Abstract
Objectives
Diabetes is associated with adverse outcomes, including death, after coronavirus disease 19 (COVID-19) infection. Beyond the lungs, Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), the etiologic agent of the COVID-19 pandemic, can infect a range of other tissues, including the kidney, potentially contributing to acute kidney injury in those with severe disease. We hypothesized that the renal abundance of angiotensin-converting enzyme (ACE) 2, the cell surface receptor for SARS-CoV-2, may be modulated by diabetes and agents that block the renin-angiotensin-aldosterone system (RAAS).
Methods
The expression of ACE 2 was examined in 49 archival kidney biopsies from patients with diabetic kidney disease and from 12 healthy, potential living allograft donors using next-generation sequencing technology (RNA Seq).
Results
Mean ACE 2 messenger RNA was increased approximately 2-fold in diabetes when compared with healthy control subjects (mean ± SD, 13.2±7.9 vs 7.7±3.6 reads per million reads, respectively; p=0.001). No difference in transcript abundance was noted between recipients and nonrecipients of agents that block the RAAS (12.2±6.7 vs 16.2±10.7 reads per million reads, respectively; p=0.25).
Conclusions
Increased ACE 2 messenger RNA in the diabetic kidney may increase the risk and/or severity of kidney infection with SARS-CoV-2 in the setting of COVID-19 disease. Further studies are needed to ascertain whether this diabetes-related overexpression is generalizable to other tissues, most notably the lungs.
Keywords: ACE 2, coronavirus, COVID-19, diabetes, kidney, renin-angiotensin-aldosterone system, RNA Seq, SARS-CoV-2
Résumé
Objectifs
Le diabète est associé à des issues défavorables, dont le décès après l’infection par la maladie à coronavirus 2019 (COVID-19, de l’anglais COronaVIrus Disease 2019). Outre les poumons, le SRAS-CoV-2, l’agent étiologique de la pandémie de la COVID-19, peut infecter de nombreux autres tissus, notamment les reins, d’où la possibilité de contribuer à l’insuffisance rénale aiguë chez les patients qui en sont atteints gravement. Nous avons posé l’hypothèse que l’excès de l’enzyme de conversion de l’angiotensine (ECA) 2 dans les reins, le récepteur à la surface des cellules sur lequel se fixe le SRAS-CoV-2, peut être modulé par le diabète et les agents qui bloquent le système rénine-angiotensine-aldostérone (SRAA).
Méthodes
Nous avons examiné l’expression de l’ECA 2 dans 49 biopsies rénales d’archives provenant de patients atteints d’une néphropathie diabétique et de 12 biopsies de tissus prélevés de donneurs vivants potentiels en bonne santé à des fins d’allogreffes à l’aide de la technologie de séquençage de nouvelle génération (RNA-Seq, de l’anglais RNA sequencing).
Résultats
L’expression moyenne de l’ARN messager de l’ECA 2 s’est révélée environ 2 fois plus élevée chez les sujets diabétiques que chez les sujets témoins en bonne santé (moyenne ± ÉT, 13,2 ± 7,9 vs 7,7 ± 3,6 lectures par million de lectures, respectivement; p = 0,001). Nous n’avons noté aucune différence dans l’excès de transcription entre les receveurs et les non-receveurs d’agents qui bloquent le SRAA (12,2 ± 6,7 vs 16,2 ± 10,7 lectures par million de lectures, respectivement; p = 0,25).
Conclusions
L’augmentation de l’ARN messager de l’ECA 2 dans les reins diabétiques peut accroître le risque ou la gravité de l’infection rénale par SRAS-CoV-2, ou les deux, dans le contexte de la COVID-19. D’autres études sont nécessaires pour vérifier si cette surexpression liée au diabète est généralisable à d’autres tissus, plus particulièrement les poumons.
Mots clés: ECA 2, coronavirus, COVID-19, diabète, reins, système rénine-angiotensine-aldostérone, RNA-Seq, SRAS-CoV-2
Key Messages.
-
•
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the etiologic agent of the coronavirus disease 19 pandemic, enters cells by binding to angiotensin-converting enzyme 2 on cell surfaces.
-
•
Beyond the lungs, the virus can infect the kidney, causing acute injury.
-
•
Angiotensin-converting enzyme 2 expression is increased approximately 2-fold in diabetic kidney disease biopsies.
Introduction
Diabetes is associated with an adverse outcome, including death, after coronavirus disease 19 (COVID-19) infection (1); however, whether it also increases susceptibility to infection is unknown. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the etiologic agent of the pandemic, binds to the cell surface ectoenzyme, angiotensin-converting enzyme (ACE) 2, the abundance of which may be a determinant of virus entry and vulnerability to infection (2). Recent studies have identified the kidney as a site of infection by SARS-CoV-2 (3), and have highlighted acute kidney injury as a common adverse outcome in patients with severe COVID-19 disease (4). Although hemodynamic imbalance and cytokine release are likely contributors to the acute loss of kidney function, the ability of SARS-CoV-2 to infect kidney cells and to induce immunologic injury and microthrombus formation (5) suggests that the virus itself may be directly involved (6).
Substantial advances have been made in the assessment of gene expression in kidney biopsies, focusing in particular on site-specific changes in glomeruli and to a lesser extent tubules (7). ACE 2, however, is highly expressed in macrophages (8) and in the microvasculature (9) that resides in the interstitial space and traditionally not subjected to selective laser capture microscopic microdissection of the kidney (7). Accordingly, we sought to examine the abundance of ACE 2 in whole rather than microdissected biopsy tissue from individuals with diabetic kidney disease.
Methods
After approval from the St. Michael’s Hospital research ethics board, our centre compiled a biobank of archived kidney biopsies from which to conduct molecular analyses. Seventy-three patients with a clinical diagnosis of diabetes and a pathologic diagnosis of diabetic nephropathy on biopsy from January 2007 to September 2016 at St. Michael’s Hospital were reviewed. All biopsies sampled the renal cortex. Of these, 49 showed only diabetic kidney disease and had adequate documentation of their clinical status and medications at the time of the biopsy with emphasis on their use of agents that block the renin-angiotensin-aldosterone system (RAAS), including ACE inhibitors, angiotensin receptor blockers (ARBs), direct renin inhibitors (DRIs) or mineralocorticoid receptor antagonists (MRAs). Glycated hemoglobin measurements within 3 months of the biopsy were available in 32 participants. Kidney biopsies from healthy control subjects were obtained from 12 potential living allograft donors.
To assess the transcriptome in these biopsies, ten 10-μm-thick sections were cut from formalin fixed paraffin-embedded kidney biopsy blocks or from fresh frozen tissue embedded in Cryomatrix (BD Biosciences, Canada). Total RNA was extracted, ribosomal RNA was removed, and complementary DNA libraries were then prepared and quantitated prior to RNA sequencing (Illumina HiSeq 3000), as previously reported (10). Abundance of ACE 2 messenger RNA was then determined and analyzed according to whether patients were receiving an agent that blocked the RAAS: ACE inhibitors, ARBs, DRIs or MRAs.
Statistical analysis
Data are expressed as mean ± SD, unless otherwise stated. The magnitude of gene expression in control and diabetic kidney disease biopsies was compared using an unpaired Student t test. Within the diabetic kidney disease group, expression levels between those receiving and not receiving treatment with an agent that blocks the RAAS were similarly compared using an unpaired Student t test. p<0.05 was considered significant.
Results
At the time of biopsy, approximately three-quarters (38 of 49) of those with diabetic kidney disease were receiving therapy with at least 1 agent that blocks the RAAS: 25 as single agent ACE inhibitor, ARB or MRA treatment with 11 prescribed dual therapy and a further 2 receiving triple ACE, ARB and MRA combination treatment (Table 1 ). As in most other sites, there were more men than women in the diabetic kidney disease group with the reverse pattern seen in the control, living allograft donors.
Table 1.
Clinical data on patients with diabetic kidney disease and healthy control subjects (potential allograft donors)
| Clinical parameters | Patients with diabetic kidney disease (n=49) | Healthy control subjects (n=12) |
|---|---|---|
| Age, years | 56±10 | 47±9 |
| % female | 33 | 92 |
| Duration of diabetes, years | 14±9 | N/A |
| HbA1c, mmol/mol, % | 66.1; 8.2%±2.3% | |
| Baseline estimated glomerular filtration rate, mL/min/1.73 m2 | 40±22 | 89±13 |
| Baseline urine albumin to creatinine ratio, mg/mmol | 337±322 | 0.8±0.9 |
| Medication usage | ||
| RAAS nonusers | 11 | N/A |
| RAAS users | 38 | |
| DRI only | 0 | |
| ACEi only | 12 | |
| ARB only | 12 | |
| MRA only | 1 | |
| DRI + ACEi | 3 | |
| DRI + ARB | 1 | |
| DRI + MRA | 0 | |
| ACEi + ARB | 5 | |
| ACEi + MRA | 1 | |
| ARB + MRA | 1 | |
| ACEi + ARB + MRA | 2 |
ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; DRI, direct renin inhibitor; HbA1C, glycated hemoglobin; MRA, mineralocorticoid receptor antagonist; N/A, not applicable; RAAS, renin-angiotensin-aldosterone system.
Note: All values are presented as mean ± SD, number of patients or as otherwise indicated.
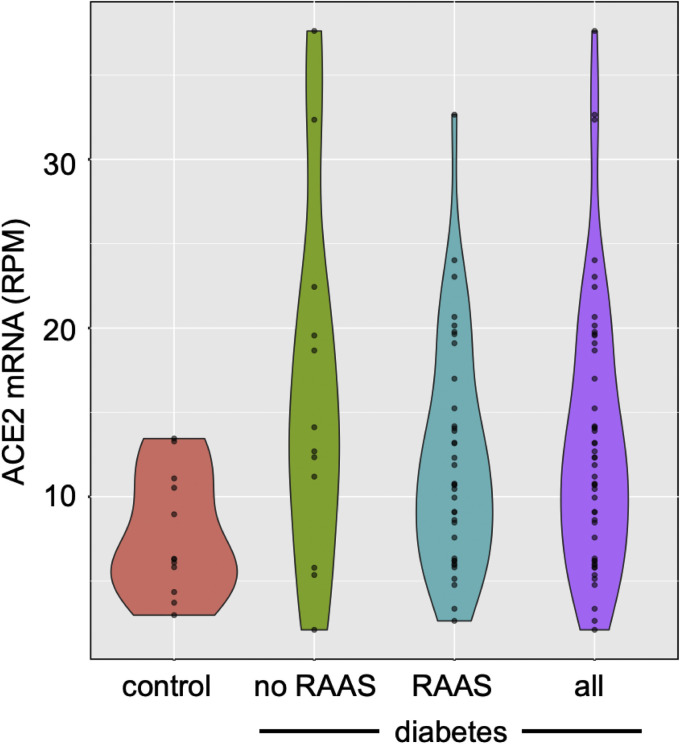
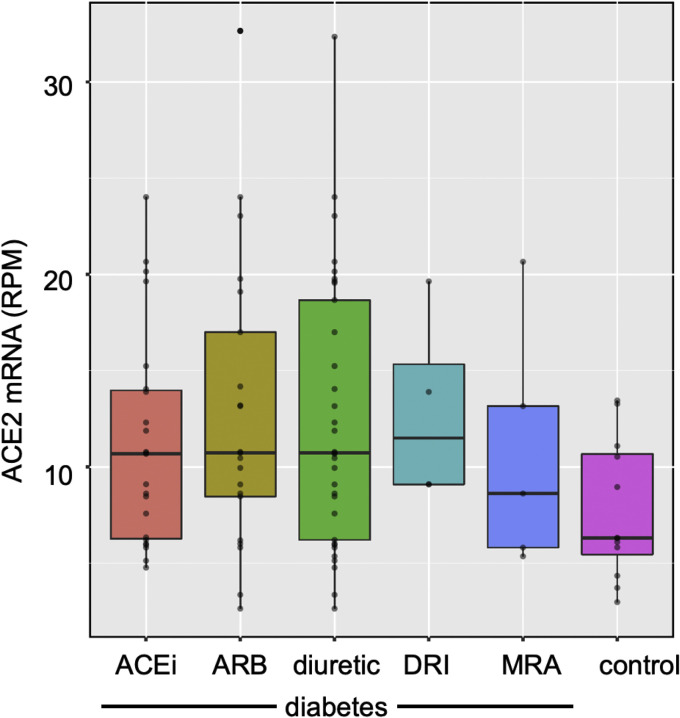
The magnitude of ACE 2 gene expression in biopsies obtained from patients with diabetic kidney disease varied widely when compared with that found in biopsies from healthy control subjects. The variability notwithstanding, mean ACE 2 messenger RNA was increased approximately 2-fold in diabetes when compared with healthy control subjects (Figure 1 , Supplementary Table 1), whereby mean expression of ACE 2 was 13.2±7.9 reads per million reads (RPMs) in biopsies from subjects with diabetic kidney disease and 7.7±3.6 RPMs in those from control subjects (p=0.001). No difference in transcript abundance was noted between recipients and nonrecipients of agents that block the RAAS (12.2±6.7 vs 16.2±10.7 RPMs, respectively; p=0.25) or among those receiving treatment with an ACE inhibitor, ARB, DRI, diuretic or MRA (Figure 2 ). Similarly, we found no relationship between ACE 2 copy number and either glycated hemoglobin (Spearman rho = 0.05; p=0.8), or between ACE 2 copy number and estimated glomerular filtration rate (ρ=0.10, p=0.5).
Figure 1.
Violin plots showing ACE2 mRNA expression as transcript union RPMs in kidney biopsies from individuals with diabetic kidney disease (purple) and living allograft donors (red) with ACE2 expression in diabetic samples also assessed according to use (blue) or not (green) of agents that block the RAAS: angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, direct renin inhibitors and mineralocorticoid receptor antagonists. ACE2, angiotensin-converting enzyme 2; mRNA, messenger RNA; RAAS, renin-angiotensin-aldosterone system; RPM, reads per million read.
Figure 2.
Box plots showing ACE2 mRNA expression as transcript union RPMs in kidney biopsies from individuals with diabetic kidney disease assessed according to use of ACEis, ARBs, DRIs, diuretics and MRAs. ACE2, angiotensin-converting enzyme 2; ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; DRI, direct renin inhibitor; MRA, mineralocorticoid receptor antagonist; mRNA, messenger RNA; RPM, reads per million read.
Discussion
An increase in ACE 2 expression in the setting of diabetic kidney disease raises the possibility that such individuals may be at higher risk of kidney infection with SARS-CoV-2 in COVID-19 disease, potentially increasing the risk of acute kidney injury and death. Furthermore, the study provides reassurance that any propensity to infection should not be exacerbated by concomitant use of agents that block the RAAS.
The main strengths of this study are the number of biopsies examined, the state-of-the-art technology used to quantify gene expression and the documentation of medications taken at the time of biopsy. We found only a single published study that compared ACE 2 messenger RNA in kidney biopsies from patients with diabetes with that of healthy control subjects using robust methodology (quantitative polymerase chain reaction) (7). In contrast with the current study of 49 biopsies, the number of biopsies in the study by Reich et al (7) was comparatively small, examining only 13 biopsies from patients with diabetes and showing a reduction in expression in diabetes. Moreover, unlike our study, the use of laser capture of proximal tubules and glomeruli in the latter study would not have fully taken into account the abundant ACE 2 expression in infiltrating cells, the postglomerular vasculature and nephron segments beyond the proximal tubule that may display differential expression in diabetes (11). Indeed, macrophages and pericytes have been shown to express high levels of ACE 2, suggesting that these cells may be particularly vulnerable to SARS-CoV-2 infection (8). In addition, the use of next-generation RNA Seq technology enabled a true measurement of ACE 2 transcript levels, as opposed to the relative levels generated by semiquantitative polymerase chain reaction or microarray-based techniques. Finally, archived remnants of formalin-fixed paraffin-embedded core biopsies have not generally been used for RNA Seq because the yield and quality of RNA from such small tissue samples has been insufficient for RNA sequencing. Our technology, developed in bladder tumours (10), overcame these limitations, enabling us to examine ACE 2 expression in small, residual amounts of clinically indicated renal biopsy samples.
In spite of the large number of biopsy specimens studied and reliance on whole rather than microdissected tissue, our study does, however, have several limitations. Most notably, the study was confined to the examination of kidney tissue and not lungs, the principal site of COVID-19 infection. Moreover, having studied only patients with biopsy-proven diabetic kidney disease, we are unable to make any inferences on whether the changes in ACE 2 messenger RNA observed in the current study apply to other organs, most notably the lungs, or if the changes in ACE 2 expression seen in our study group also apply to the kidneys of individuals with diabetes and normal kidney function. In addition, the current study which assessed gene expression in sections cut from kidney biopsies was unable to determine cell-specific patterns of expression that may have differentially affected parenchymal, vascular and inflammatory cells. Finally, although the abundance of a receptor required for SARS-CoV-2 entry was examined, many more components of the cell machinery are required for the virus to infect a cell, initiate virus replication and kill its host; although we assessed gene expression, its translation into protein was not examined.
Diabetes is a well-known risk factor not only for severe bacterial infections, but also for viral infections, such as H1N1 influenza (12). As was the case in previous human coronavirus infections, such as severe acute respiratory syndrome and Middle East respiratory syndrome, individuals with diabetes are also at higher risk of adverse outcomes with COVID-19 (13). Although this may reflect a generalized predisposition to poor outcome with infectious diseases, it may also be a consequence of an increased propensity for cellular entry and invasion by SARS-CoV-2, while noting that the current study assessed ACE 2 gene expression in the kidney and not in the lungs.
Interaction between a virus and its host cells are key determinants of infection severity. For instance, studies of a related murine coronavirus showed a dose-dependent relationship between the number of infective virus particles and the likelihood of death (14). Tissue-specific viral receptor abundance may also influence infectivity (11) and thereby contribute to the extrapulmonary manifestations of COVID-19, such as kidney and vascular disease. Consistent with this clinical observation, single cell RNA sequencing identified cells in the lung, kidney and heart as major sites of ACE 2 expression (2). Accordingly, the augmented kidney ACE 2 expression demonstrated in the present study may signify a greater propensity to renal complications of COVID-19 among individuals with diabetes.
A number of recently published observational studies concluded that the use of agents that block the RAAS increased neither the propensity to infection nor the likelihood of an adverse outcome (15, 16, 17). Although these studies did not specifically examine whether this broad conclusion also applied to individuals with diabetes, the current study does not indicate any relationship between the use of agents that block the RAAS and ACE 2 expression in the diabetic setting.
Acknowledgments
We thank Niki Dacouris, Michelle Nash, Lindita Rapi and Weiqiu Yuan for their assistance with patient data collection. We also thank the contributions of our patient partners in research: Mary Beaucage, Gwen Herrington and Dwight Sparkes. We thank the Network Biology Collaborative Centre (nbcc.lunenfeld.ca) for the RNA-Seq service, a facility supported by the Canada Foundation for Innovation, the Ontarian Government and Genome Canada and Ontario Genomics (OGI-139).
This study was funded by Can-SOLVE CKD, a SPOR grant from the Canadian Institutes of Health Research and a Transformational Grant by the Banting and Best Diabetes Centre in Toronto. R.E.G. is the Canada Research Chair in Diabetes Complications and this work was supported in part by the Canada Research Chairs’ Program. D.A.Y. is supported by a CIHR New Investigator Award and the St. Michael’s Hospital Foundation.
The study funders were not involved in the design of the study; the collection, analysis and interpretation of data or the writing of the report, nor did they impose any restrictions regarding the publication of the report.
Footnotes
To access the supplementary material accompanying this article, visit the online version of the Canadian Journal of Diabetes at www.canadianjournalofdiabetes.com.
Author Disclosures
R.E.G. reports receiving research grants to his institution from AstraZeneca and Boehringer Ingelheim; serving on advisory panels for AstraZeneca, Boehringer Ingelheim and Janssen and receiving CME speaker honoraria from AstraZeneca, Bayer, Boehringer Ingelheim and Janssen, all unrelated to the current study. He also reports being a shareholder in Certa Therapeutics, OccuRx and Fibrocor Therapeutics and is CSO of Fibrocor Therapeutics. J.L.W. and D.A.Y. are consultants and own shares in Fibrocor Therapeutics. P.S.M. was supported by a Kidney Foundation of Canada KRESCENT postdoctoral fellowship and an Eli Lilly Clinician-Scientist Trainee Fellowship in Diabetes. No other authors have any conflicts of interest to declare.
Author Contributions
R.E.G., J.L.W. and D.A.Y. designed the research. P.S.M., K.C. and L.C. acquired the data. J.L.W. and L.C. contributed to the statistical analyses. R.E.G., J.L.W., K.D.B. and D.A.Y. interpreted the data. R.E.G. drafted the manuscript. All authors reviewed and critically revised the draft for important intellectual content and approved the final manuscript to be published. J.L.W. is the guarantor of this work.
Supplementary Material
Supplementary Table 1.
Quality control meta-data for RNA Seq analysis of ACE 2 expression
| ID | ACE 2 RPM | ACE 2 count (raw read number) | Raw read count from fastqc | Mapped read numbers | Final unique read numbers | Mapping % (mapped reads/raw reads from fastqc) | Unique reads/mapped reads (%) | Unique reads/raw reads (%) | Condition |
|---|---|---|---|---|---|---|---|---|---|
| S1 | 11.88 | 1,166 | 101,465,082 | 98,099,054 | 82,972,027 | 96.682575 | 84.5798442 | 81.77397127 | DKD |
| S10 | 32.35 | 3,808 | 126,566,284 | 117,684,063 | 98,667,865 | 92.98215866 | 83.8413142 | 77.95746377 | DKD |
| S11 | 3.36 | 642 | 188,577,850 | 190,596,410 | 159,468,358 | 101.070412 | 83.6680806 | 84.56367384 | DKD |
| S12 | 5.36 | 520 | 114,244,968 | 96,992,737 | 66,805,465 | 84.89891388 | 68.8767706 | 58.47563019 | DKD |
| S13 | 6.21 | 808 | 175,045,896 | 129,964,897 | 97,959,487 | 74.24618341 | 75.3738042 | 55.96217291 | DKD |
| S14 | 6.29 | 819 | 137,316,374 | 130,057,555 | 99,596,713 | 94.71379939 | 76.5789523 | 72.53083525 | Normal Control |
| S15 | 19.77 | 2,454 | 125,973,470 | 124,069,715 | 106,369,186 | 98.48876513 | 85.7334008 | 84.43776773 | DKD |
| S17 | 2.98 | 338 | 114,739,886 | 113,279,463 | 92,896,430 | 98.72718803 | 82.0064181 | 80.96263055 | Normal Control |
| S18 | 6.32 | 797 | 131,967,904 | 125,958,052 | 98,037,370 | 95.4459745 | 77.8333488 | 74.28879828 | Normal Control |
| S19 | 18.67 | 2,234 | 125,828,736 | 119,626,461 | 92,277,030 | 95.07085965 | 77.137641 | 73.33541839 | DKD |
| S2 | 32.65 | 3,358 | 107,149,218 | 102,835,831 | 81,401,804 | 95.97441112 | 79.157044 | 75.97050685 | DKD |
| S20 | 14.05 | 1,630 | 121,341,514 | 116,008,433 | 94,539,795 | 95.6048999 | 81.4938988 | 77.91216038 | DKD |
| S21 | 10.46 | 1,168 | 114,541,108 | 111,565,723 | 87,783,595 | 97.4023431 | 78.6833022 | 76.63937999 | DKD |
| S23 | 13.16 | 1,886 | 153,494,604 | 143,258,910 | 123,152,446 | 93.33156102 | 85.964947 | 80.23242693 | DKD |
| S24 | 7.58 | 1,263 | 176,090,214 | 166,535,504 | 132,321,427 | 94.57396877 | 79.4553857 | 75.14411164 | DKD |
| S26 | 5.13 | 595 | 121,143,542 | 115,801,424 | 96,940,374 | 95.59025771 | 83.7125923 | 80.02108276 | DKD |
| S28 | 13.28 | 1,584 | 127,927,852 | 119,193,003 | 93,410,716 | 93.1720506 | 78.3692949 | 73.01827908 | Normal Control |
| S29 | 8.96 | 1,094 | 124,756,622 | 122,075,555 | 98,496,727 | 97.85096217 | 80.6850536 | 78.95110129 | Normal Control |
| S3 | 19.64 | 1,911 | 108,050,092 | 97,254,218 | 73,226,930 | 90.0084546 | 75.2943487 | 67.77127964 | DKD |
| S30 | 4.35 | 469 | 116,072,918 | 107,717,085 | 72,359,389 | 92.80122087 | 67.1754058 | 62.33959674 | Normal Control |
| S31 | 5.81 | 599 | 126,222,334 | 102,977,820 | 76,401,402 | 81.58446824 | 74.192095 | 60.52922615 | DKD |
| S32 | 37.61 | 3,643 | 103,394,492 | 96,860,281 | 85,553,782 | 93.68031036 | 88.3270017 | 82.74500928 | DKD |
| S33 | 9.95 | 1,149 | 124,113,112 | 115,461,620 | 101,243,718 | 93.02934891 | 87.6860363 | 81.57374863 | DKD |
| S34 | 6.1 | 723 | 128,575,262 | 118,359,206 | 93,275,371 | 92.05441557 | 78.8070266 | 72.5453478 | Normal Control |
| S35 | 13.45 | 1,796 | 136,947,722 | 133,454,499 | 102,748,768 | 97.44922884 | 76.9916105 | 75.02773065 | Normal Control |
| S36 | 12.31 | 1,252 | 106,491,158 | 101,699,951 | 76,515,285 | 95.50084055 | 75.2363047 | 71.85130337 | DKD |
| S38 | 6.34 | 660 | 108,149,060 | 104,020,509 | 86,927,172 | 96.1825364 | 83.5673396 | 80.37718682 | DKD |
| S4 | 9.09 | 892 | 105,342,072 | 98,043,413 | 83,559,528 | 93.07146816 | 85.2270698 | 79.3220851 | DKD |
| S40 | 22.44 | 2,083 | 102,534,852 | 92,791,329 | 80,415,714 | 90.49735499 | 86.6629618 | 78.42768818 | DKD |
| S43 | 5.82 | 717 | 129,310,302 | 123,119,602 | 91,843,518 | 95.21252375 | 74.5969907 | 71.02567744 | Normal Control |
| S47 | 10.53 | 1,301 | 125,599,084 | 123,536,436 | 97,933,170 | 98.35775235 | 79.2747251 | 77.97283776 | Normal Control |
| S48 | 11.09 | 1,358 | 129,861,432 | 122,361,656 | 88,440,726 | 94.22478569 | 72.2781375 | 68.10392018 | Normal Control |
| S49 | 19.56 | 2,664 | 140,716,858 | 136,163,725 | 113,595,350 | 96.76433011 | 83.4255599 | 80.72618421 | DKD |
| S5 | 20.66 | 2,525 | 129,277,952 | 122,208,257 | 92,915,798 | 94.53139929 | 76.0307039 | 71.87288827 | DKD |
| S50 | 8.47 | 1,178 | 145,194,772 | 138,993,857 | 122,500,834 | 95.72924361 | 88.1339914 | 84.37000335 | DKD |
| S51 | 8.62 | 1,009 | 121,066,116 | 117,037,226 | 103,152,225 | 96.67215722 | 88.1362525 | 85.20321656 | DKD |
| S52 | 5.79 | 664 | 119,156,404 | 114,491,282 | 100,216,841 | 96.08487514 | 87.53229 | 84.10529156 | DKD |
| S53 | 10.78 | 1,439 | 140,139,922 | 133,387,449 | 117,250,406 | 95.18162069 | 87.9021279 | 83.66666994 | DKD |
| S54 | 12.34 | 1,677 | 138,605,802 | 135,790,629 | 116,695,942 | 97.96893567 | 85.9381408 | 84.19268192 | DKD |
| S55 | 10.68 | 1,134 | 108,659,548 | 106,132,291 | 85,310,691 | 97.67415101 | 80.3814656 | 78.51191411 | DKD |
| S56 | 15.24 | 1,993 | 128,886,930 | 130,769,149 | 94,009,761 | 101.4603645 | 71.8898622 | 72.93971623 | DKD |
| S57 | 20.15 | 3,081 | 151,341,932 | 152,896,130 | 126,284,590 | 101.0269447 | 82.5950206 | 83.44322577 | DKD |
| S58 | 13.2 | 1,746 | 131,157,148 | 132,245,790 | 91,054,983 | 100.8300287 | 68.8528406 | 69.42433896 | DKD |
| S59 | 6.19 | 1,056 | 170,223,200 | 170,469,398 | 120,482,742 | 100.1446325 | 70.6770502 | 70.77927216 | DKD |
| S60 | 14.12 | 1,667 | 121,045,880 | 117,986,535 | 89,261,896 | 97.47257404 | 75.6543075 | 73.74220089 | DKD |
| S61 | 9.1 | 1,198 | 135,099,374 | 131,562,506 | 105,719,709 | 97.38202488 | 80.3570198 | 78.25329302 | DKD |
| S62 | 2.64 | 366 | 135,216,050 | 138,598,101 | 107,703,422 | 102.5012201 | 77.7091614 | 79.65283855 | DKD |
| S63 | 5.95 | 786 | 133,282,782 | 132,057,096 | 107,361,864 | 99.08038684 | 81.2995797 | 80.55193806 | DKD |
| S64 | 10.73 | 1,520 | 146,982,650 | 141,617,337 | 121,123,920 | 96.34969638 | 85.5290197 | 82.40695075 | DKD |
| S65 | 23.04 | 2,726 | 121,592,562 | 118,315,222 | 97,936,482 | 97.30465421 | 82.7758934 | 80.54479681 | DKD |
| S66 | 2.11 | 371 | 173,153,814 | 175,225,951 | 137,535,251 | 101.1967031 | 78.4902295 | 79.42952443 | DKD |
| S67 | 17 | 2,612 | 147,157,566 | 153,576,126 | 115,308,540 | 104.3616921 | 75.0823341 | 78.35719436 | DKD |
| S68 | 19.1 | 2,359 | 124,788,568 | 123,450,100 | 102,452,239 | 98.92741136 | 82.9908109 | 82.10066086 | DKD |
| S69 | 11.19 | 1,781 | 160,522,214 | 159,145,496 | 135,084,192 | 99.14235048 | 84.8809394 | 84.15295842 | DKD |
| S7 | 3.72 | 550 | 146,984,322 | 147,547,677 | 120,541,988 | 100.3832756 | 81.6969745 | 82.01009901 | Normal Control |
| S70 | 12.69 | 1,689 | 133,615,280 | 133,031,354 | 107,608,627 | 99.56297962 | 80.8896728 | 80.53616847 | DKD |
| S71 | 14.18 | 2,045 | 140,677,196 | 144,153,124 | 111,566,950 | 102.4708539 | 77.39475 | 79.30706125 | DKD |
| S72 | 6.01 | 1,634 | 281,300,140 | 271,680,124 | 207,399,305 | 96.58015954 | 76.3395209 | 73.72883106 | DKD |
| S73 | 4.77 | 483 | 123,833,718 | 101,246,379 | 61,657,133 | 81.75994441 | 60.8981117 | 49.79026229 | DKD |
| S8 | 24.02 | 2,450 | 103,952,142 | 101,965,133 | 79,035,117 | 98.08853482 | 77.511905 | 76.0302919 | DKD |
| S9 | 13.9 | 1,671 | 130,740,442 | 120,131,959 | 109,764,113 | 91.88584432 | 91.3696188 | 83.95574569 | DKD |
ACE 2, angiotensin-converting enzyme 2; DKD, diabetic kidney disease; ID, identification; RPM, reads per million read.
References
- 1.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020 doi: 10.1001/jama.2020.4683. (In press) [DOI] [PubMed] [Google Scholar]
- 2.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 doi: 10.1007/s11684-020-0754-0. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J Am Soc Nephrol. 2020 doi: 10.1681/ASN.2020040432. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y., Luo R., Wang K. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020 doi: 10.1016/j.kint.2020.03.005. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sise M.E., Baggett M.V., Shepard J.O., Stevens J.S., Rhee E.P. Case 17-2020: A 68-year-old man with Covid-19 and acute kidney injury. N Engl J Med. 2020;382:2147–2156. doi: 10.1056/NEJMcpc2002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020 doi: 10.1056/NEJMc2011400. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich H.N., Oudit G.Y., Penninger J.M., Scholey J.W., Herzenberg A.M. Decreased glomerular and tubular expression of ACE2 in patients with type 2 diabetes and kidney disease. Kidney Int. 2008;74:1610–1616. doi: 10.1038/ki.2008.497. [DOI] [PubMed] [Google Scholar]
- 8.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zulli A., Burrell L.M., Buxton B.F., Hare D.L. ACE2 and AT4R are present in diseased human blood vessels. Eur J Histochem. 2008;52:39–44. doi: 10.4081/1184. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Noon A.P., Aguiar Cabeza E. Next-generation RNA sequencing of archival formalin-fixed paraffin-embedded urothelial bladder cancer. Eur Urol. 2014;66:982–986. doi: 10.1016/j.eururo.2014.07.045. [DOI] [PubMed] [Google Scholar]
- 11.Pan X.W., Xu D., Zhang H., Zhou W., Wang L.H., Cui X.G. Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: A study based on single-cell transcriptome analysis. Intensive Care Med. 2020 doi: 10.1007/s00134-020-06026-1. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allard R., Leclerc P., Tremblay C., Tannenbaum T.N. Diabetes and the severity of pandemic influenza A (H1N1) infection. Diabetes Care. 2010;33:1491–1493. doi: 10.2337/dc09-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muniyappa R., Gubbi S. COVID-19 pandemic, coronaviruses, and diabetes mellitus. Am J Physiol Endocrinol Metab. 2020;318:E736–E741. doi: 10.1152/ajpendo.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Albuquerque N., Baig E., Ma X. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J Virol. 2006;80:10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds H.R., Adhikari S., Pulgarin C. Renin-angiotensin-aldosterone system inhibitors and risk of Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2008975. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancia G., Rea F., Ludergnani M., Apolone G., Corrao G. Renin-angiotensin-aldosterone system blockers and the risk of Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2006923. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. Cardiovascular disease, drug therapy, and mortality in Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007621. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]