Abstract
The rapidly spreading outbreak of COVID-19 disease is caused by the SARS-CoV-2 virus, first reported in December 2019 in Wuhan, China. As of June 17, 2020, this virus has infected over 8.2 million people but ranges in symptom severity, making it difficult to assess its overall infection rate. There is a need for rapid and accurate diagnostics to better monitor and prevent the spread of COVID-19. In this review, we present and evaluate two main types of diagnostics with FDA-EUA status for COVID-19: nucleic acid testing for detection of SARS-CoV-2 RNA, and serological assays for detection of SARS-CoV-2 specific IgG and IgM patient antibodies, along with the necessary sample preparation for accurate diagnoses. In particular, we cover and compare tests such as the CDC 2019-nCoV RT-PCR Diagnostic Panel, Cellex's qSARS-CoV-2 IgG/IgM Rapid Test, and point-of-care tests such as Abbott's ID NOW COVID-19 Test. Antibody testing is especially important in understanding the prevalence of the virus in the community and to identify those who have gained immunity. We conclude by highlighting the future of COVID-19 diagnostics, which include the need for quantitative testing and the development of emerging biosensors as point-of-care tests.
Keywords: COVID-19, SARS-CoV-2, Diagnostic testing, RT-PCR, Immunoassay, Point-of-care, FDA-EUA
Highlights
-
•
Current standard-of-care for COVID-19 diagnosis is RT-PCR, used to detect SARS-CoV-2 viral RNA.
-
•
Serological assays for detection of SARS-CoV-2 specific IgG and IgM antibodies are useful to assess virus prevalence.
-
•
Point-of-care tests for COVID-19 diagnoses can expedite results and alleviate the burden placed on healthcare providers.
-
•
Future testing strategies include developing quantitative COVID-19 diagnostic tools.
1. Introduction
A novel coronavirus disease was first reported when an outbreak of unknown respiratory illnesses occurred in Wuhan, China on December 31, 2019 (CDC, 2020h). It was quickly identified as a novel betacoronavirus, indicating a transfer of the disease from bats to humans with no clear indication of an intermediate host (Lu et al., 2020). On January 30, 2020 the World Health Organization (WHO) declared the coronavirus outbreak a public health emergency of international concern (PHEIC). On February 11, 2020 the WHO named the disease as COronaVIrus Disease 2019, or COVID-19 (WHO, 2020b), and the International Committee on Taxonomy of Viruses officially named the virus, previously the 2019-novel Coronavirus (2019-nCoV), as SARS-CoV-2 on March 2, 2020 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). The disease quickly spread throughout Southeast Asia, Europe and North America; WHO officially declared COVID-19 a pandemic on March 11, 2020 (WHO, 2020c). As of June 17, 2020, there are over 8.2 million confirmed COVID-19 cases reported world-wide (Johns Hopkins University, 2020). COVID-19 can present from mild to severe, and possibly fatal, with an increase in severity linked to age and underlying medical conditions (Guan et al., 2020).
One major problem in evaluating and monitoring the pandemic is the lack of diagnostic resources for COVID-19 (American Society of Microbiology, 2020). As the number of patients presenting with COVID-19 symptoms increase, there has been a shortage of diagnostic resources, like swabs, polymerase chain reaction (PCR) reagents, RNA isolation kits, and a growing demand for rapid, onsite diagnostics. A recent study showed that at least 35% of people are asymptomatic (CDC, 2020b), revealing an increased risk of rapid community spread and need for widespread testing. With the quick spread of COVID-19, the FDA has begun to issue Emergency Use Authorizations (EUA) to several diagnostic tests for COVID-19 (FDA, 2020a). Importantly, tests with FDA-EUA status have not received full FDA approval; rather, the authorizations for these tests are only in effect for the length of the pandemic. EUA tests for COVID-19 range from Clinical Laboratory Improvement Amendments (CLIA) certified tests to rapid diagnostics for clinical near-patient use. There has been a push towards developing point-of-care (POC) tests because the healthcare system is experiencing serious strain during the pandemic. In fact, the U.S. Department of Health and Human Service Biomedical Advanced Research and Development Authority (BARDA) granted a $13 million USD contract to Cue Health (Cue Health, 2020a, Cue Health, 2020b) and $750,000 to OraSure Technologies (OraSure Technologies, 2020) to develop portable COVID-19 diagnostic tests.
With the quickly changing landscape of available diagnostic tests for COVID-19, it is necessary for a holistic review and evaluation of diagnostic resources to be assembled. The goal of this review is to present an analysis of the current FDA-EUA diagnostic landscape for COVID-19, from patient specimen collection to commercially available diagnostic tests and future directions. We will begin our review by highlighting the structure of SARS-CoV-2 and the suspected roles and diagnostic interest of its proteins. We then cover relevant patient specimen collection techniques and sample preparation necessary prior to diagnostic testing, specifically focusing on existing viral RNA isolation methods and commercially available kits. Finally, we will move into our analysis of the current FDA-EUA diagnostic landscape, covering commercially available COVID-19 diagnostic platforms, and discussing COVID-19 immunity and how it will shape retrospective diagnostic development as well as epidemiological studies. We conclude with a discussion on necessary future works and important avenues of research.
2. SARS-CoV-2 protein structure
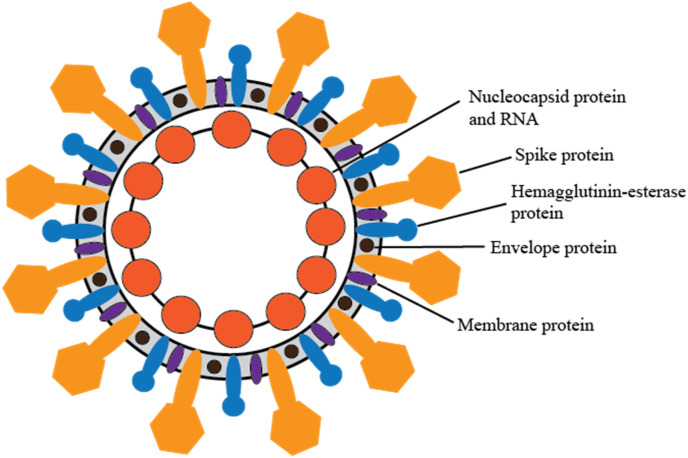
SARS-CoV-2 is composed of five proteins and a single-stranded, positive-sense RNA genome (Indwiani and Ysrafil, 2020; Fig. 1 ). A description of the proteins and their functions can be found in Table 1 . Previous studies looking at SARS-CoV have demonstrated that the strong antibody responses against the spike and the nucleocapsid proteins are of high diagnostic utility (Cheng et al., 2020).
Fig. 1.
SARS-CoV-2 Virus structure, with five main proteins. Details of protein function can be found in Table 1.
Table 1.
The five proteins composing SARS-CoV-2 (Indwiani and Ysrafil, 2020).
| Protein Name | Binding | Role |
|---|---|---|
| Spike Protein (S) | Utilizes an N-terminal signal sequence to gain access to the ER | Mediates attachment to host receptors |
| Nucleocapsid protein (N) | Binds the viral genome in a beads-on-a-string type conformation | Tethers the viral genome to replicase-transcriptase complex ;packages the encapsulated genome into viral particles |
| Envelope protein (E) | A transmembrane protein with ion channel activity | Facilitates assembly and release of the virus; involved in ion channel activity |
| Membrane protein (M) | Binds to nucleocapsid | Promotes membrane curvature |
| Hemagglutinin-esterase dimer protein (HE) | Binds sialic acids on surface glycoproteins | Thought to enhance S protein-mediated cell entry and virus spread through mucosa |
3. Specimen collection and sample preparation for diagnostic testing
The CDC has mandated that testing for SARS-CoV-2 be conducted only in consultation with a licensed healthcare provider and on persons demonstrating symptoms of COVID-19 (CDC, 2020e). The CDC has recommended nucleic acid testing of upper respiratory specimens collected by swabs. Nasopharyngeal (NP) specimens are the preferred choice for swab-based SARS-CoV-2 testing, followed by oropharyngeal (OP) specimens. As of June 2020, the CDC has allowed nasal swabs to be taken by the patient (self-swab) or health worker and used as a valid specimen for testing when NP swabs are not available. SARS-CoV-2 and its relevant biomarkers can be found in multiple specimen types besides NP, OP and nasal swabs, such as lower respiratory and blood-based specimens. Testing on alternative specimen types may be necessary depending on the goal of the test, variability of patient condition (i.e. intubation), or need for re-testing after a negative result. Here, we will cover upper respiratory, lower respiratory, and blood/serum/plasma specimens.
3.1. Clinical specimen types
3.1.1. Upper respiratory specimens
The primary goal for collecting upper respiratory specimens is to directly collect the SARS-CoV-2 virus and infected cells. Upper respiratory specimens are collected using either a swab or a wash/aspirate. Most upper respiratory swab collection techniques involve inserting a swab through either a nostril or the mouth to the desired location. Depending on the swab technique and location, the swab can be held in place, rotated, or rubbed against the surrounding tissue until sufficient specimen has been absorbed. For most upper respiratory swabs, the CDC recommends that specimens are collected with sterile flocked swabs and stored in sterile tubes containing viral transport media (VTM or UTM). For a list of CDC approved swabs, transport media, and swab techniques, see Supplementary Section 1.
Currently, NP swabs are the recommended specimen for COVID-19 diagnostics, but they must be collected by a trained healthcare worker. The current surge in the number of patients displaying COVID-19 symptoms reduces the availability of healthcare workers and the appropriate personal protective equipment necessary to perform NP or OP swabs. Considering these shortages, the CDC has approved onsite patient self-swab collection for nasal mid-turbinate swabs and anterior nares (nasal) swabs. Self-collected nasal swabs are less invasive and more comfortable compared to NP swabs collected by a healthcare worker. As the pandemic escalates, many of the specimens collected will be nasal mid-turbinate or nasal swabs. Nasopharyngeal and nasal washes/aspirates are collected using saline filled syringes or mechanical suction to collect specimens; even though they are considered acceptable specimens for COVID-19 diagnostics, the resources and time needed to perform them make them lower priority specimens to collect.
3.1.2. Lower respiratory specimens
When possible, the CDC recommends healthcare providers also take lower respiratory specimens, which can be valuable samples for diagnosing COVID-19 in severe cases (Yang et al., 2020). We discuss three types of lower respiratory specimens: sputum, tracheal aspirate and bronchoalveolar lavage (BAL) fluid. Sputum specimens should be collected from patients with deeply productive coughs, but they should never be induced. A recent study has shown that when naturally produced, sputum is a more robust specimen for diagnosis compared to throat swabs (Lin et al., 2020). Tracheal aspirates and BAL fluid specimen collection techniques involve flushing either the trachea or a small lung section with saline and aspirating it for analysis. These methods are quite invasive and should only be used if clinically indicated.
3.1.3. Whole blood, serum and plasma
Whole blood samples are collected by a healthcare provider by inserting a needle into a vein and directly collecting whole blood into a sterile tube. These samples can be stored as whole blood, serum, or plasma. In general, it is recommended that whole blood is processed into serum or plasma for storage if the sample will be analyzed at a later date. The CDC does not recommend that whole blood, serum or plasma be used as a specimen for an onsite diagnosis of COVID-19 at this time. However, whole blood is useful for conducting blood smears, looking at morphology and cell count, and examining blood cultures. After whole blood collection in a sterile tube, serum is generated after leaving whole blood at room temperature to clot. The blood is then centrifuged and the liquid supernatant, or serum, is separated from the remaining clot. For plasma collection, whole blood is collected in a sterile tube containing an anticoagulant. The blood is centrifuged, and the plasma supernatant is separated from the red blood cells and buffy coat. Serum and plasma samples can be used in serological diagnostics for epidemiological studies and recovery analysis, which will be addressed later in the review. These samples can be further processed for the detection of viral RNA in molecular diagnostics. More recently, finger prick blood drop samples collected at point-of-care or drive-through sites are used in lateral flow assays (usually immunoassays); most of these products are currently pending FDA approval. A summary of the covered swabs and extraction methods can be seen in Table 2 .
Table 2.
Clinically significant specimens collected for the detection of SARS-CoV-2, as of April 22, 2020.
| Type of Specimen | Extraction Method | Who can collect | Transport Container and Medium |
|---|---|---|---|
| Nasopharyngeal (NP) swab | A swab goes through a nostril into the nasopharynx, held, rotated and removed | Healthcare Provider Only | Sterile tubes containing 2–3 ml of viral transport media |
| Oropharyngeal (OP)/throat swab | A swab goes through the mouth to the posterior pharynx, rubbed against the pharynx and tonsillar pillars, then removed | Healthcare Provider Only | Sterile tubes containing 2–3 ml of viral transport media |
| Nasal mid-turbinate (NMT) swab | A swab goes through a nostril to the mid-turbinate, held briefly, then rotated and removed | Healthcare Provider or onsite self-swab | Transport tube containing either viral or Amies transport medium, or sterile saline |
| Anterior nares/nasal swab | A swab goes into one nostril and is rubbed against the nostril wall, then used on the second nostril | Healthcare Provider or onsite self-swab | Transport tube containing either viral or Amies transport medium, or sterile saline |
| Nasopharyngeal wash/aspirate | A tube is guided through the nose to the nasopharynx, where a Saline solution is instilled and immediately aspirated | Healthcare Provider Only | Sterile, leak-proof, screw-cap sputum collection cup or sterile dry container. |
| Sputum | The patient rinses their mouth with water and then expectorates deep cough sputum | Onsite patient collection | Sterile, leak-proof, screw-cap sputum collection cup or sterile dry container. |
| Tracheal aspirate | A catheter goes through the mouth to the trachea, where saline solution is instilled and immediately aspirated | Healthcare Provider Only | 2–3 mL in a sterile, leak-proof, screw-cap sputum collection cup or sterile dry container. |
| Bronchoalveolar lavage (BL) | A bronchoscope goes through the mouth and into the lungs, where saline solution is instilled and immediately aspirated | Healthcare Provider Only | 2–3 mL in a sterile, leak-proof, screw-cap sputum collection cup or sterile dry container. |
| Whole Blood | A needle is used for venipuncture of a viable vein, and blood is drawn out | Healthcare Provider Only | A sterile tube |
| Serum | Whole blood is drawn, left at room temperature until clotting, then centrifuged. Resulting supernatant is collected as serum | Healthcare Provider Only | A sterile tube |
| Plasma | Whole blood is drawn into a tube with anticoagulant and centrifuged; resulting supernatant is collected as plasma | Healthcare Provider Only | A sterile tube containing an anticoagulant |
3.1.4. Sample preparation for molecular diagnostics
When a specimen is analyzed using molecular diagnostics, it must first be processed into a compatible sample according to the diagnostic technique. For serological assays, whole blood, serum, or plasma samples can often be used directly. For molecular methods such as nucleic acid detection, specimens must be processed to isolate viral RNA. Current available diagnostic platforms for COVID-19 have a range of sample preparation complexity and automation. Here, we will discuss three methods of viral RNA isolation from specimens, followed by commercially available RNA extraction kits and their corresponding specimen types. The CDC has released a list of RNA isolation kits that are compatible with their SARS-CoV-2 protocol, seen in Supplementary Table S1.
3.1.5. Isolating viral RNA from a specimen
Isolating SARS-CoV-2 viral RNA from collected specimens is crucial for molecular diagnostics. After the collection of a specimen, the procedure for storage and sample preparation depend on the resources available, the time frame for sample analysis, and viral RNA isolation technique. In this section, we will discuss three methods of viral RNA isolation; more details are included in Supplementary Section 2.
Each of the three methods of viral RNA isolation can be implemented using a variety of protocols and can be purchased in kits implementing varying degrees of automation. Each of the three methods begins with cell lysis, RNAse denaturation, and protein denaturation. Cell lysis and RNAse denaturation can be achieved using chaotropic agents, while protein denaturation is often achieved by adding proteinase K. The three main methods for viral RNA isolation (Thermo Fisher Scientific, 2020b) are:
-
●Magnetic bead purification
-
○Pre-functionalized magnetic beads capture the viral RNA. External magnetic field holds the beads in place during wash and collection steps.
-
○Offers highly efficient target capture and concentration
-
○Risk of magnetic bead contamination in the isolated viral RNA solution.
-
○
-
●Spin column isolation
-
○Columns containing membranes made of silica or charged polymers trap viral RNA. Centrifugal force or vacuum is applied for wash and collection steps.
-
○Easy to implement, but requires a centrifuge or vacuum.
-
○The membranes can be clogged depending on the specimen used.
-
○
-
●Organic extraction
-
○A phenol or chloroform solution is used along with centrifugal force to aqueously separate viral RNA. Alcohol precipitation and rehydration are implemented to isolate the viral RNA.
-
○Gold-standard method for isolating viral RNA.
-
○More manually intensive when compared to other methods.
-
○
3.1.6. Commercially available RNA extraction kits
The CDC has released a list of commercially available extraction kits that can be used for sample preparation upstream of their emergency use authorization COVID-19 RT-PCR diagnostic test (CDC, 2020c). In their recommendation, the CDC highlighted primarily automated magnetic bead-based technologies from Roche, Qiagen, and bioMérieux for COVID-19 viral RNA isolation. Both manual and automated viral RNA isolation kits are normally available for purchase from major life science companies, including Roche, Qiagen, and Thermo Fisher Scientific; however, the rapid increase in COVID-19 diagnostic testing has caused a shortage of viral RNA isolation reagents. Some of the common kits used for SARS-CoV-2 RNA isolation are Qiagen's QIAamp Viral Mini Kit, Qiagen's EZ1 Virus Mini-Kit, and Roche's MagnaPure nucleic acid kit (American Society of Microbiology, 2020).
4. Current FDA-EUA diagnostic technologies for COVID-19
4.1. Current governmental and diagnostic guidelines for SARS-CoV-2
In the United States, governmental strategy to address the need for COVID-19 diagnostic tools is jointly enforced by the FDA and CDC under the Department of Health and Human Services. Operating primarily under the Public Health Service Act, they work closely to make recommendations aimed at preventing, detecting, and treating infectious disease. The CDC is called to detect and investigate diseases as well as assist the nation in implementing disease prevention tactics and health policies. Following this mission, the CDC quickly developed a nucleic acid test for the detection of SARS-CoV-2. While the FDA primarily serves as a regulatory agency for medical devices, using its EUA procedure to give emergency approval to COVID-19 diagnostic tests, it also recommends protocols for a wide range of activities related to the monitoring, diagnostics and treatment of COVID-19. The collaboration between the FDA and CDC provides guidance on the development standards, safety and use of diagnostic technologies for COVID-19 in the United State (FDA, 2020b).
The WHO's global role in monitoring, giving recommendations for best practices, and approving diagnostic tests is more or less comparable to the collaboration between the CDC and FDA in the United States. While the CDC is under the jurisdiction of the President, Congress, and the Judicial system, and is guided by internal experts, the WHO is guided by the United Nation's health ministers and gathers advice from panels of global independent experts. Similar to the FDA's EUA approval procedure, the WHO's Emergency Use Listing (EUL) validates in vitro diagnostics (IVDs) used for the detection of SARS-COV-2, focusing on IVDs most likely to be used in countries with limited resources for testing. While the FDA's EUA is meant to provide accelerated approval for all IVDs that meet requirements, the WHO's EUL prioritizes simpler products to support countries most in need (Informa, 2020).
Besides the difference in prioritization strategies of the WHO and US agencies, they have developed similar guidelines on appropriate sample types and collection, safety and sample preparation procedures, and the interpretation of diagnostic test results. However, there is a major difference between the WHO and CDC recommendations for testing methods for diagnosis of COVID-19. At this time, the WHO only recommends nucleic acid tests to be used for the diagnosis of a SARS-CoV-2 infection. The WHO has stated, based on current evidence, that point-of-care immunodiagnostic tests should only be used in research settings, and that antigen-detecting and antibody-detecting rapid diagnostic tests for patient care should not be used for diagnosis (WHO, 2020a). The WHO encourages these tests be researched and improved upon, noting their diagnostic potential and usefulness in epidemiological research.
Similar to the WHO, the CDC does not recommend the sole use of antibody testing for diagnostic purposes but does recommend it as a clinical support assessment tool. However, unlike the WHO, CDC officially recommended that all viral tests, including both nucleic acid and antigen tests, be used for the diagnosis of an infection. Both the WHO and CDC recommend testing of persons exhibiting COVID-19 symptoms, as well as close contacts of persons with positively identified infections. Additionally, they both recommend testing for a wide range of respiratory pathogens on patients’ samples suspected for COVID-19, to minimize the risk of untreated co-infection. Asymptomatic or mildly symptomatic patients are only considered for reverse transcription polymerase chain reaction (RT-PCR) testing if they have been in contact with a COVID-19 positive patient. Both organizations have released recommendations on protocols for early testing and special considerations for high population density situations and high-risk populations (CDC, 2020g).
The current standard for the diagnosis of COVID-19 is a nucleic acid test: specifically, RT-PCR for detection of SARS-CoV-2 RNA (WHO, 2020d; CDC, 2020i). In the United States, the CDC has been providing these tests to state and local public health departments, places with access to CLIA-certified laboratories. On the other hand, medical providers are seeking and obtaining tests from various commercial manufacturers who have developed tests that have received EUAs (CDC, 2020i; FDA, 2020c).
In this section, we will cover the two main types of diagnostic tests with FDA-EUA approval: nucleic acid diagnostic testing to diagnose active COVID-19 infections, and serological testing to determine COVID-19 presence in a community. The practical diagnostic considerations of RT-PCR test and serological tests are summarized in Table 3 .
Table 3.
Practical diagnostic considerations of RT-PCR test and Serological immunoassay.
| RT-PCR test | Antibody test | |
|---|---|---|
| Merit | Highly specific | Easy to use serological sample |
| Limitation | Sensitivity can suffer due to sampling errors or insufficient viral load (false negatives). Inactive virus and viral fragments could also test positive (false positives). | Generally not as accurate as RT-PCR test, with false positives and false negatives. False positives in a low prevalence population can give an exaggeration of exposure and immunity. (e.g., a specificity of 99% in a population of 1% prevalence can lead to ~50% of positive results being false.) |
| Remedy | Testing twice sequentially to improve sensitivity (e.g., a single test sensitivity of 70% would result in a 2-test sensitivity of 91%) and/or combination with chest CT scan and clinical factors | Assay validation with sufficient positive and negative sample cohorts; generally cannot be used to diagnose newly infected patients, but can be used as a screening test (Optimizing antibody test sensitivity for rule-out, optimizing specificity for rule-in) |
| Primary utility | Standard of care diagnosis of newly infected and/or active Covid-19 patients. | Screening test for stratifying newly infected patients, remotely infected patients, and asymptomatic patients; surveillance assay for seroprevalence, immunity and vaccination efficacy. |
4.2. Nucleic acid tests for viral detection of SARS-CoV-2
SARS-CoV-2 is a positive-sense, single-stranded RNA virus, with a genome of 29,881 bp in length (Lai et al., 2020). The current standard for SARS-CoV-2 diagnosis is RT-PCR. The goal of PCR is to amplify a particular gene of interest: small amounts of template DNA and primers specific to the gene of interest are cycled through heating and cooling steps, where the primers bind to and amplify the gene into millions of copies. In RT-PCR, the starting material is RNA, which is reverse-transcribed into complementary DNA (cDNA) that serves as a template in the PCR reaction. Since SARS-CoV-2 is an RNA virus, RT-PCR must be used for detection, given that RNA is the starting material.
For specific detection of SARS-CoV-2, one strategy researchers have been using are diagnostic panels consisting of primers specific to SARS-CoV-2 in RT-PCR reactions that can provide results within 2–3 h. Such tests are intended for the qualitative detection of SARS-CoV-2 viral RNA in NP and OP swab samples. Most of the available RT-PCR tests utilize oligonucleotide primers and probes selected from regions of various SARS-CoV-2 viral genes, including the nucleocapsid (N) (NeuMoDx Molecular, 2020; Luminex Molecular Diagnostics Inc, 2020; DiaSorin Molecular, 2020; Ipsum Diagnostics LLC., 2020; Thermo Fisher Scientific, 2020b; Avenillo Lab USA, 2020; PerkinElmer, 2020; Mesa Biotech Inc., 2020; Cepheid, 2020; Quest Diagnostics Infectious Disease, 2020; LabCorp. 2020), envelope (E) (Luminex Molecular Diagnostics Inc, 2020; Ortho-Clinical Diagnostics, 2020; Cepheid, 2020; QIAGEN GmbH, 2020; Roche Molecular Systems, 2020), spike (S) (Yang et al., 2020) and/or open reading frame 1 ab (ORF1ab) genes (Luminex Molecular Diagnostics Inc, 2020; Ortho-Clinical Diagnostics, 2020; Thermo Fisher Scientific, 2020a, PerkinElmer, 2020, QIAGEN GmbH, 2020; Roche Molecular Systems, 2020; DiaSorin Molecular LLC., 2020; BGI Genomics Co. Ltd., 2020; BioFire Defense, 2020; Hologic, 2020). These tests mostly utilize standard RT-PCR protocols, including cell lysis, nucleic acid extraction and purification, and multiplexed PCR amplification and detection with fluorescence signal readout. However, as many of these tests came out quickly to help initially boost COVID-19 testing, all of the tests with FDA-EUA status are still only qualitative, giving a dichotomous indication of either presence or absence of SARS-CoV-2 without quantifying viral load.
The CDC developed one of the earliest real-time RT-PCR diagnostic panels for SARS-CoV-2. The CDC's 2019-nCoV Real-Time RT-PCR Diagnostic Panel (CDC, 2020a) is a test designed to qualitatively detect two different regions of the N gene: N1 and N2, as well as the RNase P (RP) gene, from NP/OP swabs, sputum, tracheal aspirates, or bronchoalveolar lavage fluid samples. The RP gene test acts as an internal control to verify that the RT-PCR was performed correctly. The RT-PCR is designed to run on the Applied Biosystems™ 7500 Fast Dx RT-PCR Instrument, and takes about 35 minutes to complete. The analytical sensitivity of the test was determined in a series of dilution studies using characterized samples with spiked-in full-length RNA of the N gene of known titers. The lowest concentration where at least 95% of the replicates were positive was set as the limit of detection (LoD). The LoD of the CDC's 2019-nCoV Real-Time RT-PCR Diagnostic Panel was found to be 10 copies/μL. In order to evaluate its clinical performance, the CDC evaluated a total of 117 respiratory specimens collected from a handful of suspect subjects who were also tested with a composite comparator consisting of two analytically validated RT-PCR assays that targeted two unique regions of the N gene, N4 and N5. Samples that tested positive for the 2019-nCoV Real-Time RT-PCR Diagnostic Panel test as well as the composite comparator were then further investigated and confirmed to be SARS-CoV-2 positive by genetic sequencing. The CDC's 2019-nCoV Real-Time RT-PCR Diagnostic Panel has a clinical sensitivity of 100% (13/13; 95% CI: 77.2%–100%) and a clinical specificity of 100% (104/104; 95% CI: 96.4%–100%).
Having set the precedence for an RT-PCR test for SARS-CoV-2, the CDC's 2019-nCoV Real-Time RT-PCR Diagnostic Panel paved the way for other RT-PCR tests. Many of these tests were developed as standard RT-PCR kits with primers and probes for SARS-CoV-2. Like the CDC's 2019-nCoV Real-Time RT-PCR Diagnostic Panel test, these tests were developed for use on the Applied Biosystems™ 7500 Fast Dx RT-PCR Instrument (Wadsworth Center, 2020; Thermo Fisher Scientific, 2020a; Avellino Lab USA, 2020; Quidel Corporation, 2020a) or the Applied Biosystems™ 7500 Real-time PCR system (PerkinElmer, 2020; BGI Genomics Co. Ltd, 2020; Quidel Corporation, 2020a; Primerdesign Ltd., 2020; Xing et al., 2020). The PerkinElmer New Coronavirus Nucleic Acid Detection Kit reports an analytical sensitivity down to 8.3 copies/mL and 24.9 copies/mL for the ORF1ab and N gene assays, respectively (PerkinElmer, 2020), an impressive order of magnitude lower than the average LoD of ~225 copies/mL reported by other assays performed on the same system (Quest Diagnostics Infectious Disease, 2020, Ortho-Clinical Diagnostics, 2020; BGI Genomics Co. Ltd, 2020; Quidel Corporation, 2020a; Primerdesign Ltd., 2020). Assays performed on the Applied Biosystems™ 7500 Real-time PCR system can achieve lower LoDs in comparison to those performed on the Applied Biosystems™ 7500 Fast Dx RT-PCR Instrument. Average LoDs reported from assays performed on the Applied Biosystems™ 7500 Fast Dx RT-PCR Instrument are around 35,000 copies/mL (CDC, 2020a; Thermo Fisher Scientific, 2020a; Avellino Lab USA, 2020; Quidel Corporation, 2020a). This disparity in LoDs between a RT-PCR assay that is performed with faster turnaround time (~35 minutes) and a standard RT-PCR assay (~2 hours) illustrates the need for development of new technology to close this gap, such that we can perform faster assays with higher levels of analytical sensitivity. This would be advantageous during pandemics, such as the SARS-CoV-2 outbreak, to provide faster yet more accurate diagnoses.
QIAGEN GmbH's QIAstat-Dx Respiratory SARS-CoV-2 Panel (QIAGEN GmbH, 2020) is an impressive demonstration of multiplexing capabilities. This assay is performed on QIAGEN's QIAstat-Dx Analyzer 1.0, which is a fully automated system that performs all sample analysis steps including cell lysis, nucleic acid purification, master mix and reagent mixing, transfer of defined aliquots of eluate and master mix into different reaction chambers, and fluorescence detection. The assay targets two genes from SARS-CoV-2, the ORF1b and E genes, along with several other respiratory bacterial and viral infections: Adenovirus, Coronavirus 229E, Coronavirus HKU1, Coronavirus NL63, Coronavirus OC43, SARS-CoV-2, Human Metapneumovirus A + B, Influenza A, Influenza A H1, Influenza A H3, Influenza A H1N1/pdm09, Influenza B, Parainfluenza virus 1, Parainfluenza virus 2, Parainfluenza virus 3, Parainfluenza virus 4, Rhinovirus/Enterovirus, Respiratory Syncytial Virus A + B, Bordetella pertussis, Chlamydophila pneumoniae and Mycoplasma pneumoniae. The turnaround time for the test results is about 1 h and the LoD for the SARS-CoV-2 assay in this panel is 500 copies/mL. The clinical performance of the SARS-CoV-2 assay was evaluated using 30 positive nasopharyngeal swab samples and 30 negative samples. QIAGEN's QIAstat-Dx Respiratory SARS-CoV-2 Panel demonstrated a clinical sensitivity of 100% (30/30; 95% CI: 85.8%–100%) and a clinical specificity of 100% (30/30; 95% CI: 85.8%–100%). Given that patients with SARS-CoV-2 actually present with co-infections (Xing et al., 2020; Fan et al., 2020) this assay provides a better overall picture of patient condition, allowing clinicians to provide better treatment steps. Moreover, it illustrates the power of multiplexing to achieve diagnoses for many infections at the same time.
In recent years, there has been a push to make the RT-PCR testing process automated; many systems have now employed microfluidic components and magnetic affinity microsphere nucleic acid capture techniques. Three such systems currently being deployed for SARS-CoV-2 testing include the NeuMoDx™ 288 and NeuMoDx™ 96 Molecular Systems, Luminex's MAGPIX® instrument, and BD's MAX System (NeuMoDx Molecular, 2020; Luminex Molecular Diagnostics Inc, 2020; Becton and Company, 2020). Automated systems are user-friendly, only requiring users to load in the sample and press go. By limiting user interaction, automated systems minimize user error and improve assay reproducibility, eliminating the need for trained professionals. This is advantageous during pandemics such as the SARS-CoV-2 outbreak, when the number of healthcare professionals is constrained and the need for widespread testing is critical. Automated systems also use micro- and nano-scale technologies, enabling reduction of reagent and sample volumes, and making the test faster and cheaper to run.
Table 4 provides a detailed comparison of major RT-PCR tests with FDA-EUA approval. While all tests have comparable sensitivities and specificities, both Thermo Fisher Scientific's and the CDC's tests offer greater flexibility by accepting the most sample types; Thermo Fisher Scientific's test is one of the few tests that accept mid-turbinate swabs for detection. Thermo Fisher Scientific's test is also impressive as it is the only one that detects the S gene from SARS-CoV-2, which is especially important in distinguishing SARS-CoV from SARS-CoV-2. However, other tests that detect the N gene are also valuable, as this is a highly conserved (90% homology) genome domain of SARS-CoV that enables the test to have greater specificity (Dutta et al., 2020). Additionally, many of the diagnostic tests with EUA approval have the advantage of being able to process many samples at once, but a drawback to Abbott's Alinity m SARS CoV-2 Assay (Abbott Laboratories, 2020) and BD's BioGx SARS-Cov-2 Reagents for BD MAX System (Becton and Company, 2020) is that they can only process 24 samples in 2–3 hours, whereas PerkinElmer, ThermoFisher Scientific, and Roche's tests can process 96 samples in the same amount of time.
Table 4.
Comparison of molecular diagnostics for COVID-19 with FDA-EUA approval.
| Manufacturer | Test | SARS-CoV-2 Biomarkers | Sample Types Accepted | Time to Result | Sensitivity/Specificity | References |
|---|---|---|---|---|---|---|
| CDC | 2019-nCoVReal-Time RT-PCR Diagnostic Panel | N1, N2, RP | Nasopharyngeal (NP) swab, Oropharyngeal (OP) swab, Sputum, Tracheal aspirates, Bronchoalveolar lavage (BAL), Nasal aspirate | ~80 min for 1 sample | 100% (13/13)/100% (104/104) | https://www.internationalreagentresource.org/QuickLinks/COVID-19FAQ.aspx#Full%20List |
| QIAGEN | QIAstat-Dx\ RespiratorySARS-CoV-2 Panel | ORF1b, E | NP swab | ~1 h for 1 sample | 100% (30/30)/100% (30/30) | https://www.qiagen.com/us/products/diagnostics-and-clinical-research/infectious-disease/qiastat-dx-syndromic-testing/qiastat-dx-eua-us/#orderinginformation |
| PerkinElmer | PerkinElmer® New Coronavirus Nucleic Acid Detection Kit | ORF1ab and N | NP swab, OP swab | ~2 h for 96 samples | 2X LOD (100%, 20/20)4X LOD (100%, 20/20)/100% (94/94) | https://perkinelmer-appliedgenomics.com/home/products/new-coronavirus-2019-ncov-nucleic-acid-detection-kit/ |
| Thermo Fisher Scientific | TaqPath COVID-19 Combo Kit | ORF1b, S, N | NP swab, BAL, Nasal swabs, OP swabs, Nasal aspirate,Mid-turbinate swabs | 3 h for 94 samples | 100% (60/60)/100% (60/60) | https://www.thermofisher.com/us/en/home/clinical/clinical-genomics/pathogen-detection-solutions/taqpath-covid-19-diagnostic-kit.html |
| Roche | cobasSARS-CoV-2 | ORF1a/b, E | NP swab, OP swab | 3 h for 96 samples | 100% (50/50)/100% (100/100) | https://diagnostics.roche.com/us/en/products/params/cobas-sars-cov-2-test.html |
| AbbottMolecular | Alinity m SARS-CoV-2 Assay | RdRp, N | NP swabs, OP swabs, BAL | ~2 h for 24 samples | 100% (40/40)/96.5% (55/57) | https://www.molecular.abbott/us/en/products/infectious-disease/alinity-m-sars-cov-2-assay |
| Becton, Dickinson & Company (BD) | BD SARS-CoV-2 Reagents for BD MAX System | N1, N2, | NP swab, OP swab | 3 h for 24 samples | 1-2X LOD (95%, 38/40)3-5X LOD (100%, 10/10); 100% (29/29) | https://www.bd.com/en-us/offerings/capabilities/molecular-diagnostics/molecular-tests/biogx-sars-cov-2-reagents |
Some of the main customers for diagnostic tests are healthcare providers such as hospitals, urgent care facilities, and drive-through clinics that give COVID-19 diagnoses to the public. Academic institutions are also major customers of diagnostic tests, as research labs not only partner with hospitals to perform clinical studies, but also work on analytically comparing and determining the efficacy of the many FDA-EUA tests on the market. Since these nucleic acid tests are used to determine active COVID-19 diagnoses, users are required to be highly trained to ensure test accuracy; currently, these users are trained healthcare professionals and research technicians. In the future, with the advent of easy-to-use POC COVID-19 testing, more customers and users of diagnostic tests could be the patient themselves.
4.3. Immunoassay development
4.3.1. Immunity to SARS-CoV-2
It has been noted that there is deficiency in a diagnosis solely reliant on detection of viral nucleic acid, resulting in large inconsistencies or high false negative rates (Wang, Y. et al., 2020; Li and Xia, 2020). It is therefore imperative to use a combination of molecular tests and chest CT scans as well as clinical factors to provide a more accurate diagnosis (Liang, T., 2020), especially when SARS-CoV-2 RNA is below the LoD or no longer present. Serological tests are important because they provide information on patients who have been infected and already recovered, especially asymptomatic patients who were never diagnosed (Amanat et al., 2020; Vogel, 2020). Moreover, through seroprevalence studies, we can examine the growth and frequency of the infection in a community (Bendavid et al., 2020), while identifying strong responders who might be able to confer immunity to weaker responders through convalescent sera and passive antibody therapy (Casadevall and Pirofski, 2020). Understanding the SARS-CoV-2 immune response can pave the way for vaccine development and treatments, improving our understanding of correlates of protection (Okba et al., 2020).
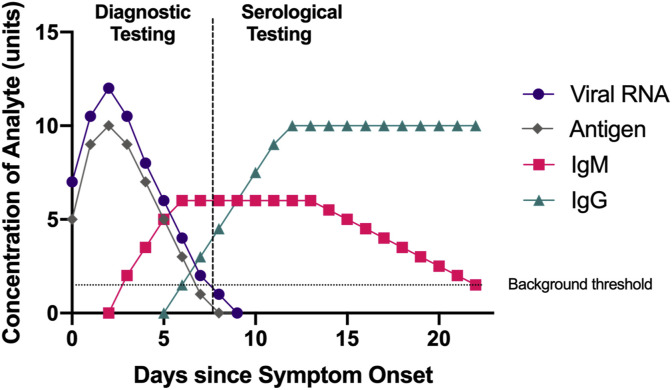
Although SARS-CoV-2 is a new virus, the immune response against it is similar to that of other pathogens: there is first an increase in immunoglobulin M (IgM), with immunoglobulin G (IgG) following after (Fig. 2 ). The timescale for when these antibodies rise is still being determined for SARS-CoV-2, but a study profiling the early SARS-CoV-2 humoral response found that the median duration of IgM detection was 5 days after symptom onset, and IgG was detected at a median of 14 days after symptom onset (Guo et al., 2020). In general, IgM is the first antibody made after infection with a new pathogen, and IgG is a more stable and longer lasting antibody present in the serum to help fight off infection. Therefore, if more antigen-specific IgG is detected in a patient's blood, it indicates a later stage of infection. For SARS-CoV-2, the IgG and IgM produced specific to the S and N proteins are of particular diagnostic interest. Some studies indicate that the S protein is more immunogenic (Amanat et al., 2020) or tends to cause a greater immune response, than the N protein, as it capable of eliciting neutralizing antibodies (Johns Hopkins Center for Health Security, 2020). However, other studies argue that the N protein is more immunogenic, as it is expressed abundantly during active infection (Dutta et al., 2020). In general, a patient with a stronger immune response has a better chance for a faster recovery, as the body actively fighting the disease.
Fig. 2.
Time course of approximate concentrations of viral RNA, antigen, and antibodies after symptom onset for a hypothetical patient with SARS-CoV-2. Diagnostic testing consists of both RT-PCR and antigen testing. While exact numbers on the duration of the antibody response are still being determined at this writing, in general, RT-PCR and antigen testing are effective to diagnose active infection when viral RNA or antigen is present. Serological assays are effective after about 5 days to detect IgM, with IgG rising afterwards (Guo et al., 2020).
Once a patient has SARS-CoV-2 specific antibodies circulating in his or her bloodstream, a natural next question is how long these antibodies remain, or how long a patient has protective immunity. To answer this, researchers have analyzed immune responses to the coronaviruses that cause the common cold, finding that protection decreases by a year or two (Tyrrell and Myint, 1996). Additionally, in examining the SARS epidemic of 2004, the specific antibodies produced against the SARS virus decrease after three years (Wu, L.-P. et al., 2007) indicating that an individual is susceptible to reinfection at that time. Research has indicated that antibodies produced against Middle East Respiratory Syndrome (MERS) decrease after 2 years (Payne et al., 2016), with more severe reactions producing stronger immune responses. A study looking at four rhesus macaques given a dose of SARS-CoV-2 discovered that when the monkeys produced neutralizing antibodies to the S protein soon after infection, they were less susceptible to reinfection (Bao et al., 2020). While this study has not been peer-reviewed, it suggests that humans with more SARS-CoV-2 specific antibodies circulating will have generated protective immunity, mitigating the risk of disease spread. Most recently, Long et al. found that the protective immune response of SARS-CoV-2 infected patients, specifically, IgG and neutralizing antibody levels, began to decrease 2–3 months after infection (Long et al., 2020). While more longitudinal studies surveying both asymptomatic and symptomatic SARS-CoV-2 patients need to be conducted to determine how long this antibody-mediated immunity will last, these preliminary studies indicate that patients who have seemingly recovered should still exercise caution and maintain good practices such as social distancing.
4.3.2. Serological tests for SARS-CoV-2
Some examples of serological tests to measure patient antibodies include rapid diagnostic tests (RDTs), enzyme-linked immunoassays (ELISAs), chemiluminescent immunoassays (CLIAs, not to be confused with CLIA acronym for Clinical Laboratory Improvement Amendments), or neutralization assays. The most common RDT is a lateral flow assay. The lateral flow immunoassay works via capillary action whereby the sample (often a finger prick blood drop) is wicked up a nitrocellulose membrane that is pre-functionalized with capture and detection antibodies, and usually gold nanoparticles or other colored nanoparticles, to generate colored lines on the membrane if the analyte of interest is present (Koczula and Gallotta, 2016). An ELISA is a plate-based assay designed to detect proteins or small molecules (British Society for Immunology, 2020). In general, an ELISA for the detection of patient antibodies is performed by first immobilizing a known capture antigen to the plate. Upon the addition of sample, patient antibodies in the sample (usually serum or plasma from venipuncture blood draw) that are specific to the capture antigen bind to the immobilized capture antigen on the plate. An enzyme-labeled detection antibody specific to any of the antibody isotypes (i.e. IgG, IgM, etc.) is added and specifically binds to the captured patient antibodies. A substrate, usually horseradish peroxidase (HRP), is added and interacts with the enzyme, causing a colorimetric change that can be measured and correlated to the presence and/or concentration of the antibody. A CLIA has the same principle as an ELISA, but are simpler tests to perform and provide larger throughput, as it has shorter incubation steps and doesn't require a reagent for stopping the enzymatic reaction (Sigma-Aldrich, 2020). CLIA tests are known to have increased sensitivity and dynamic ranges compared to ELISA tests (Monobind.Inc, 2020). The lateral flow assay, ELISA, and CLIA more frequently test for IgG and IgM antibodies; in contrast, a neutralization assay measures how many neutralizing antibodies, or those that can effectively bind to and block virus replication, are produced. Realizing the importance of measuring patient immunity to SARS-CoV-2, many companies are working to develop serological tests.
Cellex developed the first rapid antibody blood test for SARS-CoV-2 that was approved for EUA by the FDA. Cellex's qSARS-CoV-2 IgG/IgM Rapid Test is a lateral flow immunoassay (Cellex Inc., 2020) it provides results within 15–20 minutes and is used to detect patient IgG and IgM antibodies against SARS-CoV-2. The test can be used on serum, plasma, or whole blood samples. To evaluate the clinical performance of the assay, 128 SARS-CoV-2-positive patients and 250 negative control patients were tested. The clinical sensitivity of the assay was 93.8% (120/128; 95% CI: 88.2%–96.8%) and the clinical specificity of the assay was 96.0% (240/250; 95% CI: 92.8%–97.8%). The Cellex test paved the way for other lateral flow immunoassay tests to detect IgG and IgM, such as Autobio Diagnostics Anti-SARS-CoV2 Rapid Test (Autobio Diagnostics, 2020), and Chembio Diagnostic System's DPP COVID-19 IgM/IgG system, which also was approved for EUA by the FDA (Chembio Diagnostics Systems Inc, 2020). This test is advantageous in that it does not rely on visual detection for IgG/IgM detection; instead, it uses the DPP microreader for a qualitative readout, avoiding the possibility of user bias or misinterpretation. The clinical specificity of the assay is 97.6% for IgM, 96.8% for IgG, and 94.4% for IgM and IgG combined.
Ortho Clinical Diagnostics developed one of the first CLIA tests, VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack/Total Calibrator, which has been approved for EUA by the FDA (Ortho-Clinical Diagnostics, Inc., 2020). This test detects total IgG/IgM, but doesn't differentiate between the two, and takes around 50 minutes. The clinical sensitivity is 83% (30/36; 95% CI: 67.2–93.6%) and clinical specificity is 100% (400/400; 95% CI: 99.1–100.0%). Roche's technology for immunoassay detection is similar to a CLIA test: their Elecsys® system performs an electrochemiluminescent immunoassay (ECLIA) in which an electrochemical reaction initiates the main chemiluminescent reaction (Roche Diagnostics, 2020). The Elecsys® Anti-SARS-CoV-2 Test detects total antibody against N protein, and takes only 18 minutes. The clinical sensitivity is 100% ≥ 14 days post PCR confirmation (29/29; 95% CI: 88.1–100%), and the clinical specificity is 99.81% (99.65–99.91%). Similar to Roche's Elecsys® system, Bio-Rad's Platelia SARS-CoV-2 Total Ab test (Bio-Rad Laboratories, 2020) also detects total antibody against the N protein, and Abbott's SARS-CoV-2 IgG Assay (Abbott Inc, 2020).also detects antibody against the N protein, but just IgG instead of total antibody.
Table 5 provides a detailed comparison of major serological tests with FDA-EUA approval. All of the tests can be used with serum or plasma samples. Of all the tests, both Cellex's lateral flow assay and Bio-Rad's ELISA tests have comparatively lower sensitivities. Bio-Rad's test additionally has the longest turnaround time, since ELISA tests have longer incubation times compared to CLIA tests. The tests that detect total antibody against the RBD region of the S protein are especially important (Ortho-Clinical Diagnostics, 2020, DiaSorin Molecular, 2020, Siemens Medical Solutions, 2020), as it has been demonstrated that the S protein is a highly sensitive antigen for antibody detection in patients (Premkumar et al., 2020). Moreover, the RBD region of SARS-CoV-2 binds to the ACE2 receptor to enter host cells, and it is been shown that RBD-specific antibody concentrations are directly correlated with SARS-CoV-2 neutralizing antibodies in patients (ScienceDaily, 2020). These antibodies can be produced at a larger scale and can potentially be distributed as SARS-CoV-2 treatments with appropriate regulatory approval.
Table 5.
Comparision of major serological assays for COVID-19 with FDA-EUA approval.
| Manufacturer | Test | Test Type | SARS-CoV-2 Biomarkers | Time to Result | Sensitivity/Specificity | References |
|---|---|---|---|---|---|---|
| Autobio Diagnostics | Anti-SARS-CoV-2 Rapid Test | Lateral flow immunoassay | IgG and IgM only against S protein | ~15 min | 99.0% (299/302)/99.04% (309/312) | https://www.cardinalhealth.com/en/cmp/ext/med/med-lab/hardy-diagnostics-autobio-anti-sars-cov-2-rapid-test.html |
| Cellex | qSARS-CoV-2 IgG/IgM Rapid Test | Lateral flow immunoassay | IgG and IgM only against S and N proteins | ~15–20 min | 93.8% (120/128)/96% (240/250) | https://cellexcovid.com/ |
| Ortho Clinical Diagnostics | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack | Chemi-luminescent immunoassay | Total antibody against S1 protein | ~50 min | 100% (49/49)/100% (400/400) | https://www.orthoclinicaldiagnostics.com/en-us/home/ortho-covid-19-answer |
| DiaSorin | LIAISON SARS-CoV-2 S1/S2 IgG | Chemi-luminescent immunoassay | IgG against S1/S2 protein | ~35 min | 97.56% (40/41) ≥ 15 days post-symptom onset/99.3% (1082/1090) | https://www.diasorin.com/en/node/11756/ |
| Abbott Laboratories | SARS-CoV-2 IgG Assay | Chemi-luminescent microparticle immunoassay | IgG only against N protein | ~30 min | 100% (88/88) ≥ 14 days post-symptom onset/99.63% (1066/1070) | https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease/sars-cov-2 |
| Bio-Rad Laboratories | Platelia SARS-CoV-2 Total Ab assay | ELISA | Total antibody against N protein | ~100 min | 92.2% (47/51)/99.6% (684/687) | https://www.bio-rad.com/en-us/sku/72710-platelia-sars-cov-2-total-ab-assay?ID=72710 |
| Roche | Elecsys Anti-SARS-CoV-2 | Electrochemi-luminescence immunoassay | Total antibody against N protein | ~18 min | 100% (29/29) ≥14 days post-symptom onset/99.81% (5262/5272) | https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html |
| Siemens Healthcare | Atellica IM SARS-CoV-2 Total (COV2T) | Chemi-luminescent microparticle immunoassay | Total antibody against RBD of S1 protein | ~10 min | 100% (42/42) 14 days post-symptom onset/99.8% (1089/1091) | https://www.siemens-healthineers.com/en-us/laboratory-diagnostics/assays-by-diseases-conditions/infectious-disease-assays/cov2t-assay |
Currently, some of the main customers for antibody tests are healthcare providers, laboratories, and public health staff (CDC, 2020j); the tests are primarily used to evaluate populations and people who are likely to have had or have been exposed to SARS-CoV-2. Labs have used antibody tests to conduct major antibody seroprevalence studies in various counties. In the future, more community clinics, businesses, and schools might be main customers interested in mass screening and surveillance efforts to determine population prevalence. With all antibody tests, the main users are currently trained healthcare professionals and research technicians.
Like the RT-PCR assays, current immunoassays for SARS-CoV-2 are still only qualitative; they cannot be used to quantify patient antibody levels. Importantly, the results from serological tests alone should not be used to diagnose SARS-CoV-2 infection in practice. Even if high amounts of IgM are observed, indicating recent virus exposure, a standard of care molecular test should still be conducted to examine viral RNA presence. It has been shown that serological tests, when supplemented with RT-PCR for SARS-CoV-2 diagnosis, have a higher sensitivity (98.6%) than RT-PCR alone (92.2%) (Guo et al., 2020; Wang, 2020).
4.4. Point of care technologies
During a pandemic, such as the COVID-19 outbreak, it is imperative to develop and have point-of-care (POC) technologies on hand. With the number of positive cases and infections increasing at an exponential rate, mass public testing is important to rapidly identify, quarantine, and treat infected patients. Currently, the various RT-PCR tests and immunoassays are limited in availability and turnaround time in providing results back to patients. Being tied-up with large numbers of patients coming in daily, healthcare providers are cautious and stringent on limiting and choosing who gets tested. In addition, after patient sample collection, several more days are required for patient sample pickup and transport to centralized lab sites, batched patient testing, and finally result generation and reporting back to doctors for patient follow-ups. Turnaround times can therefore take up to two weeks, depending on location and demand. The inability to rapidly test large numbers of patients has been a limiting factor in preventing the spread of SARS-CoV-2, as numbers of potential positive people, many of whom have minor or no symptoms, continue to roam around untested. Therefore, it is critical to develop tests that are not limited to testing at large centralized or near-patient labs.
POC tests can help control the spread of the virus; moreover, they are essential during a pandemic because they provide a rapid and easy solution for widespread testing of the general public. POC tests are typically designed such that users can easily use the test without the need for a trained professional. They are also designed to not require complicated machinery or devices and can ideally be used in an at-home setting by consumers. Anyone and everyone can therefore be tested anywhere and everywhere. Given that POC tests provide users the ability to perform all steps of the test, from sample collection to test result readout, users can know, within minutes, whether their test result is positive or negative. This allows the user themselves to immediately act to seek professional help, instead of waiting weeks for test results.
As of June 17, 2020, there are currently four SARS-CoV-2 tests that have received EUAs for use under patient-care settings (FDA, 2020a). One of these tests is Cepheid's Xpert Xpress SARS-CoV-2 test mentioned earlier, but for use on Cepheid's GeneXpert Xpress System compact and simplified system used in physician offices and clinics. The other three tests are Abbott Diagnostic's ID NOW COVID-19 Test (Abbott Diagnostics Scarborough, 2020), Mesa Biotech's Accula SARS-CoV-2 Test (Mesa Biotech Inc., 2020), and Cue Health's Cue COVID-19 Test (Cue Health, 2020b, Cue Health, 2020a). Abbott Diagnostic's ID NOW COVID-19 Test relies on isothermal nucleic acid amplification, targeting a unique region of the RNA-dependent RNA polymerase (RdRP) gene of SARS-CoV-2. Isothermal amplification, unlike PCR, enables amplification at a constant temperature using two or three sets of primers and a polymerase with high strand displacement activity, avoiding the need for thermal cycling. To achieve comparable specificity, four different primers are used to amplify six distinct regions on the target gene. As a result, isothermal amplification can achieve higher amounts of nucleic acid copies in a shorter amount of time compared to standard PCR. The ID NOW COVID-19 Test provides results in 13 minutes or less from throat, nasal or NP swab samples, with reported analytical sensitivity of 125 copies/mL. To evaluate the clinical performance of the test, contrived NP swab samples with spiked purified viral RNA containing target SARS-CoV-2 sequences at concentrations about 2x and 5x LoD, as well as negative NP swab samples, were tested. Clinical sensitivity at 2x LoD is 100% (20/20; 83.9%–100%), and 100% (10/10; 72.3%–100%) at 5x LoD. Clinical specificity is 100% (30/30; 88.7%–100%). The ID NOW COVID-19 Test is performed on the ID NOW Instrument, which gives a simple method for mixing sample with test reagents and transferring to the test base through a cartridge.
Mesa Biotech's Accula SARS-CoV-2 Test is a combination of RT-PCR and lateral flow immunoassay (Mesa Biotech Inc, 2020). It targets the N gene of SARS-CoV-2 from nasal and throat samples. The test is performed on the Accula Dock or Silaris Dock and is relatively straightforward to use. The sample swab is dipped into a buffer vial and transferred into a test cassette, containing all reaction reagents, via a specially designed tiny bulb pipette. The test cassette sits in the dock for about 30 minutes, after which the test is completed and processed, and results can be interpreted visually. There are three lines: the internal positive process control line, the SARS-CoV-2 test line, and the internal negative process control line. The appearance of any shade of blue at the SARs-CoV-2 test line indicates a positive result. However, any appearance of blue at the negative process control line indicates an invalid test, and the test must be performed again. The reported analytical sensitivity is 200 copies/mL. To evaluate the clinical performance of the test, 30 contrived positive samples and 30 negative samples were tested, resulting in a clinical sensitivity of 100% (30/30) and clinical specificity of 100% (30/30).
The other more recent test is Cue Health's Cue COVID-19 Test, which is a rapid, portable assay that delivers results to a mobile phone in less than 25 minutes. Similar to Abbott's test, Cue's test also uses isothermal amplification on nasal swabs, but it detects the SARS-CoV-2 N gene. Additionally, Cue's disposable POC test cartridge forms a connected diagnostic platform with a mobile phone that enables a patient to have convenient access to their health information.
While all four tests have comparable sensitivity and specificity, Abbott and Cue Health's tests both use isothermal amplification and are consequently easier to use, have shorter turnaround times, and consume less power compared to Mesa Biotech and Cepheid's tests that use RT-PCR. These reasons suggest that isothermal amplification is a stronger method for POC pathogen detection compared to RT-PCR; however, it is harder to detect genes in multiplex with isothermal amplification (Lucigen, 2020). Accordingly, Cepheid's Xpert Xpress Test that uses RT-PCR has the advantage of being the only diagnostic with FDA-EUA approval for patient care settings that can detect more than one SARS-CoV-2 gene (both N2 and E), offering an additional assurance to the diagnosis. When comparing all four of the tests (Table 6 ), Cue Health's test is most promising for POC applications due to its portability, ease of use and mobile connectivity to provide patients personalized health information at their fingertips. With the growing development and availability of rapid POC tests such as Cue Health's, mass public testing can expedite a response to those who need it and prevent unnecessary spread of infections, while helping off-load the burden of healthcare providers and workers. Note that FDA-EUA is not equivalent to FDA cleared; the EUA tests usually need to be performed in CLIA certified high-complexity (H) or moderate-complexity (M) labs, or sites certified for performing CLIA-waived (W) testing, according to FDA regulation. EUA status is also temporary, so it is desirable for the EUA tests to eventually become FDA cleared under normal regulatory pathways to enable full-fledged long term usage.
Table 6.
Comparision of Major Diagnostics approved for Patient-Care settings for COVID-19 with FDA-EUA Approval.
| Manufacturer | Test | Test Type | SARS-CoV-2 Gene Biomarkers | Sample Accepted | Time to Result | Sensitivity/Specificity | References |
|---|---|---|---|---|---|---|---|
| Mesa Biotech | Accula SARS-CoV-2 Test | RT-PCR | N | Throat swab, Nasal swab | ~30 min | 100% (30/30)/100% (30/30) | https://www.mesabiotech.com/coronavirus |
| Abbott | ID NOW COVID-19 test | Isothermal DNA amplifciation | RdRp | NP swab, OP swab | <13 min | 2X LOD (100%, 20/20)5X LOD (100%, 20/20)/100% (30/30) | https://www.alere.com/en/home/product-details/id-now-covid-19.html |
| Cepheid | Xpert Xpress SARS-CoV-2 test for use on GeneXpert Xpress System | RT-PCR | N2, E | NP swab, Nasal swab, mid-turbinate swabs, OP swab | ~40 min | 100% (30/30)/100% (35/35) | https://www.cepheid.com/coronavirus |
| Cue Health | Cue COVID-19 Test | Isothermal DNA amplification | N | Nasal swabs | ~20 min | 1-2X LoD (19/20, 95%) 5X LoD (4/4, 100%) 10X LoD (4/4100%) 50X LoD (2/2100%)/30/30,100% | https://www.cuehealth.com/covid-19 |
4.5. Comparison and discussion of major commercialized diagnostic products
While there are many COVID-19 diagnostic products in the market with FDA-EUA approval, they can broadly be grouped into diagnostic tests and antibody tests. Diagnostic tests focus on nucleic acid or viral antigen detection, and are primarily utilized for active COVID-19 diagnoses. Antibody tests measure antibodies against SARS-CoV-2 in patients; these tests are utilized to glean who may still be at risk, and more broadly assess the prevalence of COVID-19 in a community. Some of the primary customers for diagnostic tests are hospitals, drive-through clinics, and academic institutions interested in providing COVID-19 diagnoses for the public; on the other hand, some of the main customers for antibody tests are healthcare providers, laboratories, public health staff, and community clinics interested in mass screening efforts to determine population prevalence.
One of the main reasons there are so many SARS-CoV-2 nucleic acid detection tests in the market is that there was never a standard protocol set in place (The Conversation US, Inc, 2020). After the CDC's initial test proved to be faulty, and test development restrictions were lifted at the end of February, many academic institutions and companies took the initiative to develop their own tests to help the country increase testing efforts. As a result, many of these tests detect different genes: while the CDC's test detects N1 and N2 regions of the N gene and the RNase P (RP) gene, Roche's cobas ® SARS-CoV-2 Test detects the ORF1 genes, and Cepheid's Xpert SARS Xpress test detects part of the envelope (E) gene. While some analyses have been conducted on comparing the performance of these tests in accurately detecting cases of COVID-19, ultimately, it has been shown that all of the kits granted an EUA status can be used for accurate diagnoses (Sethuraman et al., 2020). Some important factors to choose when considering one test over another are differences in turnaround time, test kit supply, and the population to be analyzed. For example, if aiming to detect active infections in individuals with perhaps lower viral loads, it is important to have a test with comparatively higher sensitivity in order to detect lower copy numbers of viral particles. Due to the limited availability of the testing kits, it is convenient to have molecular tests from many distributors to maintain constant supply.
There are many antibody tests circulating the market, including those that detect IgG and IgM, IgG only, and total antibody. Currently, the CDC reports that there is not a major advantage between serological assays that detect IgG specific to SARS-CoV-2, IgG and IgM specific to SARS-CoV-2, or total antibody (CDC, 2020d); however, it has also been reported that total antibody is comparatively the most sensitive serological marker, as it increases from the second week of symptom onset, whereas IgM and IgG have higher levels only in the second and third week of COVID-19 infection (Sethuraman et al., 2020). When choosing an FDA-EUA serological test, the CDC recommends that researchers choose tests with high specificity (99.5% or greater) to minimize false positive results (CDC, 2020f). Choosing a testing population with a higher likelihood of previous SARS-CoV-2 exposure also increases the positive predictive value of the test.
4.6. Cost of major commercialized diagnostic products via healthcare providers
Many of the manufacturers selling FDA-EUA approved diagnostic products only have test prices available via inquiry. However, the major customers of these diagnostic tests, such as healthcare providers (hospitals and clinical laboratories), provide cash price of testing costs on their websites. While these prices can vary drastically by provider, in general, the cost of an RT-PCR lab test is around $50-$200 (New York Times, 2020), and the cost of an antibody lab test is around $50-$150 (Sonora Quest Laboratories, 2020). However, one of the largest economic stimulus bills in the U.S., the Coronavirus Aid, Relief, and Economic Security (CARES) Act requires that group health plans and health insurances cover the cost of diagnostic tests for the detection of active SARS-CoV-2 infections, along with tests that detect antibodies against SARS-CoV-2 (Brookings Institution, 2020). It is still being debated whether full coverage should be provided by insurance only if the test is deemed medically necessary for the patient: i.e, they have presented with COVID-19 symptoms and have been referred by a medical provider (npr, 2020). In general, the clinical labs or hospitals conducting the tests will directly bill the test cost to an insurance provider; if uninsured, these costs go to a government program such as Medicare or Medicaid if the patient qualifies.
5. Future perspectives
As the COVID-19 pandemic is rapidly spreading, one major focus of future work will be continuing development of POC and home tests that do not require extensive training for operation, are easily deployable to outpatient settings and clinics, and are low-cost while still preserving the accuracy of diagnosis. POC tests have a lower barrier to implementation than lab-based tests: if FDA cleared, POC tests don't require a trained professional to operate, so users have the power to perform all the steps of the test on their own. In this way, users can know their results within minutes and seek professional help sooner, instead of waiting longer for results from a lab-based test. As we prepare for the possibility of a second wave, POC tests are more conducive to mass testing compared to lab-based tests, so we can rapidly provide accurate identification of not only symptomatic SARS-CoV-2 infected patients, but also provide detection of early infections or asymptomatic individuals. To increase the throughput and scalability of the number of tests that can be run, more POC tests should be combined with automated sample processing systems in the future, allowing more patients to get diagnoses in a timely manner.
Novel biological sensors, or biosensors, should also be developed as rapid, sensitive, and low-cost POC diagnostic devices for SARS-CoV-2 detection in the future (Nelson et al., 2020). These analytical systems consist of a transducer and immobilized biological component: the biological component recognizes a target biomarker in the sample and the transducer converts the corresponding biological signal into an electrical signal (Bhalla et al., 2016). Some common biosensors include electrochemical sensors, enzyme-based sensors, field-effect transistor (FET)-based biosensors, immunosensors, magnetic biosensors, and DNA biosensors. Biosensors have previously been used for infectious disease detection: for example, Layqah and Eissa used an electrochemical sensor with a gold-coated array of carbon electrodes to detect the spike protein of MERS-CoV in 20 minutes (Layqah and Eissa, 2019). Additionally, magnetic bionsensors such as giant magnetoresistive (GMR) sensors have been used for influenza detection: Wu et al. developed a portable GMR device that can detect influenza A nucleoprotein after the addition of magnetic nanoparticles in less than 10 minutes (Wu, K. et al., 2007). For SARS-CoV-2, FET-based biosensors have been developed to detect the SARS-CoV-2 spike protein without any sample pretreatment or labeling (Seo et al., 2020). Future biosensing devices for SARS-CoV-2 should also have limited sample processing steps, and be able to deliver quick and accurate POC diagnoses.
A future avenue also lies in developing antigen tests, which test for the presence of the SARS-CoV-2 proteins directly. Therefore, antigen tests can directly provide COVID-19 diagnoses, giving individuals a convenient way to get faster results at a lower price compared to RT-PCR assays. An immunoassay can be used for these tests, however, this time the plate is coated with antibodies specific to SARS-CoV-2, and the sample of interest is the S or N protein of SARS-CoV-2. The challenge here lies in developing and synthesizing the SARS-CoV-2 antigen-specific antibody for the test. As of June 17, 2020, Quidel has the only antigen test for SARS-CoV-2 with FDA-EUA status: their SOFIA SARS Antigen Fluorescent Immunoassay qualitatively detects the N protein in 15 minutes (Quidel Corporation, 2020b). However, as the N protein is conserved between SARS-CoV and SARS-CoV-2, Quidel's test is unable to distinguish between these two similar viruses. To eliminate this source of cross-reactivity, future antigen tests developed might target the SARS-CoV-2 S protein. Importantly, antigen-based tests can be useful as rapid diagnostic POC tests to inform duration of quarantine and social-distancing measures for COVID-19 patients with less disease severity (Cheng et al., 2020).
As the disease progresses through the population, it will be important to develop testing systems that can provide quantitative diagnoses, rather than merely qualitative ‘yes or no’ results regarding SARS-CoV-2 or IgG and IgM presence. It is imperative that we quantify the viral load to have a better sense of where a patient is in disease progression after symptom onset. Similarly, by quantifying the amount of SARS-CoV-2 specific IgG and IgM antibodies present, we can determine if a patient or a population has acquired immunity, and if so, exactly how much. This will be beneficial in identifying strong responders for providing convalescent sera.
Another future effort will focus on measuring SARS-CoV-2 infection with a host transcriptional response signature. In the past, these diagnostic profiles have been determined through a meta-analysis of publicly available data, resulting in a group of up to ten biomarker genes whose expression levels in the host are different at a specific disease state (Zhai et al., 2015; Bradley et al., 2018; Steinbrink et al., 2019). Adding on to their clinical value, these signatures have also separated viral infections from bacterial (Andres-Terre et al., 2015), correctly classified symptomatic patients without the disease from asymptomatic patients with the diseases (Andres-Terre et al., 2015), and are not confounded by gender, race, and age (Sweeney et al., 2016). Having a gene expression signature would be valuable for the detection of SARS-CoV-2 so that we can correctly distinguish patients with the disease even when the viral RNA load decreases. Moreover, even if SARS-CoV-2 mutates slightly, the gene signature should correctly classify virus presence. One group has found that there is a transcriptional response to SARS-CoV-2 that is weaker than the respiratory syncytial virus (RSV) response but stronger than the influenza response (Blanco-Melo et al., 2020). While this study has not been peer reviewed, it suggests that an antiviral response exists, paving the way for researchers to investigate further. As vaccine treatments are progressing, a SARS-CoV-2 genetic signature could also be generated to classify vaccine efficacy (Legutki and Johnston, 2013). In current vaccine efficacy trials, it takes a few months after an individual has been given the vaccine to determine and validate if he or she is mounting the appropriate immune response through antibody production (Plotkin, 2010). Instead, if there were a specific gene signature expressed a few days after a vaccine was provided, researchers would know sooner if a vaccine was effective. This could expedite vaccine trials, allowing a SARS-CoV-2 vaccine to reach the population sooner. One of the main challenges with creating such a signature is that a large population RNA-sequencing data needs to be curated first from patients with the COVID-19 disease, which will take time. Additionally, gene expression measurements involve isolating mRNA from peripheral blood mononuclear cells (PBMCs), reverse transcribing to cDNA, and amplifying the cDNA with the primers of interest. This will add on to the sample processing time, and requires a skilled technician to perform the task. Nevertheless, this is a promising quantitative approach to disease classification, and can hopefully improve the accuracy of future diagnoses.
6. Summary and conclusions
In this review, we have covered diagnostics for measuring the presence of SARS-CoV-2, which can broadly be grouped into two categories: nucleic acid detection and antibody detection. The standard of care for nucleic acid detection is RT-PCR, and this is currently being used to identify positive and negative cases of COVID-19 by testing for SARS-CoV-2 viral RNA in a patient swab sample. The most common swab types are nasopharyngeal swabs and oropharyngeal swabs. We have also covered antibody detection through serological assays, most commonly ELISA, in which SARS-CoV-2 specific IgG and IgM antibodies are detected to measure general immunity to SARS-CoV-2. This is important not only to examine the infection severity and chance for successful recovery in an individual, but also to determine if herd immunity has been reached for an overall population. Future work in this field will include quantitative testing approaches in nucleic acid and antibody/antigen assays and the development of a SARS-CoV-2 specific genetic signature. As the situation is rapidly evolving and we are learning more about this disease every day, we are more broadly advancing the field of infectious disease diagnostics.
CRediT authorship contribution statement
Neeraja Ravi: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Dana Lee Cortade: Investigation, Writing - original draft, Writing - review & editing. Elaine Ng: Investigation, Writing - original draft, Writing - review & editing. Shan X. Wang: Writing - review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements:
E.N. would like to acknowledge support from the Cancer-Translational Nanotechnology Training (Cancer-TNT) Program. D.L.K would like to acknowledge support from the Stanford Graduate Fellowship. This work is in part supported by National Institutes of Health grants R01 AI125197 and U54CA199075.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112454.
Contributor Information
Neeraja Ravi, Email: neravi@stanford.edu.
Dana L. Cortade, Email: dcortade@stanford.edu.
Elaine Ng, Email: elaineng@stanford.edu.
Shan X. Wang, Email: sxwang@stanford.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abbott Diagnostics Scarborough, Inc. ID NOW COVID-19. 2020. https://www.fda.gov/media/136525/download
- Abbott, Inc SARS-COV-IMMUNOASSAYS. 2020. https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease/sars-cov-2
- Abbott Laboratories . 2020. ALINITY m SARS-COV-2 ASSAY.https://www.molecular.abbott/us/en/products/infectious-disease/alinity-m-sars-cov-2-assay [Google Scholar]
- Amanat F., Nguyen T., Chromikova V., Strohmeier S., Stadlbauer D., Javier A., Jiang K., Asthagiri-Arunkumar G., Polanco J., Bermudez-Gonzalez M., Caplivski D., Cheng A., Kedzierska K., Vapalahti O., Hepojoki J., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020;2020 doi: 10.1101/2020.03.17.20037713. 03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Society of Microbiology . 2020. ASM Expresses Concern about Coronavirus Test Reagent Shortages.https://asm.org/Articles/Policy/2020/March/ASM-Expresses-Concern-about-Test-Reagent-Shortages [Google Scholar]
- Andres-Terre M., McGuire H.M., Pouliot Y., Bongen E., Sweeney T.E., Tato C.M., Khatri P. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory Viruses. Immunity. 2015;43(6):1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autobio Diagnostics Co, Ltd . 2020. Anti-SARS-CoV-2 Rapid Test.https://www.fda.gov/media/137364/download [Google Scholar]
- Avellino Lab USA, Inc AvellioCoV2 test. 2020. https://www.fda.gov/media/136453/download
- Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques [Preprint] Microbiology. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- Becton Dickinson, Company . 2020. BioGX SARS-CoV-2 Reagents for BD MAX System.https://www.fda.gov/media/136653/download [Google Scholar]
- Bendavid E., Mulaney B., Sood N., Shah S., Ling E., Bromley-Dulfano R., Lai C., Weissberg Z., Saavedra R., Tedrow J., Tversky D., Bogan A., Kupiec T., Eichner D., Gupta R., Ioannidis J., Bhattacharya J. COVID-19 antibody seroprevalence in santa clary county, California. medRxiv. 2020;2020 doi: 10.1101/2020.04.14.20062463. 04.14.20062463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BGI Genomics Co Ltd Real-time fluorescent RT-PCR kit for detecting SARS-2019-nCoV. 2020. https://www.fda.gov/media/136472/download
- Bhalla N., Jolly P., Formisano N., Estrela P. Introduction to biosensors. Essays Biochem. 2016;60(1):1–8. doi: 10.1042/EBC20150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bio-Rad Laboratories . 2020. Platelia SARS-CoV-2 Total Ab Assay #72710.https://www.bio-rad.com/en-us/sku/72710-platelia-sars-cov-2-total-ab-assay?ID=72710 [Google Scholar]
- BioFire Defense L. 2020. BioFire ® COVID-19 Test.www.biofiredefense.com/covid-19test [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R.A., tenOever B.R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems [Preprint] Microbiology. 2020 doi: 10.1101/2020.03.24.004655. [DOI] [Google Scholar]
- Bradley T., Ferrari G., Haynes B.F., Margolis D.M., Browne E.P. Single-cell analysis of quiescent HIV infection reveals host transcriptional profiles that regulate proviral latency. Cell Rep. 2018;25(1):107–117. doi: 10.1016/j.celrep.2018.09.020. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- British Society for Immunology . 2020. Enzyme-linked Immunosorbent Assay (ELISA)https://www.immunology.org/public-information/bitesized-immunology/experimental-techniques/enzyme-linked-immunosorbent-assay [Google Scholar]
- Brookings Institution . 2020. How the CARES Act Affects COVID-19 Test Pricing.https://www.brookings.edu/blog/usc-brookings-schaeffer-on-health-policy/2020/04/09/how-the-cares-act-affects-covid-19-test-pricing/ [Google Scholar]
- Casadevall A., Pirofski L. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.https://www.fda.gov/media/134922/download [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. COVID-19 Pandemic Planning Scenarios.https://www.cdc.gov/coronavirus/2019-ncov/hcp/planning-scenarios.html [Google Scholar]
- CDC . 2020. FAQs on COVID-19 Testing at Laboratories.https://www.cdc.gov/coronavirus/2019-ncov/lab/testing-laboratories.html [Google Scholar]
- CDC . 2020. Frequently Asked Questions about Coronavirus (COVID-19) for Laboratories.https://www.cdc.gov/coronavirus/2019-ncov/lab/faqs.html [Google Scholar]
- CDC . 2020. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-at-higher-risk.html [Google Scholar]
- CDC . 2020. Interim Guidelines for COVID-19 Antibody Testing.https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html [Google Scholar]
- CDC . 2020. Overview of Testing for SARS-CoV-2.https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html [Google Scholar]
- CDC . 2020. Situation Summary.https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/summary.html [Google Scholar]
- CDC . 2020. Testing for COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/testing.html [Google Scholar]
- CDC . 2020. Using Antibody Tests for COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests.html [Google Scholar]
- Cellex Inc qSARS-CoV-2 IgG/IgM rapid test. 2020. https://www.fda.gov/media/136625/download
- Cepheid . 2020. Xpert Xpress SARS-CoV-2 Test (POC)https://www.fda.gov/media/136315/download [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chembio Diagnostics Systems, Inc . 2020. DPP® COVID-19 IgM/IgG System.https://www.fda.gov/media/136963/download [Google Scholar]
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C. Diagnostic testing for severe acute respiratory syndrome–related coronavirus-2: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-1301. [Epub ahead of print 13 April 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue Health . 2020. The Cue Health Monitoring System.https://www.cuehealth.com/#product [Google Scholar]
- Cue Health . 2020. Cue Health Awarded $13 Million for Rapid, Portable, Point-of-Care COVID-19 Test by U.S. Department of Health and Human Services Biomedical Advanced Research and Development Authority (“BARDA”)https://www.cuehealth.com/news-listing/2020/3/26/cuehealthbardaCOVID-19 [Google Scholar]
- DiaSorin Molecular L.L.C. 2020. SimplexaTM COVID-19 Direct.https://www.fda.gov/media/136286/download [Google Scholar]
- Dutta N., Mazumdar K., Gordy J. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94(13) doi: 10.1128/jvi.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B.E., Lim K.G.E., Chong V.C.L., Chan S.S.W., Ong K.H., Kuperan P. COVID-19 and mycoplasma pneumoniae coinfection. Am. J. Hematol. 2020 doi: 10.1002/ajh.25785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2020. Emergency Use Authorizations.https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations [Google Scholar]
- FDA . 2020. MOU 225-14-017.https://www.fda.gov/about-fda/domestic-mous/mou-225-14-017#:~:text=FDA%20and%20CDC%20are%20sister,different%20statuary%20mandates%20and%20responsibilities [Google Scholar]
- FDA . 2020. FAQs on Diagnostic Testing for SARS-CoV-2.https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-diagnostic-testing-sars-cov-2 [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Wang J. Clinical Infectious Diseases; 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hologic, Inc . 2020. Panther Fusion SARS-CoV-2 Assay.https://www.fda.gov/media/136156/download [Google Scholar]
- Indwiani A., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2020;14(4) doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Informa . 2020. COVID-19 Puts the World Health Organization's Emergency Use Listing (EUL) to Work.”.https://www.mddionline.com/regulatory-and-compliance/covid-19-puts-world-health-organizations-emergency-use-listing-eul-work [Google Scholar]
- Ipsum Diagnostics LLC . 2020. COV-19 IDx Assay.https://www.fda.gov/media/136621/download [Google Scholar]
- Johns Hopkins Center for Health Security . 2020. Comparison of National RT-PCR Primers, Probes, and Protocols for SARS-Cov-2 Diagnostics.https://www.centerforhealthsecurity.org/resources/COVID-19/COVID-19-fact-sheets/200410-RT-PCR.pdf [Google Scholar]
- Johns Hopkins University . 2020. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)https://coronavirus.jhu.edu/map.html [Google Scholar]
- Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60(1):111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LabCorp . LABORATORY CORPORATION OF AMERICA; 2020. COVID-19 RT-PCR TEST.https://www.fda.gov/media/136151/download [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layqah L., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchimica Acta. 2019;186(4) doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legutki J.B., Johnston S.A. Immunosignatures can predict vaccine efficacy. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110(46):18614–18619. doi: 10.1073/pnas.1309390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;1–7 doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- Liang T. The First Affiliated Hospital, Zhejiang University School of Medicine; Hangzhou, China: 2020. Handbook of COVID-19 Prevention and Treatment. [Google Scholar]
- Lin C., Xiang J., Yan M., Li H., Huang S., Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19) Clin. Chem. Lab. Med. 2020;58(7):1089–1094. doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- Long Q., Tang X., Shi Q., Li Q., Deng H., Yuan J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucigen . 2020. Loop-Mediated Isothermal Amplification (LAMP) Primer Design and Assay Optimization.https://www.lucigen.com/docs/slide-decks/Loop-Mediated-Isothermal-Amplification-Design-Primers-Webinar-March-2018.pdf [Google Scholar]
- Luminex Molecular Diagnostics Inc . 2020. NxTAG ® CoV Extended Panel Assay Package Insert.https://www.fda.gov/media/136500/download [Google Scholar]
- Mesa Biotech Inc . 2020. Accula SARS-CoV-2 Test.https://www.fda.gov/media/136355/download [Google Scholar]
- MonobindInc . 2020. CLIA Advantages.http://www.monobind.com/Additional-Info/Assays-CLIA-Advantages [Google Scholar]
- Nelson P., Rath B., Fragkou P., Antalis E., Tsiodras S., Skevaki C. Current and future point-of-care tests for emerging and new respiratory Viruses and future perspectives. Front. Cell. and Infection Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeuMoDx Molecular I. 2020. NeuMoDxTM SARS-CoV-2 Assay INTENDED USE.https://www.fda.gov/media/136565/download [Google Scholar]
- New York Times . 2020. Most Coronavirus Tests Cost about $100. Why Did One Cost $2,315?https://www.nytimes.com/2020/06/16/upshot/coronavirus-test-cost-varies-widely.html [Google Scholar]
- npr . 2020. Insurers May Only Pay for Coronavirus Tests when They're 'Medically Necessary.https://www.npr.org/sections/health-shots/2020/06/19/880543755/insurers-may-only-pay-for-coronavirus-tests-when-theyre-medically-necessary [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Infectious Diseases (Except HIV/AIDS) 2020. SARS-CoV-2 specific antibody responses in COVID-19 patients [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- OraSure Technologies Development. 2020. https://www.orasure.com/innovation/innovation-in-development.asp
- Ortho-Clinical Diagnostics, Inc . 2020. Instructions for Use: VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total 619 9922 Reagent Pack.https://www.fda.gov/media/136967/download [Google Scholar]
- Payne D.C., Iblan I., Rha B., Alqasrawi S., Haddadin A., Al Nsour M., Alsanouri T., Ali S.S., Harcourt J., Miao C., Tamin A., Gerber S.I., Haynes L.M., Al Abdallat M.M. Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg. Infect. Dis. 2016;22(10):1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PerkinElmer I. 2020. PerkinElmer® New Coronavirus Nucleic Acid Detection Kit V 2.0.https://www.fda.gov/media/136410/download [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C., Weimer E., Scherer E., Rouphael N., Edupuganti S., Weiskopf D., Tse L., Hou Y., Margolis D., Sette A., Collins M., Schmitz J., Baric R., de Silva A. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primerdesign Ltd . 2020. Genesig ® Real-Time PCR Assay.https://www.fda.gov/media/136309/download [Google Scholar]
- QIAGEN GmbH . 2020. QIAstat-Dx ® Respiratory SARS-CoV-2 Panel Instructions for Use (Handbook)https://www.fda.gov/media/136571/download [Google Scholar]
- Quest Diagnostics Infectious Disease I. 2020. Quest SARS-CoV-2 rRT-PCR.https://www.fda.gov/media/136231/download [Google Scholar]
- Quidel Corporation . 2020. Lyra® SARS-CoV-2 Assay.https://www.fda.gov/media/136227/download [Google Scholar]
- Quidel Corporation . 2020. Sofia SARS Antigen FIA.https://www.quidel.com/immunoassays/rapid-sars-tests/sofia-sars-antigen-fia [Google Scholar]
- Roche Diagnostics . 2020. Elecsys® Anti-SARS-cov-2.https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html#productInfo [Google Scholar]
- Roche Molecular Systems . 2020. I. Cobas ® SARS-CoV-2 Qualitative Assay.https://www.fda.gov/media/136049/download [Google Scholar]
- ScienceDaily . 2020. Super-Potent Human Antibodies Protect against COVID-19 in Animal Tests: Scientists Isolate Powerful Coronavirus-Neutralizing Antibodies from COVID-19 Patients and Successfully Test in Animals.https://www.sciencedaily.com/releases/2020/06/200615140840.htm 2020. [Google Scholar]
- Seo G., Lee G., Kim M., Baek S., Choi M., Ku K., Lee C., Jun S., Park D., Kim H., Kim S., Lee J., Kim B., Park E., Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. J. Am. Med. Assoc. 2020;323(22):2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Siemens Medical Solutions . 2020. Inc. SARS-CoV-2 Total Assay.https://www.siemens-healthineers.com/en-us/laboratory-diagnostics/assays-by-diseases-conditions/infectious-disease-assays/cov2t-assay#The_Scale [Google Scholar]
- Sigma-Aldrich . CLIA/CLEIA) Development; 2020. Complete Solutions for IVD Chemiluminescent Immunoassay.https://www.sigmaaldrich.com/technical-documents/articles/ivd-immunoassay/clia-chemiluminescent-immunoassay-development.html [Google Scholar]
- Sonora Quest Laboratories . 2020. Coronavirus: what You Need to KNOW.https://www.sonoraquest.com/covid-19-information-for-patients/ [Google Scholar]
- Steinbrink J.M., Hua K., Myers R., Johnson M.D., Seidelman J., Tsalik E.L., Tsalik E.L., Alexander B.D., McClain M.T. 2885. A host transcriptional signature for accurate diagnosis of candidemia in the hospital setting. Open Forum Infectious Dis. 2019;6(Suppl. ment_2) doi: 10.1093/ofid/ofz359.163. S76–S76. [DOI] [Google Scholar]
- Sweeney T.E., Braviak L., Tato C.M., Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. The Lancet. Respir. Med. 2016;4(3):213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Conversation US Inc . 2020. There Are Many COVID-19 Tests in the US – How Are They Being Regulated?https://theconversation.com/there-are-many-covid-19-tests-in-the-us-how-are-they-being-regulated-134783 [Google Scholar]
- Thermo Fisher Scientific . 2020. TaqPath TM COVID-19 Combo Kit.https://www.fda.gov/media/136112/download [Google Scholar]
- Thermo Fisher Scientific . 2020. The Basics: RNA Isolation.https://www.thermofisher.com/us/en/home/references/ambion-tech-support/rna-isolation/general-articles/the-basics-rna-isolation.html [Google Scholar]
- Tyrrell D.A.J., Myint S.H. Coronaviruses. In: Baron S., editor. Medical Microbiology. fourth ed. University of Texas Medical Branch at Galveston; 1996. http://www.ncbi.nlm.nih.gov/books/NBK7782/ [PubMed] [Google Scholar]
- Vogel G. New blood tests for antibodies could show true scale of coronavirus pandemic. Science. 2020;80– doi: 10.1126/science.abb8028. [DOI] [Google Scholar]
- Wadsworth Center, N.Y.S. D.of P.H.CDC . 2020. New York SARS-CoV-2 Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel.https://www.fda.gov/media/135847/download [Google Scholar]
- Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J. Virol Methods. 2020;283 doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kang H., Liu X., Tong Z. Combination of RT‐qPCR testing and clinical features for diagnosis of COVID‐19 facilitates management of SARS‐CoV‐2 outbreak. J. Med. Virol. jmv. 2020;25721 doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Advice on the Use of Point-Of-Care Immunodiagnostic Tests for COVID-19.https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 [Google Scholar]
- WHO . 2020. WHO Director-General's Remarks at the Media Briefing on 2019-nCoV on 11 February 2020.https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 [Google Scholar]
- WHO . 2020. WHO Director-General's Remarks at the Media Briefing on 2019-nCoV on 11 March 2020.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 [Google Scholar]
- WHO . 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases.https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 [Google Scholar]
- Wu L.-P., Wang N.-C., Chang Y.-H., Tian X.-Y., Na D.-Y., Zhang L.-Y., Zheng L., Lan T., Wang L.-F., Liang G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13(10):1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Klein T., Krishna V., Su D., Perez A., Wang J. Portable GMR handheld platform for the detection of influenza A virus. ACS Sens. 2017;2(11):1594–1601. doi: 10.1021/acssensors.7b00432. [DOI] [PubMed] [Google Scholar]
- Xing Q., Li G., Xing Y., Chen T., Li W., Ni W., Deng K., Gao R., Chen C., Gao Y., Li Q., Yu G., Tong J., Li W., Hao G., Sun Y., Zhang A., Wu Q., Li Z., Pan S. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3550013. [DOI] [Google Scholar]
- Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li J., Yang Q., Zhao J., Zhang Z., Liu Y. 2020. Evaluating the Accuracy of Different Respiratory Specimens in the Laboratory Diagnosis and Monitoring the Viral Shedding of 2019-nCoV Infections [Preprint]. Infectious Diseases (Except HIV/AIDS) [DOI] [Google Scholar]
- Zhai Y., Franco L.M., Atmar R.L., Quarles J.M., Arden N., Bucasas K.L., Wells J.M., Niño D., Wang X., Zapata G.E., Shaw C.A., Belmont J.W., Couch R.B. Host transcriptional response to influenza and other acute respiratory viral infections – a prospective cohort study. PLoS Pathog. 2015;11(6) doi: 10.1371/journal.ppat.1004869. 8. 9, 386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.