Abstract
Background
An association between neuroinflammation and age-related neurologic disorders has been established but the molecular mechanisms and cell types involved have not been thoroughly characterized. Activity of the proinflammatory NLRP3 inflammasome is implicated in Alzheimer’s and Parkinson’s disease and our recent studies in patients suggest that dopaminergic neurons within the degenerating mesencephalon express NLRP3 throughout the progression of PD. Here, we directly test the impact of enhanced inflammasome activity in mesencephalic neurons by characterizing motor function, tissue integrity, and neuroinflammation in aging mice harboring hyperactivating mutations within the endogenous murine Nlrp3 locus, enabled only in cells expressing the dopaminergic neuron-specific Slc6a3 promoter.
Methods
We compared mice harboring inducible alleles encoding the cryopyrin-associated periodic syndrome activating mutations Nlrp3A350V and Nlrp3L351P inserted into the endogenous mouse Nlrp3 locus. Tissue specific expression was driven by breeding these animals with mice expressing Cre recombinase under the control of the dopaminergic neuron-specific Slc6a3 promoter. The experimental mice, designed to express hyperactive NLRP3 only when the endogenous mouse Nlrp3 promotor is active in dopaminergic neurons, were analyzed throughout 18 months of aging using longitudinal motor function assessments. Biochemical and histologic analyses of mesencephalic tissues were conducted in 1- and 18-month-old animals.
Results
We observed progressive and significant deficits in motor function in animals expressing Nlrp3L351P, compared with animals expressing Nlrp3WT and Nlrp3A350V. Age-dependent neuroinflammatory changes in the mesencephalon were noted in all animals. Analysis of GFAP-immunoreactive astrocytes in the substantia nigra revealed a significant increase in astrocyte number in animals expressing Nlrp3L351P compared with Nlrp3WT and Nlrp3A350V. Further analysis of Nlrp3L351P striatal tissues indicated genotype specific gliosis, elevated Il1b expression, and both morphologic and gene expression indicators of proinflammatory A1 astrocytes.
Conclusions
Dopaminergic neurons have the potential to accumulate NLRP3 inflammasome activators with age, including reactive oxygen species, dopamine metabolites, and misfolded proteins. Results indicate the Nlrp3 locus is active in dopaminergic neurons in aging mice, and that the hyperactive Nlrp3L351P allele can drive neuroinflammatory changes in association with progressive behavioral deficits. Findings suggest neuronal NLRP3 inflammasome activity may contribute to neuroinflammation observed during normal aging and the progression of neurologic disorders.
Keywords: Neuroinflammation, NLRP3 inflammasome, Astrogliosis, Cryopyrin-associated periodic syndromes
Background
Inflammasomes are a class of cytosolic multi-protein complexes whose activation results in intracellular cascades that initiate and propagate proinflammatory signaling [1]. Sustained inflammasome activation can lead to pyroptosis, a subcategory of programmed cell death [2, 3]. Inflammasome complexes are categorized based on the presence of pattern recognition receptors belonging to either the nucleotide-binding domain-like receptors (NLRs), absent in melanoma 2-like receptors (ALRs), or pyrin families [4, 5]. Upon activation, NLRP3 inflammasome proteins form a complex with ASC and pro-caspase-1, inducing maturation of caspase-1, followed by cleavage of proinflammatory cytokines including pro-interleukin-1 beta (pro-IL-1β) and pro-IL-18, and the pore-forming protein gasdermin D [6–9]. Proteolytically processed cytokines are released from the cell and can propagate inflammation in the surrounding tissue. The understanding of inflammasome biology was aided by the identification of rare familial syndromes including familial cold autoinflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS), and neonatal onset multisystem inflammatory disease (NOMID) [10, 11]. These hereditary cryopyrin-associated periodic syndromes (CAPS) are typified by chronic inflammatory symptomology caused by point mutations in the NLR family gene NLRP3, resulting in the production of a hyperactive NLRP3 protein [12, 13].
Inflammasomes are widely accepted to be key mediators of the innate immune response with many studies characterizing their activities in hematopoietic cells of the myeloid lineage, and more recent studies by our laboratory [14] and others [15–19] characterizing NLRP3 inflammasome activity in functionally related microglia of the central nervous system (CNS). Increasing evidence indicates that the inflammasome stress-sensing pathway may be utilized by a host of other cell types. NLRP3 inflammasome activity in hepatocytes is a key feature of liver disease [20], observable in animals exposed to high-fat diets [21] as well as in tissues obtained from patients with non-alcoholic fatty liver disease [22]. NLRP3 polymorphisms are associated with an elevated risk of developing Crohn’s disease [23, 24]. Subsequent studies indicate significant NLRP3 expression and inflammasome activity in intestinal epithelial cells that influence inflammatory bowel disease progression [25, 26]. Sustained NLRP3 expression was observed in distressed neurons surrounding ischemic stroke [27] as well as in primary cortical neurons in an experimental traumatic brain injury (TBI) model [28]. Expression of the inflammasome family pattern recognition receptors NLRP1 and AIM2 has also been reported in neurons in mouse and rat models respectively [29, 30]. In humans, NLRP1 expression was observed in neurons of patients with spinal cord injury [31], and in primary human neuron cultures [32]. Collectively, these data, among others, indicate that a variety of cell types utilize the inflammasomes to participate in the innate immune response.
A growing body of research has characterized the activity of intracellular inflammasomes in age-related neurodegenerative disorders based on the widely held belief that these disorders are exacerbated by chronic neuroinflammation [19, 33, 34]. The NLRP3 inflammasome is of particular interest in neurologic disorders because it can be activated by sterile cellular stressors common to many neurodegenerative diseases including proteinaceous insult [35], oxidative stress [36, 37], environmental toxicants [38], and cell death [39]. Characterizing NLRP3 in Parkinson’s disease (PD), we observed NLRP3 inflammasome activity in PD patients [40] and demonstrated that loss of Nlrp3 is neuroprotective in a toxicant-based mouse model of PD [14]. We also identified a single-nucleotide polymorphism in the NLRP3 exome sequence associated with a reduced risk of PD that we observed to negatively impact NLRP3 protein homeostasis in vitro [40]. These studies, and others [17, 41, 42], of NLRP3 activity in PD are consistent with similar studies of NLRP3 in Alzheimer’s disease (AD) [43–46], suggesting that the NLRP3 inflammasome is a core proinflammatory mediator in neurodegenerative disorders. NLRP3 activity in neurologic disorders is readily observed in microglia [47] but our studies also revealed NLRP3 expression in surviving dopaminergic (DA) neurons of the mesencephalon in post-mortem tissues obtained from PD patients [40]. Further analysis corroborated our patient data, indicating inducible expression of NLRP3 in tyrosine hydroxylase-positive differentiated Lund human mesencephalic (LUHMES) neurons in culture and evidence of NLRP3 inflammasome activity in differentiated, human SH-SY5Y cells. Our data indicating NLRP3 expression and activity in DA neurons are consistent with reports of other inflammasome-related pattern recognition receptor expression in neurons described above, yet, to our knowledge, no studies have directly tested the consequence of neuronal inflammasome activity throughout the process of aging.
To characterize the effect of enhanced NLRP3 inflammasome activity in mesencephalic neurons in the context of aging, we bred mice harboring well-characterized, inducible Nlrp3 alleles engineered to contain hyperactivating polymorphisms associated with CAPS [48] with mice expressing Cre recombinase under the control of the Slc6a3 promoter, encoding the DA neuron-specific dopamine transporter. These “knock-in” mice are designed to express hyperactive NLRP3 in a Cre-dependent manner only when the endogenous Nlrp3 promoter is active, allowing us to determine the result of enhanced NLRP3 inflammasome activity occurring exclusively when the Nlrp3 gene is active in Slc6a3-expressing neurons, throughout aging. Longitudinal behavioral analyses indicate that tissue-specific expression of the Nlrp3L351P polymorphism resulted in significant age-dependent motor deficits as compared with the Nlrp3A350V polymorphism and mice expressing the WT allele. Post-mortem studies indicate enhanced neuroinflammation specifically in tissues obtained from animals expressing Nlrp3L351P. Our collective data indicate that the Nlrp3 locus is active in DA neurons and that enhanced neuronal inflammasome activity is sufficient for the development of more widespread neuroinflammatory changes associated with motor deficits and advanced age.
Methods
Animals
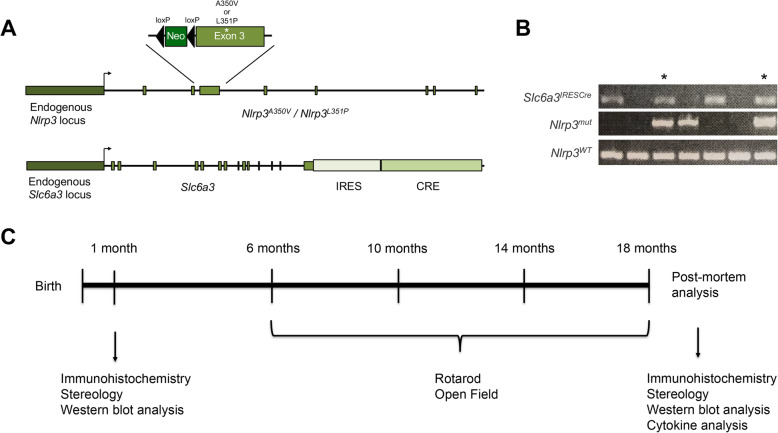
Animal experiments were approved by the Dartmouth Institutional Animal Care and Use Committee and conducted in accordance with federal guidelines pertaining to animal research. All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Dartmouth protocol entitled “Animal Models of Neurodegenerative Diseases,” protocol no. 00002117. Mice harboring Nlrp3 mutant alleles were purchased from Jackson Laboratories (Bar Harbor, ME), developed and characterized as previously described [48]. MWS-associated Nlrp3A350VneoR (stock no. 017969) and FCAS-associated Nlrp3L351PneoR (stock no. 017970) alleles were generated in the endogenous mouse Nlrp3 gene preceded by a reverse oriented neomycin cassette flanked by loxP sites. The mouse Nlrp3 allele is functionally null in both animals prior to Cre recombinase-mediated deletion of the neomycin cassette. We bred both strains to commercially available mice expressing Cre recombinase in cells expressing the dopamine transporter gene, Slc6a3IRESCre (stock no. 006660). The Slc6a3IRESCre strain is now convincingly validated and in wide use [49–51], originally characterized using the ROSA reporter line to express CRE specifically within neuronal populations of the mesencephalon [52]. This process generated mice that selectively express the hyperactive Nlrp3 alleles only when the endogenous murine Nlrp3 gene is expressed in dopaminergic neurons. Mice were heterozygous for the hyperactive Nlrp3 mutant alleles, and were engineered to retain one copy of Nlrp3WT in all cells of the animal. The breeding resulted in three distinct cohorts of animals: Nlrp3WT/Slc6a3WT, Nlrp3A350V/Slc6a3IRESCre, and Nlrp3L351P/Slc6a3IRESCre. Genotyping was conducted to validate all mice included in the study (Fig. 1b). Primers were obtained from Integrated DNA Technologies (Coralville, IA) (Table 1).
Fig. 1.
Longitudinal model of mice expressing hyperactive Nlrp3 mutations under the endogenous promotor in DA neurons. a Transgenic mice harboring Cre-driven genetic mutations in Nlrp3, either Nlrp3A350V, or Nlrp3L351P, previously associated with the cryopyrin-associated periodic syndromes, were bred with mice harboring IRES-Cre under the control of the Slc6a3 promotor. b Representative genotyping with PCR-based detection of Nlrp3WT and Nlrp3mut alleles. Mice double positive for Slc6a3IRESCre and Nlrp3mut (Nlrp3A350V or Nlrp3L351P) were included in the study as indicated by *, as well as Nlrp3WT/WT mice as controls. All mice contain one background copy of the Nlrp3WT gene. c Experimental design of longitudinal study. Histologic sections were analyzed at both 1 month and 18 months of age, and behavior analysis was conducted at the 6, 10, 14, and 18-month time points
Table 1.
Primers used for animal genotyping and gene expression analysis
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Nlrp3WT | CACCCTGCATTTTGTTGTTG | CGTGTAGCGACTGTTGAGGT |
| Nlrp3mutant | GCTACTTCCATTTGTCACGTCC | CGTGTAGCGACTGTTGAGGT |
| Slc6a3WT | TGGCTGTTGGTGTAAAGTGG | GGACAGGGACATGGTTGACT |
| Slc6a3IRESCre | TGGCTGTTGGTGTAAAGTGG | GGCCCTCACATTGCCAAAAG |
| C3 | CCAGCTCCCCATTAGCTCTG | GCACTTGCCTCTTTAGGAAGTC |
| Ggta1 | GTGAACAGCATGAGGGGTTT | GTTTTGTTGCCTCTGGGTGT |
| H2-D1 | TCCGAGATTGTAAAGCGTGAAGA | ACAGGGCAGTGCAGGGATAG |
| Il1b | CGCAGCAGCACATCAACAAGAGC | TGTCCTCATCCTGGAAGGTCCACG |
| Actb | GCACCACACCTTCTACAATGAGC | ATGTCACGCACGATTTCCC |
Gene expression analysis
To validate expression of engineered Nlrp3 alleles, DA neuron terminal-rich striatal tissues were harvested from 18-month-old animals and total mRNA was isolated using RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Total mRNA was reversed transcribed with iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Nlrp3 transcripts were amplified and sequenced with Sanger sequencing techniques conducted by Dartmouth’s Molecular Biology Shared Resource. Similar methods were utilized for real-time PCR analysis of C3, Ggta1, H2-D1, and Il1b transcripts. Analysis was completed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) with SYBR Green reagent (Bio-Rad Laboratories, Hercules, CA). All primers were obtained from Integrated DNA Technologies (Coralville, IA) (Table 1).
Behavioral analysis
Behavior was analyzed longitudinally using the rotarod and open field paradigms at 6, 10, 14, and 18 months of age as previously reported [14]. Rotarod training and testing sessions consisted of two 300-s trials separated by 15-min intervals. Recorded data for each animal captured the time each mouse spent on the rotarod, with a maximum of 300 s. Rotarod testing was performed on the fourth day following three consecutive training days. Open field testing was conducted to assess behavior and general motor activities using a computer-assisted photobeam detection system (TruScan, Thermo Fisher Scientific, Waltham, MA). Data were collected throughout the 30-min session. Measurements analyzed in this study include move time (s), rest time (s), distance moved (cm), total velocity (cm/30 min), average velocity (cm/min), and center time (s). All behavior data were analyzed with the GraphPad Prism software (San Diego, CA).
Immunohistochemistry and stereology
Immunohistochemistry and stereology experiments were performed as previously described [14]. Briefly, brain tissues were isolated and dissected. One hemisphere was submitted for sectioning and staining (Neuroscience Associates, Knoxville, TN). Sections were cut at 60 μM, stained with anti-tyrosine hydroxylase antibodies, and then counterstained with Nissl. Unbiased stereology was conducted using the optical fractionator method (Stereo Investigator, Microbrightfield, Williston, VT), counting every fourth section of the SNpC (5 sections/animal). Remaining tissues were embedded for either paraffin or cryosectioning and sliced at 5 μM. Immunohistochemical studies were performed at the Dartmouth Research Pathology Shared Resource using an automated staining system (Leica Bond Max, Buffalo Grove, IL). Immunofluorescence studies were conducted manually on paraffin sections using a sodium citrate epitope retrieval protocol. Primary antibodies used in this study include the following: ionized calcium-binding adaptor molecule 1 (IBA1) (NB100-1028, Novus Biologicals, Centennial, CO), glial fibrillary acidic protein (GFAP) (2021-05, Agilent Technologies, Santa Clara, CA), apoptosis-associated speck-like protein containing a CARD (ASC) (AL177, AdipoGen Life Science, San Diego, CA), interleukin-1 beta (IL-1β) (AF-401-NA, R&D Systems, Minneapolis, MN), and tyrosine hydroxylase (TH) (P40101, Pel-Freez Biologicals, Rogers, AR). IHC studies were conducted with either BOND polymer refined detection system (Leica Biosystems, Wetzlar, Germany) or HRP Polymer (Biocare Medical, Pacheco, CA). Images were obtained using an Olympus BX51 microscope. Confocal microscopy studies were conducted with fluorescently labeled secondary antibodies raised in donkey (Alexa Fluor 488 ab150061, Alexa Fluor 555 ab150130, Abcam, Cambridge, UK). Imaging was conducted using a Zeiss LSM 800 laser point scanning microscope under the guidance of Dartmouth’s Microscopy—Irradiation, Pre-clinical Imaging, and Microscopy Shared Resource.
SDS-PAGE and western blotting
SDS-PAGE and western blotting experiments were performed as previously described [40]. Striatal tissues were isolated and homogenized in lysis buffer (150 mM NaCl, 50 mM Tris, 1% Triton) supplemented with protease and phosphatase inhibitors. Proteins were separated using the NuPAGE system on 4–12% pre-cast gradient gels and MES/SDS running buffer (Thermo Fisher Scientific, Waltham, MA) and then were transferred to Immobilon P transfer membrane (Millipore, Burlington, MA) using a Mini Blot Module (Thermo Fisher Scientific, Waltham, MA). Blots were probed with anti-tyrosine hydroxylase (TH) antibodies (MAB318, Millipore, Burlington, MA) and anti-actin HRP-coupled antibodies (A3854, Sigma-Aldrich, St. Louis, MO). HRP-conjugated anti-mouse secondary antibodies (Millipore, Burlington, MA) were used to detect TH immunoreactivity in association with chemiluminescence-based detection, ECL-Plus (Thermo Fisher Scientific, Waltham, MA).
Statistical analysis
At least 3 biologic replicates were conducted for gene expression experiments, and data were analyzed with one-way ANOVA. All immunohistochemistry experiments were conducted by a blinded investigator, and quantified data were analyzed with one-way ANOVA or two-tailed Student’s t test as appropriate. Behavioral tests were run by an investigator blinded to genotype. Longitudinal rotarod and open field data were analyzed by two-way repeated measures ANOVA. ANOVA analyses were followed by Tukey’s post hoc multiple comparisons test when appropriate. Endpoint 18-month analysis was analyzed with one-tailed Student’s t test. Unbiased stereology was conducted by a single, blinded investigator. All statistical tests were conducted with the GraphPad Prism software (San Diego, CA).
Results
Evidence of NLRP3 inflammasome protein expression in neurons has been reported by our laboratory and others [27, 40, 53–55]; however, the impact of neuronal inflammasome activity in aging animals has not been analyzed directly. Having identified NLRP3 expression in remaining DA neurons from PD patients and in human neuronal cell lines [40], we established a genetically engineered mouse model to characterize longitudinally the effect of enhanced DA neuron-specific NLRP3 inflammasome activity throughout aging. We took advantage of DA neuron-specific Slc6a3IRESCre mice and animals originally reported by Brydges et al. [48] in which the endogenous Nlrp3 locus was modified to encode CAPS-associated activating mutations A350V or L351P under the control of Cre recombinase (Fig. 1a) (see “Methods” section). The study animals were confirmed by genotyping (Fig. 1b), and fell into three cohorts: Nlrp3WT/WT/Slc6a3IRESCre(−), Nlrp3WT/A350V/Slc6a3IRESCre(+), Nlrp3WT/L351P/Slc6a3IRESCre(+) referred to herein as Nlrp3WT, Nlrp3A350V, and Nlrp3L351P respectively. Critically, this cross generates modifications in the endogenous murine Nlrp3 locus [48], resulting in animals that will only express the hyperactive CAPS alleles when the endogenous Nlrp3 promoter is active in DA neurons.
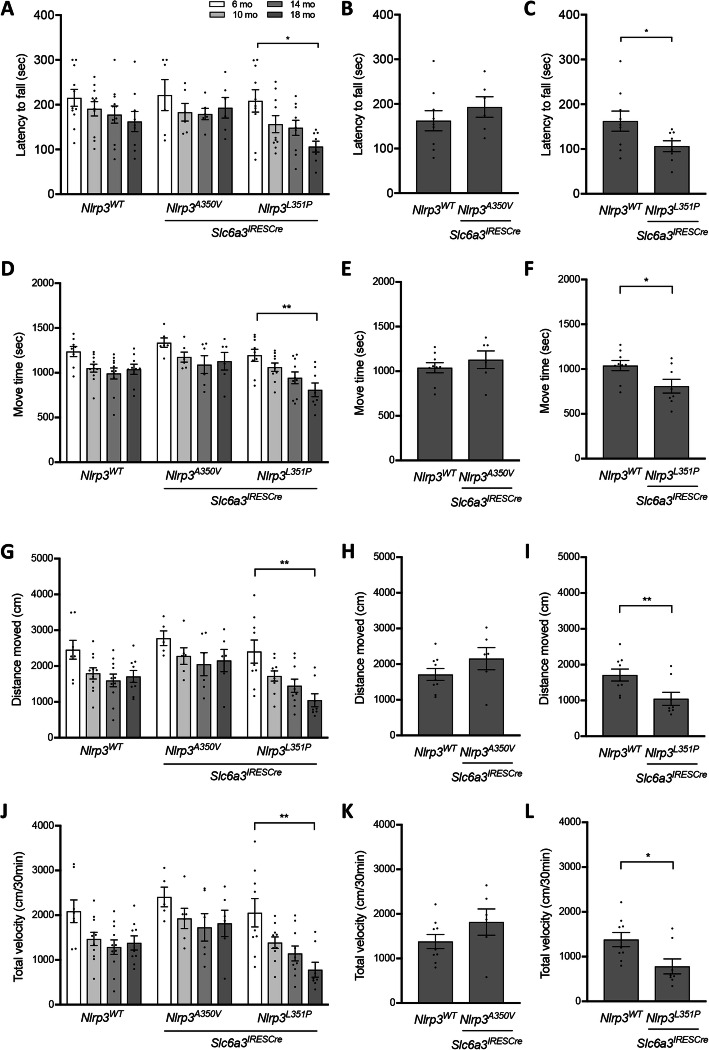
Endogenous Nlrp3 expression increases with aging in the CNS [56–59] prompting us to characterize the influence of the Nlrp3 locus in DA neurons at advanced ages (Fig. 1c). We conducted longitudinal rotarod and open field analyses comparing behavior in Nlrp3WT, Nlrp3A350V, and Nlrp3L351P expressing animals at 6, 10, 14, and 18 months of age. As expected, we observed a reduction in activity of older mice across all three genotypes (Fig. 2). Nlrp3L351P animals had a progressive decline in rotarod performance, while there was no significant change observed over the same time period in the Nlrp3WT and Nlrp3A350V expressing cohorts (Fig. 2a). Endpoint analysis further confirmed the repeated measure findings indicating that the decreased latency to fall from the rotarod was specific to the Nlrp3L351P animals (Fig. 2b, c). Open field analysis indicated that over the same 12-month time course, Nlrp3L351P animals also had significantly reduced locomotor activity, spending shorter periods of time in motion (Fig. 2d), and moving significantly shorter distances (Fig. 2g) and at reduced velocities overall (Fig. 2j). Endpoint analyses for each of these measures again confirmed the longitudinal data indicating specific deficits in Nlrp3L351P mice at 18 months of age (Fig. 2e, f, h, i, k, l). Analysis of rest time, average velocity, and time spent in the center of the open field chamber revealed a significant reduction in all genotypes throughout aging (Supplementary Figure 1). Comparing individual time points, Nlrp3L351P expressing animals spent significantly less time moving at 18 months compared to 6 months and did so with a smaller velocity (Supplementary Figures 1A, B). These behavior assays reveal that animals expressing Nlrp3L351P exhibit a significant reduction in motor function over time in the 18-month longitudinal study as compared with identically aged animals harboring Nlrp3WT and Nlrp3A350V alleles.
Fig. 2.
Behavioral analysis revealed significant reduction in motor function over time in Nlrp3L351P cohort. Behavioral analyses were conducted at 6, 10, 14, and 18-month time points in cohorts (Nlrp3WTn = 11, Nlrp3A350Vn = 6, Nlrp3L351Pn = 9). aNlrp3L351P mice had significant rotarod behavior deficits with age (two-way ANOVA, p = 0.0211 (genotype), p = 0.0052 (time); Tukey’s post hoc multiple comparisons test, * p = 0.02). b, c Endpoint analysis at 18 months shows Nlrp3L351P expressing mice, not Nlrp3A350V, were significantly impaired compared to Nlrp3WT (* p = 0.026, t test). In open field analysis, animals expressing Nlrp3L351Pd moved for significantly less time (two-way ANOVA, p = 0.0018 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0027), g moved a significantly shorter distance (two-way ANOVA, p = 0.0008 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0015), and j traveled at a significantly lower velocity (two-way ANOVA, p = 0.0008 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0020) at 18 months compared to 6 months of age. No significant changes were observed in the Nlrp3WT and Nlrp3A350V over time. Endpoint comparison at 18 months between cohorts revealed a significant decline in f move time (* p = 0.0135, t test), i distance moved (** p = 0.0085, t test), and l total velocity (* p = 0.01, t test) in animals expressing Nlrp3L351P compared to Nlrp3WT animals. e, h, k This difference was not observed in Nlrp3A350V expressing animals when compared to controls. Error bars represent s.e.m.
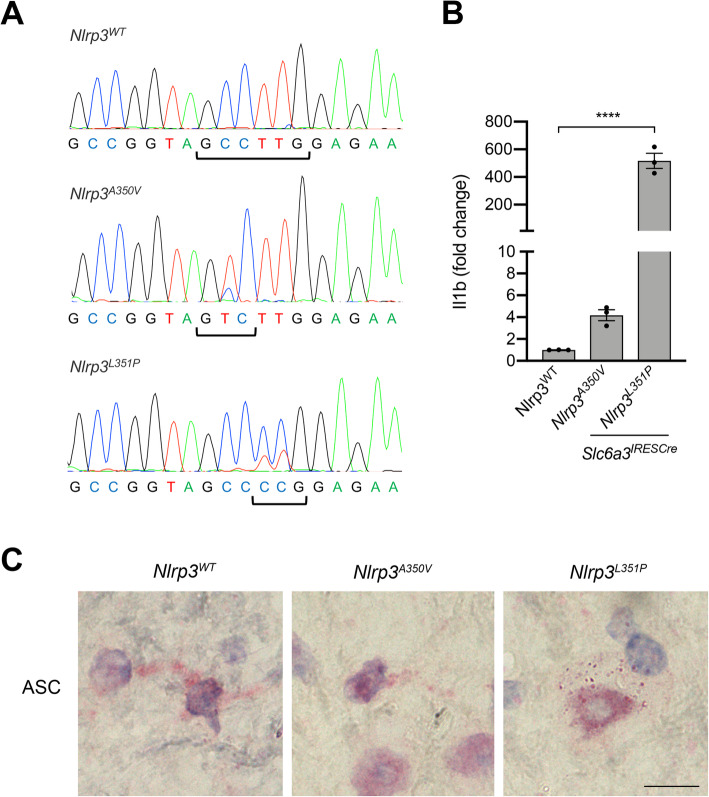
Following the observation that Nlrp3L351P animals exhibit an age-related reduction in motor function, we conducted post-mortem analysis of brain tissues to characterize NLRP3 inflammasome activity, DA neuron cell number, and neuroinflammation in our cohorts. We confirmed expression of Nlrp3 mutant transcripts using mRNA isolated from striatal tissues at 18 months of age. The hemizygous animal model is complex, harboring either wild-type Nlrp3 alleles alone or a wild-type Nlrp3 allele in combination with Nlrp3A350V or Nlrp3L351P alleles in Slc6a3-expressing cells. We isolated mRNA from dopamine fiber-rich striatal tissues and PCR amplified cDNA sequences flanking the region of Nlrp3 engineered to contain the CAPS-associated mutations. Direct sequencing of these Nlrp3 amplicons demonstrated expression of both wild-type Nlrp3 mRNA and transcripts containing Nlrp3A350V or Nlrp3L351P mutations in striatal tissues (Fig. 3a), consistent with planned hemizygosity (Fig. 1a, b), and the findings described in the original manuscript characterizing these mice [48]. To assess enhanced activity of NLRP3 in animals expressing Nlrp3A350V or Nlrp3L351P, we evaluated well-characterized and histopathologically identifiable ASC-immunoreactive specks, whose presence is indicative of inflammasome activation [60]. Immunostaining of mesencephalic sections with anti-ASC antibodies revealed distinct ASC-immunoreactive puncta in the substantia nigra pars compacta (SNpC) of animals expressing Nlrp3L351P that were not readily observed in animals expressing Nlrp3WT and Nlrp3A350V (Fig. 3c). In addition to the formation of ASC immunoreactive puncta, we measured the relative expression of Il1b, whose elevation is indicative of an inflammasome-driven proinflammatory response [1]. Real-time PCR experiments revealed elevated Il1b transcript in striatal tissues from both mutant animals, with significantly elevated expression in animals expressing Nlrp3L351P (Fig. 3b). These findings are consistent with the original report that described more severe phenotype associated with the Nlrp3L351P allele as compared with the Nlrp3A350V allele [48]. Importantly, the presence of Slc6a3-driven Nlrp3A350V and Nlrp3L351P transcripts in the aged mesencephalon is consistent with our studies in PD patients [40] indicating expression of the endogenous Nlrp3 locus in mesencephalic neurons.
Fig. 3.
Mutant alleles expressed in striatal tissues, evidence of NLRP3 inflammasome activity at 18 months of age. a mRNA was isolated from striatal tissues, reverse transcribed, amplified, and sequenced to confirm expression of Nlrp3 allele in target tissues. The alanine to valine substitution at aa350 was observed in mice expressing Nlrp3A350V (C➔ T, red peak), with evidence of the Nlrp3WT background copy expression (blue peak). The leucine to proline substitution at aa351 was observed in mice expressing Nlrp3L351P (TT ➔ CC, blue peaks), with evidence of the Nlrp3WT background copies expressed (red peaks). bIl1b mRNA transcript was reverse transcribed, amplified, and quantified with real-time PCR technologies. Nlrp3L351P mice expressed significantly more Il1b mRNA in striatal tissues compared to control animals (**** p < 0.0001, one-way ANOVA, error bars represent s.e.m.). c Histologic sections of substantia nigra were probed with anti-ASC antibodies, followed by BOND polymer detection, and imaged with a brightfield microscope. ASC puncta (red) indicative of NLRP3 inflammasome activity were observed in Nlrp3L351P mice. (60 × images, scale bar represents 10 μM)
To determine if expression of mutant Nlrp3 alleles resulting in hyperactive NLRP3 protein affected the maintenance of DA neurons throughout aging, we conducted unbiased stereology at 1- and 18-month time points in a single brain hemisphere (see “Methods” section) (Fig. 4a). One-way ANOVA analysis did not reveal significant changes in the number of TH-immunoreactive and Nissl-positive cells present in the SNpC of these tissues (Fig. 4b, c). Histologic analysis of striatal tissues immunostained with anti-TH antibodies and western blot analysis of striatal tissue homogenates revealed no significant change in TH expression (Fig. 4d–f), confirming our findings that while variable, there is no genotype-specific change in the number of DA neurons in the substantia nigra in our mouse cohorts.
Fig. 4.
Analysis of DA neuron maintenance in SNpC and striatal tissues from young and old mice. a Mesencephalic sections were stained with anti-tyrosine hydroxylase antibodies and counterstained with Nissl. TH and Nissl positive cells were counted in sections obtained from 1- and 18-month-old animals. Scale bar represents 100 μM. b Quantification of TH-immunoreactive neurons in the SNpC of animals expressing Nlrp3WT, Nlrp3A350V, and Nlrp3L351P. No significant differences in TH-immunoreactive cells were observed among cohorts. c Quantification of Nissl-positive neurons in the SNpC. No significant differences were observed with one-way ANOVA analysis. Error bars represent s.e.m. d Representative sections of striatal tissues from 1- and 18-month-old animals were sectioned and stained with anti-TH antibodies. Scale bar represents 500 μM. e Densitometric analysis of TH staining in the striatum revealed no significant differences among genotypes and ages with one-way ANOVA analysis. Error bars represent s.e.m. f Striatal tissues were homogenized and proteins were separated by SDS-PAGE. Immunoblotting with anti-TH antibodies was conducted. No significant differences in TH expression among genotypes or with age were observed. Anti-actin antibodies were used as a loading control
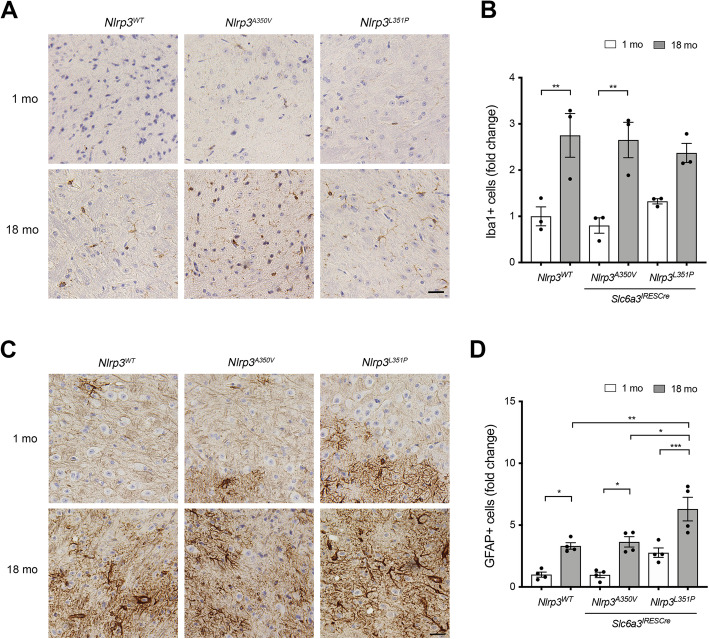
Inflammatory signaling from neurons is anticipated to be closely monitored by glial cell populations in the brain microenvironment [61, 62]. We characterized microglia and astrocytes in nigrostriatal brain tissues from 1- and 18-month-old animals to determine if neuronal expression of the mutant Nlrp3 alleles influenced glia throughout aging. Quantification of IBA1-immunoreactive microglia cells in SNpC tissues from 1- and 18-month-old animals (Fig. 5a) revealed a significant increase in cell number with age in all three genotypes (Fig. 5b). We also quantified the number of ramified and activated microglia in these tissues and observed a similar pattern with no differences between genotypes (data not shown). Staining adjacent sections with anti-GFAP antibodies to characterize astrocytes (Fig. 5c), we observed a significantly greater number of GFAP-immunoreactive astrocytes in the SNpC in 18-month-old animals compared to 1-month-old animals in all genotypes (Fig. 5d). In addition to observing an increased number of GFAP-immunoreactive astrocytes associated with aging, we also observed significantly more astroglia in 18-month-old animals expressing Nlrp3L351P compared to animals expressing Nlrp3WT and Nlrp3A350V. We and others have previously reported astrocytes to be a direct target of IL-1β [14, 63–65], the canonical proinflammatory effector cytokine of the inflammasome [7], and others have previously reported evidence of neuronal IL-1β expression [66]. Analysis of SNpC tissues was consistent with these previous reports, demonstrating easily observable IL-1β expression in TH-immunoreactive fibers (Supplementary Figure 2). While we did not observe significant nigral degeneration, these data support a model in which endogenous activity of the NLRP3 inflammasome, enhanced by the presence of the Nlrp3L351P allele, is associated with age-dependent astrogliosis and behavioral deficits.
Fig. 5.
Increased number of IBA1-immunoreactive and GFAP-immunoreactive cells in the SNpC of 18-month-old animals. a Representative histologic sections of the SNpC obtained from 1- and 18-month-old animals stained with anti-IBA1 antibodies and counterstained with BOND polymer detection. b IBA1-immunoreactive cells were quantified (10 fields/animal, n = 3). The number of IBA1-immunoreactive cells was significantly greater in 18-month-old animals compared to 1-month-old animals, indicating number of microglia in the SNpC increases with age. (Nlrp3WT ** p = 0.0094, one-way ANOVA; Nlrp3A350V ** p = 0.0041, one-way ANOVA). c Representative IHC images of SNpC tissues stained with anti-GFAP antibodies followed by BOND polymer detection. d GFAP-immunoreactive cells were quantified (10 fields/animal, n = 4). GFAP-immunoreactive cell number increases with age in all cohorts (Nlrp3WT * p = 0.03, Nlrp3A350V * p = 0.01, Nlrp3L351P *** p = 0.0008, one-way ANOVA). The number of GFAP-immunoreactive cells was significantly greater in 18-month-old Nlrp3L351P animals compared to the number of cells in 18-month-old Nlrp3WT and Nlrp3A350V animals (** p = 0.004 and * p = 0.01 respectively, one-way ANOVA). Scale bars represent 20 μM. Error bars represent s.e.m.
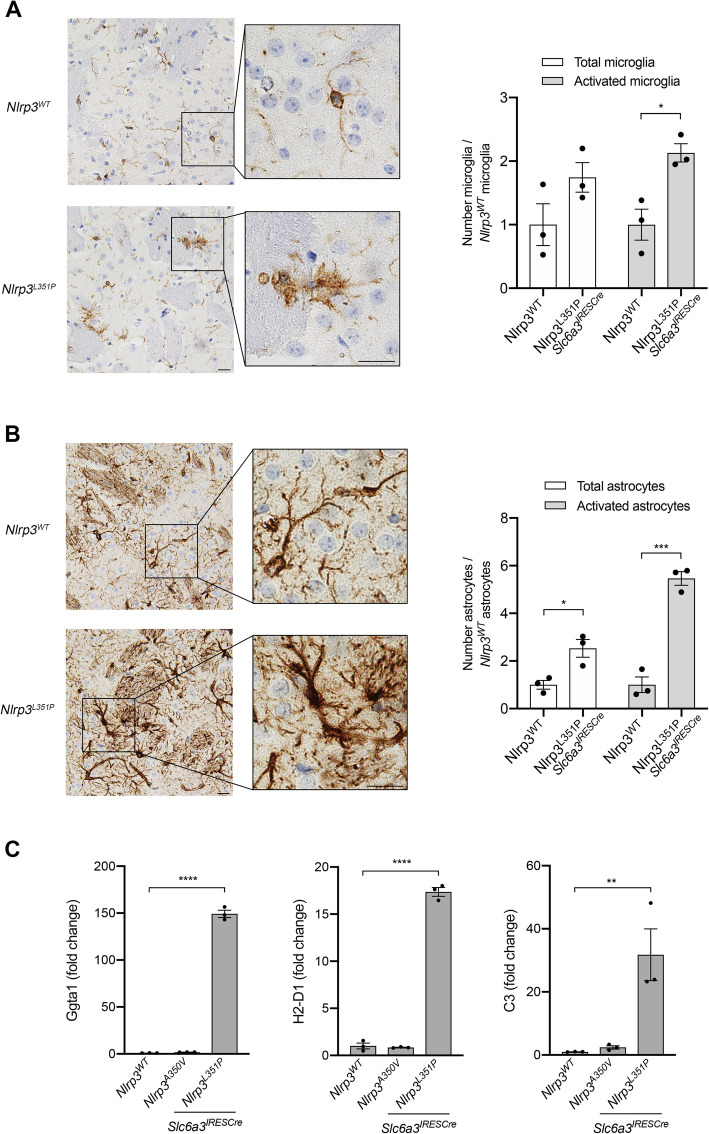
Based on our observation of IL-1β expression in neuronal fibers, we evaluated striatal tissues, reasoning that proinflammatory changes induced by DA neurons may be evident in this fiber-rich projection site. Unlike in the SNpC, microglia activation was easily observable in tissues obtained from animals expressing Nlrp3L351P (Fig. 6a, bottom panel) compared with tissues obtained from Nlrp3WT animals (top panel). Similarly, GFAP immunostaining indicated the presence of supernumerary astrocytes with a proinflammatory morphology in Nlrp3L351P animals (Fig. 6b, bottom panel), which was not observed in Nlrp3WT expressing animals (top panel). To further confirm our observation of proinflammatory changes in astroglial phenotype in both SNpC (Fig. 5c, d) and striatal tissues (Fig. 6b), we isolated mRNA from freshly frozen striatal tissues obtained from 18-month animals and measured expression of proinflammatory A1 astrocyte markers. We observed elevated expression of Ggta1, H2-D1, and C3 in animals expressing Nlrp3L351P compared to animals expressing Nlrp3WT and Nlrp3A350V (Fig. 6c). These results provide evidence of microglial activation (Fig. 6a) and the presence of proinflammatory astrocytes in animals expressing the Nlrp3L351P activating mutation in DA neurons (Fig. 6b, c), suggesting that in the aging brain, DA neurons have the capacity to initiate a widespread Nlrp3-dependent neuroinflammatory response associated with progressive loss of motor function in the absence of neuronal cell death.
Fig. 6.
Evidence of enhanced proinflammatory glial cell phenotype in the striatum of Nlrp3L351P animals. a Striatal tissue sections from 18-month-old animals were probed with anti-IBA1 antibodies followed by BOND polymer detection. Microglial activation can be visualized in animals harboring the Nlrp3L351P allele (bottom panel), compared to their WT littermates (top panel). Scale bars represent 20 μM. There are significantly more activated microglia in tissues from animals expressing Nlrp3L351P compared with tissues from Nlrp3WT animals (* p = 0.0164, t test). Error bars represent s.e.m. b Adjacent striatal sections were stained with anti-GFAP antibodies. GFAP-immunoreactive astrocytes in Nlrp3L351P animals have an increased activation phenotype (bottom panel), compared to Nlrp3WT animals (top panel). Scale bars represent 20 μM. Quantitation revealed significantly more total astrocytes (* p = 0.0217, t test) as well as significantly more activated astrocytes (*** p = 0.0005, t test) in striatal tissues from Nlrp3L351P animals compared to controls. Error bars represent s.e.m. c Expression of A1 astrocyte markers, Ggta1, H2-D1, C3, were assessed in 18-month-old animals. RNA was isolated from striatal tissues, reversed transcribed, and mRNA was measured by real-time PCR. Ggta1, H2-D1, and C3 levels were significantly elevated in Nlrp3L351P mice compared to Nlrp3WT animals (**** p < 0.0001, **** p < 0.0001, ** p = 0.009 respectively, one-way ANOVA). Error bars represent s.e.m.
Discussion
Numerous reports support a role for inflammasomes functioning in CNS microglia during normal aging and the progression of age-related neurodegenerative disorders [16, 19, 33, 34]. Our studies have been consistent with these reports [14]; however, we also unexpectedly identified NLRP3 expression in a subset of surviving DA neurons in late-stage PD patients and neuronal cell models [40]. Others report inflammasome activity in neurons [27, 53–55], but to our knowledge, no in vivo studies have examined directly the function of NLRP3 in DA neurons in the context of aging. In this study, we generated mice with the potential for DA neuron-specific NLRP3 inflammasome hyperactivity and analyzed the animals throughout aging (Fig. 1). The experiments indicate that mesencephalic neurons express the endogenous Nlrp3 gene, and that modification of the endogenous Nlrp3 locus to mimic a CAPS-associated hyperactive allele results in progressive motor dysfunction and nigrostriatal inflammation.
Among neuronal cell types, DA neurons represent an intuitive host for activity of intracellular danger sensing inflammasome signaling pathways. The processes of dopamine synthesis and metabolism induce cellular stress, including the production of dopamine quinones and intracellular ROS, a frequently reported trigger of the NLRP3 inflammasome [67, 68]. Oxidized dopamine molecules are capable of altering DNA and proteins [68], as well as inducing mitochondrial and lysosomal dysfunction, highly penetrant molecular phenotypes associated with both familial and sporadic PD [69] and reminiscent of sterile inflammatory triggers known to activate inflammasomes. These unique biologies may underlie selective vulnerability of DA neurons [70]; chronic basal levels of cellular stress are thought to negatively impact their survival throughout aging and in circumstances of toxicant-exacerbated distress. A relationship between dopamine and the NLRP3 inflammasome has been previously identified by Yan and others who report suppression of NLRP3 inflammasome-driven inflammation dependent on the DRD1 dopamine receptor in bone marrow-derived macrophages [71]. In a related report, astroglial DRD2 receptors are similarly implicated in suppression of NLRP3 inflammasome activity [72]. Critically, DA neurons express DRD2 autoreceptors that are involved in regulating the synthesis, release, and uptake of dopamine in presynaptic cells [73]. Dysregulation of these receptors is associated with multiple neurologic disorders including PD [74]. These reports highlight a regulatory pathway in which inflammasome activity is modulated by dopamine receptor activity, possibly as a means of maintaining homeostasis in microenvironments containing vulnerable neuronal populations. As animals age, dopamine receptor expression has been reported to decrease [75], which would alter the negative regulation of NLRP3-mediated inflammation. Our findings are consistent with this relationship, as we observed increased activity of the endogenous Nlrp3 locus in older animals, resulting in neuroinflammation exacerbated by the presence of the L351P NLRP3 activating mutation (Figs. 5 and 6). Findings that expression of hyperactive Nlrp3 alleles can tip the balance towards nigrostriatal inflammation and motor deficits at advanced age frame the question of whether genetic and/or environmental triggers could further upset this balance triggering sustained neurogenic inflammation capable of activating astrocytes and microglia within the nigrostriatal microenvironment.
In our study, we identified motor abnormalities specific to the L351P NLRP3 mutation in the absence of nigral cell loss. In this context, we observed changes in glial populations indicating astroglial activation in both the substantia nigra and the striatum, as well as microglial activation in striatal tissues (Figs. 5 and 6). Changes in astrocyte phenotype may help to reconcile the finding of motor deficits in animals with no significant loss of dopaminergic neurons (Figs. 2 and 4). Perisynaptic astroglia are critical for neurotransmission, forming tripartite synapses that include pre- and post-synaptic neurons [76]. Astrocyte activation, observed in conditions including stroke [77] and neurodegeneration, alters synaptic function and neurotransmission [78]. Therefore, it stands to reason that dopamine transmission in the striatum may be altered in concert with the pronounced proinflammatory astrocyte activation we observe in the Nlrp3L351P mice (Fig. 6) and thereby contribute to synaptic inefficiency and motor deficits. This possibility is supported by our finding of less severe neuroinflammatory and behavioral deficits in Nlrp3A350V animals despite similar numbers of nigral neurons (Fig. 4). Our findings are also reminiscent of PD patients who have clinical manifestations of PD based on motor symptomology, but upon imaging analysis has no observable loss of dopaminergic neurons [79]. This patient population has been referred to as “Scans Without Evidence for Dopaminergic Deficit (SWEDD).” A plausible prediction is that we have modeled the earliest stages of motor abnormalities resulting from neuronal distress associated with widespread inflammation and that analysis of animals at more advanced ages could reveal neurodegeneration.
Recently, there is an increasing appreciation for the role of astroglia in PD [80]. We observed specific changes in numbers of GFAP-immunoreactive astrocytes in the SNpC of animals expressing Nlrp3L351P, compared with animals expressing Nlrp3WT and Nlrp3A350V (Fig. 5). However, it is important to note that loss of animals in our Nlrp3A350V cohort reducing the group size to six may have impacted our statistical power. Astroglial changes were more pronounced in animals expressing Nlrp3L351P, observable in both the SNpC and the striatum, compared to phenotypically activated IBA1-immunoreactive microglia, which were only observed in the striatum (Fig. 6). To more closely examine these astrocytes, we measured mRNA expression and observed evidence of an enhanced proinflammatory A1 astrocyte signature only in Nlrp3L351P animals (Fig. 6). Previous reports of glial biology in PD patient brains report evidence of astrogliosis in the SNpC, and many of the PD genes (“PARK genes”) are known to be important in maintaining cellular processes in astrocytes, as well as in neurons [80]. The temporal function of astroglia within the processes of PD-associated neuroinflammation and neurodegeneration remains unclear. The majority of evidence suggests that astrocytes become activated subsequent to microglial activation [81]; however, it has also been shown that astrocytes can become activated in the absence of microglia-specific cytokines [82]. It is plausible that in our model, distressed DA neurons harboring elevated NLRP3 inflammasome activity communicate directly with astrocytes, consistent with the reported neuroprotective role of astroglia. We posit that at advanced ages in contexts of elevated cellular stress modeled in this study with NLRP3-activating mutations, the neuroprotective function of astrocytes is unable to persist, and ultimately the proinflammatory astroglial phenotype triggers microglial activation (Figs. 5 and 6). Our decision to analyze mice retaining a wild-type Nlrp3 allele in all cell types creates both opportunities and confounds. As a result, it is impossible to distinguish NLRP3 inflammasome activity directly related to inclusion of the hyperactivated transgene from wild-type NLRP3 inflammasome activity in glia resulting from the neuronal perturbation. For this reason, we are unable to determine with certainty that the Nlrp3L351P allele was not expressed inappropriately in glia, although we believe this to be unlikely due to previous studies utilizing this strain and our characterization of microglia and astrocytes [49–52]. Despite the shortcomings of this model system, the inclusion of an endogenous Nlrp3 allele was likely a critical factor in our discovery of an expanding neuroinflammatory phenotype resulting from expression of the Nlrp3L351P allele of neurons, a finding of importance as we work to understand the earliest stages of neuroinflammation in neurodegenerative disease. Further experiments to directly test the function of neuronal NLRP3, as well as astrocyte and microglia-specific activation in similar model systems, will more clearly characterize these important neuroglia interactions. Such studies will continue to be of high interest as they may help identify novel neuroprotective targets operating at the earliest stages of inflammatory neurodegenerative disorders.
Conclusions
Neuroinflammation is pathologically associated with many neurodegenerative disorders. We recently reported expression of NLRP3 in dopaminergic neurons in late-stage PD patient tissue [40] and have in vivo evidence that loss of Nlrp3 may prevent the progression of pathology in a toxicant model of PD [14]. Here, we developed a longitudinal mouse model to specifically determine the impact of neuronal NLRP3 activity in aging. Findings from this study conclude that aberrate NLRP3 function in DA neurons, due to the presence of Nlrp3-activating mutations associated with the CAPS diseases, results in motor function deficit and proinflammatory astrogliosis. These results are significant because they highlight complex distress-related neuroglial processes and provide a model of molecular events that exist at the transition from tissue homeostasis to progressive degenerative disease.
Supplementary information
Additional file 1: Supplementary Figure 1. Open field analysis characterizing motor function throughout longitudinal study. Behavioral analysis of mice expressing Nlrp3WT (n = 11), Nlrp3A350V (n = 6), and Nlrp3L351P (n = 9) was conducted at 6, 10, 14 and 18-month time-points. (A) Mice expressing Nlrp3L351P became significantly less mobile over time (two-way ANOVA, p = 0.0018 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0027) and (B) had a significant reduction in average velocity (two-way ANOVA, p = 0.0008 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0020). (C) Older animals of all genotypes exhibited significant changes in amount of time spent in the center of the testing box over the time-course of the longitudinal study (two-way ANOVA, p = 0.0319 (time)). Error bars represent s.e.m.
Additional file 2: Supplementary Figure 2. IL-1β protein detected in TH-immunoreactive axons. Histologic section of SNpC tissues from 18-month old animals were stained with anti-IL-1β and anti-TH antibodies. (A) Confocal microscopy of tissues from Nlrp3L351P animals revealed IL-1β protein expression in TH- immunoreactive axons (bottom panels) but not in TH-immunoreactive cell bodies (top panels). Scale bar represents 20 μM. (B) Colocalization of IL-1β and TH was observed in histologic sections from all three genotypes. Scale bar represents 10 μM.
Acknowledgements
We are grateful to the animals and shared resources at Dartmouth that made this study possible. We thank Rebecca O’Meara for supporting our IHC studies and Kenneth Orndorff for his confocal microscopy expertise. We acknowledge the Pathology and Irradiation, Pre-clinical Imaging-Irradiation, Pre-clinical Imaging, and Microscopy Shared Resource facilities at the Norris Cotton Cancer Center at Dartmouth with NCI Cancer Center Support Grant 5P30 CA023108-3.
Abbreviations
- AD
Alzheimer’s disease
- ALR
Absent in melanoma 2-like receptor
- ANOVA
Analysis of variance
- ASC
Apoptosis-associated speck-like protein containing a CARD
- CAPS
Cryopyrin-associated periodic syndromes
- CNS
Central nervous system
- DA
Dopaminergic
- FCAS
Familial cold autoinflammatory syndrome
- GFAP
Glial fibrillary acidic protein
- HRP
Horse radish peroxidase
- IBA1
Ionized calcium-binding adaptor molecule 1
- IL-1β
Interleukin-1 beta
- LUHMES
Lund human mesencephalic cells
- MWS
Muckle-Wells syndrome
- NLR
Nucleotide-binding domain-like receptor
- Nlrp3
NACHT, LRR, and PYD domains-containing protein 3
- NOMID
Neonatal onset multisystem inflammatory disease
- PARK genes
Parkinson’s associated genes
- PD
Parkinson’s disease
- ROS
Reactive oxygen species
- SNpC
Substantia nigra pars compacta
- SWEDD
Scans without evidence for dopaminergic deficit
- TH
Tyrosine hydroxylase
Authors’ contributions
KvH and MH designed the study, analyzed data, and wrote the manuscript. KvH, FA, EM, and AY conducted experiments. All authors read and approved the final manuscript.
Funding
This study was funded by a grant from the National Institute of Environmental Health Sciences (5R01ES024745-05) and a fellowship from the National Institute of Neurological Disorders and Stroke (1F31NS105245-01A1).
Availability of data and materials
Data sharing is not applicable as no datasets were generated or analyzed in this study.
Ethics approval
All animal protocols were approved by Dartmouth Institutional Animal Care and Use Committee and conducted in accordance with federal guidelines pertaining to animal research. All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Dartmouth’s protocol entitled “Animal Models of Neurodegenerative Diseases”, protocol no. 00002117.
Consent for publication
Not applicable.
Competing interests
There are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12974-020-01866-6.
References
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14(9):1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277(1):61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19(8):477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27(9):673–684. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman HM, et al. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29(3):301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann J, et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71(1):198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortimer L, et al. NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat Immunol. 2016;17(10):1176–1186. doi: 10.1038/ni.3538. [DOI] [PubMed] [Google Scholar]
- 13.Booshehri LM, Hoffman HM. CAPS and NLRP3. J Clin Immunol. 2019;39(3):277–286. doi: 10.1007/s10875-019-00638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez EM, et al. Editor’s highlight: Nlrp3 is required for inflammatory changes and nigral cell loss resulting from chronic intragastric rotenone exposure in mice. Toxicol Sci. 2017;159(1):64–75. doi: 10.1093/toxsci/kfx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman L, et al. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J Exp Med. 2017;214(5):1351–1370. doi: 10.1084/jem.20150237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E, et al. MPTP-driven NLRP3 inflammasome activation in microglia plays a central role in dopaminergic neurodegeneration. Cell Death Differ. 2019;26(2):213–228. doi: 10.1038/s41418-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar S, et al. Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson’s disease. NPJ Parkinsons Dis. 2017;3:30. doi: 10.1038/s41531-017-0032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Z, et al. Tenuigenin protects dopaminergic neurons from inflammation via suppressing NLRP3 inflammasome activation in microglia. J Neuroinflammation. 2017;14(1):256. doi: 10.1186/s12974-017-1036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heneka MT, McManus RM, Latz E. Inflammasome signalling in brain function and neurodegenerative disease. Nat Rev Neurosci. 2018;19(10):610–621. doi: 10.1038/s41583-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, et al. Pyroptosis in liver disease: new insights into disease mechanisms. Aging Dis. 2019;10(5):1094–1108. doi: 10.14336/AD.2019.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebeaupin C, et al. ER stress induces NLRP3 inflammasome activation and hepatocyte death. Cell Death Dis. 2015;6:e1879. doi: 10.1038/cddis.2015.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao L, et al. Loss-of-function CARD8 mutation causes NLRP3 inflammasome activation and Crohn’s disease. J Clin Invest. 2018;128(5):1793–1806. doi: 10.1172/JCI98642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehto S, et al. The Crohn’s disease risk factor IRGM limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Mol Cell. 2019;73(3):429–445. doi: 10.1016/j.molcel.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song-Zhao GX, et al. Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunol. 2014;7(4):763–774. doi: 10.1038/mi.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhen Y, Zhang H. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol. 2019;10:276. doi: 10.3389/fimmu.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Z, et al. Mitochondrial dysfunction induces NLRP3 inflammasome activation during cerebral ischemia/reperfusion injury. J Neuroinflammation. 2018;15(1):242. doi: 10.1186/s12974-018-1282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, et al. EK7 regulates NLRP3 inflammasome activation and neuroinflammation post-traumatic brain injury. Front Mol Neurosci. 2019;12:202. doi: 10.3389/fnmol.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Rivero Vaccari JP, et al. A molecular platform in neurons regulates inflammation after spinal cord injury. J Neurosci. 2008;28(13):3404–3414. doi: 10.1523/JNEUROSCI.0157-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adamczak SE, et al. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J Cereb Blood Flow Metab. 2014;34(4):621–629. doi: 10.1038/jcbfm.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WP, et al. Heme oxygenase-1 promotes neuron survival through down-regulation of neuronal NLRP1 expression after spinal cord injury. J Neuroinflammation. 2016;13(1):52. doi: 10.1186/s12974-016-0521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaushal V, et al. Neuronal NLRP1 inflammasome activation of Caspase-1 coordinately regulates inflammatory interleukin-1-beta production and axonal degeneration-associated Caspase-6 activation. Cell Death Differ. 2015;22(10):1676–1686. doi: 10.1038/cdd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14(7):463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 34.Voet S, et al. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol Med. 2019:11(6). [DOI] [PMC free article] [PubMed]
- 35.Codolo G, et al. Triggering of inflammasome by aggregated alpha-synuclein, an inflammatory response in synucleinopathies. PLoS One. 2013;8(1):e55375. doi: 10.1371/journal.pone.0055375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou R, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 37.Bai H, et al. Cathepsin B links oxidative stress to the activation of NLRP3 inflammasome. Exp Cell Res. 2018;362(1):180–187. doi: 10.1016/j.yexcr.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Anderson FL, et al. Inflammasomes: an emerging mechanism translating environmental toxicant exposure into neuroinflammation in Parkinson’s disease. Toxicol Sci. 2018;166(1):3–15. doi: 10.1093/toxsci/kfy219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee E, et al. MPTP-driven NLRP3 inflammasome activation in microglia plays a central role in dopaminergic neurodegeneration. Cell Death Differ. 2018. [DOI] [PMC free article] [PubMed]
- 40.von Herrmann KM, et al. NLRP3 expression in mesencephalic neurons and characterization of a rare NLRP3 polymorphism associated with decreased risk of Parkinson’s disease. NPJ Parkinsons Dis. 2018;4:24. doi: 10.1038/s41531-018-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Z, et al. The NLRP3 inflammasome is involved in the pathogenesis of Parkinson’s disease in rats. Neurochem Res. 2017;42(4):1104–1115. doi: 10.1007/s11064-017-2185-0. [DOI] [PubMed] [Google Scholar]
- 42.Gordon R, et al. Inflammasome inhibition prevents alpha-synuclein pathology and dopaminergic neurodegeneration in mice. Sci Transl Med. 2018;10:465. doi: 10.1126/scitranslmed.aah4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heneka MT, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493(7434):674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saresella M, et al. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol Neurodegener. 2016;11:23. doi: 10.1186/s13024-016-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan MS, et al. NLRP3 polymorphisms are associated with late-onset Alzheimer’s disease in Han Chinese. J Neuroimmunol. 2013;265(1-2):91–95. doi: 10.1016/j.jneuroim.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Tan MS, et al. The NLRP3 inflammasome in Alzheimer’s disease. Mol Neurobiol. 2013;48(3):875–882. doi: 10.1007/s12035-013-8475-x. [DOI] [PubMed] [Google Scholar]
- 47.Haque ME, et al. Targeting the microglial NLRP3 inflammasome and its role in Parkinson’s disease. Mov Disord. 2019. [DOI] [PubMed]
- 48.Brydges SD, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30(6):875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K, et al. Temporally restricted dopaminergic control of reward-conditioned movements. Nat Neurosci. 2020;23(2):209–216. doi: 10.1038/s41593-019-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosillo P, et al. Tsc1-mTORC1 signaling controls striatal dopamine release and cognitive flexibility. Nat Commun. 2019;10(1):5426. doi: 10.1038/s41467-019-13396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson BG, et al. RIM is essential for stimulated but not spontaneous somatodendritic dopamine release in the midbrain. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 52.Backman CM, et al. Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis. 2006;44(8):383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- 53.Zhang P, et al. Cdk5-dependent activation of neuronal inflammasomes in Parkinson’s disease. Mov Disord. 2016;31(3):366–376. doi: 10.1002/mds.26488. [DOI] [PubMed] [Google Scholar]
- 54.Fann DY, et al. Evidence that NF-kappaB and MAPK signaling promotes NLRP inflammasome activation in neurons following ischemic stroke. Mol Neurobiol. 2018;55(2):1082–1096. doi: 10.1007/s12035-017-0394-9. [DOI] [PubMed] [Google Scholar]
- 55.Liu HD, et al. Expression of the NLRP3 inflammasome in cerebral cortex after traumatic brain injury in a rat model. Neurochem Res. 2013;38(10):2072–2083. doi: 10.1007/s11064-013-1115-z. [DOI] [PubMed] [Google Scholar]
- 56.Youm YH, et al. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell Rep. 2012;1(1):56–68. doi: 10.1016/j.celrep.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youm YH, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18(4):519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spadaro O, et al. Growth hormone receptor deficiency protects against age-related NLRP3 inflammasome activation and immune senescence. Cell Rep. 2016;14(7):1571–1580. doi: 10.1016/j.celrep.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, et al. Critical role of NLRP3-caspase-1 pathway in age-dependent isoflurane-induced microglial inflammatory response and cognitive impairment. J Neuroinflammation. 2018;15(1):109. doi: 10.1186/s12974-018-1137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stutz A, et al. ASC speck formation as a readout for inflammasome activation. Methods Mol Biol. 2013;1040:91–101. doi: 10.1007/978-1-62703-523-1_8. [DOI] [PubMed] [Google Scholar]
- 61.Corrigan F, et al. Neurogenic inflammation after traumatic brain injury and its potentiation of classical inflammation. J Neuroinflammation. 2016;13(1):264. doi: 10.1186/s12974-016-0738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labzin LI, Heneka MT, Latz E. Innate immunity and neurodegeneration. Annu Rev Med. 2018;69:437–449. doi: 10.1146/annurev-med-050715-104343. [DOI] [PubMed] [Google Scholar]
- 63.Teh DBL, et al. Transcriptome analysis reveals neuroprotective aspects of human reactive astrocytes induced by interleukin 1beta. Sci Rep. 2017;7(1):13988. doi: 10.1038/s41598-017-13174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herx LM, Yong VW. Interleukin-1 beta is required for the early evolution of reactive astrogliosis following CNS lesion. J Neuropathol Exp Neurol. 2001;60(10):961–971. doi: 10.1093/jnen/60.10.961. [DOI] [PubMed] [Google Scholar]
- 65.Srinivasan D, et al. Cell type-specific interleukin-1beta signaling in the CNS. J Neurosci. 2004;24(29):6482–6488. doi: 10.1523/JNEUROSCI.5712-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huitinga I, et al. IL-1beta immunoreactive neurons in the human hypothalamus: reduced numbers in multiple sclerosis. J Neuroimmunol. 2000;107(1):8–20. doi: 10.1016/s0165-5728(00)00248-4. [DOI] [PubMed] [Google Scholar]
- 67.Zecca L, et al. Substantia nigra neuromelanin: structure, synthesis, and molecular behaviour. Mol Pathol. 2001;54(6):414–418. [PMC free article] [PubMed] [Google Scholar]
- 68.Monzani E, et al. Dopamine, oxidative stress and protein-quinone modifications in Parkinson’s and other neurodegenerative diseases. Angew Chem Int Ed Engl. 2019;58(20):6512–6527. doi: 10.1002/anie.201811122. [DOI] [PubMed] [Google Scholar]
- 69.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 70.Surmeier DJ. Determinants of dopaminergic neuron loss in Parkinson’s disease. FEBS J. 2018;285(19):3657–3668. doi: 10.1111/febs.14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan Y, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160(1-2):62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J, et al. Dopamine D2 receptor restricts astrocytic NLRP3 inflammasome activation via enhancing the interaction of beta-arrestin2 and NLRP3. Cell Death Differ. 2018;25(11):2037–2049. doi: 10.1038/s41418-018-0127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Starke K, Gothert M, Kilbinger H. Modulation of neurotransmitter release by presynaptic autoreceptors. Physiol Rev. 1989;69(3):864–989. doi: 10.1152/physrev.1989.69.3.864. [DOI] [PubMed] [Google Scholar]
- 74.Ford CP. The role of D2-autoreceptors in regulating dopamine neuron activity and transmission. Neuroscience. 2014;282:13–22. doi: 10.1016/j.neuroscience.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hemby SE, Trojanowski JQ, Ginsberg SD. Neuron-specific age-related decreases in dopamine receptor subtype mRNAs. J Comp Neurol. 2003;456(2):176–183. doi: 10.1002/cne.10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanco-Suarez E, Caldwell AL, Allen NJ. Role of astrocyte-synapse interactions in CNS disorders. J Physiol. 2017;595(6):1903–1916. doi: 10.1113/JP270988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gleichman AJ, Carmichael ST. Astrocytic therapies for neuronal repair in stroke. Neurosci Lett. 2014;565:47–52. doi: 10.1016/j.neulet.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 78.Rama Rao KV, et al. Amyloid-beta inhibits thrombospondin 1 release from cultured astrocytes: effects on synaptic protein expression. J Neuropathol Exp Neurol. 2013;72(8):735–744. doi: 10.1097/NEN.0b013e31829bd082. [DOI] [PubMed] [Google Scholar]
- 79.Nalls MA, et al. Diagnosis of Parkinson’s disease on the basis of clinical and genetic classification: a population-based modelling study. Lancet Neurol. 2015;14(10):1002–1009. doi: 10.1016/S1474-4422(15)00178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Booth HDE, Hirst WD, Wade-Martins R. The role of astrocyte dysfunction in Parkinson’s disease pathogenesis. Trends Neurosci. 2017;40(6):358–370. doi: 10.1016/j.tins.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clarke LE, et al. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A. 2018;115(8):E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. Open field analysis characterizing motor function throughout longitudinal study. Behavioral analysis of mice expressing Nlrp3WT (n = 11), Nlrp3A350V (n = 6), and Nlrp3L351P (n = 9) was conducted at 6, 10, 14 and 18-month time-points. (A) Mice expressing Nlrp3L351P became significantly less mobile over time (two-way ANOVA, p = 0.0018 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0027) and (B) had a significant reduction in average velocity (two-way ANOVA, p = 0.0008 (genotype), p < 0.0001 (time); Tukey’s post hoc multiple comparisons test, ** p = 0.0020). (C) Older animals of all genotypes exhibited significant changes in amount of time spent in the center of the testing box over the time-course of the longitudinal study (two-way ANOVA, p = 0.0319 (time)). Error bars represent s.e.m.
Additional file 2: Supplementary Figure 2. IL-1β protein detected in TH-immunoreactive axons. Histologic section of SNpC tissues from 18-month old animals were stained with anti-IL-1β and anti-TH antibodies. (A) Confocal microscopy of tissues from Nlrp3L351P animals revealed IL-1β protein expression in TH- immunoreactive axons (bottom panels) but not in TH-immunoreactive cell bodies (top panels). Scale bar represents 20 μM. (B) Colocalization of IL-1β and TH was observed in histologic sections from all three genotypes. Scale bar represents 10 μM.
Data Availability Statement
Data sharing is not applicable as no datasets were generated or analyzed in this study.