Abstract
Background
Atrial fibrillation (AF) is at least partially heritable, affecting 2–3% of the population in Europe and the USA. However, a substantial proportion of heritability is still lacking. In the present study, we aim to identify potential crucial genes associated with AF through bioinformatic analyses of public datasets.
Methods
Microarray data sets of GSE115574, GSE31821, GSE79768, GSE41177 and GSE14975 from the Gene Expression Omnibus (GEO) database were retrieved. After merging all microarray data and adjusting batch effect, differentially expressed genes (DEGs) were identified. Functional enrichment analyses based on Gene Ontology (GO) resource, Kyoto Encyclopedia of Genes and Genomes (KEGG) resource, Gene Set Enrichment Analysis (GSEA), Reactome Pathway Database and Disease Ontology (DO) were carried out. Protein-protein interaction (PPI) network was constructed using the STRING database. Combined with aforementioned significant bioinformatics information, potential crucial genes were subsequently selected. The comparative toxicogenomics database (CTD) was used to explore the interaction between potential crucial genes and AF.
Result
We identified 27 of DEGs with gene expression fold change (FC) ≥ 1.5 or ≤ 2/3 (|log2 FC| ≥ 0.58) and 5 with FC ≥ 2 or ≤ 0.5 (|log2 FC| ≥ 1) of AF patients compared with sinus rhythm controls. The most significantly enriched pathway was regulation of insulin-like growth factor transport and uptake by insulin-like growth factor binding proteins. IGFBP2, C1orf105, FHL2, CHGB, ATP1B4, IGFBP3, SLC26A9, CXCR4 and HTR2B were considered the potential crucial genes. CTD showed CXCR4, IGFBP2, IGFBP3 and FHL2 had higher scores with AF.
Conclusions
The 9 potential crucial genes, especially CXCR4, IGFBP2, IGFBP3 and FHL2, may be associated with risk of AF. Our study provided new insights of AF into genetics, molecular pathogenesis and new therapeutic targets.
Keywords: Potential crucial genes, Atrial fibrillation, Bioinformatic gene analysis, Gene expression omnibus, Biomarkers
Background
Atrial fibrillation (AF) is the most common sustained arrhythmia and is one of the major causes of stroke, heart failure, sudden death, and cardiovascular morbidity in the world [1]. The estimated number of AF patients is 34 million worldwide, and gradually increasing with the aging of the population [2]. However, the pathophysiologic mechanism underlying many AF cases remains unclear, resulting in a lack of effective treatment [3]. Only a small number of AF patients can normalize heart rhythm by catheter ablation or cardiac surgery [4]. The higher prevalence and limited treatments of AF lead to substantial public health and economic burdens [5]. Therefore, it is necessary to improve our understanding of AF pathogenesis and to develop better screening methods for AF.
Epidemiological evidence over the last decade has identified that metabolic syndrome, pre-hypertension, obesity, obstructive sleep apnea, exercise, and dietary intake of stimulants are major risk factors for AF [4, 6]. However, only a small proportion of exposed individuals eventually developed AF, suggesting that a strong genetic component might be a risk factor contributing to the susceptibility of AF. From 2007 to 2017, genetic research in European, Asian, and African-American ancestry groups have reported 17 independent signals at 14 genomic regions, such as PITX2, ZFHX3, and PRRX1, associated with AF [7, 8]. In 2018, a preliminary genome-wide association study meta-analysis including over 93,000 AF cases and more than 1 million referents identified at least 134 genetic loci significantly associated with risk of AF [9]. However, combined with the results of the genetic studies thus far, genetic variation only accounts for 42% of the heritability for AF [9]. Notably, a substantial proportion of heritability is still lacking. One potential interpretation is that unidentified genes may partially contribute to the missing heritability. Therefore, there are still many related genes to be identified, which will help us better understand the pathogenesis of AF and facilitate the discovery of novel diagnostic biomarkers or therapeutic target.
In this study, we aimed to identify the potential crucial genes for AF through Gene Expression Omnibus (GEO) database using bioinformatic methods and to analyze their expression, function and interaction.
Methods
Atrial fibrillation datasets
Raw files of 5 registered microarray data sets, including GSE115574, GSE31821, GSE79768, GSE41177 and GSE14975 (Table 1), were downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). All of these datasets were obtained from the microarray platform of Affymetrix Human Genome U133 Plus 2.0 Array [HG-U133_Plus_2]. In each data set, only human left atrial appendage (LAA) samples from AF and sinus rhythm (SR) subjects were selected, and finally 46 AF and 31 SR samples were included for subsequent analyses.
Table 1.
Characteristics of datasets in this study
| GSE series | Platform | Total | AF | SR | Country | Contributors |
|---|---|---|---|---|---|---|
| GSE115574 | GPL570 | 29 | 14 | 15 | Turkey | Deniz GC et.al |
| GSE31821 | GPL570 | 6 | 4 | 2 | France | Morel E et.al |
| GSE79768 | GPL570 | 13 | 7 | 6 | Taiwan | Tsai F et.al |
| GSE41177 | GPL570 | 19 | 16 | 3 | Taiwan | Yeh Y et.al |
| GSE14975 | GPL570 | 10 | 5 | 5 | Germany | Adam O et.al |
GSE Gene Expression Omnibus; AF Atrial Fibrillation; SR Sinus Rhythm
Data preprocessing
Series matrix files were processed with ActivePerl 5.24.2 software (https://www.activestate.com/ products/perl/) to convert the gene probe IDs to gene symbol codes. Because GSE14975 was extracted using the Affymetrix Microarray Suite 5 (MAS5) algorithm, it was log base 2 transformed. After merging all microarray data, batch effects were adjusted by the “combat” function of “sva” package of R software using empirical Bayes frameworks [10]. Finally, normalized expression values according to the “normalizeBetweenArrays” function of the package of “limma” in R software so that the expression values have similar distribution across a set of arrays [11].
Identification of differentially expressed genes (DEGs)
To assess differential expression, using the “limma” package of R software, a linear model was fitted and a simple empirical Bayes model was used to moderate standard errors [11]. A moderated t-statistic and a log-odds of differential expression was computed for each contrast for each gene. The Benjamini and Hochberg (BH) method was performed to adjust P value to reduce the false positive error. A gene was defined as a DEG between the AF and SR sample, when the adjust P value was < 0.05 and the gene expression fold change (FC) value was ≥1.5 or ≤ 2/3 (|log2 FC| ≥ 0.58), which were visualized as Volcano plots and heat map plots. “ggplot” packages of R software was applied to generate box plots for genes which have the adjusted P value < 0.05 and the gene expression FC value ≥2 or ≤ 0.5 (|log2 FC| ≥ 1).
Functional and pathway enrichment analyses of DEGs
The Gene Ontology (GO) Resource (http://geneontology.org/) is a bioinformatics tool providing a framework and set of concepts for describing the functions of gene products from all organisms [12]. Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) is a database resource integrated the information of genomes, biological pathways, diseases and chemicals [13]. Gene Set Enrichment Analysis (GSEA) (http://software.broadinstitute.org/gsea/index.jsp) is a computational method for interpreting gene expression data based on molecular signature database [14]. The Reactome Pathway Database (https://reactome.org/) is pathway annotation database collecting the biological pathways and processes in the human [15]. The Disease Ontology (DO) (http://disease-ontology.org) represents a comprehensive knowledge base of 8043 inherited, developmental and acquired human disease [16]. Before performing enrichment analysis, human genome annotation package “org.Hs.eg.db” was used to convert gene symbol codes to Entrez ID. To better understand the biological function and characteristics, R software was used to perform enrichment analysis, with the “clusterProfiler” package for Go and KEGG enrichment analyses, the “ReactomePA” package for Reactome pathway analysis and the “DOSE” package for DO enrichment analysis. The “ggplot2”, “pathview” and “graphite” packages of R software were used to visualize the plots. GO terms and KEGG maps of biological functions associated with an adjusted P value < 0.05 and Q value < 0.05 was considered to be significantly enriched. GSEA v4.0.3 was applied for GSEA analysis. Using a permutation test 1000 times, the cutoff of the significance level nominal P-value, false discovery rate (FDR) Q-value and family wise-error rate (FWER) P-value were all chosen as 0.05 for the most significant pathways related to AF.
Protein-protein interaction (PPI) network and potential crucial genes analyses
The STRING database (http:/ /string-db.org/) was performed to construct a PPI network to reveal the generic organization principles of functional cell systems and to predict protein-protein interactions [17]. The Molecular Complex Detection (MCODE) of Cytoscape was carried out to module analyze and visualize the result of PPI network. Default parameters (degree cutoff ≥2, node score cutoff ≥2, K-core ≥2, and maxi-mum depth = 100) were used. To select potential crucial genes, we synthesized above significant bioinformatics information for subsequent analyses. DEGs were considered as potential crucial genes if they met the following one of inclusion criteria: (1) Adjust P value < 0.05 and |log2FC| ≥ 1; (2) P value < 0.05, |log2FC| ≥ 0.58 and enriched in biological functions. In addition, genes with the degree of connectivity larger than 5 in the PPI network were also included.
Identification of potential crucial genes associated with AF
The comparative toxicogenomics database (CTD, http://ctdbase.org/) integrated information including chemical-gene/protein interactions, chemical-disease and gene-disease relationships to develop hypotheses related to the mechanisms of disease [18]. The association between potential crucial genes and AF risk was analyzed using the data in CTD.
Results
Identification of DEGs
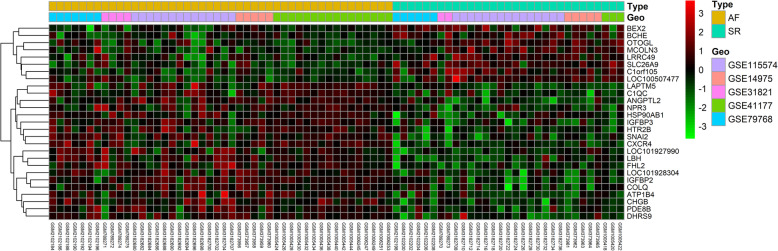
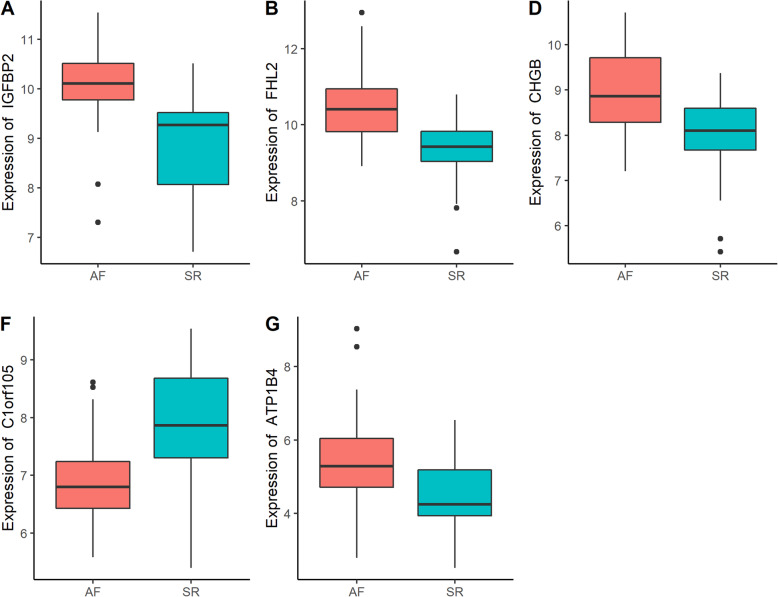
Gene expression levels of merged GEO series that have been adjusted batch effects were standardized and the results of pre- and post- standardized were presented in Supplementary Figure 1. The 54,675 probes corresponding to 21,654 genes in GSE115574, GSE31821, GSE79768, GSE41177 and GSE14975 datasets were identified and DEGs of AF were confirmed. Twenty-seven of DEGs with |log2 FC| ≥ 0.58 in LAA samples of AF patients compared with SR was identified, including 19 up-regulated genes and 8 down-regulated genes (Supplementary Table 1). Heatmap plot and Volcano plot of 27 DEGs enrolled in subsequent analyses was showed in Fig. 1 and Supplementary Figure 2. Using a screening criteria of |log2 FC| ≥ 1, there were 5 genes identified, with 4 of these genes being up-regulated and 1 down-regulated (Table 2). Boxplots for the 5 selected genes were shown in Fig. 2.
Fig. 1.
Heat map of DEGs in AF samples and SR samples. Each row represents a sample, and each column represents a single gene. Purple color represented AF samples, and blue color represented SR samples. The color scale shows the relative genes expression level in certain slide: green indicates low relative expression levels; red indicates high relative expression levels
Table 2.
The DEGs of merged data set with the use of criteria of adjust P value < 0.05 and |log2FC| ≥ 1
| Gene | Log2FC | AveExpr | t | P value | adjust P value | B |
|---|---|---|---|---|---|---|
| IGFBP2 | 1.1746 | 9.5881 | −6.0548 | < 0.0001 | 0.0006 | 8.0172 |
| C1orf105 | −1.0560 | 7.2935 | 5.6106 | < 0.0001 | 0.0011 | 6.3668 |
| FHL2 | 1.1255 | 9.9882 | −5.4755 | < 0.0001 | 0.0016 | 5.8742 |
| CHGB | 1.0836 | 8.5851 | −5.3090 | < 0.0001 | 0.0025 | 5.2745 |
| ATP1B4 | 1.0469 | 5.0220 | −4.3592 | < 0.0001 | 0.0283 | 2.0308 |
Log2FC log2 Fold Change; AveExpr Average Expression
Fig. 2.
Boxplots of gene expressions for 5 selected genes with a screening criteria of |log2 FC| ≥ 1 and adjust P value < 0.05
Functional enrichment analyses of DEGs
To further investigate the biological functions of the 27 DEGs, functional enrichment analyses were performed and results were shown in Table 3. The result of molecular function in GO revealed that two up-regulated DEGs (IGFBP2 and IGFBP3) were enriched in insulin-like growth factor I binding process (adjusted P value = 0.0168 and Q value = 0.0120) and insulin-like growth factor binding process (adjusted P value = 0.0432 and Q value = 0.0310). Using screening criteria of adjusted P value < 0.05 and Q value < 0.05, no pathway was enriched in KEGG. The ‘mineral absorption’, ‘calcium signaling pathway’ and ‘proximal tubule bicarbonate reclamation’ pathways were enriched (P values = 0.0033, 0.0331 and 0.0343 respectively). Specifically, SLC26A9 and ATP1B4 genes were enriched in mineral absorption, while two up-regulated genes (CXCR4 and HTR2B) were correlated with calcium signaling pathway. Only up-regulated ATP1B4 gene was enriched in proximal tubule bicarbonate reclamation pathway. However, these enrichments did not remain significant after multiplicity adjustment by BH.
Table 3.
Significant enriched GO terms and pathways of DEGs
| Term | Count | Genes | P value | adjust P value | Q value | |
|---|---|---|---|---|---|---|
| GO terms | ||||||
| GO:0031994 | insulin-like growth factor I binding | 2 | IGFBP2/IGFBP3 | 0.0001 | 0.0168 | 0.0121 |
| GO:0005520 | insulin-like growth factor binding | 2 | IGFBP2/IGFBP3 | 0.0006 | 0.0433 | 0.0311 |
| KEGG Pathway | ||||||
| hsa04978 | Mineral absorption | 2 | SLC26A9/ATP1B4 | 0.0033 | 0.2088 | 0.2088 |
| hsa04020 | Calcium signaling pathway | 2 | CXCR4/ HTR2B | 0.0332 | 0.2741 | 0.2741 |
| hsa04964 | Proximal tubule bicarbonate reclamation | 1 | ATP1B4 | 0.0343 | 0.2741 | 0.2741 |
| Reactome Pathway | ||||||
| R-HSA-381426 |
Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
3 | IGFBP2/ IGFBP3/ CHGB | 0.0008 | 0.0632 | 0.0435 |
GOGene Ontology; DEGs Differentially Expressed Genes
Pathway enrichment using the REACTOME database identified that three up-regulated DEGs (IGFBP2, IGFBP3 and CHGB) were enriched in Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) (P value = 0.0008 and Q value < 0.0435, Supplementary Figure 3). DO enrichment analysis revealed that DEGs were enriched in 29 biological processes (adjusted P value < 0.05 and Q value < 0.05), but not in AF associated process (Supplementary Figure 4).
GSEA was applied to test merged GEO dataset to identify functional gene sets correlated with heart. The SR > AF analysis identified five sets whose expression was correlated with AF (Supplementary Table 2). All were related to heart function (FDR Q-value < 0.05 and FWER P-value < 0.05). The sets were (1) regulation of cell growth involved in cardiac muscle cell development, (2) regulation of cardiac muscle cell differentiation, (3) cardiac muscle cell differentiation, (4) positive regulation of cardiac muscle cell differentiation, (5) physiological cardiac muscle hypertrophy. To explore whether these sets contained DEGs, we examined the leading-edge subsets for each gene set (defined above). In five leading-edge subsets, only DEG FHL2 was involved in cardiac muscle cell differentiation.
PPI network construction and potential crucial genes selection
Using the STRING platform, PPI analysis of these DEGs identified 18 nodes and 26 interactions. In addition, one significant module with 5 nodes and 9 edges was screened out via MCODE (Supplementary Figure 5a). CXCR4, IGFB P2, IGFBP3, SNAI2 and ANGPTL2 were hub nodes in module. Only CXCR4, IGFBP2 and IGFBP3 were selected for hub genes, all of which were involved in playing pivotal regulatory roles in PPI network, due to the high degree of connectivity (degree ≥5, Supplementary Figure 5b). Furthermore, after combining with the results of differential expression, enrichment analyses and PPI, IGFBP2, IGFBP3, CHGB, CXCR4, HTR2B, FHL2, C1orf105, ATP1B4 and SLC26A9 were considered potential crucial genes for further analyses.
Identification of potential crucial genes associated with AF
CTD was employed to explore the interaction between potential crucial genes and AF. As shown in Supplementary Figure 6, potential crucial genes targeting AF, left ventricular dysfunction, heart diseases and cardiovascular diseases. Inference scores in CTD reflected the association between chemical, disease and genes. The interaction results showed that CXCR4, IGFBP2, IGFBP3 and FHL2 have a higher score with AF.
Discussion
In the present study, we integrated gene expression profiles of 46 AF samples and 31 SR samples from 5 GEO datasets and analyzed the data using bioinformatics tools. A total of 27 DEGs with |log2 FC| ≥ 0.58 and 5 with |log2 FC| ≥ 1 in AF compared with SR samples were selected. Furthermore, 9 potential crucial genes (IGFBP2, IGFBP3, CHGB, CXCR4, HTR2B, FHL2, C1orf105, ATP1B4 and SLC26A9), and several important pathways, which were associated with AF risk, were identified, suggesting these may play important role in the mechanism of AF.
IGFBP2, located in chromosome 2q35, encodes the second most abundant circulating IGF-binding protein. IGFBP2, secreted by differentiating white adipocytes, regulates the functions of IGFs [19]. In the Framingham heart study, IGFBP2 was significantly associated with all-cause mortality [20]. Our study revealed that the IGFBP2 expression level was up-regulated in AF samples compared to SR samples. The enrichment analysis results of IGFBP2 were all correlated with IGFBP3.
The functional gene IGFBP3 encodes the primary carrier of IGFs (IGFBP3) in the circulation. IGFBP3 is involved in oxidative stress, atherosclerosis and left ventricular hypertrophy [21, 22]. More important, a previous study revealed low IGFBP3 serum level as an independent determinant of AF [21]. Enrichment analyses in the present study indicated that IGFBP2 and IGFBP3 were enriched in GO term of insulin-like growth factor binding and pathway of IGF transport and uptake by IGFBPs. Thus, IGFBP2 and IGFBP3 might bind and regulate the functions of IGFs through IGF transport and uptake by IGFBPs pathway, and then affect the susceptibility of AF [19].
Our study revealed that CHGB was involved in IGF transport and uptake by IGFBPs pathway including IGFBP2 and IGFBP3, and the gene expression of CHGB was higher in AF patients. Chromogranin B (CHGB) is an emerging cardiovascular biomarker, which is encoded by CHGB [23]. CHGB can regulate B-type natriuretic peptide (BNP) production through polycystin 2(PC2)-CGB-BNP signaling axis in cardiomyocyte, and integrate information from myocardial stress and neuro-endocrine activation [23, 24]. CHGB level was significantly increased in heart failure patients [23]. These findings suggested that IGF transport and uptake by IGFBPs pathway with IGFBP2, IGFBP3 and CHGB may participate in the occurrence and development of AF. A more thorough understanding of IGF transport and uptake by IGFBPs pathway in AF is necessary.
With PPI analysis, IGFBP2, IGFBP3 and CXCR4 were divided into a group according to protein-protein interactions. There was evidence that CXCR4 was overexpressed in chronic AF patients, and might contribute to the process of AF through regulating atrial fibrosis and structural remodeling [25]. In the present study, CXCR4 was also found to be potential crucial gene related to AF. KEGG pathway enrichment analysis showed that the CXCR4 and HTR2B were enriched in calcium signaling pathway, which had been extensively characterized in the role in cardiac hypertrophy and remodeling processes [26]. HTR2B is located in chromosome 2q37.1. 5-HT2B (5-hydroxytryptamine receptor 2B) receptor coded by HTR2B, is presented in the cardiovascular system, and may indirectly produce life-threatening arrhythmias and cardiodepression [27, 28]. In auricular myocytes of newborn rat, the activation of 5-HT2B enhances gap junctional intercellular communication (GJIC) in a receptor subtype-specific manner, and prolongs 5-HT exposure to alter the Cx expression pattern which associated with AF [28].
Increasing evidence demonstrated that FHL2 and its protein product has a function in cardiovascular disease [29, 30]. FHL2, located at the sarcomere, interacted with extracellular signal regulated kinase (ERK) and regulated cardiac growth, suggesting FHL2 a protective role in adrenergic-mediated cardiac hypertrophy [31, 32]. We found that FHL2 was up-regulated in AF samples compared to SR, and was involved in cardiac muscle cell differentiation. These results indicated that FHL2 might be a potential biomarker of AF.
In this study, C1orf105 and ATP1B4 had 2 fold lower and higher gene expression, respectively in AF patients than SR control. SLC26A9 and ATP1B4 were enriched in KEGG pathways of mineral absorption and proximal tubule bicarbonate reclamation. Previous study showed that a SNP on C1orf105 was associated with remodeling response to atherosclerosis [33]. Slc26a9 encodes transporters with diverse functional attributes and RT-PCR showed that Slc26a9 is detectable in heart [34]. The above evidence revealed that C1orf105, SLC26A9 and ATP1B4 were related with cardiovascular disease and might have a function in AF.
In current study, we have discussed that 9 potential crucial genes are involved in the occurrence and development of AF, suggesting these genes may serve as potential biomarkers and therapeutic targets for AF. However, the limitations of this study should be considered. Firstly, it is difficult to consider some important factors such as regions, races and age. Considering that the development of AF results from various environmental and genetic factors, some unmeasured factors including region, family history and risk factors of AF should be evaluated in further research. In addition, the potential crucial genes need further validation by RT-qPCR in clinical samples. Finally, the mechanisms in which these genes play are not completely clear. More evidence is required to find out the biological foundation.
Conclusion
Our study integrated data with relative larger sample size from multiple GEO datasets and identified 9 potential crucial genes (IGFBP2, IGFBP3, CHGB, CXCR4, HTR2B, FHL2, C1orf105, ATP1B4 and SLC26A9), and pathways using bioinformatic analyses. The exploration of potential crucial genes of AF may provide some potential aid in further identification of new biomarkers for the susceptibility of AF and useful treatment targets.
Supplementary information
Additional file 1: Figure S1. (a) Data standardization. Pre-standardization gene expression levels of each data set are presented as blue boxplots; (b) Data standardization. Post-standardization gene expression levels of each data set are presented as red boxplots.
Additional file 2: Figure S2. Volcano plot of DEGs in AF samples compared to SR samples. Red indicates the gene expression was up-regulated in AF samples compared to primary samples (adjust P value < 0.05 and |log2 FC| ≥ 0.58); Green indicates the gene expression was down-regulated in AF samples compared with primary samples (adjust P value < 0.05 and |log2 FC| ≥ 0.58); Black indicates the adjusted P value was > 0.05.
Additional file 3: Figure S3. Pathway view of Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) using the REACTOME database. IGFBP2, IGFBP3 and CHGB were enriched in the pathway.
Additional file 4: Figure S4. Bar plot of DO enrichment of DEGs. The X-axis indicates the number of genes represented in the disease.
Additional file 5: Figure S5. (a) PPI network of the DEGs and modular analysis. Yellow nodes represent DEGs in the same module. (b) Bar plot for the interaction numbers of each gene in PPI network.
Additional file 6: Figure S6. Relationship to AF diseases related to potential crucial genes based on the CTD database.
Additional file 7: Table S1. The DEGs of merged data set with the use of criteria of adjust P value < 0.05 and |log2FC| ≥ 0.58. Table S2. Summary of GSEA result with FDR Q-value < 0.05 and FWER P-value < 0.05.
Acknowledgments
Not Applicable.
Abbreviations
- AF
Atrial Fibrillation
- DEGs
Differentially Expressed Genes
- GEO
Gene Expression Omnibus
- FC
Fold Change
- LAA
Left Atrial Appendage Tissue
- SR
Sinus Rhythm
- MAS5
Affymetrix Microarray Suite 5
- BH
Benjamini and Hochberg method
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene Set Enrichment Analysis
- DO
Disease Ontology
- PPI
Protein-Protein Interaction
- MCODE
Molecular Complex Detection
- CTD
Comparative Toxicogenomics Database
- IGF
Insulin-like Growth Factor
- IGFBPs
Insulin-like Growth Factor Binding Proteins
Authors’ contributions
JZ1 and GL were responsible for study conception, design of the study, data acquisition, and analysis and interpretation of results. JZ1, XH and ZY were responsible for data acquisition. XW, YG, LL took part in the discussion of the paper. JZ1 wrote the manuscript that was reviewed and revised by GL, ZY, XH, XW, YG, LL, ZL, XC and JZ2. All authors have read and approved the manuscript.
Funding
This study was funded by research grant from the Science Foundation of Guangdong Second Provincial General Hospital (YY2018–002) and Medical Scientific Research Foundation of Guangdong Province of China (A2020453). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Microarray datasets (GSE115574, GSE31821, GSE79768, GSE41177 and GSE14975) for this study are openly available in Gene Expression Omnibus database at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115574, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31821, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79768, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41177 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14975, respectively (last accessed on 27 Dec 2019).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests, and all authors confirm its accuracy.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zebing Ye, Email: bean9350@sina.com.
Guowei Li, Email: lig28@mcmaster.ca.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-00754-5.
References
- 1.Kirchhof P, Benussi S, Kotecha D, Ahlsson A, Atar D, Casadei B, et al. 2016 ESC guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Europace. 2016;18:1609–1678. doi: 10.1093/europace/euw295. [DOI] [PubMed] [Google Scholar]
- 2.Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, et al. Worldwide epidemiology of atrial fibrillation: a global burden of disease 2010 study. Circulation. 2014;129:837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schotten U, Verheule S, Kirchhof P, Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev. 2011;91:265–325. doi: 10.1152/physrev.00031.2009. [DOI] [PubMed] [Google Scholar]
- 4.Lau DH, Schotten U, Mahajan R, Antic NA, Hatem SN, Pathak RK, et al. Novel mechanisms in the pathogenesis of atrial fibrillation: practical applications. Eur Heart J. 2016;37:1573–1581. doi: 10.1093/eurheartj/ehv375. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhof P. The future of atrial fibrillation management: integrated care and stratified therapy. Lancet. 2017;390:1873–1887. doi: 10.1016/S0140-6736(17)31072-3. [DOI] [PubMed] [Google Scholar]
- 6.Turagam MK, Velagapudi P, Kocheril AG, Alpert MA. Commonly consumed beverages in daily life: do they cause atrial fibrillation? Clin Cardiol. 2015;38:317–322. doi: 10.1002/clc.22385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res. 2014;114:1469–1482. doi: 10.1161/CIRCRESAHA.114.302225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D5D5. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi-Tope G, Vastrik I, Gopinath GR, Matthews L, Schmidt E, Gillespie M, et al. The genome knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb Symp Quant Biol. 2003;68:237–243. doi: 10.1101/sqb.2003.68.237. [DOI] [PubMed] [Google Scholar]
- 16.Schriml LM, Mitraka E, Munro J, Tauber B, Schor M, Nickle L, et al. Human disease ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res. 2019;47:D955–DD62. doi: 10.1093/nar/gky1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. The comparative Toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47:D948–DD54. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunger D, Yuen K, Ong K. Insulin-like growth factor I and impaired glucose tolerance. Horm Res. 2004;62(Suppl 1):101–107. doi: 10.1159/000080767. [DOI] [PubMed] [Google Scholar]
- 20.Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, et al. Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc. 2018;7(14):e008108. [DOI] [PMC free article] [PubMed]
- 21.Duron E, Vidal JS, Funalot B, Brunel N, Viollet C, Seux ML, et al. Insulin-like growth factor I, insulin-like growth factor binding protein 3, and atrial fibrillation in the elderly. J Gerontol A Biol Sci Med Sci. 2014;69:1025–1032. doi: 10.1093/gerona/glt206. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich N, Aumann N, Dörr M, Felix SB, Nauck M, Wallaschofski H, et al. Lack of association between insulin-like growth factor-1 or insulin-like growth factor-binding protein-3 and left ventricular hypertrophy: results of the study of health in Pomerania. J Hypertens. 2010;28:856–864. doi: 10.1097/HJH.0b013e328336274a. [DOI] [PubMed] [Google Scholar]
- 23.Heidrich FM, Melz C, Buechau MS, Pfluecke C, Quick S, Speiser U, et al. Regulation of circulating chromogranin B levels in heart failure. Biomarkers. 2018;23:78–87. doi: 10.1080/1354750X.2017.1395079. [DOI] [PubMed] [Google Scholar]
- 24.Heidrich FM, Zhang K, Estrada M, Huang Y, Giordano FJ, Ehrlich BE. Chromogranin B regulates calcium signaling, nuclear factor kappaB activity, and brain natriuretic peptide production in cardiomyocytes. Circ Res. 2008;102:1230–1238. doi: 10.1161/CIRCRESAHA.107.166033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XX, Zhang FR, Zhu JH, Xie XD, Chen JZ. Up-regulation of CXC chemokine receptor 4 expression in chronic atrial fibrillation patients with mitral valve disease may be attenuated by renin–angiotensin system blockers. J Int Med Res. 2009;37:1145–1151. doi: 10.1177/147323000903700419. [DOI] [PubMed] [Google Scholar]
- 26.Dewenter M, von der Lieth A, Katus HA, Backs J. Calcium signaling and transcriptional regulation in Cardiomyocytes. Circ Res. 2017;121:1000–1020. doi: 10.1161/CIRCRESAHA.117.310355. [DOI] [PubMed] [Google Scholar]
- 27.Kaumann AJ, Levy FO. 5-hydroxytryptamine receptors in the human cardiovascular system. Pharmacol Ther. 2006;111:674–706. doi: 10.1016/j.pharmthera.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Derangeon M, Bozon V, Defamie N, Peineau N, Bourmeyster N, Sarrouilhe D, et al. 5-HT4 and 5-HT2 receptors antagonistically influence gap junctional coupling between rat auricular myocytes. J Mol Cell Cardiol. 2010;48:220–229. doi: 10.1016/j.yjmcc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Radke MH, Polack C, Methawasin M, Fink C, Granzier HL, Gotthardt M. Deleting full length Titin versus the Titin M-band region leads to differential Mechanosignaling and cardiac phenotypes. Circulation. 2019;139:1813–1827. doi: 10.1161/CIRCULATIONAHA.118.037588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y, Bradford WH, Zhang J, Sheikh F. Four and a half LIM domain protein signaling and cardiomyopathy. Biophys Rev. 2018;10:1073–1085. doi: 10.1007/s12551-018-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell NH, Darwis D, Bueno OF, Muller JM, Schule R, Molkentin JD. Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in Cardiomyocytes. Mol Cell Biol. 2004;24:1081–1095. doi: 10.1128/MCB.24.3.1081-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz K, Schmitt JP, Vidal M, Lohse MJ. Cardiac hypertrophy: targeting Raf/MEK/ERK1/2-signaling. Int J Biochem Cell Biol. 2009;41:2351–2355. doi: 10.1016/j.biocel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Harrison SC, Zabaneh D, Asselbergs FW, Drenos F, Jones GT, Shah S, et al. A gene-centric study of common carotid artery remodelling. Atherosclerosis. 2013;226:440–446. doi: 10.1016/j.atherosclerosis.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang MH, Plata C, Zandi-Nejad K, Sindic A, Sussman CR, Mercado A, et al. Slc26a9--anion exchanger, channel and Na+ transporter. J Membr Biol. 2009;228:125–140. doi: 10.1007/s00232-009-9165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (a) Data standardization. Pre-standardization gene expression levels of each data set are presented as blue boxplots; (b) Data standardization. Post-standardization gene expression levels of each data set are presented as red boxplots.
Additional file 2: Figure S2. Volcano plot of DEGs in AF samples compared to SR samples. Red indicates the gene expression was up-regulated in AF samples compared to primary samples (adjust P value < 0.05 and |log2 FC| ≥ 0.58); Green indicates the gene expression was down-regulated in AF samples compared with primary samples (adjust P value < 0.05 and |log2 FC| ≥ 0.58); Black indicates the adjusted P value was > 0.05.
Additional file 3: Figure S3. Pathway view of Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) using the REACTOME database. IGFBP2, IGFBP3 and CHGB were enriched in the pathway.
Additional file 4: Figure S4. Bar plot of DO enrichment of DEGs. The X-axis indicates the number of genes represented in the disease.
Additional file 5: Figure S5. (a) PPI network of the DEGs and modular analysis. Yellow nodes represent DEGs in the same module. (b) Bar plot for the interaction numbers of each gene in PPI network.
Additional file 6: Figure S6. Relationship to AF diseases related to potential crucial genes based on the CTD database.
Additional file 7: Table S1. The DEGs of merged data set with the use of criteria of adjust P value < 0.05 and |log2FC| ≥ 0.58. Table S2. Summary of GSEA result with FDR Q-value < 0.05 and FWER P-value < 0.05.
Data Availability Statement
Microarray datasets (GSE115574, GSE31821, GSE79768, GSE41177 and GSE14975) for this study are openly available in Gene Expression Omnibus database at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115574, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31821, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79768, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41177 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14975, respectively (last accessed on 27 Dec 2019).