,12,19 Currently, several PIM kinase inhibitors are under investigation in clinical or pre-clinical trials. In particular, increasing evidence suggests that PIM1 targeting may enhance the treatment of FLT3-ITD-positive AML.36,39 Other studies demonstrated that PIM1 plays a critical regulatory role in the homing and migration of hematopoietic stem cells via CXCR4,36 which itself has been identified as a pivotal factor in the homing and maintenance of cancer stem cells. Therefore, PIM inhibitors may exert dual antitumor effects by targeting tumor cells and interfering with the microenvironment. Our findings suggest that PIM3 and other PIM kinases have partially overlapping and partially distinct patterns of expression and mechanisms of regulation and downstream function. Mikkers et al demonstrated the selective activation of PIM3 in Myc transgenic mice that lacked PIM1 and PIM2, and suggested that PIM3 may replace or compensate for the functions of the missing Pim kinases.40 Other researchers believe that other PIM kinases or complementary mechanisms may remain to be discovered.2 Collectively, these findings suggest that it may be more beneficial to target all the PIM genes and proteins, rather than selectively suppressing one or two family members. The development of an efficient PIM targeting strategy may rely on a better understanding of the function of each PIM kinase in AML.
Conclusion
In conclusion, our results demonstrate that PIM3 overexpression promoted AML cell proliferation by phosphorylating BAD. Moreover, we observed that the PIM3-induced regulation of chemotaxis might be mediated by the phosphorylation of CXCR4. Our findings suggest that PIM3 might represent a potential therapeutic target in AML. Our future work aims to address the exact mechanism involved in this process.
Acknowledgment
We thank the tissue bank of the Department of Hematology, West China Hospital, Sichuan University, for providing samples.
Funding Statement
This study was supported by grants to Y.W. from the National Natural Science Foundation of China (No. 81470327) and the Innovation Supporting Program of Sichuan Province, China (No. 2018RZ0137).
Abbreviations
AML, acute myeloid leukemia; PBS, phosphate-buffered saline.
Ethics Approval and Informed Consent
The patient and healthy volunteer consents were written informed consent and the study protocol was approved by the Ethical Committee of West China Hospital of Sichuan University and conformed to the Declaration of Helsinki.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23 [DOI] [PubMed] [Google Scholar]
- 2.Alvarado Y, Giles FJ, Swords RT. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5(1):81–96. doi: 10.1586/ehm.11.69 [DOI] [PubMed] [Google Scholar]
- 3.Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–1015. doi: 10.3324/haematol.2009.017079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7(1):1. doi: 10.1186/1471-2121-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondello P, Cuzzocrea S, Mian M. Pim kinases in hematological malignancies: where are we now and where are we going? J Hematol Oncol. 2014;7(1):95. doi: 10.1186/s13045-014-0095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holder SL, Abdulkadir SA. PIM1 kinase as a target in prostate cancer: roles in tumorigenesis, castration resistance, and docetaxel resistance. Curr Cancer Drug Targets. 2014;14(2):105–114. doi: 10.2174/1568009613666131126113854 [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Zhang T, Wang T, You L, Zhao Y. PIM kinases: an overview in tumors and recent advances in pancreatic cancer. Future Oncol. 2014;10(5):865–876. doi: 10.2217/fon.13.229 [DOI] [PubMed] [Google Scholar]
- 8.Chiang WF, Yen CY, Lin CN, et al. Up-regulation of a serine-threonine kinase proto-oncogene Pim-1 in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2006;35(8):740–745. doi: 10.1016/j.ijom.2006.01.027 [DOI] [PubMed] [Google Scholar]
- 9.Yan B, Yau EX, Samanta S, et al. Clinical and therapeutic relevance of PIM1 kinase in gastric cancer. Gastric Cancer. 2012;15(2):188–197. doi: 10.1007/s10120-011-0097-2 [DOI] [PubMed] [Google Scholar]
- 10.Kuo HP, Ezell SA, Hsieh S, et al. The role of PIM1 in the ibrutinib-resistant ABC subtype of diffuse large B-cell lymphoma. Am J Cancer Res. 2016;6(11):2489–2501. [PMC free article] [PubMed] [Google Scholar]
- 11.Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci U S A. 1989;86(22):8857–8861. doi: 10.1073/pnas.86.22.8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braso-Maristany F, Filosto S, Catchpole S, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22(11):1303–1313. doi: 10.1038/nm.4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173. doi: 10.1182/blood-2002-06-1677 [DOI] [PubMed] [Google Scholar]
- 14.Cohen AM, Grinblat B, Bessler H, et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45(5):951–955. doi: 10.1080/10428190310001641251 [DOI] [PubMed] [Google Scholar]
- 15.Johrer K, Obkircher M, Neureiter D, et al. Antimyeloma activity of the sesquiterpene lactone cnicin: impact on Pim-2 kinase as a novel therapeutic target. J Mol Med (Berl). 2012;90(6):681–693. doi: 10.1007/s00109-011-0848-x [DOI] [PubMed] [Google Scholar]
- 16.Hiasa M, Teramachi J, Oda A, et al. Pim-2 kinase is an important target of treatment for tumor progression and bone loss in myeloma. Leukemia. 2015;29(1):207–217. doi: 10.1038/leu.2014.147 [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Zavorotinskaya T, Dai Y, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood. 2013;122(9):1610–1620. doi: 10.1182/blood-2013-01-481457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keane NA, Reidy M, Natoni A, Raab MS, O’Dwyer M. Targeting the Pim kinases in multiple myeloma. Blood Cancer J. 2015;5(7):e325. doi: 10.1038/bcj.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013;85(5):629–643. doi: 10.1016/j.bcp.2012.09.018 [DOI] [PubMed] [Google Scholar]
- 20.Popivanova BK, Li YY, Zheng H, et al. Proto-oncogene, Pim-3 with serine/threonine kinase activity, is aberrantly expressed in human colon cancer cells and can prevent Bad-mediated apoptosis. Cancer Sci. 2007;98(3):321–328. doi: 10.1111/j.1349-7006.2007.00390.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa C, Senba M, Hashimoto T, Imaizumi A, Mori N. Expression and significance of Pim-3 kinase in adult T-cell leukemia. Eur J Haematol. 2017;99(6):495–504. doi: 10.1111/ejh.12940 [DOI] [PubMed] [Google Scholar]
- 22.Zhou ZC, Tian ZG, Yuan Z. [Expression and significance of Pim-3 gene in acute myeloid leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016;24(2):316–320. doi: 10.7534/j.issn.1009-2137.2016.02.002. Chinese. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Hou A, Gao X, et al. Lentivirus-mediated miR-23a overexpression induces trophoblast cell apoptosis through inhibiting X-linked inhibitor of apoptosis. Biomed Pharmacother. 2017;94:412–417. doi: 10.1016/j.biopha.2017.07.082 [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Yang J, Qian J, et al. PSGL-1/selectin and ICAM-1/CD18 interactions are involved in macrophage-induced drug resistance in myeloma. Leukemia. 2013;27(3):702–710. doi: 10.1038/leu.2012.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer Ther. 2019;18(1):179. doi: 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlmann A, Kipps TJ, Rassenti LZ, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br J Haematol. 2008;142(5):802–807. doi: 10.1111/j.1365-2141.2008.07261.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu XF, Li J, Vandervalk S, Wang Z, Magnuson NS, Xing PX. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. J Clin Invest. 2009;119(2):362–375. doi: 10.1172/JCI33216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boac BM, Abbasi F, Ismail-Khan R, et al. Expression of the BAD pathway is a marker of triple-negative status and poor outcome. Sci Rep. 2019;9(1):17496. doi: 10.1038/s41598-019-53695-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571(1–3):43–49. doi: 10.1016/j.febslet.2004.06.050 [DOI] [PubMed] [Google Scholar]
- 30.Li YY, Wu Y, Tsuneyama K, Baba T, Mukaida N. Essential contribution of Ets-1 to constitutive Pim-3 expression in human pancreatic cancer cells. Cancer Sci. 2009;100(3):396–404. doi: 10.1111/j.1349-7006.2008.01059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunen D, de Vries RC, Lieftink C, Beijersbergen RL, Bernards R. PIM kinases are a potential prognostic biomarker and therapeutic target in neuroblastoma. Mol Cancer Ther. 2018;17(4):849–857. doi: 10.1158/1535-7163.MCT-17-0868 [DOI] [PubMed] [Google Scholar]
- 32.de Bock CE, Demeyer S, Degryse S, et al. HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov. 2018;8(5):616–631. doi: 10.1158/2159-8290.CD-17-0583 [DOI] [PubMed] [Google Scholar]
- 33.Goldberg L, Tijssen MR, Birger Y, et al. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood. 2013;122(15):2694–2703. doi: 10.1182/blood-2013-01-477133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukaida N, Wang YY, Li YY. Roles of Pim-3, a novel survival kinase, in tumorigenesis. Cancer Sci. 2011;102(8):1437–1442. doi: 10.1111/j.1349-7006.2011.01966.x [DOI] [PubMed] [Google Scholar]
- 35.Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66(13):6741–6747. doi: 10.1158/0008-5472.CAN-05-4272 [DOI] [PubMed] [Google Scholar]
- 36.Grundler R, Brault L, Gasser C, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med. 2009;206(9):1957–1970. doi: 10.1084/jem.20082074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker S, Finter J, Forde AJ, et al. PIM kinases are essential for chronic lymphocytic leukemia cell survival (PIM2/3) and CXCR4-mediated microenvironmental interactions (PIM1). Mol Cancer Ther. 2014;13(5):1231–1245. doi: 10.1158/1535-7163.MCT-13-0575-T [DOI] [PubMed] [Google Scholar]
- 38.Asri A, Sabour J, Atashi A, Soleimani M. Homing in hematopoietic stem cells: focus on regulatory role of CXCR7 on SDF1a/CXCR4 axis. EXCLI J. 2016;15:134–143. doi: 10.17179/excli2014-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao T, Jiang N, Liao H, Shuai X, Su J, Zheng Q. The FLT3-ITD mutation and the expression of its downstream signaling intermediates STAT5 and Pim-1 are positively correlated with CXCR4 expression in patients with acute myeloid leukemia. Sci Rep. 2019;9(1):12209. doi: 10.1038/s41598-019-48687-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkers H, Allen J, Knipscheer P, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32(1):153–159. doi: 10.1038/ng950 [DOI] [PubMed] [Google Scholar]