Abstract
Cyclic GMP-AMP (cGAMP) synthase (cGAS) is a major responder to the pathogenic DNA of viruses and bacteria. Upon DNA binding, cGAS becomes enzymatically active to generate the second messenger cGAMP, leading to activation of inflammatory genes, type I interferon production, autophagy and cell death. Following genotoxic stress, cGAS can also respond to endogenous DNA, deriving from mitochondria, endogenous retroelements, and chromosomes to affect cellular signaling, secretion and cell fate decisions. However, under unperturbed conditions, signaling from self-DNA is largely, but not completely, inhibited. Here we review how endogenous DNA is exposed to cGAS, how signaling is attenuated but activated under pathological conditions, and how low-level signaling under unperturbed conditions might prime anti-pathogenic responses.
Keywords: Inflammatory signaling, cGAS, STING, self-DNA, genome integrity
cGAS – a DNA sensor for the innate immune system
In addition to the adaptive immune system, which responds to ever changing pathogen components, there is an evolutionarily older branch of the immune system known as the innate immune system [1]. Through the multifunctional and heterogeneous network of responders and effectors of the pattern recognition receptors (see Glossary), the innate immune system can respond to common pathogen components whose structures are stable over evolutionary time scales. Nucleic acids are one such component, and can potently activate innate immune signalling [2]. The presence of nucleic acids within host cells, however, raises three important questions [2]: (1) Do cells ensure that their own nucleic acids do not trigger innate immune signaling? (2) If cells disfavor activation by their own nucleic acids, how is this achieved? (3) If activation by self-DNA does occur, what are the consequences? Here we discuss this problem by using the example of the cyclic GMP-AMP (cGAMP) synthase cGAS, a major responder to DNA (Figure 1) [3]. Reviews on other innate immune DNA sensors and RNA sensors can be found elsewhere [4–6].
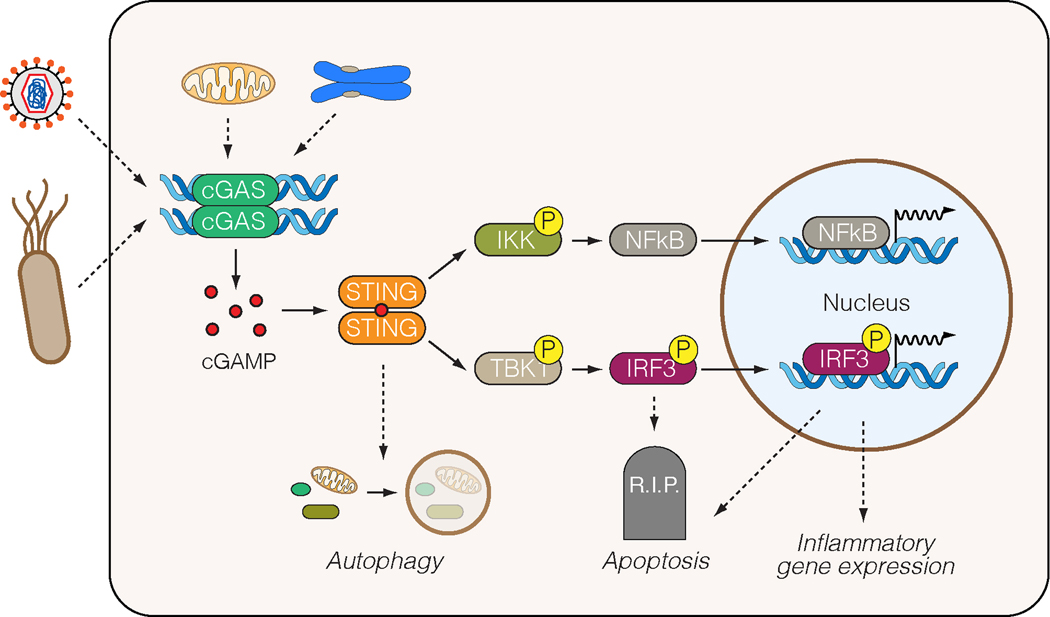
Figure 1. The cGAS pathway.
Classically, cGAS is stimulated by DNA from viruses and bacteria, but self-DNA such as chromosomal DNA or mitochondrial DNA can also stimulate it. Upon DNA binding, cGAS becomes enzymatically active and produces a second messenger, cyclic GMP-AMP (cGAMP). In turn, cGAMP binds and activates STING. This can promote autophagy, but also activates the inflammatory proteins kinases IKK and TBK1. This causes activation of NFkB and IRF3 in order to stimulate transcription of inflammatory genes. IRF3 also has a second, less well-defined activity that leads to apoptosis.
Catalytically inert on its own, cGAS only synthesizes cGAMP upon DNA binding [7]. cGAMP, a second messenger, then activates downstream signaling via an adapter, the endoplasmic reticulum (ER)-associated protein STING (stimulator of interferon genes) [7]. In turn, cGAMP-bound STING activates effector proteins, predominantly through kinase intermediaries such as TBK1 (TANK binding kinase 1) and IKK (inhibitor of nuclear factor kappa B kinase). Prominent downstream effectors include IRF3 (interferon regulatory factor 3) and NFkB (nuclear factor kappa B), which activate inflammatory gene expression and production of type I interferons. In addition, under certain circumstances, IRF3 may also have a transcription-independent function to promote apoptosis [8–10].
Upon discovery of cGAS, self-discrimination was recognized as a major question [3]. Indeed, several autoimmune diseases are connected to increased cytoplasmic DNA load, such as Aicardi-Gutieres syndrome (AGS), which can be caused by mutations in the main cytoplasmic nuclease, TREX1 (three prime repair exonuclease 1), and whose clinical manifestations in mouse models can largely be abrogated by reducing cGAS levels [7]. Thus, suppression of autoactivation of cGAS by self-DNA is critical to prevent autoimmune diseases.
Sources of self-DNA
Eukaryotes possess three major types of cellular self-DNA (Figure 2): mitochondrial DNA (mtDNA), the DNA of reverse-transcribed endogenous retroelements, and chromosomal DNA. A variation of these types of self-DNA can be present in phagocytes following phagocytosis of other cells or circulating extracellular DNA. This can lead to the intracellular presence of DNA that, in terms of the organism is self-DNA, but in terms of the concerned cell is non-self.
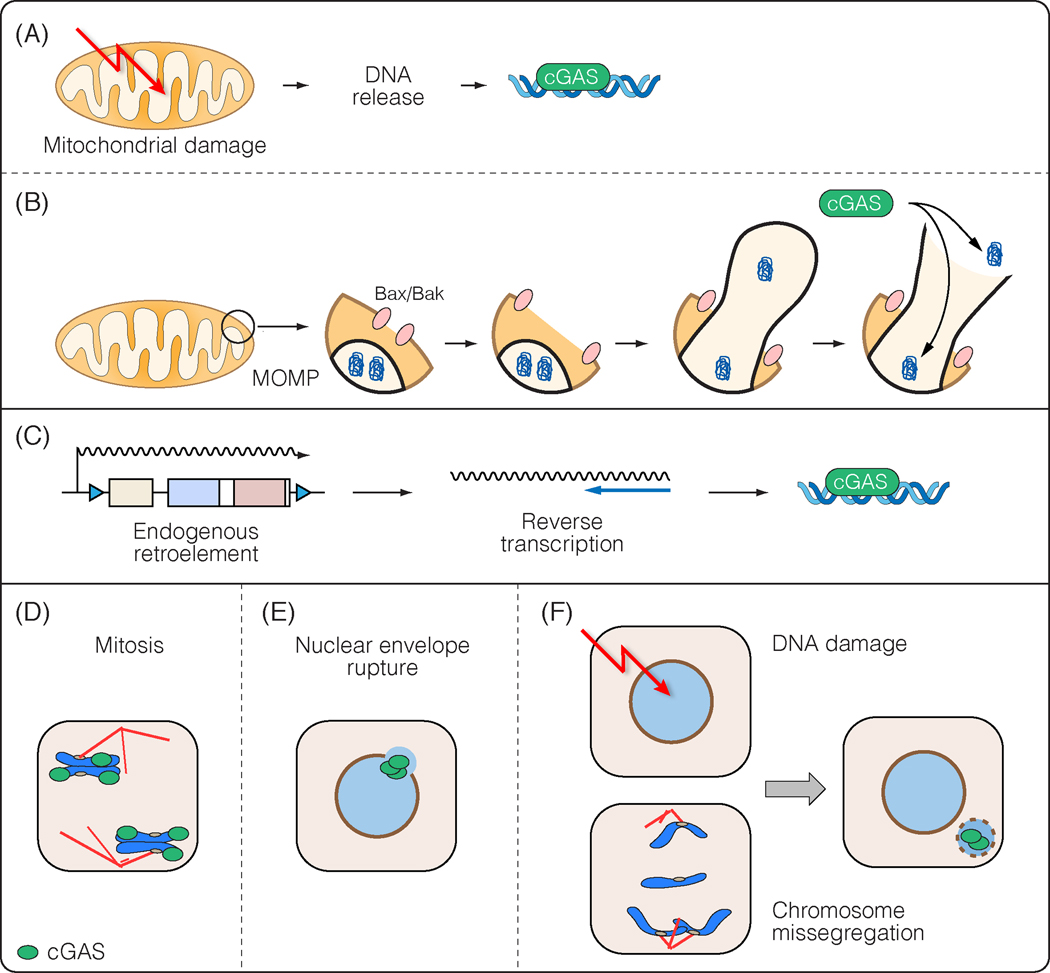
Figure 2: Sources of self-DNA for cGAS activation.
(A and B) Mitochondrial DNA. (A) Mitochondrial damage can lead to release of mitochondrial DNA (mtDNA). (B) A key event in apoptosis is mitochondrial outer membrane permeabilisation (MOMP), mediated by pores generated by the proteins Bax and Bak. These can expand to form macropores, through which the inner mitochondrial membrane can herniate. By hitherto unknown mechanisms, this can lead to release of mtDNA, and/or and influx of cGAS. (C) DNA of endogenous retroelements. If loss of silencing of endogenous retroelements occurs, their reverse transcription can lead to cytoplasmic DNA that can activate cGAS. (D – F) Chromosomal DNA. (D) Following mitotic nuclear envelope disassembly, cGAS quantitatively associates with chromosomes. (E) The nuclear envelope can sometimes break locally during interphase. This results in influx of cGAS. (F) DNA damage and chromosome missegregation can result in the formation of micronuclei, which assemble nuclear envelopes that are prone to rupture and enrichment of cGAS. Note that DNA damage may also generate other DNA fragments that may activate cGAS.
mtDNA.
Mitochondrial DNA (mtDNA) is encased within two membrane systems, the outer and inner mitochondrial membrane (OMM and IMM, respectively), where it is inaccessible to cGAS [11]. However, mitochondrial integrity can be compromised following mitochondrial damage, and during apoptosis. Damaged mitochondria are cleared in an autophagic process, mitophagy, and defects in mitophagy can lead to STING-dependent inflammatory gene induction [12]. Mitochondrial damage also occurs during infection with Dengue virus [13], suggesting a possible explanation for a conundrum: cGAS can help protect against infection by Dengue virus and other flaviviruses [13,14], despite the fact that these contain RNA genomes. Thus, cellular dysfunction during infection could lead to mtDNA release, ultimately allowing cGAS to defend against RNA viruses.
During the initiation of apoptosis, the OMM becomes permeabilized by Bax (BCL2 associated X) and Bak (BCL2 antagonist/killer). This mitochondrial outer membrane permeabilization (MOMP) [15] releases pro-apoptotic proteins from the mitochondrial inter-membrane space and ultimately results in activation of effector caspases, which rapidly promote chromosomal DNA degradation and kill the cell [15]. If caspases are inhibited while apoptosis is stimulated, mtDNA becomes accessible to cGAS [16], which will then induce inflammatory genes [16,17]. Until recently, MOMP was thought to be a point of no return in apoptosis, but careful examination of mitochondrial dynamics following weak apoptotic stimuli indicated that MOMP can also occur in a subset of mitochondria without full engagement of apoptosis, a process termed minority MOMP [18]. These mechanisms could contribute to inflammatory signaling under more physiological conditions than the dramatic manipulation of caspase inhibition under strong apoptotic stimuli. However, by definition, MOMP only affects the OMM, and MOMP can therefore not immediately explain how mitochondrial DNA is released to activate cGAS. Bax and Bak can promote large disruptions in the OMM during MOMP, facilitating IMM herniations into the cytoplasm; and these herniations are thought to be involved in mtDNA release [19,20]. Similarly, large pores can be generated by the OMM channel protein VDAC (voltage-dependent anion channel), which can also contribute to inflammatory gene expression [21]. While it has been suggested that VDAC oligomers mediate efflux of fragments of mtDNA [21], it is currently unknown how the IMM is disrupted and how mtDNA is fragmented. Perhaps, rather than large-scale efflux of mtDNA, small disruptions of the IMM might allow an influx of cGAS, which in turn might allow the generation of cGAMP and induction of inflammatory genes. Altogether, it is possible that mtDNA could be used by the host to sense a large number of cellular dysfunctions in order to activate inflammatory signaling and perhaps apoptosis.
Endogenous retroelements.
While generally silenced, endogenous retroelements can be activated under certain conditions, and may affect immune responses [22,23]. Baseline expression of retroelements can contribute to innate immune responses in cells lacking TREX1, and may therefore be relevant for autoinflammatory diseases [24]. Similar to TREX1, deficiency in RNaseH2 (ribonuclease H II), which removes RNA from DNA/RNA hybrids arising from lagging strand replication, replication errors, transcription and reverse transcription, can also lead to an increase in retroelements and cGAS signaling [25,26]. Extensive activation of endogenous retroelements has been reported during cellular senescence and ageing [27–29], and recognition of reverse transcribed retroelements by cGAS may contribute to the inflammation and pathology associated with aging [28,29]. One potential caveat is suggested by the recent finding that RNaseH2 deficiency can also cause DNA damage and micronuclei [26,30], suggesting an alternative pathway to activate cGAS in situations correlating with upregulated retroelements. Similarly, aging is associated with DNA damage [31], which could contribute to cGAS activation. Overall, cGAS signaling by endogenous retroelements is an exciting possibility, which we expect to be clarified in the future.
Chromosomal DNA.
cGAS was originally described as a cytoplasmic protein [3], with the implication that the nuclear envelope (NE) provides a major mechanism to prevent activation by chromosomal self-DNA. Recently, this concept was revised, revealing that cGAS localization is complex, and that most cell types may contain both a cytoplasmic and a nuclear pool of cGAS (Box 1). Regardless of the exact nature of cGAS subcellular localization, the NE cannot completely protect chromosomal DNA, as it disassembles during the open mitosis that occurs during metazoan cell divisions. Consistently, cGAS is sequestered onto chromosomes during mitosis [10,32,33]. In addition, the NE may break occasionally, allowing influx of cGAS [30,33–36]. Together, these findings suggest that if cGAS activation by self-DNA is limited, the NE cannot be the sole mechanism.
Box 1. The subcellular localization of cGAS.
Based on immunofluorescence microscopy and subcellular fractionation experiments, cGAS was initially postulated to be a cytoplasmic protein [3]. Since then, multiple analyses indicated that at least some cGAS could be nuclear [32,33,63,70,71,86,87], with one report even suggesting that cGAS might almost completely be nuclear [77]. These findings contrast with a limited number of studies indicating nuclear exclusion [88] or predominant association with the plasma membrane [72]. While the use of different cell types and fixation conditions, as well as possible nuclear envelope damage during biochemical fractionations could be responsible for some of the enrichment/exclusivity of cGAS within the nucleus, two conclusions are suggested by time-lapse microscopy imaging of fluorescently tagged cGAS: in interphase, newly synthesized cGAS is predominantly in the cytoplasm [10,32–35], but following mitotic nuclear envelope disassembly, cGAS is predominantly enriched on mitotic chromosomes [10,32,33,63]. Following mitosis, cGAS persists inside the nucleus for a prolonged time, with a slow appearance of cytoplasmic signal [32,33,63], together suggesting a mechanism for maintaining a pool of cGAS within the nucleus in proliferating cells [32]. Because the absence of the protein NONO (non-POU domain containing octamer binding) resulted in reduced nuclear cGAS signal [70], it is possible that NONO is required for nuclear retention of cGAS following mitosis. Overall it seems clear that cGAS cannot all be within the nucleus at all times, as cytoplasmic DNA can activate cGAS, and cGAS can be visualized on cytoplasmic DNA [3]. Furthermore, upon nuclear envelope rupture on micronuclei, an intense accumulation of cGAS occurs [30,33,36], and upon rupture of the cell’s main nucleus, dramatic enrichment of cGAS occurs close to the rupture point [89]. Similarly, many studies are largely in agreement that at least a pool of nuclear cGAS exists. The presence of cGAS within the nucleus could allow cells to respond to viruses, such as Herpesviruses and HIV, which can deliver their DNA into the nucleus, rather than the cytoplasm [70,86]. Furthermore, nuclear enrichment of cGAS might allow response to self-DNA such as following DNA damage, but potentially also during steady-state.
Irrespective of an involvement of the NE, a large body of work indicated that cGAS activation can occur following chromosomal DNA damage. Loss of a master regulator of the DNA damage response, the kinase ATM (Ataxia telangiectasia mutated), can result in cGAS-STING dependent inflammatory gene expression [37–39], although there is also one, seemingly contradictory, report suggesting that ATM can directly activate inflammatory signaling [40]. Similarly, defects in damage processing and DNA repair result in cGAS activation and inflammatory gene induction [33,41–45]. How exactly DNA damage can activate cGAS is currently unclear. One unifying possibility is that micronuclei, a common consequence of DNA damage, are involved. Micronuclei assemble defective NEs prone to rupture [46,47], and rupture of micronuclear NEs is associated with, and a requirement for, dramatic enrichment of cGAS [30,33,36,48]. For one type of DNA damage, ionizing radiation, the passage through mitosis, which generates micronuclei, is required for cGAS-dependent signaling and inflammatory gene expression [30,33]. Micronuclei that are generated following minority MOMP [18] may also contribute to stimulation of inflammatory gene expression in addition to mtDNA. Chromosomal instability, frequently observed in cancer cells, can also lead to cytoplasmic chromatin and micronuclei, and result in cGAS activation and cGAS dependent gene expression changes [49]. Sources of chromosomal instability can be a propensity for chromosome missegregation, defects in DNA replication, and chromosome fusions [50], the latter of which might functionally be equivalent to micronuclei, and can also be converted to micronuclei upon breakage [51]. However, although micronuclei generally correlate with DNA damage and situations that activate cGAS, a proper causal relationship has not been established. Furthermore, it is not clear if micronuclei generated from different sources and types of DNA are analogous, or if they differ, for example in the specific type of nuclear envelope defect. Single-cell RNA sequencing suggested that not all cells that contain micronuclei show strong induction of inflammatory genes [30], indicating that other types of aberrant DNA, or as yet undescribed regulatory mechanisms could be involved. However, a direct and quantitative comparison between cGAS-dependent gene induction in micronucleated cells with gene induction resulting from cGAS activation by viruses or cytoplasmic DNA is currently lacking.
Another chromosomal source of cytoplasmic DNA and cGAS activation can arise from recombination-mediated telomere extension, termed alternative lengthening of telomeres (ALT), which maintains functional telomeres in some cancer cells [52]. ALT cells tend to accumulate telomeric sequences in the cytoplasm, some of which may form micronuclei which can accumulate cGAS, similar to other types of micronuclei [53,54].
Self- and non-self-DNA discrimination by cGAS
cGAS contains three surfaces with which it interacts with double-stranded DNA (dsDNA) [55]. Residues within all these sites are critical for catalytic activity, and none of these regions show any obvious sequence specificity [55]. Furthermore, in contrast to the endosomal foreign DNA sensor TLR9 (Toll-like receptor 9), which uses DNA methylation patterns combined with low level sequence specificity to provide selectivity for bacterial DNA [56], no effect of DNA methylation on cGAS DNA binding or cGAMP production has so far been reported. Consistently, structural analyses exclusively indicate interactions between cGAS and the phosphodiester bond of standard B-form DNA [55,57–59]. In contrast, cGAS activation may be regulated by structural features of DNA. Activity is generally length dependent, with robust activation only observed with DNA fragments >45 bp [60,61], and smaller fragments requiring flayed ends for potent activation [60]. Length dependence may arise from the fact that cGAS is dimeric in its active form, potentially interacting with two separate DNAs [59,62]. Longer DNA is more prone to bending, allowing the two cGAS molecules of a dimer to interact with two different regions of the DNA [61]. DNA bending, as well as the stabilization of bent DNA by cGAS may support binding by other cGAS dimers, resulting in a type of oligomerization of cGAS on DNA [61]. While these findings potentially suggest that, in vivo, cGAS activation may occur more potently by DNA sequences with a base composition that is more likely to allow bending, this has not been investigated yet. Interestingly, there is one potential exception to both the sequence independence and length-dependence of cGAS activation by DNA. While shorter DNA fragments of random sequence do not induce inflammatory genes very well, transfection with 20 bp dsDNA fragments representing centromeric repeat DNA elicited a strong response [63]. However, since this phenomenon was not further investigated in an in vitro reconstituted system, it is currently unknown if this reflects direct activation of cGAS or indirect effects.
The fact that cGAS prefers bent DNA over straight DNA suggested that proteins that modify the bending status of DNA could affect cGAS activity. Indeed, in vitro, the mitochondrial nucleoid organizing protein TFAM (transcription factor A, mitochondrial), whose binding to DNA stabilizes bends, can be used to stimulate cGAS activation [61]. Similarly, cGAS activation by DNA can be stimulated by HMGB1 (high mobility group box 1), which also mediates bending [61]. This suggested that these proteins, by bending DNA and thereby supporting cGAS activation, could promote innate immune responses to self-DNA. In vitro reconstitution experiments showed that, at least in principle, addition of TFAM could potentiate cGAMP production by cGAS stimulated with DNA [61]. However, two different effects were observed in a concentration-dependent manner: at low concentrations, stimulation occurred, whereas at higher concentrations (> 1 μM), TFAM had an inhibitory effect. Within mitochondria, TFAM is thought to saturate mtDNA [11], a situation that is more likely to represent the inhibitory regime, but it is unknown what might happen if mtDNA is released during abortive apoptosis or mitochondrial damage. Limiting intracellular TFAM actually promotes cGAS-dependent induction of inflammatory genes, but this was associated with general mitochondrial dysfunction and fragility [64]. Similar to TFAM, HMGB1, depending on the concentration, can act both stimulatory and inhibitory to cGAS [61]. Intracellular concentrations of HMGB1 are estimated to be very high (~4.5 μM, only ~5 times lower than histones [65]) suggesting that, similar to TFAM, HMGB1 might be inhibitory during unperturbed situations. Genetic data point toward HMGB proteins being stimulatory to a wide range of innate immune nucleic acid sensors [66], and HMGB proteins are known to represent danger-associated molecular patterns (DAMPs), compounds released from dying or stressed cells in order to signal to neighboring cells and the immune system [67]. While it is thought that HMGB1 can directly signal via two receptors, RAGE (receptor for advanced glycation end products) and TLR4 (Toll-like receptor 4) [67], it is possible that released HMGB1, together with bound DNA, can stimulate cGAS in receiving cells. Furthermore, while the nuclear concentration of HMGB1 is very high, cytoplasmic concentration could be lower, and promote cGAS activation if it is in the stimulatory regime.
Forcing cGAS into the nucleus elicits cGAMP production, but this occurs at ~500-fold lower levels than what can be achieved by transfection of DNA into the cytoplasm [63]. Similarly, during extended mitotic arrest, when cGAS accumulates on chromosomes following nuclear envelope disassembly, phosphorylated IRF3 gradually accumulates in a manner dependent on cGAS. However, the phosphorylation level is highly attenuated when compared to cGAS activation by transfection of exogenous naked DNA [10]. Together these data indicate that chromosomal DNA is refractory to cGAS stimulation, regardless of the cell cycle stage. How might this be mediated? Chromosomal DNA is generally not found in its naked form, but is arranged into chromatin, the basic unit of which is the nucleosome, formed by ~150 bp of DNA wrapped around a histone octamer (consisting of two copies each of the histone proteins H2A, H2B, H3 and H4) [68]. Nucleosomes are unique to eukaryotes, and are not found within bacteria or viruses, the most common pathogens. Thus, they were speculated to be a signal for self-DNA [69]. Indeed, in vitro, cGAS has reduced activity on reconstituted nucleosomes or isolated cellular chromatin when compared to naked DNA [10,30,70]. Despite the lower catalytic activity on nucleosomes, cGAS actually has higher binding affinity for nucleosomes than for naked DNA [10]. This appears to be because cGAS can bind to nucleosomes by making contacts to histones, and not just DNA, in a conformation that appears to be incompatible with proper catalytic activity [10]. Because of this higher affinity for, and lower activity on, nucleosomes, nucleosomes can act as competitive inhibitors of cGAS stimulation by DNA [10]. This might provide a mechanism by which activation by nucleosome-free chromosomal DNA, such as promoter-associated DNA, is inhibited.
A number of other mechanisms have been proposed by which signaling from chromosomes can be limited. For example, it was suggested that a circular RNA specifically inhibits cGAS inside the nucleus [71]. Furthermore, cGAS was proposed to interact with phosphoinositide phosphate, resulting in quantitative sequestration at the plasma membrane [72], although this localization pattern had not been reported in other studies. If the majority of cGAS does associate with the plasma membrane, it clearly is not completely prevented from finding DNA. In particular, the dramatic and rapid enrichment of cGAS with ruptured micronuclei and chromatin close to damaged nuclear envelopes indicates that cGAS can efficiently be released. Since plasma membrane interactions appear to involve the putatively unstructured cGAS N-terminal region, which was also suggested to undergo liquid-liquid phase separation in the presence of DNA [73], DNA may remove cGAS from the plasma membrane by forcing it into phase separated condensates.
Finally, limiting the generation and half-life of the stimulatory DNA counteracts cGAS activation by self-DNA. This is indicated by the inflammatory phenotypes of diseases caused by mutations of TREX1 and DNASE2 (deoxyribonuclease II) that are caused by increases in the cytoplasmic load of chromosomally derived DNA [7,74–76], and of the inflammatory response to the loss of silencing of endogenous retroelements [25,26,28,29]. Altogether, cGAS restriction on self-DNA is a multifaceted problem, that the cell tackles at multiple levels. Recently, mutations in cGAS were identified that greatly boost cGAMP levels under uninduced conditions [77]. We expect that the identification of the mechanism by which these mutations lead to cGAS activation, and of the stimulatory DNA, will shed light on the main source of self-DNA, and on how cGAS generally prevents such activation.
Functional consequences of self-DNA induced cGAS activation
cGAS activation by self-DNA is thought to impact a number of autoinflammatory disorders, and to regulate cancer development, progression, immune infiltration and therapy response [7,78]. In tumors, as well as cancer cell lines, the cGAS pathway is frequently mutated or silenced, suggesting that it can act to limit tumor growth [79–81], although pro-tumorigenic functions have also been suggested [78]. Here, we will focus on the cellular consequences of cGAS activation by self-DNA.
While cGAS was recently suggested to respond to aberrant self-DNA by directly regulating DNA repair in a cGAMP-independent manner (Box 2), its best-defined functions following self-DNA detection involve catalytic activity and signaling via STING (Figure 3). A major role for cGAS has recently become apparent in regulating cellular senescence, a permanent cell-cycle arrest that classically occurs due to telomere attrition when cells reach their proliferative limit [31]. Because telomere attrition can be seen as a type of DNA damage, it is not surprising that other types of DNA damage can also result in senescence-like growth arrest [31], although it is currently unclear how cells decide between the various possible fates (growth, senescence, apoptosis) following DNA damage. One general aspect of senescence is an upregulation of inflammatory genes in a process known as SASP, senescence-associated secretory phenotype. Multiple studies indicated that the cGAS pathway is a major contributor to the SASP [30,32,33,36,48], although more recent investigation suggests that the SASP can occur in two waves: a cGAS-independent one followed by a second cGAS-dependent one that primarily involves type I interferons, and may be connected to the upregulation of endogenous retroelements [28]. Somewhat conflicting data have been reported on whether the cGAS pathway is also involved in other aspects of senescence. Cells appear to be less likely to undergo growth arrest during proliferative senescence when cGAS is disrupted [32,36], but, surprisingly, this effect is not as apparent with a STING disruption, indicating STING independent functions [32,36]. During DNA-damage induced senescence, the situation is less clear, and we do not yet understand how the cGAS pathway intersects with p53 and other master regulators of senescence [31,82]. Cells lacking p53 that reach a critical point in telomere attrition undergo another type of growth arrest, termed crisis [83]. Similar to senescence, cGAS activation occurs during crisis. However, rather than mediating inflammatory gene induction, cGAS promotes activation of autophagy in order to mediate cell death [83]. Although the cGAS pathway is known to regulate autophagy, similar autophagy induction was not observed in response to other types of DNA damage that were tested [83].
Box 2. A role for cGAS in DNA repair?
cGAS can be activated in response to DNA damage, raising the possibility that cGAS might – directly or indirectly – affect DNA repair. Although a limited number of recent studies investigated this idea, little consensus has emerged so far. Initial findings suggested that cGAS – exclusively cytoplasmic within the experimental setup – was specifically imported into the nucleus following DNA damage, and associated with sites of damage in order to regulate repair [88]. cGAS import was suggested to be regulated by BLK kinase (B lymphocyte kinase), hitherto not implicated in DNA damage responses, and recruitment to sites of damage was suggested to be mediated by direct interaction with γH2AX (gamma-H2AX, a DNA damage enriched histone phosphorylation) and PARP1 (poly [ADP-ribose] polymerase 1, an important DNA damage sensor and regulator of repair). cGAS depletion enhanced repair by homologous recombination (see reference [90] for a description of repair of DNA breaks), and accordingly, cGAS overexpression inhibited homologous recombination [88]. However, the authors unexpectedly did not observe a concomitant change in the competing repair pathway, non-homologous end-joining (NHEJ) [90]. In contrast, other studies did not find evidence of cGAS nuclear import following DNA damage, or association with DNA damage sites, γH2AX or PARP1 [63,87]. Further investigation of homologous recombination and NHEJ efficiencies generated evidence to suggest that cGAS inhibited both pathways [87]. In vitro reconstitution experiments suggested that cGAS interferes with the strand exchange reaction of homologous recombination [87]. However, cGAS was used at very high concentrations (>1 cGAS molecule per 20 bp of DNA), which is unlikely to represent physiological conditions (cGAS intracellular concentration is estimated to be in the nM range, and highly substoichiometric to binding sites on DNA [65]). Altogether, further investigation is required to resolve the apparent differences between these studies, and to determine if cGAS is involved in DNA repair. A possibility that needs further investigation is indirect regulation of DNA repair by modulating gene expression, autophagy [91] or interferon responses.
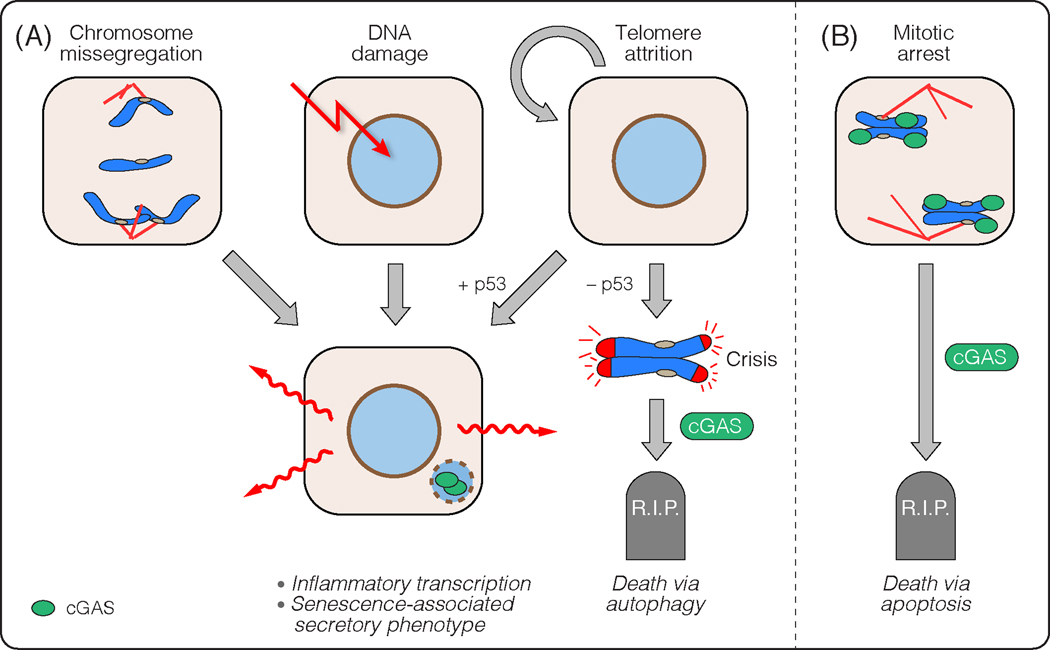
Figure 3: Consequences of cGAS activation by self-DNA.
(A) Chromosome missegregation, DNA damage, and telomere attrition can lead to induction of inflammatory genes that – at least in part – depends on cGAS. Note that a possible structure that might be responsible for cGAS activation in all these cases could be micronuclei. If telomere attrition occurs in the absence of p53, this can lead to continued proliferation, but eventually causes a cGAS-dependent upregulation of autophagy that promotes cell death. (B) During mitotic arrest, cGAS signalling builds up and eventually promotes apoptosis.
For both crisis and senescence, the precise nature of the DNA that activates cGAS is unknown, and micronuclei are frequently the favored candidates for this role [30,33,36,48,83]. Alternatively, other types of DNA fragments, reverse transcribed retroelements or mitochondrial DNA have been suggested to be involved [28,84]. Not all stimuli that generate a senescence-like growth arrest and signatures of senescence might be equal. Intuitively, it would make sense that telomere attrition might be different from ionizing radiation or mitochondrial dysfunction. Furthermore, since NE rupture appears to be a pre-requisite for interaction of cGAS with micronuclei [30], variability in NE dysfunction could modulate variability in responses.
cGAS was also connected to telomere dysfunction occurring in ALT cells [53,54]. Activation of cGAS by telomeric fragments that are a byproduct of the ALT pathway, and subsequent signaling to STING was suggested to be detrimental to cell growth [53]. This effect may be related to an anti-proliferative activity of inflammatory stress responses, or STING-dependent promotion of apoptosis [10]. Accordingly, ALT cell lines frequently show reduced expression of STING [53]. However, cGAS is lost less frequently [53]. A possible reason was suggested by another study that found evidence for cGAS-dependent activation of autophagy that positively affected cellular viability in ALT cells [54]. Thus, ALT cells might use cGAS-dependent pro-proliferative signaling, while at the same time avoiding STING-dependent anti-proliferative effects, suggesting a case of separation of function between cGAS and STING. How cGAS-dependent, pro-proliferative, autophagy in ALT might differ mechanistically from cGAS-dependent autophagy in crisis, which promotes cell death, is currently unclear.
In addition to senescence, crisis and ALT, cGAS also affects cell fate during mitotic arrest due to defective spindle assembly. During mitotic arrest, cells will eventually die in order to prevent cell division with improper spindles, which can lead to aneuploid daughter cells. cGAS is enriched on mitotic chromosomes, but downstream signaling is suppressed [10], presumably because of the poor activity on chromatin [10,70]. Nonetheless, cGAS-dependent IRF3 phosphorylation can eventually accumulate and promote apoptosis, thereby preventing cells from slipping into interphase without proper chromosome segregation [10]. Prolonged mitotic arrest is also a consequence of a number of cancer chemotherapy agents, such as taxanes. Consistently, cGAS expression was required for the taxane paclitaxel to reduce xenograft tumor growth in immunocompromised mice, and it correlated with patient survival in human non-small cell lung cancer patients [10]. Interestingly, cGAS induced apoptosis proceeds independently of transcriptional regulation by an as of yet unidentified mechanism [10].
Concluding remarks
While cGAS clearly is integral to responses to innumerable outside threats to the cell, it is intriguing that it evolved to a state where self-DNA signaling is not completely prevented. Although the organization of chromosomal DNA into nucleosomes is likely crucial for limiting cGAS, low level cGAS activity can occur even on nucleosomes [10], and forcing cGAS into the nucleus also results in some cGAMP production and inflammatory gene induction [63]. In addition to allowing cGAS to respond to abnormal self-DNA, such as damaged DNA, micronuclei, or mitotic chromosomes during mitotic arrest, low baseline stimulation of the cGAS pathway may boost anti-pathogenic gene expression, and help cells be prepared for possible future infections. This hypothesis is supported by the finding that cGAS deficient cells and mice have lower baseline expression of antipathogenic genes, respond poorer to RNA viruses, and induce inflammatory genes less well when stimulated with synthetic RNA [14]. Another potential example comes from human embryonic stem cells (hESCs), which show high baseline levels of anti-pathogenic genes [85]. In hESCs, cGAS enriches on mitotic chromosomes [10], and the rapid cell cycles of hESCs may help maintain cGAS in the nucleus after each mitosis (see Box 1), promoting expression of anti-pathogenic genes. A similar effect may also result from low level stimulation by mtDNA [17]. Overall, it appears that the cGAS pathway has evolved to strike a fine balance between preventing and allowing stimulation by self-DNA, which may break down under pathological conditions such as AGS and cancer. Future work will further establish the mechanistic basis of cGAS regulation on self-DNA and the signaling consequences (Outstanding Questions).
HIGHLIGHTS
cGAS is the major innate immune sensor of pathogenic DNA, and triggers inflammatory gene expression, autophagy, and cell death in response to detecting pathogen DNA.
cGAS can in principle also respond to all the major forms of self-DNA: mitochondrial DNA, DNA of endogenous retroelements, and chromosomal DNA.
Under unperturbed conditions, cGAS activation by self-DNA is limited. Mitochondria are separated by mitochondrial membranes, endogenous retroelements are silenced, and activation by chromosomal self-DNA is limited by non-productive binding of cGAS to nucleosomes.
In response to stress, DNA damage and cell cycle aberrations, self-DNA can eventually promote cGAS signaling, promoting inflammation, senescence and cell death.
Low-level activation of cGAS by self-DNA may boost baseline expression levels of antipathogenic genes to allow a faster response to infection.
OUTSTANDING QUESTIONS
How does cGAS signaling intersect with other innate immune pathways, and how does it intersect with cell cycle control checkpoints, and the DNA damage response?
How does chromatin inhibit cGAS signaling? Is nucleosome-mediated cGAS inhibition the only mechanism by which cGAS activity is limited on chromosomes?
What is the exact intracellular distribution of cGAS, and how is this modulated by cell type, cell cycle stage, and signaling pathways?
How is mtDNA made accessible to cGAS during MOMP? If mtDNA released into the cytoplasm, how does it traverse the inner mitochondrial membrane? And if cGAS relocates into the mitochondrial matrix, how is this mediated, and how does cGAMP find its way out?
ACKNOWLEDGMENTS
The authors thank Ryan Kim for comments on the manuscript. Work in the laboratory of H.F. is supported by the NIH (R35 GM132111).
GLOSSARY
- Alternative lengthening of telomeres (ALT):
A homologous recombination-based mechanism that solves the end-replication problem in some cancer cells that do not reactivate telomerase. In this process, the end of a telomere, which essentially resembles a DNA break, will undergo strand invasion within another telomere. Repair synthesis then leads to extension of the invading end.
- Cellular senescence:
A type of permanent cell cycle arrest. Induced when cells reach the end of their proliferative lifespan. Classically, this occurs in cells lacking telomerase when telomeres, the ends of chromosomes that lose information with every cell cycle, shrink below threshold levels. However, similar growth arrests can occur due to DNA damage.
- HMGB1 (high mobility group box 1):
A member of the high mobility group of proteins. Helps organize chromatin and regulate association of transcription factors and accessory proteins.
- Homologous recombination (HR):
A type of repair of DNA breaks that involves finding a homologous region elsewhere, and copying information to the broken ends in order to seal the break. Competes with non-homologous end-joining.
- Mitochondrial outer membrane permeabilization (MOMP):
An integral part of apoptosis. Following upstream signals, either from outside the cell (extrinsic apoptosis), or from within the cell (intrinsic apoptosis) Bax and Bak become activated. These generate pores in the outer mitochondrial membrane to release proteins that mediate the activation of caspases, proteases whose cleavage of substrates results in cell death.
- Mitophagy:
The clearance of dysfunctional or damaged mitochondria by a specialized autophagy pathway.
- Non-homologous end-joining (NHEJ):
A type of repair of DNA breaks that essentially glues the two broken ends back together. Competes with homologous recombination.
- Senescence-associated secretory phenotype (SASP):
The upregulation of inflammatory genes that is a hallmark of senescent cells. This may signal to neighboring cells and tissues, and indirectly cause some of the pathology of aging.
- Pattern recognition receptors:
Germ line encoded intracellular or extracellular receptors that detect specific components of pathogens and activate aspects of innate immune signaling in response.
- TFAM (mitochondrial transcription factor A):
The major protein factor that organizes mitochondrial DNA into nucleoids.
Footnotes
DISCLAIMER STATEMENT
We declare that we have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Brubaker SW et al. (2015) Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stetson DB and Medzhitov R (2006) Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 [DOI] [PubMed] [Google Scholar]
- 3.Sun L et al. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J and Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 [DOI] [PubMed] [Google Scholar]
- 5.Khan S et al. (2019) Cytosolic Nucleic Acid Sensors in Inflammatory and Autoimmune Disorders. Int Rev Cell Mol Biol 344, 215–253 [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki T and Kawai T. (2019) Discrimination Between Self and Non-Self-Nucleic Acids by the Innate Immune System. Int Rev Cell Mol Biol 344, 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ablasser A and Chen ZJ (2019) cGAS in action: Expanding roles in immunity and inflammation. Science 363, eaat8657 [DOI] [PubMed] [Google Scholar]
- 8.Tang C-HA et al. (2016) Agonist-Mediated Activation of STING Induces Apoptosis in Malignant B Cells. Cancer Res 76, 2137–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay S and Sen GC (2017) RIG-I-like receptor-induced IRF3 mediated pathway of apoptosis (RIPA): a new antiviral pathway. Protein Cell 8, 165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zierhut C et al. (2019) The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 178, 302–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonekamp NA and Larsson N-G (2018) SnapShot: Mitochondrial Nucleoid. Cell 172, 388–388.e1 [DOI] [PubMed] [Google Scholar]
- 12.Sliter DA et al. (2018) Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Aguirre S et al. (2017) Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2, 17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW et al. (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legrand AJ et al. (2019) The Diversification of Cell Death and Immunity: Memento Mori. Mol Cell 76, 232–242 [DOI] [PubMed] [Google Scholar]
- 16.White MJ et al. (2014) Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159, 1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rongvaux A et al. (2014) Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichim G et al. (2015) Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol Cell 57, 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McArthur K et al. (2018) BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359, eaao6047 [DOI] [PubMed] [Google Scholar]
- 20.Riley JS et al. (2018) Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J 37, e99238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J et al. (2019) VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366, 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowe HM and Trono D. (2011) Dynamic control of endogenous retroviruses during development. Virology 411, 273–287 [DOI] [PubMed] [Google Scholar]
- 23.Kassiotis G and Stoye JP (2016) Immune responses to endogenous retroelements: taking the bad with the good. Nat. Rev. Immunol. 16, 207–219 [DOI] [PubMed] [Google Scholar]
- 24.Stetson DB et al. (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokatayev V et al. (2016) RNase H2 catalytic core Aicardi-Goutières syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med 213, 329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartsch K et al. (2017) Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26, 3960–3972 [DOI] [PubMed] [Google Scholar]
- 27.De Cecco M et al. (2013) Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cecco M et al. (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon M et al. (2019) LINE1 Derepression in Aged Wild-Type and SIRT6- Deficient Mice Drives Inflammation. Cell Metab. 29, 871–885.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie KJ et al. (2017) cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Deursen JM (2014) The role of senescent cells in ageing. Nature 509, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H et al. (2017) cGAS is essential for cellular senescence. Proceedings of the National Academy of Sciences 114, E4612–E4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding SM et al. (2017) Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denais CM et al. (2016) Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raab M et al. (2016) ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 [DOI] [PubMed] [Google Scholar]
- 36.Glück S et al. (2017) Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 19, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Härtlova A et al. (2015) DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 [DOI] [PubMed] [Google Scholar]
- 38.Quek H et al. (2017) Rats with a missense mutation in Atm display neuroinflammation and neurodegeneration subsequent to accumulation of cytosolic DNA following unrepaired DNA damage. J. Leukoc. Biol. 101, 927–947 [DOI] [PubMed] [Google Scholar]
- 39.Song X et al. (2019) Accumulation of Cytoplasmic DNA Due to ATM Deficiency Activates the Microglial Viral Response System with Neurotoxic Consequences. J. Neurosci. 39, 6378–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Q et al. (2015) DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Reports 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erdal E et al. (2017) A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev 31, 353–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coquel F et al. (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61 [DOI] [PubMed] [Google Scholar]
- 43.Reisländer T et al. (2019) BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat Commun 10, 3143–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heijink AM et al. (2019) BRCA2 deficiency instigates cGAS-mediated inflammatory signaling and confers sensitivity to tumor necrosis factor-alpha-mediated cytotoxicity. Nat Commun 10, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gratia M et al. (2019) Bloom syndrome protein restrains innate immune sensing of micronuclei by cGAS. J Exp Med 216, 1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crasta K et al. (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatch EM et al. (2013) Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dou Z et al. (2017) Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakhoum SF et al. (2018) Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-David U and Amon A. (2020) Context is everything: aneuploidy in cancer. Nat Rev Genet 21, 44–62 [DOI] [PubMed] [Google Scholar]
- 51.Umbreit NT et al. (2020) Mechanisms generating cancer genome complexity from a single cell division error. Science 368, eaba0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dilley RL and Greenberg RA (2015) ALTernative Telomere Maintenance and Cancer. Trends Cancer 1, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y-A et al. (2017) Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat Struct Mol Biol 24, 1124–1131 [DOI] [PubMed] [Google Scholar]
- 54.Barroso-González J et al. (2019) RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol Cell 76, 11–26.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie W et al. (2019) Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proceedings of the National Academy of Sciences 116, 11946–11955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemmi H et al. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 57.Gao P et al. (2013) Cyclic [G(2’,5’)pA(3“,5”)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Civril F et al. (2013) Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X et al. (2013) Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herzner A-M et al. (2015) Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16, 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreeva L et al. (2017) cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 [DOI] [PubMed] [Google Scholar]
- 62.Zhang X et al. (2014) The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Reports 6, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gentili M et al. (2019) The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Reports 26, 2377–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West AP et al. (2015) Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hein MY et al. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 [DOI] [PubMed] [Google Scholar]
- 66.Yanai H et al. (2009) HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103 [DOI] [PubMed] [Google Scholar]
- 67.Bianchi ME et al. (2017) High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol. Rev. 280, 74–82 [DOI] [PubMed] [Google Scholar]
- 68.Luger K et al. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 69.Zierhut C and Funabiki H. (2015) Nucleosome functions in spindle assembly and nuclear envelope formation. Bioessays 37, 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lahaye X et al. (2018) NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell 175, 488–501 [DOI] [PubMed] [Google Scholar]
- 71.Xia P et al. (2018) A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 48, 688–701.e7 [DOI] [PubMed] [Google Scholar]
- 72.Barnett KC et al. (2019) Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell 176, 1432–1446.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du M and Chen ZJ (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao D et al. (2015) Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences 112, E5699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Motani K et al. (2015) DNA-Mediated Cyclic GMP-AMP Synthase-Dependent and - Independent Regulation of Innate Immune Responses. J Immunol 194, 4914–4923 [DOI] [PubMed] [Google Scholar]
- 76.Takahashi A et al. (2018) Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat Commun 9, 1249–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volkman HE et al. (2019) Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife 8, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon J and Bakhoum SF (2020) The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov 10, 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia T et al. (2016) Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Reports 14, 282–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia T et al. (2016) Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res 76, 6747–6759 [DOI] [PubMed] [Google Scholar]
- 81.Konno H et al. (2018) Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 33, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang C et al. (2015) The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612–aaa5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nassour J et al. (2019) Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vizioli MG et al. (2020) Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev 34, 428–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu X et al. (2018) Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 172, 423–438.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orzalli MH et al. (2015) cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proceedings of the National Academy of Sciences 112, E1773–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang H et al. (2019) Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J 38, e102718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H et al. (2018) Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 339, 786. [DOI] [PubMed] [Google Scholar]
- 89.Young AM et al. (2020) BAF facilitates interphase nuclear envelope repair through recruitment of nuclear transmembrane proteins. bioRxiv 39, 2020.01.10.902296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mehta A and Haber JE (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6, a016428–a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gui X et al. (2019) Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]