Abstract
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) has rapidly caused a global pandemic associated with a novel respiratory infection: coronavirus disease-19 (COVID-19). Angiotensin-converting enzyme-2 (ACE2) is necessary to facilitate SARS-CoV-2 infection, but—owing to its essential metabolic roles—it may be difficult to target it in therapies. Transmembrane protease serine 2 (TMPRSS2), which interacts with ACE2, may be a better candidate for targeted therapies. Using publicly available expression data, we show that both ACE2 and TMPRSS2 are expressed in many host tissues, including lung. The highest expression of ACE2 is found in the testes, whereas the prostate displays the highest expression of TMPRSS2. Given the increased severity of disease among older men with SARS-CoV-2 infection, we address the potential roles of ACE2 and TMPRSS2 in their contribution to the sex differences in severity of disease. We show that expression levels of ACE2 and TMPRSS2 are overall comparable between men and women in multiple tissues, suggesting that differences in the expression levels of TMPRSS2 and ACE2 in the lung and other non–sex-specific tissues may not explain the gender disparities in severity of SARS CoV-2. However, given their instrumental roles for SARS-CoV-2 infection and their pleiotropic expression, targeting the activity and expression levels of TMPRSS2 is a rational approach to treat COVID-19.
Abbreviations and Acronyms: ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease-19; GTEx, genotype-tissue expression; SARS-CoV-2, severe acute respiratory syndrome-coronavirus 2; TMPRSS2, transmembrane protease serine 2
In December 2019, a novel member of the Coronaviridae family, severe acute respiratory syndrome (SARS)-coronavirus 2 (SARS-CoV-2), has spread rapidly throughout the world, probably from Wuhan, Hubei province, China, resulting in an infectious respiratory infection: coronavirus disease-19 (COVID-19).1, 2, 3 As of May 1, 2020, more than 3.2 million persons are confirmed to have contracted SARS-CoV-2, and nearly 250,000 have died from this global pandemic.4 This global public health emergency requires a rapid and effective response to mitigate COVID-19–related morbidity and mortality.2 Elucidating the mechanism of SARS-CoV-2 infection is necessary for the rational design of therapeutics, the use of novel or repurposed therapeutics, development of an effective vaccine, and understanding the clinical course of COVID-19.
Infections by the SARS coronaviruses—SARS-CoV and SARS-CoV-2—are dependent on host proteins angiotensin-converting enzyme 2 (ACE2) receptor, which has been the subject of multiple investigations in the literature.5 , 6 However, viral entry requires not only binding to the ACE2 receptor but also priming of the virus’s spike (S) protein by the transmembrane protease serine 2 (TMPRSS2) by cleavage of the S proteins at the S1/S2 and S2 sites. This cleavage step is necessary for the virus-host cell membrane fusion and cell entry.6 , 7 One striking observation made across most countries is the increased severity of disease among older men with SARS-CoV-2 infection,8 although there might also be differences in infectivity. Although early data of 140 patients with SARS-CoV-2 from Wuhan, China, demonstrated a nearly equal male to female ratio for infection,9 subsequent studies of hospitalized and deceased patients have identified an increased male prevalence from approximately 55% to 85% in China,1 , 10, 11, 12, 13, 14, 15, 16 to 82% in Italy,17 and approximately 63% in New York City of hospitalized patients.18 It is interesting that this skewed sex difference in disease severity was also reported in studies of respiratory infections caused by related coronaviruses, SARS-CoV and Middle East respiratory syndrome (MERS)-CoV, suggesting a potentially shared underlying mechanism responsible for the high disease severity among infected men.19, 20, 21, 22 Host susceptibility to severe COVID-19 disease is also associated with older age, hypertension, heart failure, chronic kidney disease, diabetes, obesity, and the use of ACE inhibitors.22, 23, 24 Although gender can be associated with these additional comorbidities, male sex remains an independent variable associated with severe COVID-19 infection in multiple studies.22 , 23. It is possible that expression differences in ACE2 and TMPRSS2 explain the increased severity of disease among men. ACE2 is located at Xp22.2, within the nonpseudoautosomal portion of the X chromosome (15,561,033-15,602,069, GRCh38). It displays incomplete X-chromosome inactivation and shows a male-biased expression pattern in several tissues.25 TMPRSS2 is located at 21q22.3, within chromosome 21 (41,464,305-41,508,158, GRCh38), and its expression is modulated by androgen signaling via multiple androgen receptor elements upstream of the gene’s transcriptional start site.26 , 27 In this study, we evaluate the expression levels of both ACE2 and TMPRSS2 in many host tissues, including lung. Given the increased severity of disease among older men with SARS-CoV-2 infection, we also address the potential roles of ACE2 and TMPRSS2 in their contribution to the sex differences in severity of disease.
Methods
ACE2 and TMPRSS2 Gene Expression Analyses
ACE2 and TMPRSS2 expression data were obtained directly from the genotype-tissue expression (GTEx) project (https://gtexportal.org) or from the Human Protein Atlas GTEx data (RNAseq based on RSEMv1.2.22 (v7). Expression from all tissue samples available were plotted using the box plots available from the GTExPortal website with plots shown as median and 25th and 75th percentiles and dots displayed as outliers if they are above or below 1.5 times the interquartile range. Comparison of ACE2 and TMPRSS2 expression was performed using GTEx tool multiGeneQueryPage.
Statistical Analyses
From the Human Protein Atlas GTEx data (RNAseq based on RSEMv1.2.22 (v7), box plots were created using SPSS Software (SPSS Statistics, IBM, Chicago, Illinois) and extreme outliers in each type of sample were identified (data not shown). If extreme outliers were present in the sample population, the significance of the difference was calculated using the Mood’s median test. For all other samples without extreme outliers, Kruskal–Wallis was used to calculate the significance.
TMPRSS2 Allele Frequencies
Allele frequency of 2 missense variants rs75603675 and rs12329760 in TMPRSS2 gene were calculated using the Geography of Genetic Variants Browser.28
Results
Expression of ACE2 and TMPRSS2 in Multiple Tissues
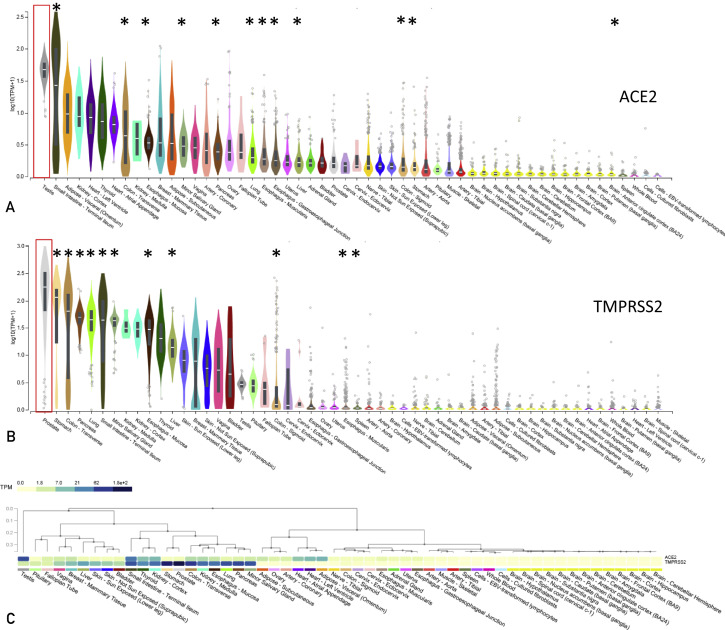
Given the necessity of ACE2 and TMPRSS2 genes for SARS CoV-2 infection, we evaluated their expression in human tissues using data from the GTEx project (https://gtexportal.org). Expression of ACE2 and TMPRSS2 was detectable in multiple tissues. As the common symptoms of COVID-19 involve cough, sore throat, gastrointestinal issues, anosmia, and dysgeusia,14 , 29 we analyzed the levels of ACE2 and TMPRSS2 in tissues associated with these sequelae. Both ACE2 and TMPRSS2 were expressed in the lung, with expression levels of TMPRSS2 higher than ACE2 (Figure 1 ). Tissues associated with the gastrointestinal system with elevated ACE2 expression include small intestine (terminal ileum), colon (transverse), esophagus (mucosa), minor salivary gland and pancreas, esophagus, liver, colon (sigmoid) and stomach (Figure 1A). TMPRSS2 expression was also detected in gastrointestinal tissues including stomach, colon (transverse), pancreas, small intestine (terminal ileum), minor salivary gland, esophagus (mucosa), liver, and colon (sigmoid) (Figure 1B). Some patients with COVID-19 experience anosmia and dysgeusia, findings that may be a result of olfactory nerve abnormalities.29 Although GTEx does not have expression data specifically from olfactory tissues, the overall expression levels of both ACE2 and TMPRSS2 appear low in other nervous system tissues. The highest expression of ACE2 was observed in the testes, and the prostate displayed the highest expression of TMPRSS2 (Figure 1). Although the overall expression of ACE2 and TMPRSS2 varied in each tissue, multiple tissues expressed both ACE2 and TMPRSS2 (Figure 1C). However, some tissues—such as adipose, heart, artery, and ovary—had high ACE2 and low TMPRSS2 expression, whereas the prostate, stomach, bladder, skin, liver, and pituitary had high TMPRSS2 and low ACE2 expression (Figure 1C). Although the pathophysiology of SARS-CoV-2 is not completely understood, and these data alone cannot establish causality between infection and ACE2 or TMPRSS2 expression, they do suggest that additional tissues other than the lung—including, but not limited to, kidney, testes, and skin—may also be infected by SARS-CoV-2. This observation may explain the pleiotropic effects of SARS-CoV-2 infection but should be confirmed in vitro and in vivo. 30, 31, 32
Figure 1.
ACE2 and TMPRSS2 gene expression data. The data were obtained directly from the Genotype-Tissue Expression (GTEx) Project (https://gtexportal.org). Samples were sorted based on the median expression on a log scale using transcripts per million (TPM) unit. ACE2 (A) and TMPRSS2 (B) expression from all tissue samples available were plotted using the box plots available from the GTExPortal website with plots shown as median and 25th and 75th percentiles and dots displayed as outliers if they are above or below 1.5 times the interquartile range. Red boxes show the testes and prostate expression of ACE2 and TMPRSS2, respectively. Tissues associated with common COVID-19 symptoms are marked with an asterisk∗. In (C), a comparison of ACE2 and TMPRSS2 expression using GTEX tool multiGeneQueryPage.
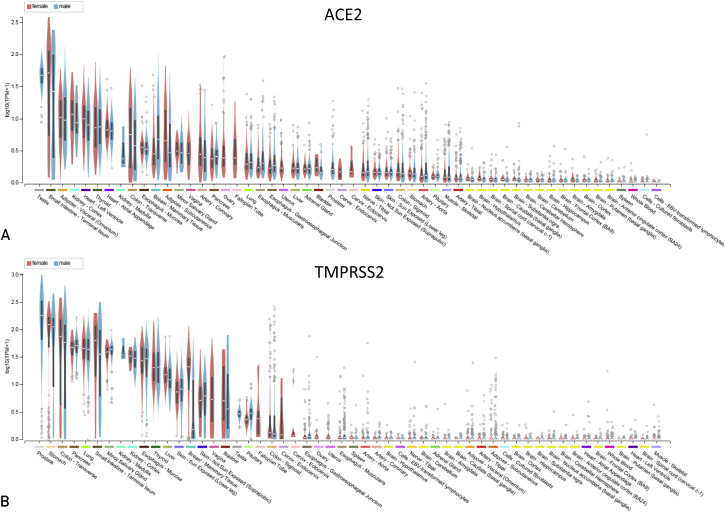
Expression of ACE2 and TMPRSS2 is Similar in Men and Women
Although the frequency of men and women testing positive for SARS-CoV-2 may be similar, men are more likely to die (60% men vs 40% women) following the infection compared with women, an observation that does not appear to be geographically specific (Table 1 ). To evaluate whether ACE2 or TMPRSS2 contribute to the sex disparity in severity of disease, we analyzed their sex differential gene expression using the GTEx data that were available directly through the GTEx portal and also through the Human Protein Atlas (https://proteinatlas.org) (Figure 2 and Table 2 ). Focusing on the tissues with the highest expression of ACE2 and TMPRSS2, only the subcutaneous adipose tissue showed a significant difference between sex, with men displaying lower median ACE2 expression compared with women (Table 2) (men 1.3 transcripts per million [TPM] vs women 2.2 TPM, P value <0.0001). In some tissues, a higher numerical median expression of ACE2 (lung: men 0.7 vs women 0.6; thyroid gland: men 4.4 vs women 3.4; heart atrial appendage: men 3.6 vs 3.4 women; and visceral adipose: men 5.5 vs women 5.4 TPM) and TMPRSS2 (colon transverse: men 104.1 vs women 90.8; pancreas: men 52.7 vs women 51.0; salivary gland: men 46.3 vs women 42.5; and esophagus: men 32.7 vs women 28.8 TPM) was observed (Table 2); however, these findings were not considered significant. No significant differences in ACE2 or TMPRSS2 expression were observed in lung tissue (Table 2).
Table 1.
Sex Disaggregated Information From 32 Countries With 982, 274 Total Cases As Of May 2, 2020
| Country | Total cases | Total cases |
Total cases |
Cases |
Cases |
Total deaths |
Total deaths |
Total deaths |
Deaths |
Deaths |
Deaths |
Deaths |
Deaths confirmed+ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (M) | (F) | (% M) | (% F) | (M) | (F) | (% M) | (% F) | confirmed + (% M) | confirmed + (% F) | (M:F ratio) | |||
| The Netherlands | 38,365 | 14,579 | 23,786 | 38 | 62 | 4566 | 2603 | 1963 | 57 | 43 | 17.9 | 8.3 | 2.2 |
| Finland | 4740 | 2275 | 2465 | 48 | 52 | 145 | 75 | 70 | 52 | 48 | 3.3 | 2.8 | 1.2 |
| Denmark | 8851 | 3717 | 5134 | 42 | 58 | 434 | 247 | 187 | 57 | 43 | 6.7 | 3.6 | 1.8 |
| Greece | 2324 | 1278 | 1046 | 55 | 45 | 132 | 100 | 32 | 76 | 24 | 7.8 | 3.0 | 2.6 |
| Italy | 176,716 | 86,591 | 90,125 | 49 | 51 | 23,164 | 14,593 | 8571 | 63 | 37 | 16.9 | 9.5 | 1.8 |
| Republic of Ireland | 19,666 | 8260 | 11,406 | 42 | 58 | 903 | 479 | 424 | 53 | 47 | 5.8 | 3.7 | 1.6 |
| Spain | 204,856 | 90,137 | 114,719 | 44 | 56 | 15,853 | 9195 | 6658 | 58 | 42 | 10.2 | 5.8 | 1.8 |
| Switzerland | 29,376 | 13,513 | 15,863 | 46 | 54 | 1408 | 817 | 591 | 58 | 42 | 6.0 | 3.7 | 1.6 |
| Belgium | 47,682 | 17,642 | 30,040 | 37 | 63 | 5286 | 2696 | 2590 | 51 | 49 | 15.3 | 8.6 | 1.8 |
| Germany | 156,337 | 75,042 | 81,295 | 48 | 52 | 5908 | 3368 | 2540 | 57 | 43 | 4.5 | 3.1 | 1.4 |
| Portugal | 24,322 | 9972 | 14,350 | 41 | 59 | 948 | 465 | 483 | 49 | 51 | 4.7 | 3.4 | 1.4 |
| Austria | 15,314 | 7504 | 7810 | 49 | 51 | 550 | 308 | 242 | 56 | 44 | 4.1 | 3.1 | 1.3 |
| Sweden | 19,621 | 8829 | 10,792 | 45 | 55 | 2355 | 1342 | 1013 | 57 | 43 | 15.2 | 9.4 | 1.6 |
| Northern Ireland | 2724 | 1117 | 1607 | 41 | 59 | 206 | 115 | 91 | 56 | 44 | 10.3 | 5.6 | 1.8 |
| Norway | 7605 | 3726 | 3879 | 49 | 51 | 195 | 107 | 88 | 55 | 45 | 2.9 | 2.3 | 1.3 |
| Romania | 11,313 | 5091 | 6222 | 45 | 55 | 619 | 396 | 223 | 64 | 36 | 7.8 | 3.6 | 2.2 |
| Ukraine | 9410 | 4140 | 5270 | 44 | 56 | 239 | 131 | 108 | 55 | 45 | 3.2 | 2.0 | 1.6 |
| Luxembourg | 3741 | 1908 | 1833 | 51 | 49 | 89 | 50 | 39 | 56 | 44 | 2.6 | 2.1 | 1.2 |
| Europe | 782,963 | 355,322 | 427,641 | 45 | 55 | 63,000 | 37,087 | 25,913 | 59 | 41 | 10.4 | 6.1 | 1.7 |
| Dominican Republic | 6293 | 3398 | 2895 | 54 | 46 | 282 | 220 | 62 | 78 | 22 | 6.5 | 2.1 | 3.0 |
| Peru | 28,699 | 17,793 | 10,906 | 62 | 38 | 782 | 555 | 227 | 71 | 29 | 3.1 | 2.1 | 1.5 |
| Mexico | 15,529 | 9007 | 6522 | 58 | 42 | 1434 | 975 | 459 | 68 | 32 | 10.8 | 7.0 | 1.5 |
| Colombia | 5597 | 2910 | 2687 | 52 | 48 | 253 | 157 | 96 | 62 | 38 | 5.4 | 3.6 | 1.5 |
| Ecuador | 15,728 | 8650 | 7078 | 55 | 45 | 871 | 592 | 279 | 68 | 32 | 6.8 | 3.9 | 1.7 |
| Canada | 26,638 | 11,721 | 14,917 | 44 | 56 | 1067 | 534 | 534 | 50 | 50 | 4.6 | 3.6 | 1.3 |
| Argentina | 3892 | 1946 | 1946 | 50 | 50 | 192 | 125 | 67 | 65 | 35 | 6.4 | 3.5 | 1.9 |
| Americas | 102,376 | 55,426 | 46,950 | 54 | 46 | 4881 | 3158 | 1723 | 65 | 35 | 5.7 | 3.7 | 1.6 |
| China | 55,924 | 28,521 | 27,403 | 51 | 49 | 2114 | 1353 | 761 | 64 | 36 | 4.7 | 2.8 | 1.7 |
| South Korea | 10,752 | 4301 | 6451 | 40 | 60 | 244 | 127 | 117 | 52 | 48 | 3.0 | 1.8 | 1.6 |
| Australia | 6713 | 3357 | 3357 | 50 | 50 | 84 | 51 | 33 | 61 | 39 | 1.5 | 1.0 | 1.6 |
| Philippines | 7955 | 4296 | 3659 | 54 | 46 | 530 | 350 | 180 | 66 | 34 | 8.1 | 4.9 | 1.7 |
| Indonesia | 9511 | 5611 | 3900 | 59 | 41 | 773 | 526 | 247 | 68 | 32 | 9.4 | 6.3 | 1.5 |
| Thailand | 2938 | 1616 | 1322 | 55 | 45 | 52 | 40 | 12 | 77 | 23 | 2.5 | 0.9 | 2.7 |
| Asia | 93,793 | 47,702 | 46,091 | 51 | 49 | 3797 | 2447 | 1350 | 64 | 36 | 5.1 | 2.9 | 1.8 |
| South Africa | 3142 | 1414 | 1728 | 45 | 55 | 43 | 24 | 19 | 56 | 44 | 1.7 | 1.1 | 1.6 |
| Total | 982,274 | 459,863 | 522,411 | 47 | 53 | 71,721 | 42,716 | 29,005 | 60 | 40 | 9.3 | 5.6 | 1.7 |
The data were obtained from the Global Health 505033 (http://globalhealth5050.org/covid19) tracker. Data from Iran, India, and Pakistan are not indicated in table. Regions were color-coded as follows: Europe (purple), Americas (green), Asia (orange), and Africa (blue).
Confirmed + = confirmed positive
Figure 2.
Sex differences in TMPRSS2 and ACE2 expression data. The data were obtained directly from the Genotype-Tissue Expression (GTEx) Project (https://gtexportal.org). Female subjects (pink) and male subjects (blue) were arranged based on sorting using the median expression on a log scale using transcripts per million (TPM) unit. TMPRSS2 (A) or ACE2 (B) expression from all tissues available were plotted using the box plots available from the GTExPortal website with plots shown as median and 25th and 75th percentiles with dots displayed as outliers if they are above or below 1.5 times the interquartile range.
Table 2.
Median ACE2 and TMPRSS2 TPM Expression Values.
| TMPRSS2 | Male median TPM | Female median TPM | P value |
|---|---|---|---|
| Stomach | 109.4 | 132.5 | 0.309 |
| Colon-transverse | 104.1 | 90.8 | 0.460 |
| Small intestine | 63.6 | 86.1 | 0.186 |
| Lung | 48.2 | 49.5 | 0.244 |
| Pancreas | 52.7 | 51.0 | 0.436 |
| Salivary gland | 46.3 | 42.5 | 0.228 |
| Esophagus | 32.7 | 28.8 | 0.649 |
| Kidney | 31.6 | 31.1 | 0.876 |
| Thyroid | 21.6 | 21.1 | 0.921 |
| Liver | 12.2 | 14.6 | 0.284 |
| ACE2 | Male median TPM | Female median TPM | P value |
|---|---|---|---|
| Small intestine | 38.0 | 50.7 | 0.385 |
| Adipose tissue subcutaneous | 1.3 | 2.2 | <0.0001 |
| Adipose tissue visceral | 5.5 | 5.4 | 0.971 |
| Thyroid gland | 4.4 | 3.4 | 0.059 |
| Kidney | 4.7 | 8.8 | 0.104 |
| Heart atrial appendage | 3.6 | 3.4 | 0.563 |
| Heart left ventricle | 4.7 | 5.7 | 0.104 |
| Colon-sigmoid | 0.2 | 0.2 | 0.769 |
| Colon-transverse | 3.3 | 4.0 | 0.460 |
| Lung | 0.7 | 0.6 | 0.574 |
| Salivary gland | 1.5 | 1.6 | 0.636 |
The data were obtained from The Human Protein Atlas GTEx data (RNAseq based on RSEMv1.2.22 [v7]) and sorted from the highest expressing tissues from male and female patients (all age groups included). Box plots were created using SPSS Software (IBM, Chicago, Illinois), and extreme outliers in each type of sample were identified. If extreme outliers were present in the sample population, the significance of the difference was calculated using the Mood’s median test. For all other samples without extreme outliers, Kruskal–Wallis was used to calculate the significance.
TPM = transcripts per million.
Discussion
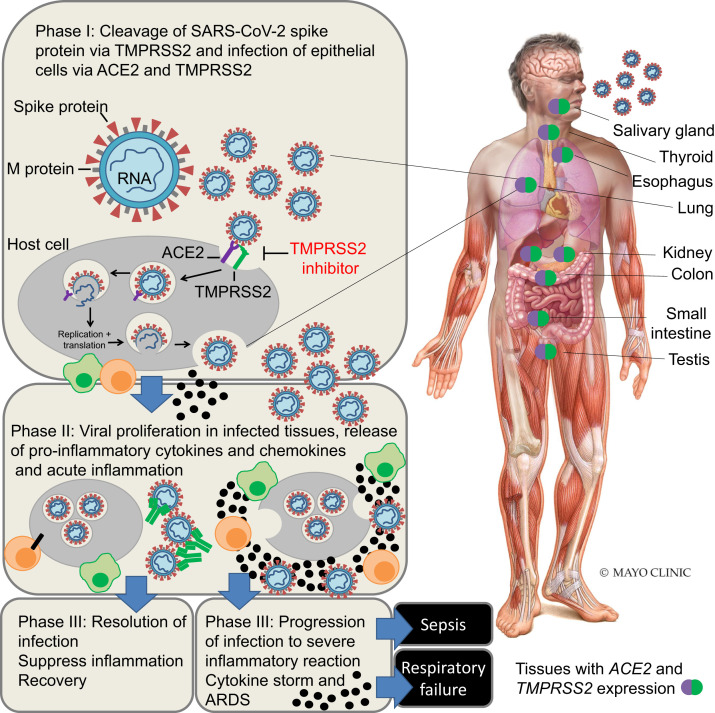
The rapid development of novel approaches or repurposing of existing therapies is critical in mitigating the coronavirus pandemic. Because of the centrality of ACE2 and TMPRSS2 as host-expressing genes necessary for SARS-CoV-2 infection and subsequent immune reactions34 (Figure 3 ), it is reasonable to develop agents that target their proteins or inhibit their activity.35 Therapies that target ACE2 received some focus in the literature,36 and the use of decoy ACE2 receptors could have promise.37 However, directly antagonizing human ACE2 may be challenging owing to the essential regulatory role of ACE2 in heart function, as demonstrated by the severe cardiac defects observed in ACE2 knockout (KO) mice.38 Furthermore, there are potentially beneficial effects of ACE2 that occur because of the degradation of angiotensin I and angiotensin II by ACE2. By contrast, TMPRSS2 KO mice lack an observable phenotype and appear healthy, suggesting that TMPRSS2 is nonessential for mouse reproduction, development, and growth.39 Further, TMPRSS2 KO mice had reduced infection and virus spread within the airway and less severe immune response due to SARS-CoV and the related MERS-CoV viruses, demonstrating the critical role of TMPRSS2 in coronavirus infection.40
Figure 3.
Schematic of SARS-CoV-2 infection of host tissue and disease pathogenesis. SARS-CoV-2 infects host cells (primarily epithelial cells) that express the host receptor, ACE2 and TMPRSS2, resulting in phase I of infection. In phase II, viral proliferation occurs in infected cells and this results in a local immune response, release of cytokines and chemokines (black circles), attraction of macrophages (green cell) and T cells (orange cell) to infected cells, and activation of further adaptive immune responses. In most cases, there is a healthy immune response, and infected cells are eliminated and further viral infection can be blocked by neutralizing antibodies (green). In this phase III of infection, there is a reduction of virus spread, resolution of infection, suppression of inflammation with limited tissue injury, and eventual recovery. However, in some cases of phase III of infection, the viral infection may lead to increased production of proinflammatory cytokines, resulting in a cytokine storm, causing multiorgan damage and acute respiratory distress syndrome (ARDS).
Inhibition of TMPRSS2 activity using camostat mesylate in human lung cells in vitro has a demonstrated efficacy against SARS-CoV-2 infection.6 Therefore, this agent appears to be a logical therapeutic approach, as it is already used in Japan to treat pancreatitis.41 Given the strong preclinical support of repurposing camostat mesylate for SARS-CoV infection, clinical trials evaluating camostat mesylate alone or in combination with hydroxychloroquine have begun in Europe (https://clinicaltrials.gov/ct2/show/NCT04321096 and https://clinicaltrials.gov/ct2/show/NCT04338906), and we are actively pursuing them in the United States. In addition, another TMPRSS2 inhibitor—nafamostat—may also have clinical utility against SARS-CoV-2 infection.42
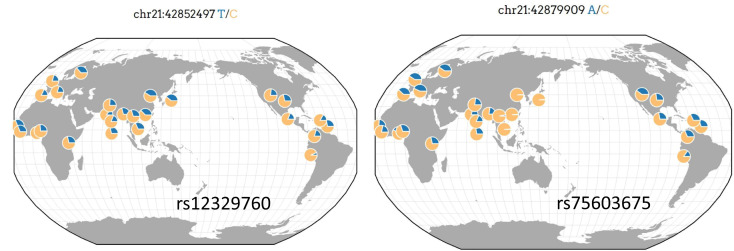
An alternative strategy is to inhibit the androgen receptor (AR). The TMPRSS2 expression is modulated by androgen signaling via multiple androgen receptor elements upstream of the gene’s transcriptional start site.26 , 27 As shown in Figure 1B, and by others, TMPRSS2 is highly expressed in prostate epithelium.26 Consistent with this, an aberrant fusion of TMPRSS2 with ERG or with other partner oncogenes (ETV1, ETV4, ETV5) is a common feature in prostate cancer.43 As it is well documented that TMPRSS2 is an androgen-responsive gene, a novel approach to SARS-CoV-2 infection may be to reduce the expression of TMPRSS2 by inhibition of AR signaling through the use of androgen deprivation therapy (ADT) or antiandrogens, which are standard treatments for prostate cancer.44 Although preclinical studies are necessary to evaluate this novel approach to SARS-CoV-2 infection, it has the benefit that the use and low toxicity profile of AR-directed therapies have been well established in prostate cancer studies.44 Targeting TMPRSS2 protease activity through protease inhibitors or indirectly through ADT requires an understanding of the functional polymorphisms present in the gene. Of the 13 common markers (minor allele frequency 0.5) within TMPRSS2, there are 2 missense variants (rs75603675; c.23G>T p.Gly8Val and rs12329760; c.589G>A p.Val197Met) whose frequencies vary by ancestry and geography (Figure 4 ). Targeting TMPRSS2 protease activity through protease inhibitors or indirectly through ADT will require additional studies to determine the role of rs75603675 and rs12329760 on TMPRSS2 expression, protease activity, and in response to protease inhibition.
Figure 4.
Allele frequency of 2 missense variants rs12329760 and rs75603675 in TMPRSS2 gene. The allele frequencies were calculated using the Geography of Genetic Variants Browser.28 Genomic positions in GRCh37.
Our observations are consistent with previous studies evaluating TMPRSS2 expression in lung tissue using GTEx data,45 although—owing to the limitation of the data—they do not consider age, menopausal status, or ancestry. Although a very slight increase in TMPRSS2 expression was observed in the bronchial epithelial cells of male patients compared with female patients, using GSE66499 microarray data,45 it is unclear whether this small increase in TMPRSS2 expression explains the increase in severity of disease in men. Our results are also consistent with previous studies supporting no significant differences in ACE2 expression in lung tissue in association with age (>60 and <60 years), race (white and Asian), and sex using RNAseq, microarray, and GTEx datasets.46 Combined, these findings suggest that differences in the expression levels of TMPRSS2 and ACE2 in the lung and other non–sex-specific tissues likely do not explain the gender disparities in severity of SARS CoV-2. However, the observation of high ACE2 expression in the testes is an intriguing observation, and a recent study hypothesized that the testes could serve as a reservoir for SARS-CoV-2.30 However, the expression of genes in the testes has been associated with “leaky expression,” without evidence of a functional consequence of the expressed genes.47 Therefore, the expression of ACE2 in the testes warrants additional investigation, including demonstration of SARS-CoV-2 in semen. Additional studies are also needed to evaluate whether androgens (or even estrogens) contribute to sex-associated severity of disease independently of ACE2 or TMPRSS2 functions.
Conclusion
SARS-CoV-2 has rapidly caused a global pandemic. Both ACE2 and TMPRSS2 are expressed in many host tissues, but the expression levels of ACE2 and TMPRSS2 are overall comparable between men and women in multiple tissues. This suggests that expression differences of TMPRSS2 and ACE2 in the lung and other non–sex-specific tissues may not explain the gender disparities in severity of SARS CoV-2. However, targeting the activity and expression levels of TMPRSS2 is a rational approach to treat COVID-19 and should be explored further.
Acknowledgments
The genotype-tissue expression (GTEx) project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2) from The Human Protein Atlas GTEx data (RNAseq based on RSEMv1.2.22 [v7]) on April 15, 2020.
Footnotes
Potential Competing Interests: Dr Fonseca has served as a consultant for Amgen, BMS, Celgene, Takeda, Bayer, Janssen, Novartis, Pharmacyclics, Sanofi, Merck, Juno, Kite, Aduro, OncoTracker, GSK, and AbbVie. He has served on scientific advisory boards for Adaptive Biotechnologies and OncoTracker. Dr Elhaik has served as a consultant for DNA Diagnostics Center. The remaining authors report no competing interests.
Supplemental Online Material
References
- 1.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.J., Dong X., Cao Y.Y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan W.J., Ni Z.Y., Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Y., Tu L., Zhu P. Clinical features of 85 fatal cases of COVID-19 from Wuhan: a retrospective observational study. Am J Respir Crit Care Med. 2020;201(11):1372–1379. doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J., Tong Z., Guan X., Du B., Qiu H. Clinical characteristics of patients who died of coronavirus disease 2019 in China. JAMA Netw Open. 2020;3(5):e208147. doi: 10.1001/jamanetworkopen.2020.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasselli G., Zangrillo A., Zanella A. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrilli C.M., Jones S.A., Yang J. Factors associated with hospitalization and critical illness among 4,103 patients with COVID-19 disease in New York City. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin J.-M., Bai P., He W. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alghamdi I.G., Hussain, Almalki S.S., Alghamdi M.S., Alghamdi M.M., El-Sheemy M.A. The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int J Gen Med. 2014;7:417–423. doi: 10.2147/IJGM.S67061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. 2017;198(10):4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badawi A., Ryoo S.G. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int J Infect Dis. 2016;49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y., Yu X., Zhao H., Wang H., Zhao R., Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24(1):108. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler D.J., Mozsary C., Meydan C. Host, viral, and environmental transcriptome profiles of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) bioRxiv. 2020 doi: 10.1101/2020.04.20.048066. [DOI] [Google Scholar]
- 25.Tukiainen T., Villani A.C., Yen A. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550(7675):244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin B., Ferguson C., White J.T. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180–4184. [PubMed] [Google Scholar]
- 27.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcus J.H., Novembre J. Visualizing the geography of genetic variants. Bioinformatics. 2017;33(4):594–595. doi: 10.1093/bioinformatics/btw643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xydakis M.S., Dehgani-Mobaraki P., Holbrook E.H. Smell and taste dysfunction in patients with COVID-19. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shastri A., Wheat J., Agrawal S. Delayed clearance of SARS-CoV2 in male compared to female patients: high ACE2 expression in testes suggests possible existence of gender-specific viral reservoirs. medRxiv. 2020 [Google Scholar]
- 31.Marzano A.V., Genovese G., Fabbrocini G. Varicella-like exanthem as a specific COVID-19-associated skin manifestation: multicenter case series of 22 patients. J Am Acad Dermatol. 2020;83(1):280–285. doi: 10.1016/j.jaad.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su H., Yang M., Wan C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98(1):219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Global Health5050. http://globalhealth5050.org/covid19
- 34.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020;10(6):779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N Engl J Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteil V., Kwon H., Prado P. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crackower M.A., Sarao R., Oudit G.Y. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 39.Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006;26(3):965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93(6):e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibo J., Ito T., Kawabe K. Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity. Lab Invest. 2005;85(1):75–89. doi: 10.1038/labinvest.3700203. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto M., Matsuyama S., Li X. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linn D.E., Penney K.L., Bronson R.T., Mucci L.A., Li Z. Deletion of interstitial genes between TMPRSS2 and ERG promotes prostate cancer progression. Cancer Res. 2016;76(7):1869–1881. doi: 10.1158/0008-5472.CAN-15-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucas J.M., Heinlein C., Kim T. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4(11):1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. medRxiv. 2020 doi: 10.18632/aging.103415. https://www.org/10.1101/2020.03.30.20047878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai G. medRxiv; 2020. Bulk and single-cell transcriptomics identify tobacco-use disparity in lung gene expression of ACE2, the receptor of 2019-nCov. [DOI] [Google Scholar]
- 47.Kryuchkova-Mostacci N., Robinson-Rechavi M. Tissue-specific evolution of protein coding genes in human and mouse. PLoS One. 2015;10(6):e0131673. doi: 10.1371/journal.pone.0131673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.