Abstract
This paper presents an updated and comprehensive review on the different methods used for detection and quantification of viruses in wastewater treatment systems. The analysis of viability of viruses in wastewater and sludge is another thrust of this review.
Recent studies have mostly focused on determining the abundance and diversity of viruses in wastewater influents, in samples from primary, secondary, and tertiary treatment stages, and in final effluents. A few studies have also examined the occurrence and diversity of viruses in raw and digested sludge samples. Recent efforts to improve efficiency of virus detection and quantification methods in the complex wastewater and sludge matrices are highlighted in this review.
A summary and a detailed comparison of the pre-treatment methods that have been utilized for wastewater and sludge samples are also presented. The role of metagenomics or sequencing analysis in monitoring wastewater systems to predict disease outbreaks, to conduct public health surveillance, to assess the efficiency of existing treatment systems in virus removal, and to re-evaluate current regulations regarding pathogenic viruses in wastewater is discussed in this paper. Challenges and future perspectives in the detection of viruses, including emerging and newly emerged viruses such as the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), in wastewater systems are discussed in this review.
Abbreviations: ASTM, American Society for Testing Materials; CER, Cation exchange resin; cDNA, Complementary DNA; cPCR, Conventional Polymerase Chain Reaction; COVID-19, Coronavirus disease 2019; CPE, Cytopathic effects; DNA, Deoxyribonucleic Acid; EV, Enterovirus; EVE, Enzymatic virus elution; ELISA, Enzyme Linked Immunosorbent Assay; EFM, Epifluorescence Microscopy; FCM, Flow Cytometry; GUI, Graphical user interface; HAV, Hepatitis A virus; HFUFS, Hollow fibre ultrafiltration system; HAdV, Human adenovirus; HAdV2, Human adenovirus 2; HAdV 40, Human adenovirus 40; HEV, Hepatitis E virus; HSV1, Herpes Simplex Virus Type 1; HHV, Human herpesvirus; IFA, Immunofluorescence Assay; ICC-PCR, Integrated Cell Culture-Polymerase Chain Reaction; ICC-RT-qPCR, Integrated Cell Culture-real time RT-PCR; JCPyV, JC Polyomavirus; HPV, Human papillomavirus; HPyV, Human polyomavirus; HTtV, Human Torque teno virus; μPAD, Microfluidic paper analytic device; MMTV, Mouse mammary tumour virus; MHV, Murine Hepatitis Virus; NGS, Next Generation Sequencing; NoV, Norovirus; NoV GI, Norovirus GI; NoV GII, Norovirus GII; PMMV, Pepper Mild Mottle Virus; PTA, Phosphotungstic acid; PEG, Polyethylene Glycol; PCR, Polymerase Chain Reaction; PV, Poliovirus; PV1, Poliovirus 1; φ6, Pseudomonas phage; PFGE, Pulsed-Field Gel Electrophoresis; qPCR, Real-Time Polymerase Chain Reaction; RT-PCR, Reverse Transcription-Polymerase Chain Reaction; RT-qPCR, Real Time RT-PCR; RNA, Ribonucleic Acid; RoV, Rotavirus; RoV-A, Rotavirus A; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; TEM, Transmission Electron Microscopy; VIRADEL, Virus Adsorption-Elution
Keywords: Virus detection, Virus concentration, Sequencing, SARS-CoV-2, Sludge, Emerging viruses
Graphical abstract
1. Introduction
Viruses are ubiquitous and persistent in raw wastewater and treated wastewater as well as in the receiving water bodies (Fumian et al., 2010). One of the main sources of viruses, including viral pathogens in wastewater is the human fecal matter, particularly that from infected persons (Gerba et al., 2017; Hellmér et al., 2014a; Symonds et al., 2009). Sewage systems receive enteric viruses excreted by infected individuals. An infected person sheds 105 to 1012 viral particles per gram of fecal matter (Gerba, 2000). In addition to human pathogenic viruses, waterborne viruses that originate from food production, animal husbandry, seasonal surface runoff and other sources are present in wastewater (Corsi et al., 2014). The bodies that receive treated wastewater are oftentimes used for recreational activities and agriculture, and as a source of raw water for drinking water production (Corsi et al., 2014; Dias et al., 2019). Effluents from wastewater treatment plants (WWTPs) are widely used for irrigation and for aquifer recharge (Gerba et al., 2017). The presence of potentially pathogenic viruses in wastewater is of concern since it can pose risks to human health (Carducci et al., 2009; Naddeo and Liu, 2020). Sludges from wastewater treatment plants are also utilized for agricultural applications as soil conditioner or fertilizer (Andreadakis et al., 2002; Bibby and Peccia, 2013; Grobelak et al., 2019). Aside from the concerns related to reuse of wastewater and sludge, exposure of WWTP workers to viral pathogens is a potential risk. Aeration facilities in WWTPs generate and diffuse bioaerosols containing chemicals and microorganisms including viruses that are typically non-waterborne (Pasalari et al., 2019; Wang et al., 2020b). Aerosols are also emitted during the handling of sludge and land application of biosolids (Bibby and Peccia, 2013).
2. Occurrence of viruses in wastewater and sludge
The abundance and diversity of pathogenic viruses in wastewater has been shown to reflect the pattern of infection in human population (Fumian et al., 2010; Montazeri et al., 2015). Adenovirus (HAdV), rotavirus (RoV), hepatitis A virus (HAV), and other enteric viruses, such as noroviruses (NoV), coxsackievirus, echovirus, reovirus and astrovirus are some of the principal human pathogens viruses transmissible via water media. Enteric viruses cause waterborne diseases such as diarrhea in children and in adults (Pietruchinski et al., 2006) and are associated with other disease outbreaks (Thongprachum et al., 2018). Enteric viruses may lead to symptoms notably nausea, vomiting, and fever (Bishop and Kirkwood, 2008). In addition to diarrhea and other gastrointestinal diseases, some enteric viruses have been related to respiratory diseases such as bronchiolitis (Kocwa-Haluch, 2001).
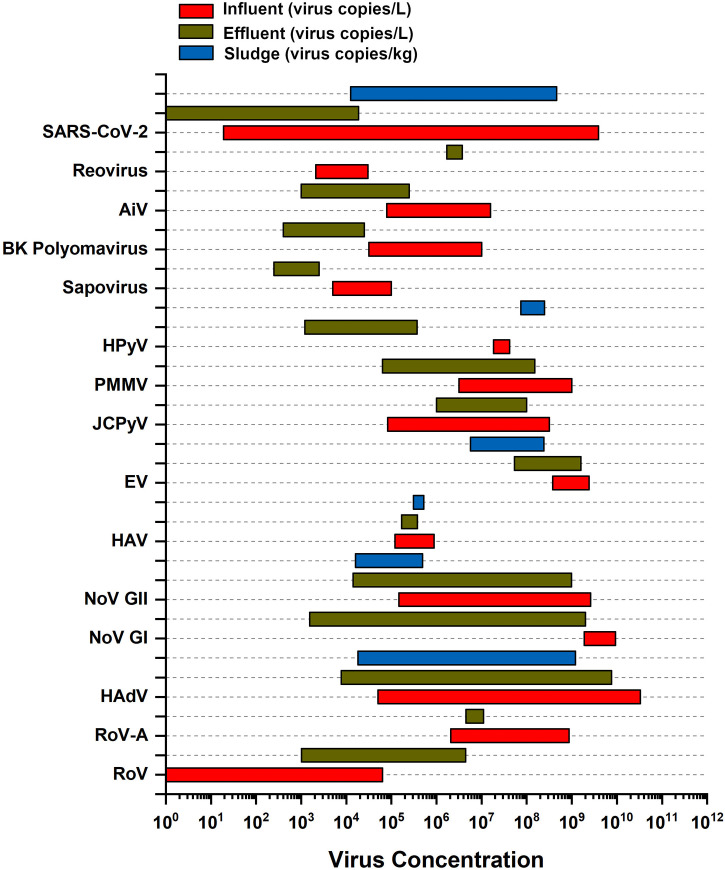
The aims of quantification of viruses in wastewater include: i) the determination of the level of risk associated to waterborne diffusion of viruses, ii) the evaluation of the efficiency of disinfection as measure of control of virus, iii) the surveillance on the extent of diffusion of viruses in a population (Barcelo, 2020). These objectives can be met by determining both the concentration of virus and its infectivity. Virus requires a host species for replication therefore, in absence of the latter, the concentration of a virus in an environment can be constant or progressively reduced by inactivation. Table 1 and Fig. 1 report literature data concerning the range of concentrations and viability of representative viruses that can populate wastewater both continually (e.g., enteric viruses) and sporadically (e.g., coronaviruses during an epidemic). It can be observed that WWTP processes are not completely effective in the reduction of viral genomes' concentrations of most viruses in wastewater (Table 1). For a complete information on the presence of viruses in water, as well as in other environments, their viability and corresponding potential infectivity should also be investigated. This information is an essential input for determining pathways of virus transmission, quantifying the extent of disease risk, and identifying interventions through the use Quantitative Microbial Risk Assessment (QMRA) and Infectious Disease Transmission Modeling (IDTM) (Brouwer et al., 2018).
Table 1.
Occurrence and viability of viruses in wastewater and sludge.
| Virus |
Concentration |
Viability |
||||||
|---|---|---|---|---|---|---|---|---|
| (size; typology) | WWTP | Influent (genome copies/L) | Effluent (genome copies/L) | Sludge (genome copies/kg of dry solids) | Reference | Sample matrix | Number samples with infectious virus/total number of samples | Reference |
| HEV (27–34 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | 7.80 × 103 to 5.80 × 108 | 4.00 × 103 to 1.00 × 104 | ⁎⁎ |
Izopet et al., 2019⁎ Di Profio et al., 2019; Masclaux et al., 2013; Iaconelli et al., 2020; Beyer et al., 2020 |
Effluent | 30/30 | Baez et al., 2017 |
| RoV (70 nm; non-enveloped, linear, segmented, positive-sense dsRNA virus) |
Municipal | 1.00 × 101 to 6.31 × 104 | 1.01 × 103 to 4.47 × 106 | 8.00 × 103 to 8.00 × 105b (primary sludge) |
Drancourt, 2017⁎ He et al., 2011 Osuolale and Okoh, 2017; Qiu et al., 2018; Prado et al., 2014 |
Effluent pre-UV | 3/51 | Qiu et al., 2018 |
| Effluent post-UV | 4/50 | Qiu et al., 2018 | ||||||
| Activated sludge | 3/12 | Schlindwein et al., 2010 | ||||||
| RoV-A (70 nm; non-enveloped, linear, segmented, positive-sense, dsRNA virus) |
Hospital | 2.08 × 106 to 8.7 × 108 | 4.50 × 106 to 1.10 × 107 9.12 × 106 to 3.72 × 107a |
⁎⁎ |
Drancourt, 2017⁎ Prado et al., 2011; Prado et al., 2019 |
⁎⁎ | – | |
| HAdV (70–90 nm; non-enveloped, linear, non-segmented, dsDNA virus) |
Municipal | 5.00 × 104 to 3.30 × 1010 | 7.76 × 103 to 7.60 × 109 5.50 × 104 to 7.59 × 105a |
1.80 × 104 to 1.20 × 109b (Primary sludge) 9.40 × 109 to 3.90 × 1011 (Thickened sludge) 9.10 × 107 to 6.90 × 109 (Anaerobically digested sludge) |
Payne, 2017⁎ Calgua et al., 2011; Qiu et al., 2018; Schlindwein et al., 2010; La Rosa et al., 2020; Prado et al., 2019; Sidhu et al., 2018; Prado et al., 2014; Wong et al., 2010 |
Effluent | 15/16 | Thompson et al., 2003 |
| 6/12 | Li et al., 2010 | |||||||
| Secondary effluent | 4/12 | Li et al., 2010 | ||||||
| Effluent pre-UV | 16/51 | Qiu et al., 2018 | ||||||
| 8/12 | Schlindwein et al., 2010 | |||||||
| Effluent post-UV | 10/50 | Qiu et al., 2018 | ||||||
| Activated sludge | 12/12 | Schlindwein et al., 2010 | ||||||
| Thickened sludge | 2/2 | Wong et al., 2010 | ||||||
| Anaerobically digested sludge | 1/8 | Wong et al., 2010 | ||||||
| Hospital | 1.70 × 105 to 2.30 × 107 | 6.60 × 104 to 2.90 × 106 | ⁎⁎ | Prado et al., 2011 | ⁎⁎ | ⁎⁎ | – | |
| NoV GI (27–35 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | 1.90 × 109 to 9.30 × 109 | 1.55 × 103 to 2.00 × 109 | 5.00 × 107 (Anaerobically digested sludge) |
Cunliffe et al., 2014⁎ Qiu et al., 2018; La Rosa et al., 2020; Wong et al., 2010 |
⁎⁎ | ⁎⁎ | – |
| Nov GII (27–35 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | 1.48 × 105 to 2.60 × 109 | 1.41 × 104 to 9.90 × 108 | 1.60 × 104 to 4.90 × 105b (Primary sludge) 1.50 × 108 (Anaerobically digested sludge) |
Cunliffe et al., 2014⁎ Masclaux et al., 2013; Qiu et al., 2018; La Rosa et al., 2020; Prado et al., 2014; Wong et al., 2010 |
⁎⁎ | ⁎⁎ | – |
| Hospital | 0.00 to 1.80 × 106 |
3.20 × 104 to 9.60 × 107 | ⁎⁎ | Prado et al., 2011 | ⁎⁎ | ⁎⁎ | – | |
| HAV (27–32 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | 1.20 × 105 to 8.90 × 105 | 1.70 × 105 to 3.80 × 105 | Activated sludge: 3.10 × 105 to 5.20 × 105b |
Rasche et al., 2019⁎ Schlindwein et al., 2010; Villar et al., 2007; Prado et al., 2014 |
Effluent | 8/12 | Schlindwein et al., 2010 |
| Activated Sludge | 2/12 | Schlindwein et al., 2010 | ||||||
| EV (27–30 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | 3.80 × 108 to 2.40 × 109 | 5.40 × 107 to 1.60 × 109 | 5.47 × 107 to 1.15 × 108 (Primary sludge) 5.66 × 106 to 1.44 × 107 (Activated Sludge) 1.44 × 107 to 2.40 × 108 (Thickened Sludge) 2.60 × 107 to 7.60 × 107 (Anaerobically digested sludge) |
Rajtar & Majek, 2008⁎ Schvoerer et al., 2001; Monpoeho et al., 2000; Wong et al., 2010 |
Effluent | 9/25 | La Rosa et al., 2020 |
| Effluent Pre-UV | 17/51 | Qiu et al., 2018 | ||||||
| Effluent Post-UV | 7/50 | Qiu et al., 2018 | ||||||
| Thickened Sludge | 2/2 | Wong et al., 2010 | ||||||
| Anaerobically Digested Sludge | 1/8 | Wong et al., 2010 | ||||||
| JCPyV (40 nm; non-enveloped, circular, non-segmented dsDNA virus) |
Municipal | 8.33 × 104 to 3.20 × 108 | 1.00 × 106 to 1.00 × 108 2.82 × 105 to 7.76 × 106a |
Zhang⁎⁎ |
Nelson et al., 2012⁎ Prado et al., 2019; Fumian et al., 2010; Bofill-Mas et al., 2010 |
⁎⁎ | ⁎⁎ | ⁎⁎ |
| PMMV (310 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal Ponds | 3.16 × 106 to 1.00 × 109 | 6.31 × 104 to 1.52 × 108 | ⁎⁎ |
Symonds et al., 2014; Schmitz et al., 2016; Kitajima et al., 2018⁎ |
Effluent | Infectivity confirmed by inoculating effluent with test plants | Bačnik et al., 2020 |
| HPyV (40–50 nm; non-enveloped, circular, non-segmented dsDNA virus) |
Municipal | 1.83 × 107 to 4.17 × 107 | 1.20 × 103 to 3.70 × 105 | 7.40 × 107 to 2.50108 (Anaerobically digested sludge) |
De Gascun & Carr, 2013⁎ McQuaig et al., 2009; Wong et al., 2010 |
⁎⁎ | ⁎⁎ | ⁎⁎ |
| Sapovirus (41–46 nm; non-enveloped, linear, non-segmented positive-sense, ssRNA virus) |
Municipal | 5.01 × 103 to 1.00 × 105 | 2.50 × 102 to 2.51 × 103 | ⁎⁎ |
D’Souza, 2015⁎ Schmitz et al., 2016 |
⁎⁎ | ⁎⁎ | ⁎⁎ |
| BK Polyomavirus (40–44 nm; non-enveloped, circular, non-segmented dsDNA virus ) |
Municipal | 3.16 × 104 to 1.00 × 107 | 0.40 × 103 to 2.51 × 104 | ⁎⁎ |
Ambalathingal et al., 2017⁎ Schmitz et al., 2016 |
⁎⁎ | ⁎⁎ | ⁎⁎ |
| AiV (30 nm; non-enveloped, linear, segmented, positive-sense ssRNA virus) |
Municipal | 7.90 × 104 to 1.58 × 107 | 1.00 × 103 to 2.51 × 105 | ⁎⁎ |
D’Souza, 2015⁎ Schmitz et al., 2016; Kitajima et al. (2014) |
⁎⁎ | ⁎⁎ | ⁎⁎ |
| Parechovirus (28 nm; non-enveloped, linear, non-segmented, positive-sense ssRNA virus) |
Municipal | ⁎⁎ | ⁎⁎ | 2.51 × 107 to 2.51 × 108 (Anaerobically digested sludge) |
Olijve et al., 2018; ⁎ Bibby and Peccia, 2013 |
Sludge | 1/307 | Hamparian et al., 1985 |
| Reovirus (85 nm; non-enveloped, linear, segmented, positive-sense dsRNA virus) |
Municipal | 2.10 × 103 to 3.00 × 104 (in titer per litre of sewage) |
1.70 × 106 to 3.72 × 106 0.00 to 6.03 × 106c |
⁎⁎ |
Betancourt & Gerba, 2016⁎ Ridinger et al., 1982 |
Effluent pre-UV | 47/51 | Qiu et al., 2018 |
| Effluent post-UV | 24/50 | Qiu et al., 2018 | ||||||
| Raw Sludge | 12/15 | Gallagher and Margolin, 2007 | ||||||
| Anaerobically digested sludge | 1/9 | Gallagher and Margolin, 2007 | ||||||
| SARS-CoV-1 (150–200 nm; enveloped, linear, non-segmented positive-sense, ssRNA virus) |
Hospital sewage | ⁎⁎ | ⁎⁎ | ⁎⁎ | Wang et al., 2005b | Hospital sewage seeded with virus | 1/1 | Wang et al., 2005b |
| Hospital sewage before disinfection with chlorine | 0/10 | |||||||
| SARS-CoV-2 (60–140 nm; enveloped, linear, non-segmented positive-sense, ssRNA virus) |
Municipal | 1.90 × 101 to 3.00 × 106 | 2.40 × 103 to 2.51 × 105a |
1.25 × 104 to 4.6 × 108 (Primary Sludge) 1.17 × 104 to 4.02 × 104 (Waste Activated Sludge) |
Zhu et al., 2020; ⁎ See Table 4 for complete list of references |
Influent | 0/8 | Rimoldi et al., 2020 |
| Effluent | 0/4 | |||||||
| Hospital | 0.00 (after primary disinfection tank before septic tank) |
0.50 × 100 to 1.87 × 104 (after septic tank with disinfection) |
Zhang et al., 2020 | ⁎⁎ | ⁎⁎ | ⁎⁎ | ||
Notes:
dsDNA, double-stranded DNA
ssRNA, single-stranded DNA
Reference for virus size and structure.
No available quantitative (concentration) or viability data of virus in specified sample matrix in previous studies.
Secondary Treatment Effluent.
Expressed in genome copies/L.
Before UV Treatment.
Fig. 1.
Occurrence of representative viruses in wastewater.
Enteric viruses are non-enveloped viruses with an enhanced resistance. Some enteric viruses such RoV have been shown to be resistant to UV disinfection. The resistance is attributed to its structure, specifically to the presence of a double-stranded RNA and three-layered capsid protein (Li et al., 2009). On the contrary, enveloped viruses are more susceptible to inactivation in wastewater. The presence of solvents, detergents and disinfectant in wastewater rapidly compromise the lipidic viral envelope or the surface protein. Severe Acute Respiratory Syndrome (SARS) coronavirus (SARS-CoV-1), an enveloped virus, was found to be persistent in raw sewage at 20 °C for 2 days and 14 days at 4 °C (Wang et al., 2005b, Wang et al., 2005c). A limited number of studies have shown that the viability of SARS coronaviruses has not been observed in effluent from WWTPs (see Table 1). SARS-CoV-1 has also been reported by Wang et al. (2005c) to be inactivated by free chlorine with concentration greater than 0.5 mg/L and by chlorine dioxide with concentration greater than 2.19 mg/L. Mechanisms of removal of viruses from WWTP include adsorption of viruses on larger aggregated particles that are separated from wastewater by sedimentation (Verbyla and Mihelcic, 2015), retention by membrane and biofilm layers, predation and enzymatic breakdown in membrane bioreactors (Chaudhry et al., 2015) , and inactivation by disinfection processes such as UV, chlorination, and ozonation (Xagoraraki et al., 2014). As discussed, the resistance of the viruses to inactivation mechanisms is influenced by the viral structure. Double-stranded viruses, including reovirus, HAdV and RoV, have been shown to be generally more resistant to UV radiation than single-stranded ones (Calgua et al., 2014; Harris et al., 1987; Li et al., 2009). This behavior of the double-stranded viruses is attributed to the capability to repair their genomes during replication in the host cells (Beck et al., 2014; Calgua et al., 2014). In addition to the influence of the viral structure to inactivation, some WWTP processes also reduce the rate of inactivation of viruses. Although adsorption of viruses to solids contributes to their removal from wastewater, this also provides a degree of protection of the viruses from inactivation (Hejkal et al., 1981).
This paper aims to present and compare the currently developed methods of detection and quantification of viruses in wastewater. It also aims to present a review of the different pre-treatment steps needed to increase the efficiency of detection and quantification methods of viruses in wastewater and associated products. In this paper, particular attention is paid to the detection and quantification of the recently emerged SARS-CoV-2. This virus belongs to the Coronaviridae family, characterized by a protein capsid, containing the viral RNA, protected by a bilipidic envelope with embedded spike proteins. This virus can be transmitted by inhalation of infected respiratory particles (Chan et al., 2020), though other potential routes of transmission have been postulated to be important, notably via fomites (van Doremalen et al., 2020), ocular surface (Lu et al., 2020), and fecal–oral route (Wu et al., 2020a; Xiao et al., 2020; Yeo et al., 2020). SARS-CoV-2 has been reported to affect the human gastroenteric tract (Ding and Liang, 2020) and the presence of the viral RNA was detected in fecal samples (Wu et al., 2020a; Xiao et al., 2020) and wastewater conveyed to WWTPs (Ahmed et al., 2020; La Rosa et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Wu et al., 2020b; Wurtzer et al., 2020).
Therefore, the analysis of wastewater constitutes a powerful tool for surveillance of the propagation of diseases associated with pathogenic viruses. The presence of these different pathways highlights the importance of the control and removal of viruses in wastewater treatment. Correspondingly, the efficiency of treatment systems to remove viruses must be determined based on their quantification and identification. The presence of human pathogenic viruses in wastewater not only poses a specific sanitary risk, but it also provides a reliable indicator of the extent of circulation of the viruses in the population.
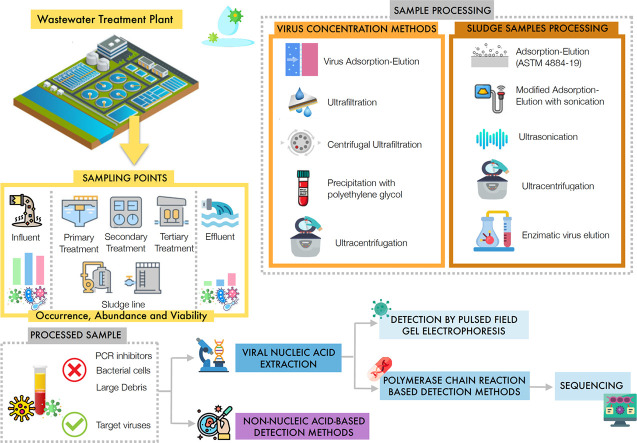
3. Methods of pre-treatment of wastewater samples
The accuracy of detection of viruses depends on the sample volume, nucleic acid extraction yield (for nucleic acid-based methods) and purity (Bofill-Mas and Rusiñol, 2020; Haramoto et al., 2018; Hryniszyn et al., 2013; Sidhu et al., 2013). A study conducted by Hjelmsø et al. (2017) showed that both the concentration of nucleic acids and the nature of the methods used for the extraction significantly influence the results of viral metagenomic analyses, particularly those of viral community composition, viral specificity, and viral pathogen detection. This means that the methods for concentration, nucleic acid extraction, and detection must be chosen appropriately.
This section presents a summary of concentration methods of wastewater samples, pre-treatment methods for sludge samples, and nucleic acid extraction methods.
3.1. Concentration methods
The processing of a sample collected for detection and quantification of viruses depends on the type of sample matrix. Influents generally have higher concentration of viruses than other environmental samples (Haramoto et al., 2018). Raw wastewater samples have higher turbidity (Falman et al., 2019; Qiu et al., 2016; Sidhu et al., 2013), higher suspended solids (Prado et al., 2019) and higher organic matter concentrations (Falman et al., 2019) than other environmental water samples. In addition, influent wastewaters have high concentrations of humic acids and heavy metals, that may interfere with the molecular methods of assaying for viruses (Prado et al., 2019). On the other hand, sludges are very heterogeneous sample matrices, in which viruses tend to be adsorbed on the surface of the flocs (Symonds et al., 2014). These characteristics affect the accuracy of the detection of viruses in these samples. This necessitates the use of concentration steps, some of which consist of primary and secondary concentration methods. It should be noted that molecular methods for detection and quantification of viruses do not provide complete information on the infectivity of viruses present in water media. Those methods determine the presence of molecular fragments (DNA or RNA) of the viruses. The viability of a virus can be determined through the cytopathogenic effect of the infected sample in suitable cell lineages that act as a host species for the virus. Therefore, for the reliable determination of this parameter it is important to preserve the viability of the virus during the sampling, the handling and the treatment of the wastewater sample. A summary and comparison of different types of concentration methods used in processing samples for detection of viruses from wastewater treatment plants are presented in the following discussion and summarized in Table 2 .
Table 2.
Methods of concentration of viruses in wastewater samples.
| Method | Sample Volume (L) | Advantages | Disadvantages | References |
|---|---|---|---|---|
| VIRADEL | 0.50–400 | Reduces amount of PCR inhibitors | Higher sample volume needed Requires pre-conditioning of samples (when using electronegative membranes) Multiple steps lead to viral loss |
Falman et al., 2019; Kuo et al., 2010; Masclaux et al., 2013; O’Brien et al., 2017; Osuolale and Okoh, 2017; Prado et al., 2011; Soto-Beltran et al., 2013 |
| Ultrafiltration | 1–10 | May be used for simultaneous concentration of viruses and other microbes | Clogging of filters when sample is of high turbidity (except for tangential ultrafiltration) Slow filtration rate |
Cashdollar and Wymer, 2013; Jahne et al., 2020; Morales-Morales et al., 2003; Sidhu et al., 2018 |
| Centrifugal ultrafiltration | 0.01–0.10 | Lower sample volume needed Reduces amount of PCR inhibitors Rapid and simpler method Easier processing of multiple samples |
Small pore size may result to clogging of filters High turbidity samples may need pre-filtration |
Nordgren et al., 2009; Sidhu et al., 2013 |
| Precipitation with PEG | 0.40–1 | Higher efficiency in concentrating RNA viruses | Concentrates enzymatic inhibitors (PCR inhibitors) |
Amdiouni et al., 2012; Ibrahim et al., 2017; Masclaux et al., 2013; Strubbia et al., 2019 |
3.2. Virus adsorption-elution method
Viruses can be removed from water matrices through adsorption onto a membrane filter and subsequent recovery by elution: a process named as virus adsorption-elution (VIRADEL). Metcalf (1961) first reported the recovery of influenza virus from aqueous suspensions through adsorption on a membrane filter. Wallis and Melnick (1967) examined the concentration of enteroviruses by dilution in salt solution to allow adsorption to membrane filters, and subsequent elution. In the current VIRADEL method, the samples pass through a membrane filter that may have an electronegative or electropositive charge. The viruses are retained on the surface of the filter due to electrostatic attraction between viral particle and membrane surface (Farkas et al., 2018). The VIRADEL method was developed for water samples with low turbidity (Farkas et al., 2018). Modifications to this method are necessary for samples with high turbidity, such as influent wastewaters. Prado et al. (2011) pre-filtered influent hospital wastewater samples before electronegative membrane filtration. VIRADEL method has been widely used for large volumes of sample to be processed, notably river samples (Hamza et al., 2009; Maunula et al., 2012), tap water samples (Hill et al., 2009; Polaczyk et al., 2007), groundwater samples (Lee et al., 2011), and coastal waters (Fong et al., 2005; Goyal and Gerba, 1983; Katayama et al., 2002). Wastewater sample volumes ranging from 0.5 to 400 L were used in recent studies that applied VIRADEL as primary concentration method (Falman et al., 2019; Kuo et al., 2010; Masclaux et al., 2013; Osuolale and Okoh, 2017; Prado et al., 2011; Soto-Beltran et al., 2013). However, the processing of large volumes of samples leads to the concentration of PCR inhibitors. The volume of sample to be treated must be adequate to ensure a final high viral content, keeping low that of PCR inhibitors, in order to facilitate the subsequent molecular amplification techniques of the nucleic material of the virus (Qiu et al., 2016). This consequently improves the quality of viral DNA or RNA that will be extracted in the downstream processes. Aside from the difficulties associated to the handling of large volumes of samples, another disadvantage of VIRADEL is the number of steps, including: i) pre-filtration, ii) membrane adsorption, iii) elution, resulting to prolonged processing time and loss in yield of recovered viruses.(Qiu et al., 2016)
3.2.1. Electronegative filtration
The outer surface of non-enveloped viruses is composed of capsid proteins whose isoelectric point is typically below the pH of wastewater. Therefore, this class of viruses has a net surface negative electrical charge (Hamza et al., 2009; Mayer et al., 2015). The use of electronegative membranes, common in VIRADEL methods, requires the lowering of the pH prior to filtration in order to induce the protonation of the protein capsid and the resultant inversion of the electrical charge of the viral surface (Hamza et al., 2009). The positively charged viruses can then be adsorbed onto the surface of a negatively charged membrane (Cashdollar and Wymer, 2013; Hamza et al., 2009). The addition of salts, such as magnesium or aluminum chlorides improves the adsorption of the viruses on the negatively charged-membrane (Hamza et al., 2009; Haramoto et al., 2009; Masclaux et al., 2013; Osuolale and Okoh, 2017). The first report on the adjustment of pH of suspension medium to 5.0 for optimum adsorption of poliovirus (PV) to membrane filters was made by Wallis and Melnick (1967). Hamza et al. (2014) used a negatively charged membrane of 0.45 μm pore size for adsorption of human enteric viruses from sewage. The sample's pH was adjusted to 3.5 to modify the charge of the viruses and enhance the attraction to the electronegative membrane. Prado et al. (2011) concentrated effluent and pre-filtered influent wastewater samples with gastroenteric and HAV using a negatively charged membrane.
The recovery of the viruses through elution is done after the adsorption to the electronegative membrane. In some methods, the elution step is eliminated, and the nucleic acids of the viruses are directly extracted from the filter (Ahmed et al., 2015). This latter method has been used to concentrate and directly extract nucleic acids of JC Polyomavirus (JCPyV) and BK Polyomavirus (McQuaig et al., 2009) and other enteric viruses (Ahmed et al., 2015) in environmental water samples.
3.2.2. Electropositive filtration
Electropositive membrane filters can be alternatively used in VIRADEL methods. On the basis of the electrical charge of virus suitable for adsorption on membrane filters, the adjustment of the sample pH to reverse the virus surface charge is not necessary when using electropositive membranes (Cashdollar and Wymer, 2013). Soto-Beltran et al. (2013) examined the effectiveness of two electropositive filters in the concentration of PV from a WWTP's effluent (prior to chlorination stage). In that study, two methods of concentration using NanoCeram filter and 1MDS filter were compared in terms of virus recovery. Overall efficiencies of these methods were 57% for NanoCeram and 23% for 1MDS filter methods. The Virosorb 1MDS filter is made of an electropositive glass and cellulose medium (Polaczyk et al., 2007; Soto-Beltran et al., 2013). The NanoCeram filter is made of nanoalumina fibers, which makes it electropositive (Karim et al., 2009; Soto-Beltran et al., 2013).
3.2.3. Virus elution from electronegative or electropositive filters
After being retained by the filters, the viruses must be recovered through elution from the collection substrate (Pepper and Gerba, 2015). For negative filter membranes, a common elution solution is an alkaline solution of beef extract in glycine buffer solution (Hamza et al., 2009). This solution is passed through the filter and results in the desorption of the retained viruses from the filter. The alkaline pH restores the native negative charge of the virus, resulting in an increased repulsion and subsequent desorption of the viruses from the filter membrane. In addition, the additive organic matter present in the buffer solution competes with the viruses for adsorption to the filter facilitating its removal (Pepper and Gerba, 2015).
Other elution buffer solutions used include glycine/NaOH solution (Di Bonito et al., 2017; Miura et al., 2011), sodium polyphosphate (Soto-Beltran et al., 2013) and skimmed milk (Cupples et al., 2010) to recover viruses from environmental samples. These elution buffer solutions are in the range of 70 mL to 1000 mL in volume (Falman et al., 2019; Hamza et al., 2009; Kuo et al., 2010; Prado et al., 2011; Soto-Beltran et al., 2013).
3.2.4. Secondary concentration
After elution of the viruses mobilized from the retaining membrane, the eluate frequently needs to undergo further re-concentration. The aim of this step is to reduce as much as possible the sample volume prior to nucleic acid extraction steps (Pepper and Gerba, 2015). In a study by Falman et al. (2019), the efficiency in recovery of PVs in wastewater was evaluated for several secondary concentration methods. Adsorption using an electropositive filter and elution with beef extract was used as a primary concentration method. The reported results showed that optimized flocculation with skimmed milk resulted in the efficient recovery of poliovirus type 1 (PV1) with yield of 106 ± 25% while the yield with Polyethylene glycol (PEG) precipitation was 59 ± 19%.
3.3. Ultrafiltration methods
This method relies on the principle of size-exclusion using membrane filters that have pore sizes that are smaller than the viral particles (Morales-Morales et al., 2003; Olszewski et al., 2005).
In a study conducted by Sidhu et al. (2018), the concentration of influent and effluent wastewater treatment samples was done using a hollow fibre ultrafiltration system (HFUFS), followed by another filtration step with a 100 K molecular weight cut-off filter. Results showed that the recovery of the HAdV was 37% from influent samples and 67% from effluent samples. A study by Prado et al. (2019) on the detection of enteric viruses from wastewater treatment plant samples used ultrafiltration as a primary concentration step, followed by celite-based secondary concentration. One disadvantage of this method is the small pore size of filters, leading to clogging of filters when samples have high turbidity. Grassi et al. (2010) concentrated raw and treated wastewater samples to detect RoV using tangential flow ultrafiltration. The tangential flow ultrafiltration method is generally suitable for high turbidity samples since clogging is prevented because the water flow is parallel to the membrane surface (Farkas et al., 2018).
3.4. Centrifugal ultrafiltration method
Qiu et al. (2016) developed a one-step centrifugal ultrafiltration method to concentrate human enteric viruses from wastewater. This concentration technique makes use of size exclusion through a centrifugal filter, Centricon Plus-70, with a molecular weight cut-off (MWCO) of 30 kDa. Compared to the 10 L volume of sample needed in VIRADEL, this rapid concentration method used a lower input volume of 100 mL. The reported results showed that the centrifugal ultrafiltration method resulted in an efficient recovery that was comparable to that of the VIRADEL method. In another study by Sidhu et al. (2013), centrifugal ultrafiltration was used to concentrate HAdV from 10 mL of primary wastewater samples.
These latter studies utilized the input sample volume that was smaller compared to the amount needed using the VIRADEL technique. This allows for easier collection of samples, simpler processing, and shorter filtration time. This also allows processing of multiple samples simultaneously.(Qiu et al., 2016)
Centrifugal ultrafiltration does not require any pH adjustments before filtration and after elution, which is necessary in the electronegative filtration methods, thus maintaining the stability of viruses sensitive to pH variations.
3.5. Direct precipitation with polyethylene glycol
Polyethylene glycol (PEG) is an inert and biocompatible polymer frequently applied for precipitation of proteins. PEG acts as an “inert solvent sponge” (Atha and Ingham, 1981) sequestrating water molecules from the solvation layer surrounding the proteins of the viral capsid, enhancing the virus-virus interactions and resulting in the precipitation. Early applications of precipitation with PEG method to recover viruses from environmental samples include release of solids-associated viruses from wastewater and sludge samples (Wellings et al., 1976) and their use to concentrate viruses from shellfish, natural fresh and estuarine water, and freshwater sediment samples (Lewis and Metcalf, 1988). Masclaux et al. (2013) utilized precipitation with PEG to concentrate Hepatitis E viruses in influent and effluent samples from municipal WWTPs. Another study by Strubbia et al. (2019) applied PEG to concentrate NoV from raw sewage samples. In the same study a modified PEG method (Pyro-PEG) was utilized, applying elution with sodium pyrophosphate and sonication prior the PEG precipitation step. An increased number of norovirus GI (NoV GI) and norovirus GII (NoV GII) concentrations in the treated wastewater samples was observed using Pyro-PEG in comparison to conventional PEG method. This increase in detection is attributed to the sonication step which released viral particles from organic matter and reduced the number of bacteria in the sample.
The application of PEG in the study of Amdiouni et al. (2012) resulted to a high efficiency in the recovery of RNA viruses. This is useful for the concentration of viruses from wastewater samples since several RNA viruses are present in wastewater (Adriaenssens et al., 2018; Ng et al., 2012).
As a drawback, PEG may result to non-selective precipitation because it induces the precipitation of various proteins, such as enzymes, that can interfere and even inhibit the subsequent detection of viral genome by PCR amplification methods (Masclaux et al., 2013; Shieh et al., 1995).
3.6. Skimmed milk flocculation
Skimmed milk flocculation, a concentration method first developed to recover adenoviruses from seawater Calgua et al. (2008) is also used to recover viruses from wastewater samples. This method relies on the physical processes including i) adsorption of viruses to pre-flocculated skimmed milk proteins, ii) sedimentation of the flocs with adsorbed viruses, and iii) dissolution of sediments using a phosphate buffer solution. The combination of elution with glycine buffer and the skimmed milk flocculation was successfully used by Calgua et al. (2013) to recover HAdV, JCPyV, and NoVGII from raw urban sewage samples. Assis et al. (2018) applied the skimmed milk flocculation for the recovery of HAdV and RoV from WWTP effluent samples. The latter study revealed that higher recovery rates of HAdV and RoV were obtained by eliminating the initial centrifugation step and by doubling the concentration of skimmed milk. The elimination of centrifugal step was done because of low solids in the treated effluent.
An advantage of this concentration method is that it does not require special equipment and reduced number of processing steps, allowing simultaneous concentration of a large number of samples (Calgua et al., 2008).
3.7. Pre-treatment of sludge samples
Viruses tend to be adsorbed on sludge flocs via electrostatic and hydrophobic interactions. Because sludge is a highly aggregated matrix, viruses must first be desorbed from the sludge through pre-treatment methods.(Pepper and Gerba, 2015)
The traditional method used for pre-treatment of sludge samples is based on the American Society for Testing Materials (ASTM) Standard 4994-19. It involves the concentration of viruses to sludge flocs by adsorption and subsequent elution of viruses. In this method, AlCl3 solution is added to the sample and the pH is adjusted to the value of 3.5 to achieve the optimum adsorption of viruses on the sludge flocs. The mixture is then subjected to centrifugation for the separation of the liquid phase. Solids are washed with an eluent solution to desorb the viruses, and the eluate is subjected to another centrifugation step. The pelleted solids are discarded and the supernatant is passed through a 0.22 μm filter to remove bacteria and other particles (ASTM D449-19, 2019; Pepper and Gerba, 2015; Xagoraraki et al., 2014).
Wu and Liu (2009) used a modified method where sonication (50 kHz, 100 W) of the sample was conducted before the last centrifugation step. This pre-treatment step was conducted prior to the direct counting of viral particles by epifluorescence microscopy (EFM). The effect of sonication on the recovery was examined by varying the duration of sonication. The viral count after one minute of sonication was found to be significantly higher than that obtained without sonication. The sonication enhanced the desorption of viruses from the solid sludge and the release of viruses from their host cells. However, sonication carried out for longer than a minute led to lower viral counts, which may be attributed to possible damage to viral particles. Ultrasonication of activated sludge samples was applied by Ma et al. (2013) to separate viruses from the sample matrix, and subsequently increased the efficiency of flow cytometry (FCM), which requires a sample free of aggregated flocs. Aside from sonication, Brown et al. (2015) used Tween 80, a non-ionic surfactant and emulsifier, and sodium pyrophosphate, an ionic dispersant, to enhance the dislodgement of viruses from the sludge matrix, demonstrating an increased efficiency of virus counting with FCM.
Ultracentrifugation was also examined and compared to the beef extract elution method in recovering enteric viruses from primary sewage sludge samples. However, both methods resulted in less than 7.5% mean recovery of HAdV, Rotavirus A (RoV-A), NoV GII and HAV from the sludge. These results suggested that the applied concentration methods were unable to reduce the presence of the inhibitors that affected the subsequent PCR-based detections (Prado et al., 2014).
Enzymatic virus elution (EVE), a pre-treatment step that minimizes the content of PCR inhibitors in sludge samples, was studied by Sano et al. (2003). Instead of the commonly used 10% beef extract as eluant, a solution containing hydrolytic enzymes (10 g/L) and a cation exchange resin (CER), at a concentration of 10 milliequivalents per liter (meq/L), was added to the sample pellets after the first centrifugation step. Hydrolytic enzymes used by Sano et al. (2003) were lysozyme, carboxylesterase, chymotrypsin, and papain. Multivalent cations present in the sample can enhance the adsorption of viruses to sludge flocs. The addition of CER sequestrates these cations, preventing the re-sorption of the viruses onto particle of sludge samples. In the study by Sano et al. (2003), the efficiency of RT-PCR was increased by using the EVE method instead of the conventional elution with 10% beef extract. The overall increase in virus recovery efficiency (from 7% to 31%) resulting from the combined EVE method and RT-PCR was ascribed to: i) the reduction of the concentration of PCR inhibitors from the sludge, i.e. peptides decomposed by the additive hydrolytic enzymes, ii) the capture of multivalent cations by the cation exchange resin; iii) the elimination of beef extract eluant, which is a source of further components which inhibit PCR amplification of viral genome (Hata et al., 2011; Rock et al., 2010).
Elution with beef extract and elution with glycine were compared by Rock et al. (2010) in terms of the reduction of PCR inhibitors present in biosolids or treated sewage sludges. Excitation-emission spectroscopy results showed that the natural organic matter levels were higher in the samples eluted with beef extract than in samples eluted with glycine. Lower organic matter levels led to higher amplification efficiency of PV genome by RT-qPCR.
3.8. Nucleic acid extraction methods
In the PCR-based methods, the target sequence of the genome material of the virus is amplified. Viruses have genome materials that are either RNA or DNA (Artika et al., 2020). The viral genome materials may be classified as single-stranded or double-stranded. Nucleic acid strands have different polarities, whether positive (+) or negative (−). The viral genome structures are either linear or circular. Viruses also have segmented or complete genomes (Guttman, 2013; Murphy, 1988; O'Carroll and Rein, 2016).
As previously discussed, the PCR-based amplification starts with template DNA sample. In the case of RNA viruses, RNA is reverse transcribed to complementary DNA (cDNA), which becomes the starting material. The quality and purity of these biomacromolecules affect the efficiency of the amplification and quantification methods.
The isolation and purification of DNA/RNA proceeds in these steps: lysis, purification, and recovery. Different types of DNA extraction methods include boiling, column method, magnetic beads, and use of FTA cards (Barbosa et al., 2016).
In the recent studies investigating presence of viruses in wastewater samples, commercially available DNA and RNA kits have been extensively used. The most common DNA extraction kits rely on the use of a column that has silica-based membranes (Barbosa et al., 2016). The binding of DNA to silica in the presence of chaotropic salt sodium iodide (NaI) was first examined in the study of Vogelstein and Gillespie (1979) to extract DNA from agarose gel. Boom et al. (1990) also developed the method that made of use of silica, in the presence of the chaotropic agent guanidinium thiocyanate, to purify nucleic acids from human serum and urine.
The column extraction method is classified as a solid phase-DNA extraction method. In this method, the sample undergoes lysis prior to column loading. Lysate is loaded to the column in the presence of chaotropic salts such as guanidine hydrochloride, guanidine isothiocyanate, sodium iodide, and sodium perchlorate. These salts allow the nucleic acids to selectively adsorb to the silica support, through disrupting their affinity to water. Purification comes after adsorption through washing with a reagent. The last step involves elution of the nucleic acids from the column using a buffer solution.(Barbosa et al., 2016; Butler, 2010)
Examples of the kits with silica-based membranes that were used to extract viral nucleic acids from wastewater samples include QIAamp DNA Blood Mini Kit (Hamza et al., 2014), QIAamp DNA Blood Maxi Extraction Kit (Jahne et al., 2020), QIAamp Viral RNA Mini Kit (Calgua et al., 2011; Masclaux et al., 2013; Prado et al., 2011, Prado et al., 2019), Qiagen DNeasy Blood and Tissue kit (Sidhu et al., 2018), and AllPrep DNA/RNA Mini Kit (Symonds et al., 2014).
Automatic extractors have been used by Ibrahim et al. (2017) and Di Bonito et al. (2017) to extract nucleic acids of viruses from influent and effluent wastewater samples. Most of the automated extractors rely on the use of magnetic beads which bind the nucleic acids, leaving impurities in the solution. Elution is conducted to recover the DNA bound on the beads (Barbosa et al., 2016). An advantage of using automatic extractors is their high throughput and lower variability of analytical results (Dundas et al., 2008).
4. Detection methods of viruses in wastewater
There have been numerous studies on the detection of viruses in different water matrices, including: surface water treated for drinking water purposes (Wang et al., 2020b), bottled drinking water (Martin-Latil et al., 2012), well water (Emelko et al., 2019; Ji et al., 2020), seawater (Canh et al., 2019), irrigation water (Rusiñol et al., 2020), surface water (Canh et al., 2019; Guo et al., 2018; Hata et al., 2014; Khalenkov et al., 2008) and wastewater (Di Bonito et al., 2017; Ibrahim et al., 2017; Jahne et al., 2020; Masclaux et al., 2013; Prado et al., 2019; Qiu et al., 2016; Symonds et al., 2014).
Different sampling points in the examined treatment plants have been used in these studies, as shown in Table 3 . Samples were taken from influents, primary settling tanks, secondary and tertiary treatment effluents, and final effluents. Some of these studies examined the fate of viruses inside WWTPs by examining samples originating from secondary treatment steps (Ibrahim et al., 2017; Prado et al., 2011, Prado et al., 2019; Symonds et al., 2014). Other studies collected samples prior to and after tertiary treatment to determine its effectiveness in the context of the removal of viruses (Prado et al., 2011; Qiu et al., 2018). Aside from the presence of viruses in wastewater, the occurrence of viruses in the sludges produced in the examined treatment operations has been studied (Brown et al., 2015; Symonds et al., 2014; Wu and Liu, 2009). Few studies have investigated the presence and abundance of viruses in the ambient air from WWTPs (Brisebois et al., 2018; Masclaux et al., 2014; Pasalari et al., 2019).
Table 3.
Detection and quantification of viruses in wastewater and sludge samples.a
| Wastewater sample | Viruses | Concentration/pre-treatment method | Nucleic acid extraction | Virus detection/quantification | Reference |
|---|---|---|---|---|---|
| Wastewater treatment samples: (a) Effluent of natural oxidizing pond (b) Effluent of rotating biodisk |
AiV Genotype B | Beef extract and AlCl3 method followed by precipitation with PEG | Automatic extractor NucliSENS® EasyMag™ | RT-PCR | Ibrahim et al., 2017 |
| Raw sewage | Oncogenic viruses: HPV HPyV HHV MMTV |
Elution with glycine | Automatic extractor NucliSENS® EasyMag™ | Combined multiplex PCR and bead-based Luminex technology | Di Bonito et al., 2017 |
| Influent wastewater | HAdV EV RoV NoV GI NoV GII |
VIRADEL | QIAamp DNA Blood Mini Kit | Combined real time PCR (qPCR) and multiplex Luminex xMAP assay | Hamza et al., 2014 |
| Combined wastewater; Graywater | HAdV NoV GI NoV GII |
Ultrafiltration followed by elution with sodium polyphosphate solution | QIAamp DNA Blood Maxi Extraction Kit | (a) For HAdV: qPCR and TaqMan assay; Digital droplet PCR (ddPCR) (b) For Nov GI and NoV GII: RT-qPCR; RT-ddPCR |
Jahne et al., 2020 |
| Influent wastewater | PV1 | Primary: Filtration with electropositive filter and elution with beef extract and glycine Secondary: (a) beef extract-Celite (b) ViroCap flat disc filter (c) concentrating Pipette (d) PEG/NaCl precipitation (e) skimmed-milk flocculation |
– | – | Falman et al., 2019 |
| (a) Influent municipal wastewater; (b) Effluent municipal wastewater |
HEV HAdV 40 Nov GII Porcine Adenovirus |
(a) VIRADEL (b) Precipitation with PEG |
QIAamp Viral RNA Mini Kit | RT-PCR | Masclaux et al., 2013 |
| Effluent | EV RoV |
Adsorption-elution method (Mg-method and Al-method) | ZR Viral RNA kit | RT-qPCR | Osuolale and Okoh, 2017 |
| Hospital wastewater treatment plant samples: (a) Influent (b) Effluent of sedimentation tank (c) Final effluent (after chlorination) |
RoV-A HAdV NoV GI NoV GII HAV |
Adsorption-elution using electronegative membrane | QIAamp Viral RNA Kit | (a) Conventional PCR, RT-PCR (b) qPCR |
Prado et al., 2011 |
| Municipal wastewater treatment samples: (a) Pre-UV treatment (b) Post-UV treatment |
NoV RoV Sapovirus Astrovirus JC Virus HAdV EV Reovirus |
Adsorption-elution using NanoCeram disc filters | MagaZorb total RNA Prep kit | qPCR | Qiu et al., 2018 |
| Municipal wastewater treatment samples: (a) Influent (b) Effluent of Secondary Treatment (c) Reclaimed water |
HAdV JCPyV RoV-A |
(a) For Influent and Secondary Effluent: Concentration with Celite (b) Reclaimed water: Ultrafiltration |
QIAmp Viral RNA Mini Kit | RT-qPCR | Prado et al., 2019 |
| Wastewater treatment samples: (a) Influent (b) Effluent |
HAdV HPyV HTtV |
Ultrafiltration | Qiagen DNeasy Blood and Tissue kit | qPCR | Sidhu et al., 2018 |
| Wastewater treatment samples: (a) Influent (b) Effluent of water treatment ponds (c) Effluent of UASB reactor (d) Sludge from water treatment pond (e) Sludge from UASB reactor |
NoV RoV PMMV EV |
Sludge samples: ASTM 4994-19 Wastewater samples: (a) For EV: Dilution with beef extract solution, centrifugation and filtration through protein-treated filter (b) For q-PCR: Adsorption- elution |
AllPrep DNA/RNA Mini Kit | (a) For NoV, RoV, PMMV: RT-qPCR (b) For EV: Cell Culture |
Symonds et al., 2014 |
| Municipal wastewater treatment samples: (a) Influent (b) Sludge |
Virus type not determined (quantification only) | Elution with solutions (a) Beef extract (b) Sodium pyrophosphate (c) Glycine (d) Potassium citrate (e) Lysozyme |
– | (a) Epifluorescence Microscopy (EFM) using SYBR Green I as stain (b) Transmission Electronic Microscopy (TEM) (a) Pulsed-field Gel Electrophoresis |
Wu and Liu, 2009 |
| Influent wastewater | HAdV JCPyV |
Concentration with Phosphate-buffered saline (PBS) solution | For qPCR: QIAamp Viral RNA mini Kit | (a) Immunofluorescence assay (b) Plaque assay (c) Tissue culture infectious dose-50 (d) qPCR |
Calgua et al., 2011 |
| Influent wastewater Primary settled wastewater Secondary settled wastewater Activated sludge Effluent |
Virus type not determined (quantification only) | Ultrasonication | – | Flow cytometry (FCM) | Ma et al., 2013 |
| Activated sludge | Virus type not determined (quantification only) | Addition of dispersants Tween 80, sodium pyrophosphate, and sodium cholate; Ultrasonication |
– | Flow cytometry | Brown et al., 2015 |
| Activated sludge mixed liquor | Virus type not determined (quantification and determination of viral size distribution only) | Sonication | PFGE Method | PFGE | Otawa et al., 2007 |
| Sludge samples (influent and effluent of anaerobic digesters) | Enteroviruses Coronavirus HKU1, Klassevirus, Cosavirus Parechovirus |
Precipitation with PEG | Qiagen Viral RNA extraction kit | PCR; High-throughput Sequencing |
Bibby and Peccia, 2013 |
| Sludge sewage samples | Enteric viruses | EVE | Sepa-gene RV-R | RT-PCR | Sano et al., 2003 |
| Sewage sludge samples | HAdV RoV-A NoV GII HAV |
Ultracentrifugation; Beef Extract Elution |
QIAamp Viral RNA Kit | RT-qPCR qPCR |
Prado et al., 2014 |
| Primary sludge samples Activated sludge samples Thickened sludge samples |
EV | Beef Extract elution with sonication; Precipitation with PEG |
RNeasy plant mini kit | RT-PCR | Monpoeho et al., 2000 |
| Raw sludge samples Treated sludge samples |
Mammalian orthoreovirus | Adsorption elution; Organic Flocculation |
QIAamp Viral RNA Kit | ICC-RT-qPCR; Plaque Assay |
Gallagher and Margolin, 2007 |
Note: aData for SARS-CoV-2 are reported in Table 4.
As discussed, the sample processing methods affect the efficiency of subsequent detection methods. The choice of a specific method for detection or quantification also depends on the ease of cultivation of the viruses in laboratory conditions (Ibrahim et al., 2017). Specific characteristics of the viruses affect the concentration and detection methods to be applied to the samples being examined. Wastewater and sludge samples have both a variety of DNA or RNA viruses (Bibby and Peccia, 2013; Ng et al., 2012). Viruses are also classified according to their structures as enveloped and non-enveloped viruses. Non-enveloped viruses are characterized by the encapsulation of their nucleic acids by capsid proteins. Enveloped viruses have an additional lipid bilayer membrane that surrounds the capsid proteins (Lucas and Knipe, 2002; Ye et al., 2016a). Disruption of the lipid bilayers by sample processing methods can lead to lower recoveries, affecting subsequent detection (Ye et al., 2016a). This is of particular interest for investigations related to coronaviruses (CoVs) (Casanova et al., 2009; Geller et al., 2012; La Rosa et al., 2012; Wang et al., 2005b).
Methods for the detection and quantification of viruses found in wastewater include epifluorescence microscopy, transmission electronic microscopy, pulsed-field gel electrophoresis, immunofluorescence assay, flow cytometry, traditional cell-culture, and molecular methods. Main features of these methods are summarized in this section.
It is noted that these detection methods provide different information regarding the presence of viruses in wastewater and sludge samples. As presented in the following discussions, some methods provide qualitative data and others quantitative data. Molecular methods for the quantification of a virus are based on the determination of the number of selected segments of the genetic material of the virus. Thus, the virus can be detected even if inactivated, i.e. when the viral capsid or envelope is compromised, and even when the genetic material is fragmented. Cell-culture based methods and immunological methods are used for analysis of viability of viruses.
4.1. Transmission electron microscopy (TEM)
Viruses are typically too small for detection using optical microscopy. Electron microscopy enables the inspection of these small particles (Laue, 2010). One of the earliest methods used to quantify viruses is TEM, an imaging technique with nanometer-scale resolution. This method allows the quantification, and identification and classification of viruses according to morphology.(Roingeard et al., 2019)
A basic methodology of TEM is negative staining. In negative staining, the virus particles are adsorbed on a pre-treated specimen support. After adsorption, the particles are stained with heavy metals, commonly uranyl acetate and phosphotungstic acid (PTA). After the staining and drying steps, the samples are analyzed under the electron microscope. Information from negative staining includes virus count, and virus sizes and structures (Laue, 2010).
Wu and Liu (2009) examined the morphology of viruses present in activated sludge and anaerobic digestion sludge samples from a municipal wastewater treatment plant using TEM. The observed morphologies were then used to identify viral particles according to a protocol by the International Committee on Taxonomy of Virus guidelines. TEM results showed that the sludge samples had numerous morphological types, indicating a diverse viral community.
TEM may be applied to identify viruses in emerging infectious diseases since viruses' morphologies are known to be stable even after the mutation of their nucleic acids (Laue, 2010). However, a limitation of the TEM is that it is highly selective to host-specific infectious virus, leading to a viral count result lower than the actual population (Brown et al., 2015). The evaluation of very large number of samples using TEM is difficult since it requires the expertise of highly trained personnel and sophisticated equipment while TEM analyses cannot be automated at present (Barreto-Vieira and Barth, 2015).
4.2. Nucleic acid staining with fluorescent dyes
This virus quantification method makes use of highly fluorescent nucleic acid dyes (Wen et al., 2004). Water samples are passed through a filter, commonly with a pore size of 0.22 μm (Otawa et al., 2007; Wu and Liu, 2009). The nucleic acids in the virus particles are then stained by a fluorescent dye. Staining with this type of dye allows the formation of fluorescent dots with dimensions larger than actual virus particles. The formation of fluorescent dots occurs when the fluorescent dye bound with nucleic acids are put under excitation. When the sample is viewed under an epifluorescence microscope, the fluorescent dots can be counted as the virus particles (Ortmann and Suttle, 2009).
One of the common dyes used in the recent relevant studies is SYBR Green I whose blue emission is observed using an epifluorescence microscope (Otawa et al., 2007). Counting of viruses from influent, effluent, and sludge samples was done by Wu and Liu (2009) using epifluorescence microscopy with SYBR Green I. Otawa et al. (2007) also employed this direct count method, using SYBR Green I as staining agent, to enumerate viruses in mixed liquor activated sludge samples.
One advantage of this method is that the stained virus particles can be counted even at lower magnifications thus obviating the need to use TEM instrumentation (Ortmann and Suttle, 2009). Another advantage of EFM is that it can be used to count viruses that are not cultivable in laboratories. Aside from this, through the use DNase treatment, the EFM method can differentiate virus particles with nucleic acids from virus-like particles without nucleic acids. However, a limitation of this method is that it has a low efficiency in detecting RNA viruses or single-stranded DNA viruses (Forterre et al., 2013).
4.3. Flow cytometry (FCM)
The combination of the use of fluorescent nucleic acid-specific dyes and flow cytometry has been applied for the quantification of viruses in wastewater samples.
In FCM, samples containing viruses are diluted with buffer solutions and stained with fluorescent dyes. These samples are then injected into the flow cytometer. The hydrodynamics effect of the surrounding sheath fluid allows the virus particles to enter a stream in a single file. The individual particles intersect with a beam of monochromatic light, commonly from an argon-ion laser. The scattering and fluorescence produced by interactions of each particle with the incident laser beam is collected by detectors and analysed as the scatter and fluorescence intensity, respectively (Brown and Wittwer, 2000).
In the FCM approach, the samples are stained with a fluorescent dye, which binds selectively to DNA or RNA. The intensity of fluorescence of the DNA/dye and RNA/dye complexes is therefore correlated with the DNA/RNA content of the sample (Adan et al., 2017; Brown and Wittwer, 2000). Advantages of FCM are its notably high accuracy and high speed of quantification (Ma et al., 2013).
Ma et al. (2013) utilized FCM to determine the abundance of viruses in influent, primary and secondary settled water, activated sludge and effluent samples from a municipal wastewater plant. Comparing viral counts determined by FCM and EFM showed a higher sensitivity and higher speed of quantification using FCM. Brown et al. (2015) quantified viruses from activated sludge samples using FCM. In that study, the FCM method was shown to have a higher sensitivity than that of TEM by a factor of 2.7.
4.4. In situ fluorescence
The fluorescence-based analyses discussed in the preceding paragraphs can only be performed in the specialized laboratories. Pollard (2012) presented the conceptual design of a fluorescence instrument for in-situ and/or online monitoring of abundance of viruses in varying water matrices including effluent wastewater. The proposed portable instrument has an inline filter to remove bacteria from the water sample. A mixing coil is positioned after the filter where DNase and the fluorescent probe SYBR-Gold are added to the sample to form the DNA/RNA-SYBR viral complex. This mixture is directed to a reverse osmosis (RO) filter that concentrates the viruses. The RO concentrate, which contains stained viral particles, passes through a unit that measures the fluorescence signal. On the other hand, the permeate becomes the “background fluorescence”, which is subtracted from the sample viral fluorescence. Although the proposed instrument is still at a conceptual stage, tests of this method showed that the EEM fluorescence intensity has a linear correlation (r2 = 0.97) with the viral count.
4.5. Immunofluorescence assay (IFA)
One advantage of IFA over other quantification methods is that it can be used to analyse the infectivity of viruses (Calgua et al., 2011). In this method, a sample from an infected cell culture is adsorbed on a microscope slide. The viral protein antigen is detected by sequentially incubating the sample fixed on the slide with a specific antibody and a fluorescent chemical-conjugated secondary antibody that recognizes the former. Under optical excitation, the fluorophore-conjugated antibody fluoresces. When viewed under a fluorescent microscope, the antigen-antibody complex appears as a fluorescent particle (Dowd et al., 2009; Im et al., 2019).
This method was successfully used by Calgua et al. (2011) to quantify HAdV and JCPyV in raw sewage entering wastewater treatment plants. The reported results showed that the sensitivity of IFA is an order of magnitude higher than those of the other cell culture methods used in the study. The viability of the human adenovirus 2 (HAdV 2) found in activated sludge samples and effluent samples from a wastewater treatment plant was also verified by Schlindwein et al. (2010) using IFA.
4.6. Enzyme linked immunosorbent assay (ELISA)
ELISA is a method that detects the presence of microbial antigens in various matrices (Boonham et al., 2014). It relies on the principle of antigen binding to its specific antibody and eliciting a change in color or fluorescence due to the resultant enzyme activity. The first step of the process is the binding of antigen at a specific antibody immobilized on a surface, commonly in a set of 96-well microtiter plates. A second enzyme-linked antibody, specific for the same antigen, is utilized to form an antibody-antigen-antibody sandwich. The enzyme-coupled antibody reacts with a substrate that changes color when modified by the enzyme. The change in color or fluorescence is correlated with the concentration of the probed antigens in the sample.(Gan and Patel, 2013)
Kargar et al. (2013) utilized ELISA to detect RoV-A in influent and effluent samples of urban and hospital sewage disposal systems. This detection allowed determining the efficiencies of removal of RoV from urban and hospital wastewater treatment systems.
4.7. Pulsed-field gel electrophoresis (PFGE)
The PFGE method makes use of a pulsating electric field to enable the separation of high molecular weight DNA fragments according to their molecular sizes (Nassonova, 2008; Díez et al., 2000).
In this method, alternating electric fields generated between two separate electrodes cause the molecules to re-orient periodically to align to the imposed electric field (Le Tang et al., 2017). The ability of DNA molecules to re-orient themselves and respond to the imposed modulated electric field depends on their molecular sizes and charges. It takes a shorter time for the smaller molecules to re-orient by migrating through the pores of a gel matrix towards the new anodes. On the other hand, larger DNA molecules take a longer time to re-orient. The larger DNA molecules that migrate slower than the set pulse time tend to migrate as one band through the gel matrix (Le Tang et al., 2017; Lopez-Canovas et al., 2019).
The band pattern formed by a viral community serves as its fingerprint. The number of the formed bands gives an estimate of the number of different viruses (diversity) in a sample (Díez et al., 2000).
Otawa et al. (2007) applied the PFGE method to determine the diversity of viruses in activated sludge samples from 14 wastewater treatment plants. The method revealed that the prevailing sizes of viral DNAs in the samples were in the range of 40 to 70 kb. Based on the similarity of the band patterns observed in the PFGE data, the PFGE method was able to demonstrate that the detected viral species were common to the different activated sludge wastewater treatment plants.
Wu and Liu (2009) used PFGE in their study concerned with the diversity of the viruses in influent, primary settling tank, effluent, and sludge samples of a municipal wastewater treatment plant. Their experimental results showed that the activated sludge, anaerobic digestion sludge, and effluent had the highest number of bands. Similar band patterns were observed between the influent and primary settling tank samples. The PFGE method revealed that the dominant sizes of viral DNAs were in the ranges of 30 to 80 kb and 200–350 kb.
4.8. Molecular methods
Molecular methods for the detection and quantification of viruses are based on nucleic acid amplification. This section discusses the different methods of viral nucleic acid amplification, and subsequent detection and analysis.
4.8.1. Conventional polymerase chain reaction (cPCR)
PCR is an amplification method which relies on the principle of cloning a DNA fragment from a DNA extract through in vitro replication (Kadri, 2020). In the case of detection of viruses, this method amplifies or clones a fragment of the viral genome (Rodriguez et al., 2009). A pair of oligonucleotides (primers), which define the starting site of DNA polymerase, is used to flank the DNA fragment that is to be replicated. Each of the oligonucleotide is designed to become attached to a certain edge of the DNA target, depending on its given sequence (Kadri, 2020). Amplification through PCR makes use of an thermostable enzyme, commonly the Taq Polymerase, to synthesize new DNA strands from existing strands in the DNA extract (Lo and Allen, 2006). PCR proceeds in three steps: denaturation or separation of double strands of DNA at temperature >90 °C, annealing of primers at temperature close to their melting temperature (Tm) , and elongation at ≥72 °C (Kadri, 2020; Mackay, 2002). Repeating these three steps produces multiple copies of DNA sequences (Lo and Allen, 2006). Two-step PCR, which combines annealing and extension, has also been used leading to reduction of time of amplification (Girones et al., 1993; Jin et al., 2014).
After the amplification step, the PCR products are subjected to agarose gel electrophoresis (Mackay, 2002; Rodriguez et al., 2009) for their qualitative analysis.
4.8.2. Reverse transcription-polymerase chain reaction (RT-PCR)
In the case of RNA viruses, the amplification of the nucleic acid starts with the production of DNA from RNA using a reverse transcriptase enzyme (Cobo, 2012). In this method, the produced DNA is the cDNA which becomes the new template that is amplified using the PCR method described in Section 3.8.1.
Reverse Transcription allows creating cDNAs from RNA viruses (Schinazi et al., 2013). RNA viruses present in wastewater include, among many others, NoV, Aichi virus (AiV), parechovirus, HAV, hepatitis E virus (HEV), astrovirus, RoV and CoVs (Hellmér et al., 2014b).
Ibrahim et al. (2017) utilized the RT-PCR technique to amplify nucleic acids of AiVs, which have RNA genomes, in wastewater samples. Amplification with RT-PCR enabled the analysis of AiVs which are not easily cultivable in the laboratory. The RT-PCR method was also used to amplify the nucleic acids of HEVs, which are positive-strand RNA viruses, in influent and effluent samples of wastewater treatment plants (Di Profio et al., 2019; Masclaux et al., 2013). Osuolale and Okoh (2017) successfully used RT-PCR to amplify the nucleic acids of the RNA viruses enterovirus (EV) and RoV, in wastewater effluent samples. In this study, the RoVs were subjected to heating prior RT-PCR to separate its double-stranded RNA.
RT-PCR was used by Monpoeho et al. (2000) to determine the abundance of EV in primary sludge, activated sludge and thickened sludge samples. The results showed that the largest viral concentration was present in the primary sludge samples. Smaller concentrations were found in the activated sludge samples, followed by the thickened sludge samples.
RT-PCRs are conducted using random, oligo (dT) or specific primers (Newby et al., 2009; Rodriguez et al., 2009). The use random primers will result in non-specific production of cDNA from the existing RNA array in the sample. Oligo (dT) primers hybridize the polyadenine tails in mRNAs (Jalali et al., 2017; Rodriguez et al., 2009). On the other hand, specific primers amplify a region of interest in the viral genome (Rodriguez et al., 2009). The choice of primers affects the result of the amplification method. This is relevant to the detection of emerging viruses in wastewater.
4.8.3. Real-time polymerase chain reaction (qPCR)
The qPCR is a method that allows simultaneous PCR amplification of nucleic acids and detection of the products (Cobo, 2012). It is classified as a quantitative method because it allows quantification of the target sequences, in contrast to the conventional PCR method, which provides qualitative data through gel electrophoresis (Kadri, 2020). Aside from providing quantitative data, qPCR also displays high sensitivity and is not affected by limiting concentrations of reagents (Watzinger et al., 2006).
In the qPCR method, PCR products are labelled by binding with fluorescent dyes or fluorogenic probes (Arya et al., 2005). A Real-Time thermocycler is used to monitor the fluorescence emission during the PCR amplification (Arya et al., 2005; Cobo, 2012). The principle of quantification is based on the correlation between the intensity of fluorescence emission and the amount of amplification of the product of PCR after each cycle (Kadri, 2020).
RT-qPCR uses the same principle as qPCR except that it starts with RNA material instead of DNA, requiring a reverse transcription step (RT) prior to that of qPCR. Another advantage of the qPCR or RT-qPCR over the conventional method is that it eliminates the step of the agarose gel electrophoresis after the amplification step.
Qiu et al. (2018) utilized qPCR to detect and quantify HAdV, JCPyV, and astrovirus in samples collected before and after UV treatment in a wastewater treatment plant. In the same study, RT-qPCR was used to detect and quantify NoV GII, reovirus, EV, RoV, and sapovirus.
Prado et al. (2011) used qPCR and RT-qPCR to detect viruses in hospital wastewater effluents. The latter study demonstrated that the use of RT-qPCR in detecting NoV was more efficient than that of the conventional RT-PCR. This was attributed to the possibility of amplifying shorter fragments in RT-qPCR than was achieved in conventional RT-PCR. Kong et al., also reported that shorter PCR products, or amplicons, are generally amplified more effectively than long ones. However, this hypothesis contrasts with results of other studies, in which there were no sensitivity differences between conventional RT-PCR and RT-qPCR based on the length of the amplicons (Bastien et al., 2008). Therefore, other factors could contribute to the variations, for instance the specificity of the primers, their accessibility to the region to be amplified, and the protocols used (Ferreira et al., 2009).
In a study by Wong et al. (2010), quantification by RT-qPCR showed no significant difference between enteric virus levels in dewatered sludge and sludge after mesophilic anaerobic digestion treatment. This showed that anaerobic digestion may not be efficient for the inactivation of viruses in wastewater sludge.
RT-qPCR was used to detect and quantify SARS-CoV-2 in wastewater samples of Massachusetts (Wu et al., 2020b). Results of these measurements gave a value of 1.04 × 103 genome copies per liter of sample. While additional WWTPs in other areas should be tested further, the results showed that RT-qPCR data from wastewater samples could be used to estimate the viral burden in the affected population. RT-qPCR was also applied to detect and quantify SARS-CoV-2 in raw wastewater samples of Paris, France (Wurtzer et al., 2020). Although the infectivity of the viruses in the examined samples was not yet ascertained by this method, these RT-qPCR results are highly important because they can be interpreted to indicate that contamination of the wastewaters in Paris had existed even before the onset of the rapid increase of the Coronavirus disease 2019 (COVID-19) cases in the affected population. These studies demonstrate the potential role of RT-qPCR in the development of the Wastewater Based Epidemiology (WBE) approach that can be applied to investigate viral disease outbreaks in communities (Ahmed et al., 2020; Xagoraraki et al., 2014).
It should be noted that a limitation of the qPCR and RT-qPCR methods, as in the conventional PCR and RT-PCR, is that they detect both infectious and non-infectious viruses. Still, the data of these methods are useful in evaluating the efficiency of disinfection treatments, since they detect viruses that cannot be effectively detected in cell culture-based methods. These PCR-based methods may be used to detect viruses, such as RoV and NoV, that are difficult to propagate in cell cultures (Qiu et al., 2018).
Recently, combined azo dye pre-treatment and qPCR has been applied even for the quantification of damaged viruses, overcoming the difficulties associated to the culturing of some viruses for the determination of the viability (Leifels et al., 2020).
4.8.4. Multiplex PCR
Multiplex PCR allows simultaneous detection of different viruses present in a single sample using more than one set of primers in one reaction (Newby et al., 2009). Multiplex qPCR also allows detection of different specific viruses using more than one fluorescent reporter (Haramoto et al., 2018).
Hamza et al. (2014) used Luminex assay, a novel multiplex PCR-based assay, to simultaneously detect human enteric viruses present in wastewater influents. This approach makes use of the Luminex XMap technology with microspheres that are uniquely labelled with variable amounts of 2–3 different fluorophores to obtain up to 500 spectrally distinct microsphere sets. In this method oligonucleotide probes specific for desired DNA sequences are coupled to the beads and the PCR products obtained using biotin-labeled primers are hybridized with probes. Addition of phycoerytrin-streptovidin complex that interacts with biotinylated sequences allows detecting the presence of amplicons on the beads by fluorescence emission. The microspheres enter an analyzer that makes use of the principle of flow cytometry. The beads are identified after they pass through lasers of different wavelengths and the spectral response from each bead is associated with a single oligonucleotide probe. The Luminex analyzer also measures the fluorescence emitted by the phycoerythrin to quantify the amount of bound nucleic acids (Dunbar and Hoffmeyer, 2013; Hamza et al., 2014).
The results reported in Hamza et al. (2014) showed that the multiplex assay was as sensitive as qPCR in the detection of several enteric viruses in wastewater samples. Di Bonito et al. (2017) also successfully used the multiplex PCR bead-based technology (Luminex) to detect oncogenic viruses in untreated wastewater. This class of viruses, that are related to human cancer, include mucosal and cutaneous human papillomavirus (HPV), human polyomavirus (HPyV), human herpesvirus (HHV) and mouse mammary tumor virus (MMTV).
The multiplex qPCR has been shown to have a similar sensitivity to singleplex PCR in the detection of RoV-A in raw sewage samples (Fumian et al., 2010).
Aside from the multiplex assay's sensitivity, another advantage of this method is its speed associated with the simultaneous detection principle that it employs. It also requires lower reagent cost and labor compared to singleplex PCR-based assays.(Dunbar and Hoffmeyer, 2013; Fumian et al., 2010; Hamza et al., 2014)
4.8.5. Sequencing
Aside from quantification, PCR products may also be subjected to DNA sequencing. DNA sequencing is the process of determining the specific order of nucleotides for a segment of DNA (Artika et al., 2020).
The most common sequencing method is based on the dideoxy or chain-termination method, or the Sanger method (Clark et al., 2019; Hoy, 2019). In this method, DNA polymerase randomly produces multiple partial copies of the DNA template that have varying lengths. The lengths of each subset DNA copies differ by one nucleotide, resulting to each copy ending in a different nucleotide position. Each final nucleotide is labelled with a fluorescent tag, with a different color for ddATP, ddGTP, ddCTP and ddTTP. The DNA copies undergo capillary electrophoresis where they are separated by size, with the smaller fragments forming the fastest moving bands. A fluorescent detector records the fluorescence emitted by the final nucleotides of each fragment. The sequence of the template DNA is revealed by the diagram of fluorescent peaks produced (Clark et al., 2019; Hoy, 2019).
Recent studies concerned with the detection of viruses in wastewater have applied the Sanger method using commercially available sequencing kits such as the BigDye Terminator Cycle (Di Profio et al., 2019; Hellmér et al., 2014b; Ibrahim et al., 2017; Montazeri et al., 2015; Prado et al., 2011). Most of the sequencing analyses used in these and similar studies are now automated to increase speed and sensitivity of the process (Artika et al., 2020).
Direct sequencing was conducted by Ibrahim et al. (2017) to identify the genotype of AiVs found in the secondary effluent of a pilot wastewater treatment plant. The sequence analysis revealed that the strains of AiV found in the wastewater samples were the same strains found in patients with acute gastroenteritis.
Hellmér et al., 2014a, Hellmér et al., 2014b used the sequencing analysis to investigate whether the identification of pathogenic viruses in sewage can be indicative of an epidemic outbreak. Wastewater samples from Gothenburg, Sweden were subjected to analysis by RT-qPCR and sequence analysis. The reported results showed that two strains of the HAV detected in sewage samples were the same strains found in fecal samples of infected patients in Gothenburg. The strains were also found to be the same strains associated to an outbreak in the region at the time the sewage samples were collected.
In a similar study (Di Profio et al., 2019), wastewater samples from Abruzzo Region in Southern Italy were collected to monitor the prevalence of HEV. Results of sequencing analyses showed the presence of the HEV Genotype 3 strains, which were previously detected in animal and human hosts in the region. Although further exploration of these findings is needed, the result of this study suggests that the high number of people infected with HEV in the region may be due to transmission from animals to humans.
Partial sequencing was conducted by Prado et al. (2011) to determine the diversity of viruses in samples from a hospital wastewater treatment plant located in Rio de Janeiro, Brazil. Results of this study showed that HAdV serotypes 40 and 41 detected in the effluent samples were serotypes that are associated to gastroenteritis cases in children in Rio de Janeiro and Salvador, Brazil.
High-throughput sequencing has been recently applied in studies investigating the prevalence and diversity of viruses in sewage samples. The Next Generation Sequencing (NGS) technique allows a larger number of samples to be analyzed in a single run (Hoy, 2019; Martínez-Puchol et al., 2020). In this type of sequencing, the following steps are involved: producing millions of copies of DNA fragments, and conducting and recording millions of sequences simultaneously (Clark et al., 2019; Hoy, 2019). This parallel sequencing of a large number of samples reduces turnaround time and cost of sequencing (Hoy, 2019; Martínez-Puchol et al., 2020).
Fumian et al. (2019) successfully used NGS to determine Norovirus GII genotypes found in influent, primary effluent, and secondary effluent from a wastewater treatment plant in Rio de Janeiro, Brazil. Results demonstrated the prevalence of 6 major genotypes of NoV GII in the wastewater samples collected for a period of one year. These genetic data were compared to the corresponding clinical data as part of a surveillance of the circulation of pandemic and epidemic norovirus strains in the examined area. Two strains of noroviruses, GII.5 and GII.7, were found to be prevalent in the wastewater samples but were not detected in the clinical samples. This indicates that these strains may lead to asymptomatic or mild infections, which are not reported.
NGS techniques were used by Martínez-Puchol et al. (2020) to determine the distribution of human pathogenic viruses in three wastewater treatment plants in Barcelona City, Spain. Sequencing analyses showed that the three wastewater treatment plants had similar distribution of types of HAdV, HPV and HEV types despite the differences in the location, and volume of wastewater treated.
Raw and digested sludge samples from wastewater treatment plants in the United States of America were tested using DNA sequencing to assess infectious risks related to reuse of sludge as agricultural soil amendment (Bibby and Peccia, 2013). NGS showed that aside from the viruses normally associated with gastrointestinal infection, the examined sludge samples had an abundance of viruses associated with respiratory diseases. These viruses include HAdV Types B and C, Bocavirus, and Coronavirus HKU1, which may be transmitted by airborne route (droplet spread) or by fomites. This poses an additional concern since aerosols are known to be released during application of stabilized sludge on agricultural land. This indicates the presence of another possible pathway of transmission of viruses related to human disease by exposure to sludge from wastewater treatment. Current regulations of pathogens in sludge include the monitoring of levels of enteroviruses and fecal coliforms. The abundance of emerging viruses other than EV in sludge may indicate a need to review the regulations.
Recent metagenomic analyses of samples from WWTPs have revealed that a large number of viral genomes cannot be recognized using information from public sequence databases. Tamaki et al. (2012) found that 85–90% of viral sequences derived from tropical WWTP samples were unclassified. Similar results were obtained by Alhamlan et al. (2013) during the analysis of the viral community of a dairy lagoon wastewater, where 81–83% of derived sequences were unrecognizable according to the GenBank database. Around 37% of sequences derived from different WWTP samples studied by Ng et al. (2012) were unclassifiable by BLASTx against the GenBank viral database. These results show that further work using sequencing is needed to fully identify novel viral species in WWTP systems.
The latter studies show the role of sequencing analysis in the ongoing effort to identify the diversity of strains of viruses in wastewater and wastewater sludge samples. The use of this approach to monitor wastewater systems may serve as an early warning method to detect possible disease outbreaks (Hellmér et al., 2014b) and as a tool for public health surveillance (Fernandez-Cassi et al., 2018). It may also be used as a basis for re-evaluation of regulatory requirements related to pathogens in wastewater systems (Bibby and Peccia, 2013; Montazeri et al., 2015).
4.8.6. Integrated cell culture- polymerase chain reaction (ICC-PCR)
4.8.6.1. Cell culture
Traditional cell culture methods use cytopathic effects (CPE) for measuring infectious viruses (Qiu et al., 2018). In this approach, cells are cultivated under controlled conditions and infected with viruses, which need host cells to replicate. The identification of presence of viruses is observed through CPEs that are visible under light microscopy. CPEs are manifested as deterioration of monolayer cells that have been infected by a virus (Dilnessa and Zeleke, 2017).
The observed CPEs and the rates of CPE appearance are used to identify viruses. Destruction of the monolayer cell, in which the infected cells shrink easily and detach from the glass, is typically associated to EV. Focal degeneration of infected cells is typical of herpesviruses. Enlargement and formation of grape-like clusters are associated with HAdV. The appearance of altered areas, referred to as inclusion bodies, in stained cells is also a characteristic type of cytopathic effects observed for HAdV and herpesviruses (Suchman and Blair, 2007).
Aside from the long period needed to obtain results, another limitation of the traditional cell culture is that some viruses do not grow in cell cultures (Dilnessa and Zeleke, 2017; Reynolds, 2004). Another complication is that some viruses such as RoV and HAV do not give visible cytopathogenic results and are therefore not detected by conventional cell culture method. It is also not a highly specific method since multiple types of viruses can be supported by a single cell line (Reynolds, 2004).
4.8.6.2. ICC-PCR
The combination of cell culture and PCR was developed to address the limitations of each method when used individually (Rodriguez et al., 2009). In the ICC-PCR method, the samples with viruses are introduced to cell culture media and incubated for 2–3 days. Samples of cell culture with viruses are subjected to freezing, which causes lysis of the cells. The virus particles are released from their host cells when lysis occurs. After this, PCR-based amplification is applied to the lysate (Newby et al., 2009).
The approach of integrated cell culture-real time PCR (ICC-qPCR) was used by Qiu et al. (2018) to evaluate the efficiency of UV disinfection in a wastewater treatment plant. The results showed that the ICC-qPCR method was more efficient than qPCR in detecting reovirus. This study also demonstrated that RoV was rarely detected by the ICC-qPCR method. This was attributed to the insufficient rate of RoV propagation in cell culture in the study.
Chowdhary et al. (2005) utilized ICC-PCR to detect PVs in wastewater from sewage treatment plants. Since PVs are RNA viruses, the extracted viral RNAs were subjected to reverse transcription before amplification. ICC-PCR provided results in 4–5 days while traditional cell culture took 18 days.
The viability of HAdV in raw sewage, secondary effluent, and reclaimed water was assessed by Prado et al. (2019) using ICC-qPCR. Using this method showed that a significant amount of HAdV was viable even after undergoing the activated sludge wastewater treatment.
Integrated cell culture- real time RT-PCR (ICC-RT-qPCR) was shown to be more efficient than traditional plaque assay in detecting the presence of mammalian orthoreovirus in raw and treated sludge samples (Gallagher and Margolin, 2007).
The latter studies showed that in terms of virus inactivation, the ICC-PCR method is useful in assessing the efficiency of different stages of wastewater treatment in virus removal. However, ICC-PCR has its limitations such as the non-detection of some viruses that are not cultivable in cell culture. It should also be noted that it is designed to detect specific types of viruses depending on the PCR primers used. It is also therefore prone to false negatives in the detection of other viruses present in the sample. Because of this, some studies still use the traditional cell culture along with ICC-PCR (Qiu et al., 2018).
4.9. Process control viruses
The efficiency of the methods used to concentrate, detect and quantify viruses is monitored through process controls (Blanco Fernández et al., 2017; Hennechart-Collette et al., 2015). Process control viruses are either added to samples prior to their concentration, to concentrated samples prior to nucleic acid extraction, or to nucleic acid extracts prior to the amplification and detection (Haramoto et al., 2018). The chosen process control viruses must have similar morphology as the target viruses, must not be naturally found in the samples and must be easily quantified by nucleic acid-based assays and infectivity assays (Blanco Fernández et al., 2017; Diez-Valcarce et al., 2011). Murine norovirus has been used as a process control for NoV because of the former's similarity in the morphology and genetic structure to the target virus (Hennechart-Collette et al., 2015). Murine norovirus was used by Kitajima et al. (2014) as a process control virus to determine the efficiencies of viral nucleic acid extraction and RT-qPCR in a study on the detection and quantification of viruses, including NoVs, in wastewater. Bacteriophage PP7 was utilized by Fumian et al. (2010) as a process control to determine the efficiency of concentration of RoV-A from wastewater samples. Mengovirus strain vMC0 has been used as a process control virus to assess the efficiency of concentration, nucleic acid extraction, and detection of enteric viruses from water samples (Coudray-Meunier et al., 2015; Teixeira et al., 2020). Farkas et al. (2018) utilized mengovirus strain vMC0 as a process control to assess the efficiency of different concentration methods in the recovery of enteric viruses from wastewater samples. Mengovirus has a similar structure as HAV (Coudray-Meunier et al., 2015), and was utilized as a process control to asses viral RNA extraction and PCR efficiencies in a study on the detection of HAV from WWTPs (Ouardani et al., 2016). However, the study of Petterson et al. (2015) showed that there is a low correlation between the recoveries of spiked mengovirus, and the enteric viruses HAdV, NoV GI, and NoV GII from surface water samples. This result indicated that there is a need for further exploration of process control viruses that would be as close as possible to the structure and behavior of the target viruses.
5. Challenges and perspectives
5.1. Concentration methods of wastewater for assay of emerging viruses: coronaviruses and SARS-CoV-2
Since the identification of coronaviruses in the mid-1960s, several novel viruses, including SARS-CoV-1 (2002, China), Middle East respiratory syndrome–CoV (MERS-CoV, 2012, Saudi Arabia), and SARS-CoV-2 (2019, China), have emerged. Up to now, seven human CoVs have been identified. CoVs are enveloped single-stranded RNA viruses with sizes ranging from 60 to 220 nm. The viral protein capsid is covered by a lipid bilayer membrane that contains proteins or glycoproteins and crown-like spikes on the surface. Contrarily to “simple” non-enveloped RNA viruses, CoVs are less stable in wastewater. SARS-CoV-1 has been shown to persist up to 2 days at room temperature, both in domestic and hospital sewages, and more than 14 days at 4 °C (Wang et al., 2005b, Wang et al., 2005c). These considerations pertinent to the stability of CoVs, should be taken into account during the sampling and storage of wastewater samples, as well as during the treatment of wastewater for the detection, quantification and particularly in the determination of the viability of these viruses.
Methods for concentration of CoVs in water media were recently examined by La Rosa et al. (2020). Enveloped viruses such as CoVs are sensitive to pH variations, with their optimal stability at slight acidic pH of 6–6.5 (Sattar et al., 2009). Nevertheless, SARS-CoV-2 demonstrated high stability in a wide range of pH values (Chin et al., 2020). These should be considered in the case of VIRADEL recovery of CoVs where virus elution from electronegative or electropositive membranes is performed at strong alkaline or acid pH, respectively. Five studies have examined the efficiency of recovery of CoVs: the recovery of bovine enteric CoV (Collomb et al., 1986), SARS-CoV-1 (Wang et al., 2005a), murine hepatitis virus (MHV)(Ye et al., 2016b), bovine CoV(Abd-Elmaksoud et al., 2014), and transmissible gastroenteritis virus (TGEV) (Blanco et al., 2019). Adsorption on glass wool and glass powder (Abd-Elmaksoud et al., 2014; Blanco et al., 2019; Collomb et al., 1986) or on silica gel compounded with aluminum hydroxide (Wang et al., 2005a), followed by elution with neutral (Wang et al., 2005a) or alkaline (Abd-Elmaksoud et al., 2014; Blanco et al., 2019; Collomb et al., 1986) buffers were some of the techniques adopted for the concentration of the above mentioned CoVs. PEG precipitation, ultracentrifugation and ultrafiltration were compared by Ye et al. (2016b) in the context of the recovery efficiency of MHV. Ultrafiltration was demonstrated the most efficient method with 25.1% of recovery of MHV. Blanco et al. (2019) applying virus adsorption onto glass wool with subsequent elution with alkaline buffer and PEG precipitation, explored on the recovery of TGEV, effects of pH, contact time and composition of eluent. An adsorption of 42.7% and the complete removal of virus from the glass wool adsorbent was obtained by overnight elution with glycine/beef extract buffer at pH of 11.0 in presence of TWEEN 80 (0.3%).
Recent results of the investigation of the detection and quantification of SARS-CoV-2 are summarized in Table 4 . Most of those studies have been published as pre-print articles and not yet subjected to peer-review process at the moment of writing of this review. Therefore, the information reported in these publications should be considered with caution. In absence of a general protocol for recovery of CoVs and of SARS-CoV-2 from wastewater, different procedures of sampling, sample storage and concentration, and virus detection have been applied (Table 4, Table 5 ).
Table 4.
Detection and quantification of SARS-CoV-2 in wastewater and sludge.
| Reference; Location of WWTPs |
Sample type; Sampling mode; Volume; Storage temperature |
Sample pre-treatment | Concentration method | PCR inhibitor treatment | RNA extraction | Concentration in genome copies/L |
Viability test results, number samples with infectious virus/total number of samples | Detection/quantization/viability analysis methods | |
|---|---|---|---|---|---|---|---|---|---|
| Influent | Effluent | ||||||||
|
Medema et al., 2020; The Netherlands |
Municipal wastewater; Composite sampling (24 h); 250 mL; 4 °C |
Centrifugation | Centrifugal filtration (Centricon Plus-70, MWCO of 10 kDa) | – | RNeasy PowerMicrobe (Qiagen) and Biomerieux Nulisens Kit (Biomerieux) in combination with semi-automated KingFisher mL purification system (Thermo Scientific) |
a | a | a | RT-PCR: (one step) EvoScript RNA Probes Master (Roche); Indirect evaluation by F-specific RNA phages assay |
| (Wu et al., 2020b, Wu et al., 2020a) Massachusetts, USA |
Municipal wastewater; Composite sampling (24 h); n.r.; 4 °C |
Pasteurization (60 °C, 90 min) and filtration (0.2 μm pore size) |
PEG/NaCl, centrifugation | – | TRIzol | 1.04 × 103 | a | a | RT-PCR Reverse transcriptase, NEB RT-qPCR; TaqMan fast advanced master mix, Thermofisher |
|
Nemudryi et al., 2020; Montana, USA |
Municipal wastewater; Composite sampling (24 h); 500 mL; n.r. |
Filtration (5 μm and 0.45 μm pore size) | Centrifugal filtration (Corning Spin-X UF, MWCO of 100 kDa) | – | RNeasy Mini Kit (Qiagen) | 104 | a | a | RT-qPCR (one-step) TaqPath™ 1-Step RT-qPCR Master Mix, CG (ThermoFisher); RT-PCR SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific); Q5 High-Fidelity DNA Polymerase (New England Biolabs) |
|
Ahmed et al., 2020 Australia |
Municipal wastewater; Composite sampling (24 h) and grab sampling; 100–200 mL; 4 °C |
Filtration (0.45 μm pore size) | a. Electronegative membrane filtration (HAWP09000, Merk; pore size of 0.45 μm) b. Centrifugal filtration (Centricon Plus-70, MWCO of 10 kDa) |
– | RNeasy PowerWater Kit; RNeasy PowerMicrobiome (Qiagen) | 1.90 × 101 to 1.2 × 102 |
a | a | RT-qPCR (one-step) iTaq™ Universal Probes One-Step Reaction Mix (Bio-Rad) |
|
Wurtzer et al., 2020; France |
Municipal wastewater; n.r.; 11 mL; n.r. |
– | Ultracentrifugation | PCR Inhibitor removal resin (Zymo Research) | PowerFecal Pro kit with a QIAsymphony automated extractor (QIAGEN) | 5.00 × 104 to 3.00 × 106 |
a | a | RT-qPCR |
| Wang et al., 2020b, Wang et al., 2020a; China | Hospital sewage; n.r.; n.r.; n.r. |
– | – | – | SARS-CoV-2 nucleic acid detection kit (Shanghai Berger Medical Technology Co., China) | a | a | 0/6 | RT-PCR; Viability in Vero-E6 cells |
| (Randazzo et al., 2020); Spain |
Municipal wastewater; Grab sampling 200 mL; 4 °C |
pH Adjustment at 6; | Precipitation with AlCl3, centrifugation; elution with beef extract (3%, pH 7.4), centrifugation and resuspension in PBS | – | Nucleospin RNA virus Kit (Macherey-Nagel) | 1.48 × 105 to 3.90 × 105 |
Secondary Effluent: 2.51 × 105 Tertiary Effluent: No detection |
a | PrimeScript™ One Step RT-PCR Kit; RT-qPCR diagnostic panel assays (US CDC, 2019-nCoV RUO Kit and the positive control 2019-nCoV_N_Positive Control by Integrated DNA Technologies). |
|
La Rosa et al., 2020; Italy |
Municipal wastewater; Grab sampling; 250 mL; −20 °C |
Pasteurization (57 °C, 30 min) |
PEG-Dextran precipitation | One step PCR Inhibitor removal kit (Zymo Research) | NucliSENS miniMAG semi-automated extraction system (bioMerieux,) | a | a | a | RT-PCR SuperScript III/IV Reverse Transcriptase (ThermoFisher Scientific); Kit Platinum™ SuperFi™ Green PCR Master Mix, Thermo), |
|
Rimoldi et al., 2020; Italy |
Municipal wastewater; Grab sampling; 500 mL; Specific temperature not reported (samples were under refrigeration) |
Filtration (0.7 μm and 0.2 μm nominal pore size) | Not carried out | – | QIAMP VIRAL RNA mini kit (Qiagen, Hilden, Germany) | a | a | Influent: 0/8 Effluent: 0/4 |
Real-Time RT-PCR Viability in Vero E6 cells |
|
Bar Or et al., 2020; Israel |
Municipal wastewater; Composite sampling (24 h); 0.25–1 L; −20 °C or − 80 °C |
Centrifugation | a. PEG or alum precipitation, centrifugation, b. Centrifugal filtration (Amicon, MWCO of 30 kDa) |
– | RNA extraction kit (RNeasy mini kit- QIAGEN and EasyMAG-bioMerieux, France) | Only Cycle Treshold (Ct) numbers were given (i.e. number of cycles required for the fluorescent signal to cross the threshold) |
Only Cycle Treshold (Ct) numbers were given (i.e. number of cycles required for the fluorescent signal to cross the threshold) |
a | RT-qPCR |
| Haramoto et al., 2020 | Municipal wastewater; Grab sampling; Influent: 200 mL Secondary Effluent: 5000 mL; n.r. |
– | Electronegative membrane-vortex method; Adsorption direct RNA extraction method |
– | RNeasy PowerWater Kit (Qiagen) | Not detected | 2.4 × 103 | a | RT-qPCR Nested PCR |
| Zhang et al., 2020 | Municipal wastewater; Grab sampling; 2.0 L; 4 °C |
Centrifugation | PEG 6000/NaCl precipitation | EZ1 virus Mini kit (Qiagen, Germany) |
Not detected (After primary disinfection tank before septic tank) |
0.5 × 103 to 18.7 × 103 (After septic tank with disinfection with sodium hypochlorite) |
a | RT-qPCR | |
| (Alpaslan Kocamemi et al., 2020); Turkey |
Municipal wastewater sludge; Grab sampling; n.r.; n.r. |
Centrifugation; filtration (0.45 and 0.2 μm nominal pore size); pH Adjustment at 7.0–7.2 | PEG 8000/centrifugation | n.r. | Roche MagNA pure LC total nucleic acid isolation kit using Roche MagNA pure LC system (Penzberg, Germany) | Primary Sludge: 1.25 × 104 Waste Activated Sludge: 1.17 × 104 to 4.02 × 104 |
a | RT-qPCR | |
| Peccia et al., 2020 | Municipal wastewater sludge; Grab sampling; 2.5 mL; -80 °C |
Not carried out | Not carried out (Direct addition of sludge to RNA extraction kit) |
n.r. | RNeasey PowerSoil Total RNA kit, Qiagen | Primary Sludge: 1.7 × 106 to 4.6 x 108 |
a | RT-qPCR | |
Note: n.r. Not reported
No available quantitative (concentration) or viability data of virus in wastewater or sludge
Table 5.
Main primers and probes used in molecular detection and quantification of SARS-CoV-2 in wastewater.
| Study | Oligonucleotide name | Oligonucleotide Sequencea (5′ to 3′) | Target genes | Reference |
|---|---|---|---|---|
|
Ahmed et al., 2020; April 18, 2020 |
N_Sarbeco_F | CACATTGGCACCCGCAATC | Nucleocapsid protein (N) | (Corman et al., 2020) |
| N_Sarbeco_R | GAGGAACGAGAAGAGGCTTG | |||
| N_Sarbeco_P | FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ | |||
| NIID_2019-nCOV_N_F2 | AAATTTTGGGGACCAGGAAC | Nucleocapsid protein (N) | (Shirato et al., 2020) | |
| NIID_2019-nCOV_N_R2 | TGGCAGCTGTGTAGGTCAAC | |||
| NIID_2019-nCOV_N_R2ver3 | TGGCACCTGTGTAGGTCAAC | |||
| NIID_2019-nCOV_N_P2 | FAM-ATGTCGCGCATTGGCATGGA-BHQ | |||
|
La Rosa et al., 2020; May 7, 2020 |
2274 - CO-FW1 | GTGCTAAACCACCGCCTG | ORF1ab | (La Rosa et al., 2020) |
| 2275 - CO-REV1 | CAGATCATGGTTGCTTTGTAGGT | |||
| 2276 - CO-FW2 | CGCCTGGAGATCAATTTAAACAC | |||
| 2277 - CO-REV2 | ACCTGTAAAACCCCATTGTTGA | |||
| WuhanCoV-spk1-f | TTGGCAAAATTCAAGACTCACTTT | Spike protein gene (S). | (Shirato et al., 2020) | |
| WuhanCoV-spk2-r | TGTGGTTCATAAAAATTCCTTTGTG | |||
| NIID_WH-1_F24381 | TCAAGACTCACTTTCTTCCAC | |||
| NIID_WH-1_R24873 | ATTTGAAACAAAGACACCTTCAC | |||
| RdRP_SARSr-F2 RdRP_SARSr-R1 RdRP_SARSr-P2 |
GTGARATGGTCATGTGTGGCGG CARATGTTAAASACACTATTAGCATA FAM-CAGGTGGAACCTCATCAGGAGATGCBHQ1 |
RdRP gene | (Corman et al., 2020) | |
|
Wurtzer et al., 2020; April 17, 2020 Alpaslan Kocamemi et al., 2020 May 16, 2020 |
RdRp_SARSr-F | GTGARATGGTCATGTGTGGCG | RNA-dependent RNA polymerase gene (RdRp) | (Corman et al., 2020) |
| RdRp_SARSr-R | CARATGTTAAASACACTATTAGCATA | |||
| RdRP_SARSr-P1 | FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ | (Corman et al., 2020) | ||
| RdRp_SARSr-P2 | FAM-CAGGTGGAACCTCATCAGGAGATGC-BBQ | |||
|
Medema et al., 2020 March 30, 2020 Wu et al., 2020 April 7, 2020 Randazzo et al., 2020 April 28, 2020 Peccia et al., 2020a June 12, 2020 |
2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | Nucleocapsid protein (N) | Centers for Disease Control and Prevention, CDC (US 2020) |
| 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | |||
| 2019-nCoV_N1-P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | |||
| 2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | |||
| 2019-nCoV_N2-F | GCGCGACATTCCGAAGAA | |||
| 2019-nCoV_N2-F | FAM-ACAATTTGCCCCCAGCGCTTCAG- BHQ1 | |||
| 2019-nCoV_N3-F | GGGAGCCTTGAATACACCAAAA | |||
| 2019-nCoV_N3-F | TGTAGCACGATTGCAGCATTG | |||
| 2019-nCoV_N3-F | FAM-AYCACATTGGCACCCGCAATCCTG- BHQ1 | |||
|
Medema et al., 2020 March 30, 2020 Wurtzer et al., 2020 April 17, 2020 |
E_Sarbeco_F | ACAGGTACGTTAATAGTTAATAGCGT | Envelope protein gene (E) | (Corman et al., 2020) |
| E_Sarbeco_R | ATATTGCAGCAGTACGCACACA | |||
| E_Sarbeco_P | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ | |||
| F. Wu et al., 2020 April 7, 2020 |
S-RPA-Forward_v1 | GAAATTAATACGACTCACTATAGGGAGGT TTCAAACTTTACTTGCTTTACATAGA |
Spike protein gene (S) | (Zhang et al., n.d.) |
| S-RPA-Reverse_v1 | TCCTAGGTTGAAGATAACCCACATAATAAG | |||
|
Nemudryi et al., 2020 April 20, 2020 |
N1 forward and reverse | sequences not available | Nucleocapsid protein (N) | 2019-nCoV CDC EUA Kit-IDT#10006606 |
| N2 forward and reverse | sequences not available | Nucleocapsid protein (N) | ||
|
Zhang et al., 2020 May 14,2020 |
CCDC-ORF1, forward primer Reverse primer Fluorescence probe |
CCCTGTGGGTTTTACACTTAA ACGATTGTGCATCAGCTGA FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 |
ORFlab | Not indicated |
| CCDC-N, forward primer Reverse primer Fluorescence probe |
GGGGAACTTCTCCTGCTAGAAT CAGACATTTTGCTCTCAAGCTG FAM-TTGCTGCTGCTTGACAGATT-TAMRA |
Nucleocapsid protein (N) | Not indicated | |
|
Haramoto et al., 2020 20 June 2020 |
N_Sarbeco_F1 | CACATTGGCACCCGCAATC | Nucleocapsid protein (N) | (Corman et al., 2020) |
| N_Sarbeco_R1 | GAGGAACGAGAAGAGGCTTG | |||
| N_Sarbeco_P1 | FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ | |||
| NIID_2019-nCOV_N_F2 | AAATTTTGGGGACCAGGAAC | Nucleocapsid protein (N) | (Shirato et al., 2020) | |
| NIID_2019-nCOV_N_R2ver3 | TGGCACCTGTGTAGGTCAAC | |||
| NIID_2019-nCOV_N_P2 | FAM-ATGTCGCGCATTGGCATGGA-BHQ1 | |||
| 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | Nucleocapsid protein (N) | Centers for Disease Control and Prevention, CDC (US 2020) | |
| 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | |||
| 2019-nCoV_N1-P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | |||
| 2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | |||
| 2019-nCoV_N2-F | GCGCGACATTCCGAAGAA |
Note: FAM: 6-carboxyfluorescein; BBQ: blackberry quencher; BHQ (and BHQ1): Black Hole Quencher 1
N1 and N2 primer sets only
Currently, scarce information is available on the efficiency of recovery of SARS-CoV-2 from wastewater. This requires investigations on the determination of the concentration of SARS-CoV-2 in wastewater model samples spiked with a known number of virus copies. As most of the existing concentration methods have been used to recover non-enveloped human enteric viruses, further research must be conducted to evaluate the efficiency of existing concentration methods in recovering enveloped viruses, such as the SARS-CoV-2 (Ahmed et al., 2020). As previously discussed, enveloped viruses have a structure that is different from non-enveloped viruses. Enveloped viruses tend to be sensitive to some organic solvents used in concentration methods (La Rosa et al., 2020; Ye et al., 2016a). A related study by Ye et al., 2016a, Ye et al., 2016b compared recovery efficiencies of different methods used to concentrate enveloped viruses from wastewater. The latter study showed that using ultracentrifugation at 100,000g for an hour resulted in lower recoveries of murine hepatitis virus (MHV) and Pseudomonas phage (φ6). The lower recovery efficiencies were attributed to the strong centrifugal forces that may have disrupted the lipid bilayer membranes of the enveloped viruses. In the same study, an optimized ultrafiltration method, with a pre-filtration step using a 0.22 μm filter, resulted in relatively higher recoveries of enveloped viruses. On the other hand, PEG precipitation showed low recoveries of the model enveloped viruses. Percentage recoveries of SARS-CoV-2 have not been determined due to handling and facility restrictions related to biosafety (Ahmed et al., 2020). Further studies need to be carried out to determine the efficiencies of alternative concentration methods applicable to recover SARS-CoV-2 viruses from wastewater and in sludge samples. Control experiments on the efficiency of recovery of viruses, from SARS-CoV-2 infected wastewater, were performed by determining the concentration of F-specific RNA phages according to ISO 10705 (Medema et al., 2020), or by spiking mengovirus vMC0 (University of Valencia, Spanish Type Culture Collection, CECT 100000) in wastewater samples according to ISO 15216-2:2017 (Randazzo et al., 2020). Haramoto et al. (2020) utilized the electronegative membrane-vortex method to concentrate SARS-CoV-2 from wastewater samples and evaluated the efficiency of recovery using pepper mild mottle virus (PMMV). Medema et al. (2020) suggested the spiking of other human CoVs, such as CoV 229E, to the sewage samples as a reliable internal standard for the determination of the efficiency of recovery of CoVs from wastewater. SARS-CoV-1 is most proximal to the novel SARS-CoV-2, among the viruses investigated in wastewater. Wang et al. (2005a) reported an average SARS-CoV-1 recovery efficiency of 1% by applying adsorption of virus on silica gel compounded with aluminum hydroxide and elution with a buffer at pH of 7.4.
Modes of sampling and sample storage can affect the detection and viability of CoVs in wastewater. As discussed, SARS-CoV-1 infectivity persisted in municipal and hospital wastewater for 2 days at room temperature and for more than 14 days at 4 °C. On the other hand, to the best of the authors' knowledge, information is lacking on the effect of freezing on viability and efficiency of recovery of CoVs from wastewater. Composite and grab samples from WWTPs were analyzed for detection of SARS-CoV-2 (Table 4). In some investigations, debris and particulate were removed by centrifugation or filtration or by a combination of these techniques prior to concentration of the wastewater samples. The nature of these treatments can affect the final virus concentration since CoVs tend to be adsorbed onto organic matter and can even be protected by suspended solids (Gundy et al., 2008). For safe handling of samples, pasteurization at 57 or 60 °C can be applied for inactivation of viruses and other pathogens present in wastewater (La Rosa et al., 2020; Wu et al., 2020b). Several techniques for secondary concentration of the water sample, or combination of them, were used, including: i) centrifugal filtration (Ahmed et al., 2020; Bar Or et al., 2020; Medema et al., 2020; Wu et al., 2020b), ii) ultracentrifugation (Wurtzer et al., 2020), iii) precipitation with PEG (Bar Or et al., 2020; La Rosa et al., 2020; Wu et al., 2020b; Zhang et al., 2020), aluminum chloride (Randazzo et al., 2020) or alum (Bar Or et al., 2020); iv) electronegative membrane filtration (Ahmed et al., 2020; Haramoto et al., 2020).
5.2. Detection and quantification of SARS-CoV-2 in wastewater and sludge
The investigations on SARS-CoV-2 in wastewater and WWTPs exploit the methodological principles applied to other pathogenic and non-pathogenic viruses, applying some modifications necessary for adaption to the characteristics of this new strain of coronavirus.
Here, we briefly describe the molecular materials and procedures adopted so far by different authors for the study of SARS-CoV-2 in wastewater samples. Table 5 reports the main sequences of primers and fluorescent probes used.
Medema et al. (2020) used either of two different commercially systems, RNeasy Power Microbiome Kit (Qiagen) and Nuclisens kit (Biomerieux), in combination with the semi-automated KingFisher mL purification system (ThermoScientific) to extract RNA from viral particles in samples from WWTPs in The Netherlands. For identification, one-step RT-qPCR method (EvoScript RNA Probes Master, Roche) was used with primers specific for three regions (N1−N3) of the nucleocapsid protein gene (N) and envelope protein gene (E). A quantitative culture assay for F-specific RNA phage was also conducted in this study to indirectly evaluate the efficiency of recovery of SARS-CoV-2 by the purification and concentration treatment steps. The effect of these sample processing steps to the viability of the viruses was also examined.
Ahmed et al. (2020) extracted RNA, using the RNeasy PowerWater Kit and RNeasy PowerMicrobiome Kit (Qiagen), from the ultra-filtered sample or from the electronegative membrane. For specific detection and quantification, RT-qPCR was used with the primers N_Sarbeco and NIID_2019-nCOV_N, both of which are specific for the nucleocapsid protein gene. One-step kit RT-qPCR was adopted, where reverse transcription and qPCR occurred in the same reaction well.
La Rosa et al. (2020) assessed the presence of SARS-CoV-2 in Italian WWTP by means of RT-PCR, using two groups of specific primers for SARS-CoV-2: a newly developed set targeting ORF1ab (La Rosa et al., 2020) and a published set, designed for the pharyngeal swab, specific for the spike (S) protein gene. In this study, broad-range primers, which were designed before the detection of the new strain of coronavirus, were also tested, amplifying a conserved region of the ORF1ab of Coronavirinae members. However, these primers did not give specific signals due to some nucleotide differences in this region found with subsequent sequencing of SARS-CoV-2.
Wurtzer et al. (2020) investigated the presence of viruses in wastewater samples collected from 3 major WWTPs in Paris, using the RT-qPCR with primers targeting the envelope protein gene (E) and RNA-dependent RNA polymerase gene (RdRP). After concentration by ultracentrifugation, the viral genome was extracted using an optimized protocol from the commercial PowerFecal Pro kit on a QIAsymphony extractor (Qiagen).
Nemudryi et al. (2020) detected SARS-CoV-2 in concentrated wastewater samples from WWTP of Bozeman (USA) by one step RT-qPCR. The primers, supplied by the commercially 2019-nCoV EUC CDC kit, were specific for two regions (N1, N2) of the nucleocapsid protein gene. RNA extraction was carried out via the commercially RNeasy mini kit (Qiagen). Nemudryi et al. (2020) also evaluated the phylogenetic relationship of the isolated SARS-CoV-2 genome with the other sequenced worldwide, using 10 published primer pairs (Table 5) to amplify with non-quantitative RT-PCR and sequence some polymorphic regions dispersed in the genome.
Wu et al. (2020b) preliminarily detected the presence of SARS-CoV-2 in PEG-precipitated wastewater samples from a WWTP of Massachusetts (USA) with classical PCR and published primers directed towards the (S) protein gene spike. After detection, RT-qPCR was used to quantify the viral titer, with primers targeting N1, N2 and N3 loci of the nucleocapsid protein gene (N) and published by US, CDC. Viral RNA was extracted with Trizol (Thermofisher).
The concentration of SARS-CoV-2 in raw wastewater obtained by recent studies varies from 101 to 106 genome copies/L (Table 4). Concentrations of SARS-CoV-2 in secondary effluent range from 103 to 105 genome copies/L (Haramoto et al., 2020; Randazzo et al., 2020). SARS-CoV-2 was also present at a concentration of 103 to 104 genome copies/L in the effluent from the septic tanks of Fangcang hospital in Wuhan, Hubei, China (Zhang et al., 2020). The obtained concentration of the virus in primary sludge samples is 104 to 108 genome copies/L (Alpaslan Kocamemi et al., 2020; Peccia et al., 2020). This concentration in primary sludge samples is slightly higher than that in the wastewater samples. These results are in accordance with the tendency of enveloped viruses to adsorb to the solid-fraction of wastewater (Ye et al., 2016a), resulting to their accumulation in the wastewater sludge. This is important in the study of the emerging viruses, such as SARS-CoV-2, since the concentrated sludge sample can provide information on the viruses present in wastewater and in corresponding affected communities.
As of this time of writing of the review, only two studies have performed analysis of viability of SARS-CoV-2 in wastewater. Wang et al. (2020a) conducted the analysis by inoculation of the virus on Vero-E6 cells. No viable virus was detected in all of the influent and effluent hospital wastewater samples. Rimoldi et al. (2020) performed the same viability test of SARS-CoV-2 in municipal wastewater samples, in which no viable virus was also detected. The study of Zhang et al. (2020) revealed that SARS-CoV-2 genomes were not detected in the influent of a hospital septic tank (after primary disinfection tank) but were detected in the effluent at a concentration of 0.50 × 103 to 1.87 × 104 genome copies/L after disinfection with sodium hypochlorite. The viral genomes in the septic tank effluent were detected when the free chlorine concentration has declined to non-detectable level. The absence in the influent and presence in the effluent suggested the release of embedded viruses in the stool particles, which provided protection from the treatment process. Although the latter study did not perform a viability analysis, the results indicate the need for further studies to confirm that the sewage or wastewater treatment systems are not SARS-CoV-2 transmission pathways.
5.3. Detection of viruses from aerosols in wastewater systems
A limited number of studies have shown that wastewater systems emit bioaerosols that have significant concentrations of viruses such as RoV, NoV, HAdV, HAV, and Herpes Simplex Virus Type 1 (HSV1) (Brisebois et al., 2018; Masclaux et al., 2014; Pasalari et al., 2019). Viruses in aerosols pose a risk to the WWTP or sewage workers who are exposed to these viruses via the inhalation or contamination by contact with deposited aerosols on surfaces (Masclaux et al., 2014). Highly resistant viruses, such as RoV and NoV, that are present in the local atmosphere of a WWTP can also pose risks to the residents living near the facility (Pasalari et al., 2019).
Although no evidence has so far emerged concerning the emission of SARS-CoV-2 to the ambient air at WWTPs or sewage systems, future studies on this potential exposure pathway need to be conducted to assess possible risks to workers and affected residents. The study of van Doremalen et al. (2020) examined the stability of SARS-CoV-2 in aerosols. Although the experimental condition used in that study had only a 3 h duration, it showed that SARS-CoV-2 remained viable in aerosols. The study also compared the viability of SARS-CoV-2 and SARS-CoV-1 in aerosols. It showed that both viruses have similar stabilities. SARS-CoV-1 virus was previously reported to be transmitted through aerosols in sewage systems of a residential building in Hong Kong, leading to a disease outbreak in the examined area (Chim et al., 2003; Hung, 2003).
5.4. Development of rapid detection methods
Regulations related to wastewater treatment do not commonly require routine monitoring of viruses before wastewater discharge to receiving bodies of water. However, several studies have shown that pathogenic viruses remain abundant in wastewater and sludge even after treatment. Monitoring of viruses is not widely conducted in wastewater treatment plants because of the cost, time and complexity involved in conducting detection methods. The need for rapid, highly accurate, and sensitive methods at optimum cost arises.
The possibility of use of in situ monitoring tools is to be further explored in this context. The use of biosensors to detect viruses in environmental samples has been previously studied but there is only a limited number of studies on application of this approach to detect viruses in wastewater. Chung et al. (2019) utilized a microfluidic paper analytic device (μPAD) to detect NoV in environmental water samples including reclaimed wastewater. In the latter study, NoV viruses were detected without prior sample concentration and nucleic acid amplifications. The NoV were captured by the μPAD, where antibody-conjugated fluorescent submicron particles were also added. The use of μPAD relies on the correlation between the virus concentration and the number of aggregated particles formed by antibody-antigen binding. Additional visualization methods were developed in the latter study to make the detection method suitable to field applications. The quantity of NoV in the samples was determined using a smart-phone fluorescence microscope and a mobile graphical user interface (GUI) application. However, this method can only be used to detect specific viruses whose detection depends on the antibody used. The use of biosensors such as μPAD is a potential tool for rapid in detection of viruses in environmental water samples such as wastewater and is still in the nascent stage.
Along with the molecular detection methods, tools such as paper-based devices for rapid, in situ, and sensitive detection can possibly provide timely information about the circulation of emerging pathogenic viruses such as SARS-CoV-2 in wastewater and in affected communities (Mao et al., 2020).
6. Conclusions
There is a wide range of detection and quantification methods that have been used to detect and quantify viruses in wastewater and sludge samples. Molecular methods have been extensively used in the recent studies for specific and sensitive detection of viruses in wastewater. However, it is notable that there are instances when the use of at least two detection methods proved helpful in monitoring the abundance and diversity of viruses in wastewater and sludge. This is to ensure that wider range of viruses found in samples are detected and quantified. Analysis of viability of viruses is also needed for a complete information regarding their occurrence in wastewater and sludge samples.
Pre-treatment methods must be chosen according to the characteristics of the wastewater or sludge sample matrices. In applying these techniques, methods to reduce effects of inhibitors that are originally present in the wastewater or sludge samples, or in the reagents must be considered. Recent developments that resulted in increasing efficiency of detection of viruses in sludge samples focused on their release from the highly aggregated sludge matrix. Addition of optimized ultrasonication step to the traditional adsorption-elution method, and the use of dispersants and enzymes to break down sludge flocs are the current developments in the sludge sample processing methods.
High-throughput sequencing has been used in the comprehensive studies of wastewater treatment systems, allowing for a large number of samples to be processed simultaneously. This approach has been shown to be important in studying the fate of viruses during and after wastewater and sludge treatment processes, in evaluating efficiencies of the treatment systems, and in identifying paths of transmission of human diseases through exposure to wastewater or sludge.
Recent studies have illustrated the importance of data of molecular methods for studying the previous, ongoing and possible disease outbreaks. Detection and quantification methods developed to ascertain the occurrence and identities of viruses in wastewater clearly play an important role in using the WBE approach in studying the circulation of infectious viruses such as SARS-CoV-2 in exposed communities.
Research needs identified in this review include further validation of the processing methods for wastewater samples, with a specific focus on ascertaining concentrations of emerging viruses such as SARS-CoV-2 using seeded samples. Further validation of assays that use primers specific to SARS-CoV-2 in wastewater matrices should also be conducted. Further studies should also include SARS-CoV-2 infectivity assays of influent, effluent and sludge samples. Future research on the presence of viruses in aerosols emitted from wastewater or sewage systems must also be conducted.
The development of rapid, and sensitive methods could also be considered to complement the molecular methods to provide timely information about presence and abundance of viruses in wastewater samples, as part of routine monitoring or as part of a WBE approach to investigate ongoing or emerging disease outbreaks.
CRediT authorship contribution statement
Mary Vermi Aizza Corpuz: Data curation, Writing - original draft, Writing - review & editing. Antonio Buonerba: Conceptualization, Data curation, Writing - original draft, Writing - review & editing. Giovanni Vigliotta: Data curation, Writing - original draft, Writing - review & editing. Tiziano Zarra: Writing - review & editing. Florencio Ballesteros: Writing - review & editing. Pietro Campiglia: Writing - review & editing. Vincenzo Belgiorno: Writing – original draft, Writing - review & editing, Funding acquisition. Gregory Korshin: Writing - original draft, Writing - review & editing. Vincenzo Naddeo: Conceptualization, Project administration, Supervision, Funding acquisition, Validation, Writing - original draft, Writing - review & editing.
Declaration of competing interest
On behalf of all authors, I declare that there is no conflict of interest. All authors have NO affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest in the subject matter or materials discussed in this manuscript.
Acknowledgements
The authors would like to express their sincere gratitude to the support from i) Sanitary Environmental Engineering Division (SEED) and grants (FARB projects) from University of Salerno coordinated by prof. V. Naddeo; ii) Inter-university centre for prediction and prevention of major hazards (Consorzio inter-Universitario per la previsione e la prevenzione dei Grandi Rischi, C.U.G.RI.); iii) University of the Philippines Diliman and the Engineering Research and Development for Technology (ERDT) through the Department of Science and Technology Philippines. The authors would also like to acknowledge the help of Carlo Cabreros and James Almalvez from University of the Philippines Diliman and Fabiano Castrogiovanni from University of Salerno for providing writing assistance.
Editor: Dr. Damia Barcelo
References
- Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food Environ. Virol. 2014;6:253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adan A., Alizada G., Kiraz Y., Baran Y., Nalbant A. Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- Adriaenssens E.M., Farkas K., Harrison C., Jones D.L., Allison H.E., McCarthy A.J. vol. 3. 2018. Viromic Analysis of Wastewater Input to a River Catchment Reveals a Diverse Assemblage of RNA Viruses. mSystems. e00025-18, /msystems/3/3/msys.00025-18.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P.S., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81:2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhamlan F.S., Ederer M.M., Brown C.J., Coats E.R., Crawford R.L. Metagenomics-based analysis of viral communities in dairy lagoon wastewater. J. Microbiol. Methods. 2013;92:183–188. doi: 10.1016/j.mimet.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Alpaslan Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in istanbul wastewater treatment plant sludges (preprint) Infect. Dis. Ther. 2020 doi: 10.1101/2020.05.12.20099358. (except HIV/AIDS) [DOI] [Google Scholar]
- Ambalathingal G.R., Francis R.S., Smyth M.J., Smith C., Khanna R. BK Polyomavirus: Clinical Aspects, Immune Regulation, and Emerging Therapies. Clin. Microbiol. Rev. 2017;30:503–528. doi: 10.1128/CMR.00074-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdiouni H., Maunula L., Hajjami K., Faouzi A., Soukri A., Nourlil J. Recovery comparison of two virus concentration methods from wastewater using cell culture and real-time PCR. Curr. Microbiol. 2012;65:432–437. doi: 10.1007/s00284-012-0174-8. [DOI] [PubMed] [Google Scholar]
- Andreadakis A.D., Mamais D., Gavalaki E., Kampylafka S. Sludge utilisation in agriculture: possibilities and prospects in Greece. Water Sci. Technol. 2002;46:231–238. doi: 10.2166/wst.2002.0340. [DOI] [PubMed] [Google Scholar]
- Artika I.M., Wiyatno A., Ma’roef C.N. Pathogenic viruses: Molecular detection and characterization. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya M., Shergill I.S., Williamson M., Gommersall L., Arya N., Patel H.R. Basic principles of real-time quantitative PCR. Expert. Rev. Mol. Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- Assis A.S.F., Fumian T.M., Miagostovich M.P., Drumond B.P., da Rosa e Silva M.L. Adenovirus and rotavirus recovery from a treated effluent through an optimized skimmed-milk flocculation method. Environ. Sci. Pollut. Res. 2018;25:17025–17032. doi: 10.1007/s11356-018-1873-x. [DOI] [PubMed] [Google Scholar]
- ASTM D449-19 . ASTM International; West Conshohocken, PA: 2019. Standard Practice for Recovery of Viruses From Wastewater Sludges. [DOI] [Google Scholar]
- Atha D.H., Ingham K.C. Mechanism of precipitation of proteins by polyethylene glycols. J. Biol. Chem. 1981;256:12108–12117. [PubMed] [Google Scholar]
- Bačnik K., Kutnjak D., Pecman A., Mehle N., Tušek Žnidarič M., Gutiérrez Aguirre I., et al. Viromics and infectivity analysis reveal the release of infective plant viruses from wastewater into the environment. Water Res. 2020;177:115628. doi: 10.1016/j.watres.2020.115628. [DOI] [PubMed] [Google Scholar]
- Baez P.A., Lopez M.C., Duque-Jaramillo A., Pelaez D., Molina F., Navas M.-C. First evidence of the Hepatitis E virus in environmental waters in Colombia. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E.F., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. 2020. Regressing SARS-CoV-2 Sewage Measurements Onto COVID-19 Burden in the Population: A Proof-of-Concept for Quantitative Environmental Surveillance. medRxiv 2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C., Nogueira S., Gadanho M., Chaves S. Molecular Microbial Diagnostic Methods. Elsevier; 2016. DNA extraction: finding the most suitable method; pp. 135–154. [DOI] [Google Scholar]
- Barcelo D. An environmental and health perspective for COVID-19 outbreak: meteorology and air quality influence, sewage epidemiology indicator, hospitals disinfection, drug therapies and recommendations. J. Environ. Chem. Eng. 2020;8 doi: 10.1016/j.jece.2020.104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Vieira D.F., Barth O.M. In: Microbiology in Agriculture and Human Health. Shah M.M., editor. InTech; 2015. Negative and positive staining in transmission electron microscopy for virus diagnosis. [DOI] [Google Scholar]
- Bastien P., Procop G.W., Reischl U. Quantitative real-time PCR is not more sensitive than “conventional” PCR. J. Clin. Microbiol. 2008;46:1897–1900. doi: 10.1128/JCM.02258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S.E., Rodriguez R.A., Linden K.G., Hargy T.M., Larason T.C., Wright H.B. Wavelength dependent UV inactivation and DNA damage of adenovirus as measured by cell culture infectivity and long range quantitative PCR. Environ. Sci. Technol. 2014;48:591–598. doi: 10.1021/es403850b. [DOI] [PubMed] [Google Scholar]
- Betancourt W.Q., Gerba C.P. Rethinking the Significance of Reovirus in Water and Wastewater. Food Environ. Virol. 2016;8:161–173. doi: 10.1007/s12560-016-9250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S., Szewzyk R., Gnirss R., Johne R., Selinka H.-C. Detection and Characterization of Hepatitis E Virus Genotype 3 in Wastewater and Urban Surface Waters in Germany. Food Environ. Virol. 2020;12(2):137–147. doi: 10.1007/s12560-020-09424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R.F., Kirkwood C.D. Encyclopedia of Virology. Elsevier Ltd; 2008. Enteric viruses; pp. 116–123. [Google Scholar]
- Blanco Fernández M.D., Barrios M.E., Cammarata R.V., Torres C., Taboga O.A., Mbayed V.A. Comparison of internal process control viruses for detection of food and waterborne viruses. Appl. Microbiol. Biotechnol. 2017;101:4289–4298. doi: 10.1007/s00253-017-8244-2. [DOI] [PubMed] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Rodriguez-Manzano J., Calgua B., Carratala A., Girones R. Newly described human polyomaviruses Merkel Cell, KI and WU are present in urban sewage and may represent potential environmental contaminants. Virol. J. 2010;7:141. doi: 10.1186/1743-422X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/JCM.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., Kreuze J., Winter S., van der Vlugt R., Bergervoet J., Tomlinson J., Mumford R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Brisebois E., Veillette M., Dion-Dupont V., Lavoie J., Corbeil J., Culley A., Duchaine C. Human viral pathogens are pervasive in wastewater treatment center aerosols. J. Environ. Sci. 2018;67:45–53. doi: 10.1016/j.jes.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer A.F., Masters N.B., Eisenberg J.N.S. Quantitative microbial risk assessment and infectious disease transmission modeling of waterborne enteric pathogens. Curr. Environ. Health Rpt. 2018;5:293–304. doi: 10.1007/s40572-018-0196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Wittwer C. Flow cytometry: principles and clinical applications in hematology. Clin. Chem. 2000;46:1221–1229. doi: 10.1093/clinchem/46.8.1221. [DOI] [PubMed] [Google Scholar]
- Brown M.R., Camézuli S., Davenport R.J., Petelenz-Kurdziel E., Øvreås L., Curtis T.P. Flow cytometric quantification of viruses in activated sludge. Water Res. 2015;68:414–422. doi: 10.1016/j.watres.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Butler J.M. Fundamentals of Forensic Typing. Elsevier; 2010. DNA extraction.pdf; pp. 99–109. [Google Scholar]
- Calgua B., Mengewein A., Grunert A., Bofill-Mas S., Clemente-Casares P., Hundesa A., Wyn-Jones A.P., López-Pila J.M., Girones R. Development and application of a one-step low cost procedure to concentrateviruses from seawater samples. J. Virol. Methods. 2008;153:79–83. doi: 10.1016/j.jviromet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Calgua B., Barardi C.R.M., Bofill-Mas S., Rodriguez-Manzano J., Girones R. Detection and quantitation of infectious human adenoviruses and JC polyomaviruses in water by immunofluorescence assay. J. Virol. Methods. 2011;171:1–7. doi: 10.1016/j.jviromet.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Calgua B., Rodriguez-Manzano J., Hundesa A., Suñen E., Calvo M., Bofill-Mas S., Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods. 2013;187:215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Calgua B., Carratalà A., Guerrero-Latorre L., de Abreu Corrêa A., Kohn T., Sommer R., Girones R. UVC inactivation of dsDNA and ssRNA viruses in water: UV fluences and a qPCR-based approach to evaluate decay on viral infectivity. Food Environ. Virol. 2014;6:260–268. doi: 10.1007/s12560-014-9157-1. [DOI] [PubMed] [Google Scholar]
- Canh V.D., Osawa H., Inoue K., Kasuga I., Takizawa S., Furumai H., Katayama H. Ferrihydrite treatment to mitigate inhibition of RT-qPCR virus detection from large-volume environmental water samples. J. Virol. Methods. 2019;263:60–67. doi: 10.1016/j.jviromet.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Carducci A., Battistini R., Rovini E., Verani M. Viral removal by wastewater treatment: monitoring of indicators and pathogens. Food Environ. Virol. 2009;1:85–91. doi: 10.1007/s12560-009-9013-x. [DOI] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Poon V.K.-M., Chan W.-M., Ip J.D., Cai J.-P., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry R.M., Nelson K.L., Drewes J.E. Mechanisms of pathogenic virus removal in a full-scale membrane bioreactor. Environ. Sci. Technol. 2015;49:2815–2822. doi: 10.1021/es505332n. [DOI] [PubMed] [Google Scholar]
- Chim S., Tsui S., Chan K., Au T., Hung E., Tong Y., Chiu R., Ng E., Chan P., Chu C., Sung J., Tam J., Fung K., Waye M., Lee C., Yuen K., Lo Y. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362:1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary R., Shukla A., Datta T., Dhole T.N. Rapid detection of sewage sample polioviruses by integrated cell culture polymerase chain reaction. Arch. Environ. Occup. Health. 2005;60:223–228. doi: 10.3200/AEOH.60.4.223-228. [DOI] [PubMed] [Google Scholar]
- Chung S., Breshears L.E., Perea S., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.-Y. Smartphone-based paper microfluidic particulometry of norovirus from environmental water samples at the single copy level. ACS Omega. 2019;4:11180–11188. doi: 10.1021/acsomega.9b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.P., Pazdernik N.J., McGehee M.R. Molecular Biology. Elsevier; 2019. DNA sequencing; pp. 240–269. [DOI] [Google Scholar]
- Cobo F. Application of molecular diagnostic techniques for viral testing. TOVJ. 2012;5:104–114. doi: 10.2174/1874357901206010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collomb J., Laporte J., Vautherot J.F., Schwartzbrod L. Research on coronaviruses in water. I. Adsorption and elution of the coronavirus on glass powder. Virologie. 1986;37:95–105. [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi S.R., Borchardt M.A., Spencer S.K., Hughes P.E., Baldwin A.K. Human and bovine viruses in the Milwaukee River watershed: hydrologically relevant representation and relations with environmental variables. Sci. Total Environ. 2014;490:849–860. doi: 10.1016/j.scitotenv.2014.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray-Meunier C., Fraisse A., Martin-Latil S., Guillier L., Delannoy S., Fach P., Perelle S. A comparative study of digital RT-PCR and RT-qPCR for quantification of Hepatitis A virus and Norovirus in lettuce and water samples. Int. J. Food Microbiol. 2015;201:17–26. doi: 10.1016/j.ijfoodmicro.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Cunliffe N.A., Glass R.I., Nakagomi O. Manson’s Tropical Infectious Diseases. Elsevier; 2014. Rotavirus and Other Viral Diarrhoea; pp. 207–214.e3. [Google Scholar]
- Cupples A.M., Rose J.B., Xagoraraki I. In: Environmental Microbiology. Mitchell R., Gu J.-D., editors. John Wiley & Sons, Inc; Hoboken, New Jersey: 2010. New molecular methods for detection of waterborne pathogens. [Google Scholar]
- D’Souza D.H. Advances in Microbial Food Safety. Elsevier; 2015. Update on foodborne viruses: types, concentration and sampling methods; pp. 102–116. [DOI] [Google Scholar]
- De Gascun C.F., Carr M.J. Human Polyomavirus Reactivation: Disease Pathogenesis and Treatment Approaches. Clin. Dev. Immunol. 2013;2013:1–27. doi: 10.1155/2013/373579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonito P., Iaconelli M., Gheit T., Tommasino M., Della Libera S., Bonadonna L., La Rosa G. Detection of oncogenic viruses in water environments by a Luminex-based multiplex platform for high throughput screening of infectious agents. Water Res. 2017;123:549–555. doi: 10.1016/j.watres.2017.06.088. [DOI] [PubMed] [Google Scholar]
- Di Profio F., Melegari I., Palombieri A., Sarchese V., Arbuatti A., Fruci P., Marsilio F., Martella V., Di Martino B. High prevalence of hepatitis E virus in raw sewage in Southern Italy. Virus Res. 2019;272 doi: 10.1016/j.virusres.2019.197710. [DOI] [PubMed] [Google Scholar]
- Dias E., Ebdon J., Taylor H. Estimating the concentration of viral pathogens and indicator organisms in the final effluent of wastewater treatment processes using stochastic modelling. Microbial Risk Anal. 2019;11:47–56. doi: 10.1016/j.mran.2018.08.003. [DOI] [Google Scholar]
- Díez B., Antón J., Guixa-Boixereu N., Pedrós-Alió C., Rodríguez-Valera3 F. Pulsed-field gel electrophoresis analysis of virus assemblages present in a hypersaline environment. Int. Microbiol. 2000;3:159–164. [PubMed] [Google Scholar]
- Diez-Valcarce M., Cook N., Hernández M., Rodríguez-Lázaro D. Analytical application of a sample process control in detection of foodborne viruses. Food Anal. Methods. 2011;4:614–618. doi: 10.1007/s12161-011-9262-9. [DOI] [Google Scholar]
- Dilnessa T., Zeleke H. Cell Culture, Cytopathic Effect and Immunofluorescence Diagnosis of Viral Infection. J. Microbiol. Mod. Tech. 2017;2 doi: 10.15744/2575-5498.2.102. [DOI] [Google Scholar]
- Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission: a COVID-19 virological and clinical review. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.052. S0016508520305710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S.E., Halonen M.J., Maier R.M. In: Environmental Microbiology. Maier R.M., Pepper I.L., Gerba C.P., editors. Elsevier Inc; 2009. Immunological methods. [Google Scholar]
- Drancourt M. Acute Diarrhea. Infect. Dis. 2017:335–340.e2. [Google Scholar]
- Dunbar S.A., Hoffmeyer M.R. The Immunoassay Handbook. Elsevier; 2013. Microsphere-based multiplex immunoassays: development and applications using Luminex® xMAP® technology; pp. 919–927. [DOI] [Google Scholar]
- Dundas N., Leos N.K., Mitui M., Revell P., Rogers B.B. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diag. 2008;10:311–316. doi: 10.2353/jmoldx.2008.070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelko M.B., Schmidt P.J., Borchardt M.A. Confirming the need for virus disinfection in municipal subsurface drinking water supplies. Water Res. 2019;157:356–364. doi: 10.1016/j.watres.2019.03.057. [DOI] [PubMed] [Google Scholar]
- Falman J.C., Fagnant-Sperati C.S., Kossik A.L., Boyle D.S., Meschke J.S. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food Environ. Virol. 2019;11:20–31. doi: 10.1007/s12560-018-09364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., McDonald J., Malham S., Jones D. Two-step concentration of complex water samples for the detection of viruses. MPs. 2018;1:35. doi: 10.3390/mps1030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Timoneda N., Martínez-Puchol S., Rusiñol M., Rodriguez-Manzano J., Figuerola N., Bofill-Mas S., Abril J.F., Girones R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018;618:870–880. doi: 10.1016/j.scitotenv.2017.08.249. [DOI] [PubMed] [Google Scholar]
- Ferreira F.F.M., Guimarães F.R., Fumian T.M., Victoria M., Vieira C.B., Luz S., Shubo T., Leite J.P.G., Miagostovich M.P. Environmental dissemination of group A rotavirus: P-type, G-type and subgroup characterization. Water Sci. Technol. 2009;60:633–642. doi: 10.2166/wst.2009.413. [DOI] [PubMed] [Google Scholar]
- Fong T.-T., Griffin D.W., Lipp E.K. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. AEM. 2005;71:2070–2078. doi: 10.1128/AEM.71.4.2070-2078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P., Soler N., Krupovic M., Marguet E., Ackermann H.-W. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013;21:1–5. doi: 10.1016/j.tim.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Leite J.P.G., Castello A.A., Gaggero A., Caillou M.S.L. de, Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Fioretti J.M., Lun J.H., dos Santos I.A.L., White P.A., Miagostovich M.P. Detection of norovirus epidemic genotypes in raw sewage using next generation sequencing. Environ. Int. 2019;123:282–291. doi: 10.1016/j.envint.2018.11.054. [DOI] [PubMed] [Google Scholar]
- Gallagher E.M., Margolin A.B. Development of an integrated cell culture—real-time RT-PCR assay for detection of reovirus in biosolids. J. Virol. Methods. 2007;139:195–202. doi: 10.1016/j.jviromet.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Gan S.D., Patel K.R. Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Investig. Dermatol. 2013;133:1–3. doi: 10.1038/jid.2013.287. [DOI] [PubMed] [Google Scholar]
- Geller C., Varbanov M., Duval R.E. Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses. 2012;4:3044–3068. doi: 10.3390/v4113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerba C.P. Assessment of enteric pathogen shedding by bathers during recreational activity and its impact on water quality. Quant. Microbiol. 2000;2:55–68. [Google Scholar]
- Gerba C.P., Betancourt W.Q., Kitajima M. How much reduction of virus is needed for recycled water: a continuous changing need for assessment? Water Res. 2017;108:25–31. doi: 10.1016/j.watres.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girones R., Allard A., Wadell G., Jofre J. Application of PCR to the detection of adenoviruses in polluted waters. Water Sci. Technol. 1993;27:235–241. doi: 10.2166/wst.1993.0352. [DOI] [Google Scholar]
- Goyal S.M., Gerba C.P. Viradel method for detection of rota virus from seawater. J. Virol. Methods. 1983;7:279–285. doi: 10.1016/0166-0934(83)90080-0. [DOI] [PubMed] [Google Scholar]
- Grassi T., Bagordo F., Idolo A., Lugoli F., Gabutti G., De Donno A. Rotavirus detection in environmental water samples by tangential flow ultrafiltration and RT-nested PCR. Environ. Monit. Assess. 2010;164:199–205. doi: 10.1007/s10661-009-0885-x. [DOI] [PubMed] [Google Scholar]
- Grobelak A., Czerwińska K., Murtaś A. Industrial and Municipal Sludge. Elsevier; 2019. General considerations on sludge disposal, industrial and municipal sludge; pp. 135–153. [DOI] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2008;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Guo X., Wang S., Zhao C., Li J., Zhong J. An integrated cell absorption process and quantitative PCR assay for the detection of the infectious virus in water. Sci. Total Environ. 2018;635:964–971. doi: 10.1016/j.scitotenv.2018.04.223. [DOI] [PubMed] [Google Scholar]
- Guttman B. Brenner’s Encyclopedia of Genetics. Elsevier; 2013. Virus; pp. 291–294. [DOI] [Google Scholar]
- Hamparian V.V., Ottolenghi A.C., Hughes J.H. Enteroviruses in sludge: multiyear experience with four wastewater treatment plants. Appl. Environ. Microbiol. 1985;50:280–286. doi: 10.1128/aem.50.2.280-286.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Stang A., Sure K., Überla K., Wilhelm M. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 2009;43:2657–2668. doi: 10.1016/j.watres.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Wilhelm M. Development of a Luminex assay for the simultaneous detection of human enteric viruses in sewage and river water. J. Virol. Methods. 2014;204:65–72. doi: 10.1016/j.jviromet.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Utagawa E., Ohgaki S. Recovery of human norovirus from water by virus concentration methods. J. Virol. Methods. 2009;160:206–209. doi: 10.1016/j.jviromet.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G.D., Adams V.D., Sorensen D.L., Curtis M.S. Ultraviolet inactivation of selected bacteria and viruses with photoreactivation of the bacteria. Water Res. 1987;21:687–692. doi: 10.1016/0043-1354(87)90080-7. [DOI] [Google Scholar]
- Hata A., Katayama H., Kitajima M., Visvanathan C., Nol C., Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl. Environ. Microbiol. 2011;77:4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kojima K., Sano S., Kasuga I., Kitajima M., Furumai H. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci. Total Environ. 2014;468–469:757–763. doi: 10.1016/j.scitotenv.2013.08.093. [DOI] [PubMed] [Google Scholar]
- He X.Q., Cheng L., Zhang D.Y., Xie X.M., Wang D.H., Wang Z. One-year monthly survey of rotavirus, astrovirus and norovirus in three sewage treatment plants (STPs) in Beijing, China and associated health risk assessment. Water Sci. Technol. 2011;64:1202–1210. doi: 10.2166/wst.2011.080. [DOI] [PubMed] [Google Scholar]
- Hejkal T.W., Wellings F.M., Lewis A.L., LaRock P.A. Distribution of viruses associated with particles in waste water. Appl. Environ. Microbiol. 1981;41:628–634. doi: 10.1128/AEM.41.3.628-634.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennechart-Collette C., Martin-Latil S., Guillier L., Perelle S. Determination of which virus to use as a process control when testing for the presence of hepatitis A virus and norovirus in food and water. Int. J. Food Microbiol. 2015;202:57–65. doi: 10.1016/j.ijfoodmicro.2015.02.029. [DOI] [PubMed] [Google Scholar]
- Hill V.R., Polaczyk A.L., Kahler A.M., Cromeans T.L., Hahn D., Amburgey J.E. Comparison of hollow-fiber ultrafiltration to the USEPA VIRADEL technique and USEPA method 1623. J. Environ. Qual. 2009;38:822–825. doi: 10.2134/jeq2008.0152. [DOI] [PubMed] [Google Scholar]
- Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy M.A. Insect Molecular Genetics. Elsevier; 2019. DNA Sequencing and the evolution of the “-omics”; pp. 223–262. [DOI] [Google Scholar]
- Hryniszyn A., Skonieczna M., Wiszniowski J. Methods for detection of viruses in water and wastewater. AiM. 2013;03:442–449. doi: 10.4236/aim.2013.35060. [DOI] [Google Scholar]
- Hung L.S. The SARS epidemic in Hong Kong: what lessons have we learned? J. R. Soc. Med. 2003;96:374–378. doi: 10.1258/jrsm.96.8.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconelli M., Bonanno Ferraro G., Mancini P., Suffredini E., Veneri C., Ciccaglione A.R., Bruni R., Della Libera S., Bignami F., Brambilla M., De Medici D., Brandtner D., Schembri P., D’Amato S., La Rosa G. Nine-Year Nationwide Environmental Surveillance of Hepatitis E Virus in Urban Wastewaters in Italy (2011–2019) Int. J. Env. Res. Pub. He. 2020;17(6):2059. doi: 10.3390/ijerph17062059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim C., Hammami S., Mejri S., Mehri I., Pothier P., Hassen A. Detection of Aichi virus genotype B in two lines of wastewater treatment processes. Microb. Pathog. 2017;109:305–312. doi: 10.1016/j.micpath.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Im K., Mareninov S., Diaz M.F.P., Yong W.H. In: Biobanking, Methods in Molecular Biology. Yong W.H., editor. Springer New York; New York, NY: 2019. An introduction to performing immunofluorescence staining; pp. 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izopet J., Tremeaux P., Marion O., Migueres M., Capelli N., Chapuy-Regaud S., Mansuy J.-M., Abravanel F., Kamar N., Lhomme S. Hepatitis E virus infections in Europe. J. Clin. Virol. 2019;120:20–26. doi: 10.1016/j.jcv.2019.09.004. [DOI] [PubMed] [Google Scholar]
- Jahne M.A., Brinkman N.E., Keely S.P., Zimmerman B.D., Wheaton E.A., Garland J.L. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: Implications for onsite reuse. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali Mehdi, Zaborowska J., Morteza Jalali. Basic Science Methods for Clinical Researchers. Elsevier; 2017. The polymerase chain reaction; pp. 1–18. [DOI] [Google Scholar]
- Ji P., Aw T.G., Van Bonn W., Rose J.B. Evaluation of a portable nanopore-based sequencer for detection of viruses in water. J. Virol. Methods. 2020;278 doi: 10.1016/j.jviromet.2019.113805. [DOI] [PubMed] [Google Scholar]
- Jin H.X., Seo S.B., Lee H.Y., Cho S., Ge J., King J., Budowle B., Lee S.D. Differences of PCR efficiency between two-step PCR and standard three-step PCR protocols in short tandem repeat amplification. Aust. J. Forensic Sci. 2014;46:80–90. doi: 10.1080/00450618.2013.788681. [DOI] [Google Scholar]
- Kadri K. In: Synthetic Biology - New Interdisciplinary Science. L. Nagpal M., Boldura O.-M., Baltă C., Enany S., editors. IntechOpen; 2020. Polymerase chain reaction (PCR): principle and applications. [DOI] [Google Scholar]
- Kargar M., Javdani N., Najafi A., Tahamtan Y. First molecular detection of group A rotavirus in urban and hospital sewage systems by nested-RT PCR in Shiraz, Iran. J. Environ. Health Sci. Eng. 2013;11:4. doi: 10.1186/2052-336X-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M.R., Rhodes E.R., Brinkman N., Wymer L., Fout G.S. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 2009;75:2393–2399. doi: 10.1128/AEM.00922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Shimasaki A., Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. AEM. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalenkov A., Laver W.G., Webster R.G. Detection and isolation of H5N1 influenza virus from large volumes of natural water. J. Virol. Methods. 2008;149:180–183. doi: 10.1016/j.jviromet.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes — Identification of potential viral indicators. Sci. Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. Npj Clean Water. 2018;1:19. [Google Scholar]
- Kocwa-Haluch R. Waterborne enteroviruses as a hazard for human health. Pol. J. Environ. Stud. 2001;10:485–487. [Google Scholar]
- Kuo D.H.-W., Simmons F.J., Blair S., Hart E., Rose J.B., Xagoraraki I. Assessment of human adenovirus removal in a full-scale membrane bioreactor treating municipal wastewater. Water Res. 2010;44:1520–1530. doi: 10.1016/j.watres.2009.10.039. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., della Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanita. 2012;48:397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue M. Methods in Cell Biology. Elsevier; 2010. Electron microscopy of viruses; pp. 1–20. [DOI] [PubMed] [Google Scholar]
- Lee H., Kim M., Paik S.-Y., Lee C.H., Jheong W.-H., Kim J., Ko G. Evaluation of electropositive filtration for recovering norovirus in water. J. Water Health. 2011;9:27–36. doi: 10.2166/wh.2010.190. [DOI] [PubMed] [Google Scholar]
- Leifels M., Dan C., Sozzi E., Shoults D.C., Wuertz S., Mongkolsuk S., Sirikanchana K. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications; a systematic review (preprint) Occup. Environ. Health. 2020 doi: 10.1101/2020.05.08.20095364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/AEM.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Gu A.Z., He M., Shi H.-C., Yang W. UV inactivation and resistance of rotavirus evaluated by integrated cell culture and real-time RT-PCR assay. Water Res. 2009;43:3261–3269. doi: 10.1016/j.watres.2009.03.044. [DOI] [PubMed] [Google Scholar]
- Li D., He M., Jiang S.C. Detection of Infectious Adenoviruses in Environmental Waters by Fluorescence-Activated Cell Sorting Assay. Appl. Environ. Microbiol. 2010;76:1442–1448. doi: 10.1128/AEM.01937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.M.D., Allen C.K.C. Clinical Applications of PCR. Humana Press; New Jersey: 2006. Introduction to the polymerase chain reaction; pp. 177–194. [DOI] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. S246812532030087X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Canovas L., Martinez Benitez M.B., Herrera Isidron J.A., Flores Soto E. Pulsed field gel electrophoresis: past, present, and future. Anal. Biochem. 2019;573:17–29. doi: 10.1016/j.ab.2019.02.020. [DOI] [PubMed] [Google Scholar]
- Lu C., Liu X., Jia Z. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W., Knipe D.M. Viral capsids and envelopes: structure and function. Encycloped. Life Sci. 2002;7 [Google Scholar]
- Ma L., Mao G., Liu J., Yu H., Gao G., Wang Y. Rapid quantification of bacteria and viruses in influent, settled water, activated sludge and effluent from a wastewater treatment plant using flow cytometry. Water Sci. Technol. 2013;68:1763–1769. doi: 10.2166/wst.2013.426. [DOI] [PubMed] [Google Scholar]
- Mackay I.M. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Martínez-Puchol S., Rusiñol M., Fernández-Cassi X., Timoneda N., Itarte M., Andrés C., Antón A., Abril J.F., Girones R., Bofill-Mas S. Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens. Sci. Total Environ. 2020;713 doi: 10.1016/j.scitotenv.2020.136604. [DOI] [PubMed] [Google Scholar]
- Martin-Latil S., Hennechart-Collette C., Guillier L., Perelle S. Duplex RT-qPCR for the detection of hepatitis E virus in water, using a process control. Int. J. Food Microbiol. 2012;157:167–173. doi: 10.1016/j.ijfoodmicro.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Masclaux F.G., Hotz P., Friedli D., Savova-Bianchi D., Oppliger A. High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Res. 2013;47:5101–5109. doi: 10.1016/j.watres.2013.05.050. [DOI] [PubMed] [Google Scholar]
- Masclaux F.G., Hotz P., Gashi D., Savova-Bianchi D., Oppliger A. Assessment of airborne virus contamination in wastewater treatment plants. Environ. Res. 2014;133:260–265. doi: 10.1016/j.envres.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Maunula L., Söderberg K., Vahtera H., Vuorilehto V.-P., von Bonsdorff C.-H., Valtari M., Laakso T., Lahti K. Presence of human noro- and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J. Water Health. 2012;10:87–99. doi: 10.2166/wh.2011.095. [DOI] [PubMed] [Google Scholar]
- Mayer B.K., Yang Y., Gerrity D.W., Abbaszadegan M. The impact of capsid proteins on virus removal and inactivation during water treatment processes. Microbiol. Insights. 2015;8s2 doi: 10.4137/MBI.S31441. MBI.S31441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S.M., Scott T.M., Lukasik J.O., Paul J.H., Harwood V.J. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. AEM. 2009;75:3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Metcalf T.G. Use of membrane filters to facilitate the recovery of virus from aqueous suspensions1. Appl. Microbiol. 1961;9:376–379. doi: 10.1128/AEM.9.5.376-379.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Masago Y., Sano D., Omura T. Development of an effective method for recovery of viral genomic RNA from environmental silty sediments for quantitative molecular detection. Appl. Environ. Microbiol. 2011;77:3975–3981. doi: 10.1128/AEM.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monpoeho S., Dehée A., Mignotte B., Schwartzbrod L., Marechal V., Nicolas J.-C., Billaudel S., Férré V. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. BioTechniques. 2000;29:88–93. doi: 10.2144/00291st03. [DOI] [PubMed] [Google Scholar]
- Montazeri N., Goettert D., Achberger E.C., Johnson C.N., Prinyawiwatkul W., Janes M.E. Pathogenic enteric viruses and microbial indicators during secondary treatment of municipal wastewater. Appl. Environ. Microbiol. 2015;81:6436–6445. doi: 10.1128/AEM.01218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Morales H.A., Vidal G., Olszewski J., Rock C.M., Dasgupta D., Oshima K.H., Smith G.B. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 2003;69:4098–4102. doi: 10.1128/AEM.69.7.4098-4102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F.A. Laboratory Diagnosis of Infectious Diseases Principles and Practice. Springer New York; New York, NY: 1988. Virus taxonomy and nomenclature; pp. 153–176. [DOI] [Google Scholar]
- Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci.: Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- Nassonova E.S. Pulsed field gel electrophoresis: theory, instruments and application. Cell Tiss. Biol. 2008;2:557–565. doi: 10.1134/S1990519X08060011. [DOI] [PubMed] [Google Scholar]
- Nelson C.D.S., Derdowski A., Maginnis M.S., O’Hara B.A., Atwood W.J. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology. 2012;428:30–40. doi: 10.1016/j.virol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater (preprint) Epidemiology. 2020 doi: 10.1101/2020.04.15.20066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby D.T., Marlowe E.M., Maier R.M. In: Environmental Microbiology. Maier R.M., Pepper I.L., Gerba C.P., editors. Elsevier Inc; 2009. Nucleic acid–based methods of analysis; p. 42. [Google Scholar]
- Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P.-E. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res. 2009;43:1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- O’Brien E., Munir M., Marsh T., Heran M., Lesage G., Tarabara V.V., Xagoraraki I. Diversity of DNA viruses in effluents of membrane bioreactors in Traverse City, MI (USA) and La Grande Motte (France) Water Res. 2017;111:338–345. doi: 10.1016/j.watres.2017.01.014. [DOI] [PubMed] [Google Scholar]
- O’Carroll I.P., Rein A. Encyclopedia of Cell Biology. Elsevier; 2016. Viral nucleic acids; pp. 517–524. [DOI] [Google Scholar]
- Olijve L., Jennings L., Walls T. Human Parechovirus: an Increasingly Recognized Cause of Sepsis-Like Illness in Young Infants. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski J., Winona L., Oshima K.H. vol. 51. 2005. Comparison of 2 Ultrafiltration Systems for the Concentration of Seeded Viruses From Environmental Waters; p. 10. [DOI] [PubMed] [Google Scholar]
- Ortmann A.C., Suttle C.A. In: Bacteriophages, Methods in Molecular Biology. Clokie M.R.J., Kropinski A.M., editors. Humana Press; Totowa, NJ: 2009. Determination of virus abundance by epifluorescence microscopy; pp. 87–95. [DOI] [PubMed] [Google Scholar]
- Osuolale O., Okoh A. Human enteric bacteria and viruses in five wastewater treatment plants in the Eastern Cape, South Africa. J. Inf. Public Health. 2017;10:541–547. doi: 10.1016/j.jiph.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Otawa K., Lee S.H., Yamazoe A., Onuki M., Satoh H., Mino T. Abundance, diversity, and dynamics of viruses on microorganisms in activated sludge processes. Microb. Ecol. 2007;53:143–152. doi: 10.1007/s00248-006-9150-9. [DOI] [PubMed] [Google Scholar]
- Ouardani I., Turki S., Aouni M., Romalde J.L. Detection and molecular characterization of hepatitis A virus from Tunisian wastewater treatment plants with different secondary treatments. Appl. Environ. Microbiol. 2016;82:3834–3845. doi: 10.1128/AEM.00619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasalari H., Ataei-Pirkooh A., Aminikhah M., Jafari A.J., Farzadkia M. Assessment of airborne enteric viruses emitted from wastewater treatment plant: atmospheric dispersion model, quantitative microbial risk assessment, disease burden. Environ. Pollut. 2019;253:464–473. doi: 10.1016/j.envpol.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Payne S. Viruses. Elsevier; 2017. Family Adenoviridae; pp. 261–267. [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Omer S.B. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics (preprint) Epidemiology. 2020 doi: 10.1101/2020.05.19.20105999. [DOI] [Google Scholar]
- Pepper I.L., Gerba C.P. In: Environmental Microbiology. Pepper I.L., Gerba C.P., Gentry T.J., editors. Elsevier; 2015. Environmental sample collection and processing; pp. 157–175. [DOI] [Google Scholar]
- Petterson S., Grøndahl-Rosado R., Nilsen V., Myrmel M., Robertson L.J. Variability in the recovery of a virus concentration procedure in water: implications for QMRA. Water Res. 2015;87:79–86. doi: 10.1016/j.watres.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Pietruchinski E., Benati F., Lauretti F., Kisielius J., Ueda M., Volotão E.M., Soares C.C., Hoshino Y., Linhares R.E.C., Nozawa C., Santos N. Rotavirus diarrhea in children and adults in a southern city of Brazil in 2003: distribution of G/P types and finding of a rare G12 strain. J. Med. Virol. 2006;78:1241–1249. doi: 10.1002/jmv.20686. [DOI] [PubMed] [Google Scholar]
- Polaczyk A.L., Roberts J.M., Hill V.R. Evaluation of 1MDS electropositive microfilters for simultaneous recovery of multiple microbe classes from tap water. J. Microbiol. Methods. 2007;68:260–266. doi: 10.1016/j.mimet.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Pollard P.C. Fluorescence instrument for in situ monitoring of viral abundance in water, wastewater and recycled water. J. Virol. Methods. 2012;181:97–102. doi: 10.1016/j.jviromet.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Prado T., Silva D.M., Guilayn W.C., Rose T.L., Gaspar A.M.C., Miagostovich M.P. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 2011;45:1287–1297. doi: 10.1016/j.watres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Prado T., Gaspar A.M.C., Miagostovich M.P. Detection of enteric viruses in activated sludge by feasible concentration methods. Braz. J. Microbiol. 2014;45:343–349. doi: 10.1590/S1517-83822014000100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., de Castro Bruni A., Barbosa M.R.F., Garcia S.C., de Jesus Melo A.M., Sato M.I.Z. Performance of wastewater reclamation systems in enteric virus removal. Sci. Total Environ. 2019;678:33–42. doi: 10.1016/j.scitotenv.2019.04.435. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Lee B.E., Ruecker N.J., Neumann N., Ashbolt N., Pang X. A one-step centrifugal ultrafiltration method to concentrate enteric viruses from wastewater. J. Virol. Methods. 2016;237:150–153. doi: 10.1016/j.jviromet.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Ferrando E.C., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area (preprint) Occup. Environ. Health. 2020 doi: 10.1101/2020.04.22.20075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajtar B., Majek M. Enteroviruses In Water Environment – A Potential Threat To Public Health. Ann. Agric. Environ. Med. 2008;15:199–203. [PubMed] [Google Scholar]
- Rasche A., Sander A.-L., Corman V.M., Drexler J.F. Evolutionary biology of human hepatitis viruses. J. Hepatol. 2019;70:501–520. doi: 10.1016/j.jhep.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K.A. Public Health Microbiology. Humana Press; New Jersey: 2004. Integrated cell culture/PCR for detection of enteric viruses in environmental samples; pp. 069–078. [DOI] [PubMed] [Google Scholar]
- Ridinger D.N., Spendlove R.S., Barnett B.B., George D.B., Roth C. Evaluation of Cell Lines and Immunofluorescence and Plaque Assay Procedures for Quantifying Reoviruses in Sewaget. Appl. Env. Microbiol. 1982;43:7. doi: 10.1128/aem.43.4.740-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers (preprint) Epidemiology. 2020 doi: 10.1101/2020.05.01.20086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C., Alum A., Abbaszadegan M. PCR inhibitor levels in concentrates of biosolid samples predicted by a new method based on excitation-emission matrix spectroscopy. Appl. Environ. Microbiol. 2010;76:8102–8109. doi: 10.1128/AEM.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.A., Pepper I.L., Gerba C.P. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 2009;75:297–307. doi: 10.1128/AEM.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard P., Raynal P.-I., Eymieux S., Blanchard E. Virus detection by transmission electron microscopy: still useful for diagnosis and a plus for biosafety. Rev. Med. Virol. 2019;29 doi: 10.1002/rmv.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Timoneda N., Fernández-Cassi X., Pérez-Cataluña A., Fernández-Bravo A., Moreno-Mesonero L., Moreno Y., Alonso J.L., Figueras M.J., Abril J.F., Bofill-Mas S., Girones R. Metagenomic analysis of viruses, bacteria and protozoa in irrigation water. Int. J. Hyg. Environ. Health. 2020;224 doi: 10.1016/j.ijheh.2019.113440. [DOI] [PubMed] [Google Scholar]
- Sano D., Fukushi K., Yoshida Y., Omura T. Detection of enteric viruses in municipal sewage sludge by a combination of the enzymatic virus elution method and RT-PCR. Water Res. 2003;37:3490–3498. doi: 10.1016/S0043-1354(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Sattar S.A., Springthorpe V.S., Karim Y., Loro P. Chemical disinfection of non-porous inanimate surfaces experimentally contaminated with four human pathogenic viruses. Epidemiol. Infect. 2009;102:493–505. doi: 10.1017/S0950268800030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinazi R.F., Kohler J.J., Kim B. Brenner’s Encyclopedia of Genetics. Elsevier; 2013. Reverse transcription; p. 224. [DOI] [Google Scholar]
- Schlindwein A.D., Rigotto C., Simões C.M.O., Barardi C.R.M. Detection of enteric viruses in sewage sludge and treated wastewater effluent. Water Sci. Technol. 2010;61:537–544. doi: 10.2166/wst.2010.845. [DOI] [PubMed] [Google Scholar]
- Schmitz B.W., Kitajima M., Campillo M.E., Gerba C.P., Pepper I.L. Virus Reduction during Advanced Bardenpho and Conventional Wastewater Treatment Processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- Schvoerer E., Ventura M., Dubos O., Cazaux G., Serceau R., Gournier N., Dubois V., Caminade P., Fleury H.J.A., Lafon M.-E. Qualitative and quantitative molecular detection of enteroviruses in water from bathing areas and from a sewage treatment plant. Res. Microbiol. 2001;152:179–186. doi: 10.1016/s0923-2508(01)01190-1. [DOI] [PubMed] [Google Scholar]
- Shieh Y.-S.C., Wait D., Tai L., Sobsey M.D. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods. 1995;54:51–66. doi: 10.1016/0166-0934(95)00025-P. [DOI] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of Genetic Diagnostic Methods for Novel Coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.061. (advpub) [DOI] [PubMed] [Google Scholar]
- Sidhu J.P.S., Ahmed W., Toze S. Sensitive detection of human adenovirus from small volume of primary wastewater samples by quantitative PCR. J. Virol. Methods. 2013;187:395–400. doi: 10.1016/j.jviromet.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Sidhu J.P.S., Sena K., Hodgers L., Palmer A., Toze S. Comparative enteric viruses and coliphage removal during wastewater treatment processes in a sub-tropical environment. Sci. Total Environ. 2018;616–617:669–677. doi: 10.1016/j.scitotenv.2017.10.265. [DOI] [PubMed] [Google Scholar]
- Soto-Beltran M., Ikner L.A., Bright K.R. Effectiveness of poliovirus concentration and recovery from treated wastewater by two electropositive filter methods. Food Environ. Virol. 2013;5:91–96. doi: 10.1007/s12560-013-9104-6. [DOI] [PubMed] [Google Scholar]
- Strubbia S., Schaeffer J., Oude Munnink B.B., Besnard A., Phan M.V.T., Nieuwenhuijse D.F., de Graaf M., Schapendonk C.M.E., Wacrenier C., Cotten M., Koopmans M.P.G., Le Guyader F.S. Metavirome sequencing to evaluate norovirus diversity in sewage and related bioaccumulated oysters. Front. Microbiol. 2019;10:2394. doi: 10.3389/fmicb.2019.02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchman E., Blair C. 2007. Cythopatic Effects_Protocol.pdf. [Google Scholar]
- Symonds E.M., Griffin D.W., Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl. Environ. Microbiol. 2009;75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Verbyla M.E., Lukasik J.O., Kafle R.C., Breitbart M., Mihelcic J.R. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Tamaki H., Zhang R., Angly F.E., Nakamura S., Hong P.-Y., Yasunaga T., Kamagata Y., Liu W.-T. Metagenomic analysis of DNA viruses in a wastewater treatment plant in tropical climate: viral metagenomics in a WWTP. Environ. Microbiol. 2012;14:441–452. doi: 10.1111/j.1462-2920.2011.02630.x. [DOI] [PubMed] [Google Scholar]
- Tang Le, Wang Chun-Xiao, Liu Shu-Lin. Reference Module in Life Sciences. Elsevier; 2017. Pulsed field gel electrophoresis ☆. p. B9780128096338070000. [DOI] [Google Scholar]
- Teixeira P., Costa S., Brown B., Silva S., Rodrigues R., Valério E. Quantitative PCR detection of enteric viruses in wastewater and environmental water sources by the Lisbon municipality: a case study. Water. 2020;12:544. doi: 10.3390/w12020544. [DOI] [Google Scholar]
- Thompson S.S., Jackson J.L., Suva-Castillo M., Yanko W.A., El Jack Z., Kuo J., Chen C.-L., Williams F.P., Schnurr D.P. Detection of Infectious Human Adenoviruses in Tertiary-Treated and Ultraviolet-Disinfected Wastewater. Water Environ. Res. 2003;75:163–170. doi: 10.2175/106143003x140944. [DOI] [PubMed] [Google Scholar]
- Thongprachum A., Fujimoto T., Takanashi S., Saito H., Okitsu S., Shimizu H., Khamrin P., Maneekarn N., Hayakawa S., Ushijima H. Detection of nineteen enteric viruses in raw sewage in Japan. Infect. Genet. Evol. 2018;63:17–23. doi: 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla M.E., Mihelcic J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015;71:107–124. doi: 10.1016/j.watres.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Villar L.M., de Paula V.S., Diniz-Mendes L., Guimarães F.R., Ferreira F.F.M., Shubo T.C., Miagostovich M.P., Lampe E., Gaspar A.M.C. Molecular detection of hepatitis A virus in urban sewage in Rio de Janeiro, Brazil. Lett. Appl. Microbiol. 2007;45:168–173. doi: 10.1111/j.1472-765X.2007.02164.x. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis C., Melnick J.L. Concentration of enteroviruses on membrane filters. J. Virol. 1967;1:472–477. doi: 10.1128/JVI.1.3.472-477.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Guo T.-K., Zhen B., Kong Q.-X., Yi B., Li Z., Song N., Jin M., Wu X.-M., Xiao W.-J., Zhu X.-M., Gu C.-Q., Yin J., Wei W., Yao W., Liu C., Li J.-F., Ou G.-R., Wang M.-N., Fang T.-Y., Wang G.-J., Qiu Y.-H., Wu H.-H., Chao F.-H., Li J.-W. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005;11:4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Guo T.-K., Zhen B., Kong Q.-X., Yi B., Li Z., Song N., Jin M., Xiao W.-J., Zhu X.-M., Gu C.-Q., Yin J., Wei W., Yao W., Liu C., Li J.-F., Ou G.-R., Wang M.-N., Fang T.-Y., Wang G.-J., Qiu Y.-H., Wu H.-H., Chao F.-H., Li J.-W. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital. J. Virol. Methods. 2005;128:156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., Si B., Guo B.-Z., Liu C., Ou G.-R., Wang M.-N., Fang T.-Y., Chao F.-H., Li J.-W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Kjellberg I., Sikora P., Rydberg H., Lindh M., Bergstedt O., Norder H. Hepatitis E virus genotype 3 strains and a plethora of other viruses detected in raw and still in tap water. Water Res. 2020;168 doi: 10.1016/j.watres.2019.115141. [DOI] [PubMed] [Google Scholar]
- Watzinger F., Ebner K., Lion T. Detection and monitoring of virus infectionsby real-time PCR. Mol. Asp. Med. 2006;27:254–298. doi: 10.1016/j.mam.2005.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings F.M., Lewis A.L., Mountain C.W. Demonstration of solids-associated virus in wastewater and sludge. Appl. Environ. Microbiol. 1976;31:354–358. doi: 10.1128/AEM.31.3.354-358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K., Ortmann A.C., Suttle C.A. Accurate estimation of viral abundance by epifluorescence microscopy. Appl. Environ. Microbiol. 2004;70:3862–3867. doi: 10.1128/AEM.70.7.3862-3867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Onan B.M., Xagoraraki I. Quantification of enteric viruses, pathogen indicators, and salmonella bacteria in class B anaerobically digested biosolids by culture and molecular methods. AEM. 2010;76:6441–6448. doi: 10.1128/AEM.02685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Liu W.-T. Determination of virus abundance, diversity and distribution in a municipal wastewater treatment plant. Water Res. 2009;43:1101–1109. doi: 10.1016/j.watres.2008.11.039. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases (preprint) Infect. Dis. Ther. 2020 doi: 10.1101/2020.04.05.20051540. (except HIV/AIDS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters (preprint) Epidemiology. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., Yin Z., Svambayev Z. Fate of viruses in water systems. J. Environ. Eng. 2014;140 doi: 10.1061/(ASCE)EE.1943-7870.0000827. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020:140445. doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Abudayyeh, O.O., Gootenberg, J.S., n.d. A protocol for detection of COVID-19 using CRISPR diagnostics 8.
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]