Abstract
The coronavirus disease 2019 (COVID-19) pandemic poses a current world-wide public health threat. However, little is known about its hallmarks compared to other infectious diseases. Here, we report the single-cell transcriptional landscape of longitudinally collected peripheral blood mononuclear cells (PBMCs) in both COVID-19- and influenza A virus (IAV)-infected patients. We observed increase of plasma cells in both COVID-19 and IAV patients and XIAP associated factor 1 (XAF1)-, tumor necrosis factor (TNF)-, and FAS-induced T cell apoptosis in COVID-19 patients. Further analyses revealed distinct signaling pathways activated in COVID-19 (STAT1 and IRF3) versus IAV (STAT3 and NFκB) patients and substantial differences in the expression of key factors. These factors include relatively increase of interleukin (IL)6R and IL6ST expression in COVID-19 patients but similarly increased IL-6 concentrations compared to IAV patients, supporting the clinical observations of increased proinflammatory cytokines in COVID-19 patients. Thus, we provide the landscape of PBMCs and unveil distinct immune response pathways in COVID-19 and IAV patients.
Keywords: COVID-19, influenza, scRNA-seq, apoptosis, IL-6R
Graphical Abstract
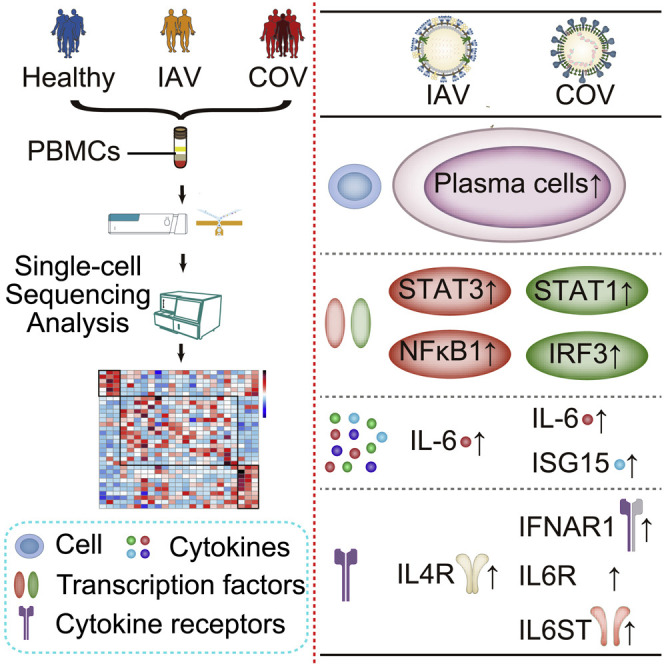
Highlights
-
•
We generated a single-cell atlas of PBMCs in both COVID-19 and influenza patients
-
•
Plasma cells increase significantly in both COVID-19 and influenza patients
-
•
COVID-19 is featured with XAF1-, TNF-, and FAS-induced T cell apoptosis
-
•
COVID-19 activates distinct pathway (STAT1/IRF3) versus influenza (STAT3/NFκB)
COVID-19 and influenza are both respiratory infections with cytokine release syndrome. Zhu et al. use single-cell RNA sequencing of longitudinally collected PBMCs in both patients to reveal distinct immune response landscapes of the two diseases and identify virus-specific cell composition and immune response pathways.
Introduction
A novel coronavirus (CoV), designated severe acute respiratory syndrome (SARS)-CoV-2, underlies a pandemic infection, coronavirus disease 2019 (COVID-19), which rapidly spread globally and has been proclaimed a severe public health emergency of international concern (PHEIC) by the World Health Organization (WHO) (Wang et al., 2020a). By June 30, 2020, more than 200 countries and territories reported COVID-19 infections, together comprising 10,185,374 confirmed cases and 503,862 deaths (data from WHO, the Centers for Disease Control and Prevention [CDC], and the European Centre for Disease Prevention and Control [ECDC] are at https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports).
Common respiratory viruses include CoV, influenza A virus (IAV), influenza B virus, respiratory syncytial virus, parainfluenza virus, adenovirus, etc. CoV belongs to the virus family Coronaviridae, which is an enveloped, nonsegmented, single-stranded positive-sense RNA virus (Zhao et al., 2017). SARS-CoV-2 is a β-coronavirus, like Middle East respiratory syndrome (MERS)-CoV and SARS-CoV, both of which are also suggested to have originated in bats (Zhou et al., 2020b). Evidence supports that the cellular receptor angiotensin-converting enzyme 2 (ACE2) and the serine protease for SARS-CoV entry, transmembrane serine protease 2 (TMPRSS2) (Li et al., 2003), are key mediators for SARS-CoV-2 host cell entry (Wang et al., 2020b). It has been shown that ACE2 is expressed not only in the respiratory system but in a range of organs, tissues, and cell types (Xu et al., 2020a), indicating that viral infection can rapidly spread throughout the body as disease progresses.
SARS-CoV first emerged in China in 2002–2003, and MERS-CoV was first reported in Saudi Arabia in 2012, with mortality rates around 10% (8,098 cases and 774 deaths; data from WHO) and 34.4% (2,494 cases and 858 deaths; data from WHO) of SARS-CoV and MERS-CoV, respectively (Wu et al., 2020). In particular, based on the data collected from WHO (up to March 24, 2020), 4.9% of SARS-CoV-2 cases are fatal (823,626 cases and 40,598 deaths), lower than that of MERS-CoV and SARS-CoV (Liu et al., 2017). Flu season occurs annually, and influenza symptoms are similar to respiratory diseases caused by CoVs. According to annual estimates of the burden of seasonal influenza in the United States, the influenza viruses have caused an estimated 9,200,000–35,600,000 illnesses, 139,000–708,000 hospitalizations, and 4,000–20,000 deaths from pneumonia and influenza and 12,000–56,000 deaths from respiratory and circulatory symptoms (data from 2010–2011 to 2015–2016 influenza seasons) with a mortality rate of 0.04%–0.83% (Rolfes et al., 2018).
SARS-CoV-2 infection diagnostics include pneumonia detection using computed tomography (CT) scans and viral RNA detection (extracted and tested by real-time RT-PCR with SARS-CoV-2-specific primers and probes) in throat swab samples, secretions acquired from the lower respiratory tract, peripheral blood, or feces. Patients with mild symptoms present with fever, cough, myalgia or fatigue, and sputum production, though infected individuals are sometimes asymptomatic. Such mild symptoms rarely include intestinal signs and symptoms (Huang et al., 2020).
After COVID-19 diagnosis, other symptoms can be detected by routine blood examination. The neutrophils in 38% of COVID-19 patients were above the normal range, while the hemoglobin in 51% COVID-19 patients lies below the normal range, according to research at the Jinyintan Hospital in Wuhan, China. In addition, lymphocyte levels decreased in 35% of patients (Chen et al., 2020), suggesting possible dysfunctional cell-mediated immunity in COVID-19 patients.
In addition to acute respiratory distress syndrome (ARDS), virally driven hyperinflammation is another major cause of mortality (Huang et al., 2014). Increased proinflammatory cytokine or chemokine responses even initiated viral sepsis and overwhelming systemic inflammation, contributing to cytokine storm syndromes (CSSs) that include acute inflammatory-induced lung injury and development of pneumonitis, ARDS and respiratory failure resulting in shock, hemodynamic instability, multiple organ dysfunction, and even death. It has been reported that interleukin-6 (IL-6) concentrations and ferritin increase with illness deterioration in non-survivors compared with survivors within a subgroup of patients with COVID-19 (Zhou et al., 2020a). Furthermore, we recently reported that during the acute phase, a group of proinflammatory cytokines was upregulated in lung injury (Murray score) in severe patients. Importantly, these cytokines can be used as biomarkers to predict disease severity after SARS-CoV-2 infection (Liu et al., 2020).
Although there is accumulating clinical data regarding blood cell indices, underlying molecular mechanisms have yet to be clarified. Here, we report the transcriptome dynamics of peripheral blood mononuclear cells (PBMCs) from patients with COVID-19, comparing these to profiles in IAV patients and control, healthy donors. We examined the landscape and features of these infections by integrating single-cell RNA sequencing (scRNA-seq) with clinical symptoms. We observed an increased proportion of plasma cells, as well as a reduction of lymphocytes in the clinic. Further analyses suggest that XAF1-, tumor necrosis factor (TNF)-, and Fas-induced apoptosis may confer this reduction. Furthermore, distinct from that in IAV patients, expression of IL6R and IL6ST was upregulated in COVID-19 patients, which synergistically promotes increased proinflammatory cytokines during pathogenesis. We also discovered that several interferon (IFN)-stimulated genes (ISGs; including ISG15, IFI44, IFI44L, and RSAD2) were specifically upregulated in PBMCs from COVID-19 patients, enhancing antiviral and immune modulatory functions after viral infection.
Results
Single-Cell Transcriptional Landscape of PBMCs from COVID-19 and IAV Patients
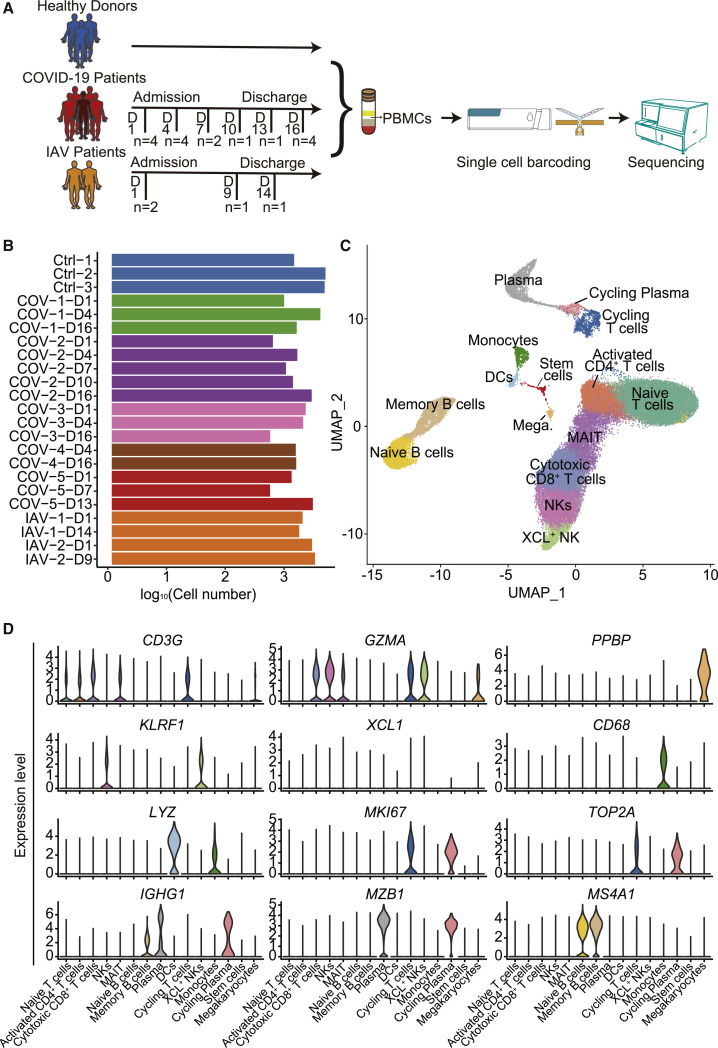
To investigate the pathogenesis and mechanism of SARS-CoV-2 infections in COVID-19 patients, blood samples were collected from three healthy controls, two IAV-infected patients, and five COVID-19 patients, including four patients (COV-1 to COV-4) with uncomplicated disease courses and one patient (COV-5) that subsequently progressed to severe disease (Figure 1 A; Table S1). Seasonal IAV-infected patients were chosen as controls because both SARS-CoV-2 and IAV are contagious RNA viruses, and both caused respiratory tract infection. However, COVID-19 has distinct clinical signatures compared to IAV infections, including high morbidity and mortality. COVID-19 patients were enrolled within 5–10 days of symptom onset based on positive nucleic acid testing results, and the day when PBMCs were first collected was named day 1. Time points corresponding to disease onset and sample collection are listed in Table S1. PBMCs were also collected at subsequent time points as shown in Figure 1A. After filtering out cells with low quality, we obtained transcriptome datasets from 46,022 cells with an average of 2,000 cells for each participant at each time point (Figure 1B). To uncover immune cell populations in COVID-19, we performed unsupervised clustering and obtained 15 cell populations (Figure 1C). Immunocytes such as T cells, B cells, monocytes, natural killer (NK) cells, DCs, stem cells, and megakaryocytes were identified based on the expression of classic cell-type markers (Figure 1C). With this approach, five populations were annotated as T cells, including naive T cells (CD3+CCR7+GZMA−), activated CD4+ T cells (CD3+CD4+IL7R+), cytotoxic CD8+ T cells (CD3+CD8+GZMA+), mucosal associated invariant T cells (MAIT cells; CD3+SLC4A10+), and cycling T cells (CD3+MKI67+); four populations were annotated as B cells, including naive B cells (MS4A1+IGHG1−), plasma cells (MZB1+IGHG1+), cycling plasma cells (MZB1+IGHG1+MKI67+), and memory B cells (MS4A1+IGHG1+); two were annotated as NK cells (KLRF1+); and one population was labeled DCs (CD1C+LYZ+) and monocytes (LYZ+CD68+) (Figures 1D and S1A). Most of the clusters consisted of cells from multiple patients, indicating common immune traits among patients. In addition, PBMC samples from patients did not express ACE2 and TMPRSS2 receptors and did not exhibit viral reads, indicating that SARS-CoV-2 may not infect PBMCs (Figure S1B).
Figure 1.
Single-Cell Gene Expression Profiling of Immune Cells Derived from PBMCs of the Participants
(A) Schematic outline of the study design. 10 subjects, including three healthy donors, five COVID-19 patients, and two IAV-infected patients were included in this study.
(B) Bar plot shows the log10 transformed cell number of each sample for every donor at different time points. Blue represents three healthy donors, orange represents two IAV-infected patients, and five COVID-19 patients are displayed using five different colors.
(C) The clustering result of 46,022 cells from ten donors. Each point represents one single cell, colored according to cell type. Mega., Megakaryocytes.
(D) Expression levels of cell typing genes in cell type clusters. CD3G indicates T cells, KLRF1 and XCL1 indicate NKs, MS4A1 indicates B cells, IGHG1 and MZB1 indicate plasma cells, CD68 indicates monocytes, LYZ indicates DCs, MKI67 and TOP2A indicate cycling T cells, GZMA indicates cytotoxic CD8+ T cells and NKs, and PPBP indicates megakaryocytes.
Increased Plasma Cells in PBMCs from COVID-19 and IAV Patients
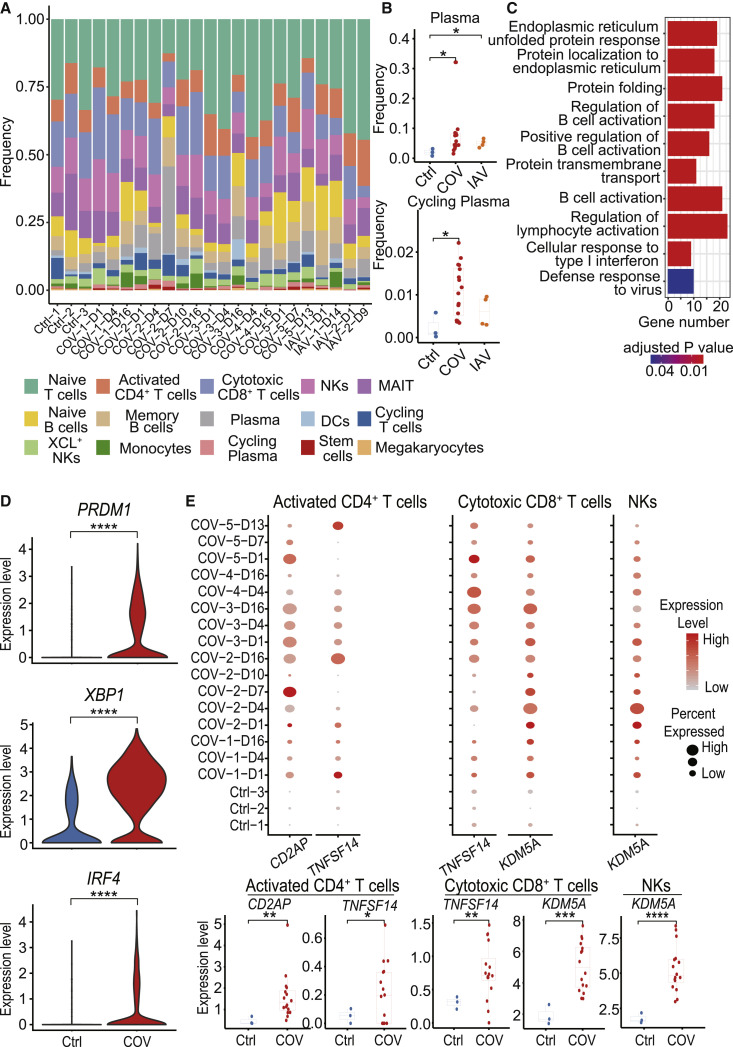
The general patterns of PBMC populations were comparable across patients (Figure 2 A; Table S2). The proportion of plasma cells and cycling plasma cells were increased significantly in both COVID-19- and IAV-infected patients (Figures 2B and S2), and there was no difference between these two types of virus infection. The increased plasma cells may induce protective neutralizing antibodies to prevent viruses infecting cells. As expected, when examining the functions of upregulated genes in B cells of COVID-19 patients compared with healthy donors, protein complex assembly and protein-transport-related pathways were especially enriched, which may be because a large number of proteins are synthesized during this process (Figure 2C). B-cell-activation-related pathways were also enriched (Figure 2C), and representative genes include PRDM1, XBP1, and IRF4 (Figure 2D). The identity and function of plasma cells are dependent upon the transcription factors PRDM1, XBP1, and IRF4 (Ochiai et al., 2013; Shaffer et al., 2004). PRDM1 has a central role in determining and shaping the secretory arm of mature B cell differentiation and in promoting immunoglobulin (Ig) synthesis. XBP1 is a positively acting transcription factor in the CREB-ATF family that is expressed at a large amounts in plasma cells and is crucial for increasing protein synthesis in plasma cells (Shaffer et al., 2004). IRF4 regulates immunoglobulin class-switch recombination, and sustained and higher concentrations of IRF4 are known to promote the generation of plasma cells (Ochiai et al., 2013). Following increased plasma cells and expression of relevant transcription factors, we found that the expression of CD2AP on activated CD4+ T cells was elevated compared with healthy controls (Figure 2E). The adaptor molecule CD2AP in CD4+ T cells modulates the differentiation of follicular helper T cells and improves protective antibody responses in viral infection (Raju et al., 2018). In addition to supporting plasma cell function, TNFSF14 expression was also increased in activated CD4+ T cells and cytotoxic CD8+ T cells. These factors promote T cell activation as well as T cell recruitment to tissues from peripheral blood. KDM5A, which encodes an H3K4me3 demethylase that is required for NK cell and T cell activation, is upregulated in NK cells and cytotoxic CD8+ T cells of COVID-19 (Figure 2E). Taken together, these results demonstrated that elevated plasma cells and increased activation of T cells and NK cells in COVID-19 patients may contribute to defense against the virus.
Figure 2.
Dynamic Composition and Functional Changes in Immune Cells during SARS-CoV-2 Infection
(A) The cell-type frequency in each sample. Bars are colored by cell types.
(B) Differences in plasma and cycling plasma proportion among samples from healthy donors (Ctrl) (n = 3), COVID-19 patients (COV) (n = 16), and IAV patients (IAV) (n = 4). Student’s t test was applied to test the significance of the difference. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Enriched GO terms for upregulated genes in COVID-19 patients compared to healthy controls in B cells.
(D) The differential expression levels of B cell activation-related genes PRDM1, XBP1, and IRF4 between healthy donors (Ctrl) and COVID-19 patients (COV) in plasma cells.
(E) The expression levels of T cell activation related genes in activated CD4+ T cells, cytotoxic CD8+ T cells, and NKs in samples from healthy donors and COVID-19 patients. In the upper panel, the color of each dot represents expression levels of the gene, while the dot size represents the fraction of cells expressing the gene in the specific cell type. In the lower panel, the difference between healthy donors (Ctrl) (n = 3) and COVID-19 patients (COV) (n = 16) were tested using the Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
IFN Response and Lymphocyte Apoptosis in COVID-19 Patients
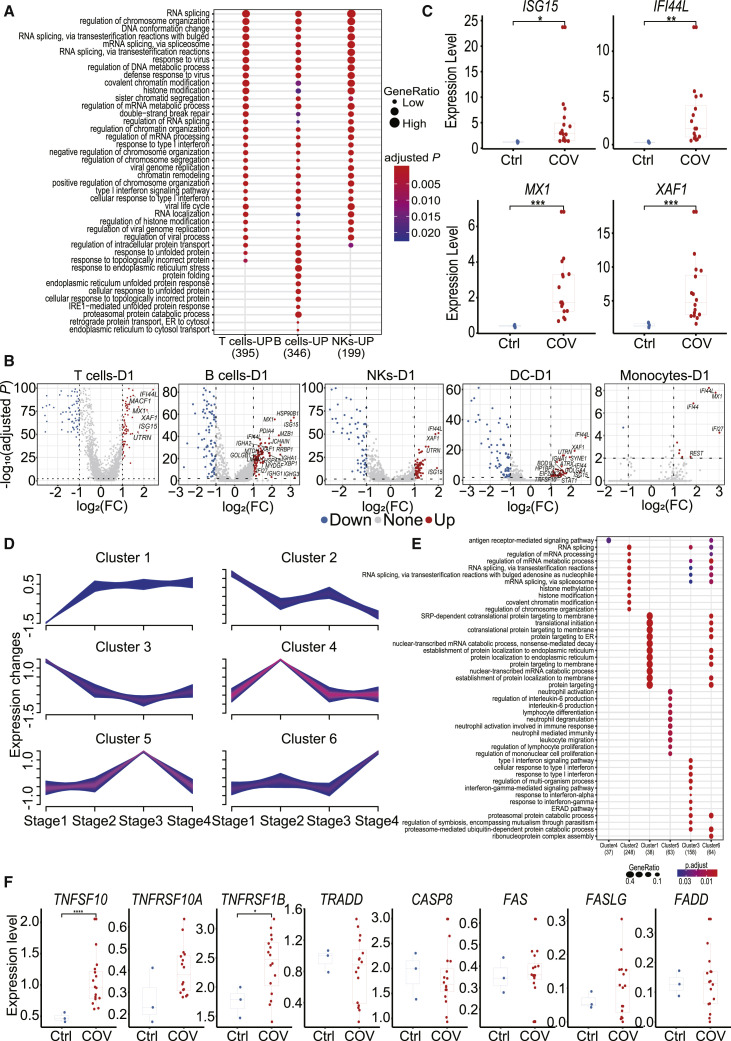
Gene Ontology (GO) analyses were performed to gain insight into functions of different cell subsets between COVID-19 patients and healthy controls. Genes in the group “response to type I IFN signaling” were enriched in T, B, and NK cell subsets of D1 and D4, but not D16 samples (Figures 3 A and S3A), consistent with the concept that the IFN response is essential to the response triggered by viral infection. Consistently, “defense response to virus signaling” genes were also enriched in T, B, and NK cells of all five COVID-19 patients on D1 and D4, but not D16 (Figures 3A and S3A), implying an ongoing immune response against SARS-CoV-2 virus. Furthermore, endoplasm- and protein-unfolding-related pathways were especially enriched in B cells at all the three time points (D1, D4, and D16) (Figures 3A and S3A), which may be due to a higher proportion of plasma cells in B cell clusters because high demand of protein synthesis was required during antibody production. Other signaling pathways, such as “regulation of chromosome organization,” “DNA conformation change,” etc. in SARS-CoV-2 infection were also upregulated. The roles of these genes will need further investigation.
Figure 3.
Analysis of IFN Response- and Apoptosis-Associated Genes in COVID-19 Patients
(A) The top 20 enriched biological processes by GO analysis in day 1 samples from COVID-19 patients compared to healthy controls in different cell populations. Dot color indicates the statistical significance of the enrichment (p), and dot size represents gene ratio annotated to each term.
(B) The differentially expressed genes in day 1 samples from COVID-19 patients compared to healthy controls in different cell subsets. Red dots represent genes upregulated in COVID-19 patients (adjusted p < 0.01 and fold Change (FC) ≥ 2), while blue dots represent downregulated genes in COVID-19 patients (adjusted p < 0.01 and FC ≤ 0.5). Genes with log2(FC) ≥ 1.5 were labeled by gene symbols.
(C) The gene expression of ISG15, IFI44L, MX1, and XAF1 in healthy donors (Ctrl) (n = 3) and COVID-19 patients (COV) (n = 16). Student’s t test was applied to test the significance of the difference. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Genes clustered by their expression pattern along the progression of the disease by the mfuzz R package.
(E) The top 10 enriched biological processes in each cluster of genes as revealed by GO analysis.
(F) The difference in expression levels of apoptosis-associated genes between COVID-19 patients (COV) (n = 5) and healthy controls (Ctrl) (n = 3) in T cells. Student’s t test was applied. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Differentially expressed genes (DEGs) in these transcriptomic profiles were then compared between COVID-19 patients and healthy controls. ISGs, which are vital to early viral control (Schoggins and Rice, 2011), were identified in patients infected with SARS-CoV-2 on D1 and D4 (Figures 3B and S3B), in line with the enrichment for “response to type I IFN signaling” pathways in our GO analysis (Figures 3A and S3A). Among these ISGs, ISG15, IFI44L, MX1, and X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) were upregulated in T, B, NK, and DC cell subsets (Figures 3B and S3B). The expression of these four genes was also much higher in COVID-19 patients compared to healthy controls at the bulk level (Figure 3C). We then examined transcription dynamics of these genes during the disease process. To achieve this, we divided the disease processes of COVID-19 patients from symptom onset to discharge into four stages (Table S3). We identified six time-dependent expression patterns (Figure 3D) and investigated their biological significance (Figure 3E). Cluster 3 contained 158 genes that had decreased expression levels over time. The functions of these genes were significantly enriched in biological processes associated with IFN responses, indicating that the transcriptional regulation of these genes is dynamic and that they were activated at early time points and deactivated at late time points (Figure 3E). Cluster 1 contained 38 genes whose expression levels were elevated from stage 2. GO enrichment analysis showed that their functions were significantly enriched in translation and protein-synthesis-related pathways. This is consistent with the timing of antibody production (Thevarajan et al., 2020), since a large amount of proteins synthesis occurs during this process (Figure 3E).
Severe patient (COV-5) had a stronger response to IFNa upon SARS-CoV-2 infection than the mild patients did. In addition, the expression of ISG15, IFI44L, MX1, and XAF1 was higher at earlier time points of disease progression and then decreased at later phases individually (Figure S3C), illustrating their dynamic responses to interferons.
The ubiquitin-like protein ISG15, IFI44L, and MX1 all have roles in the antiviral response (Perng and Lenschow, 2018), while XAF1 participates in pro-apoptotic responses and has been reported to form a positive feedback loop with IRF-1 that drives apoptosis under stress (Jeong et al., 2018). TP53-mediated apoptosis was also enhanced by XAF1 via post-translational modification (Zou et al., 2012).Therefore, expression of genes linked to XAF1-mediated apoptosis, including IRF1, TP53, BCL2L11, and CASP3, was analyzed (Figure S3D). Expression of IRF1, TP53, and CASP3 was increased in T, B, and NK cell subsets in COVID-19 patients compared to controls, while BCL2L11 exhibited different patterns in different cell subsets. In addition to the XAF1 related apoptosis pathway, expression of genes in other apoptosis-linked pathways, including TNF and Fas pathways (Elmore, 2007), was examined in both COVID-19 patients and healthy controls (Figures 3F and S3E). The expression of TNFSF10 (TRAIL) and its receptor TNFRSF10A were increased in T cells from COVID-19 patients relative to healthy controls. Other TNF path members, including TNFRSF1B, were also relatively upregulated in COVID-19 patients. As for the FAS path, the expression of FAS, FASLG, and FADD were upregulated in T cells of COVID-19 patients, though not significantly (Figure 3F). In B and NK cell subsets, TNFSF10 and FADD were notably increased in COVID-19 patients with other genes increased mildly, except that Fas in B cells and TRADD in NK cells were subtly decreased (Figure S3E). Taken together, we find that upregulated genes relevant to the XAF1, TNF, and Fas pathways may lead to increased T cell apoptosis in COVID-19 patients.
Immune Molecular Signatures of COVID-19 Patients Compared to IAV Patients
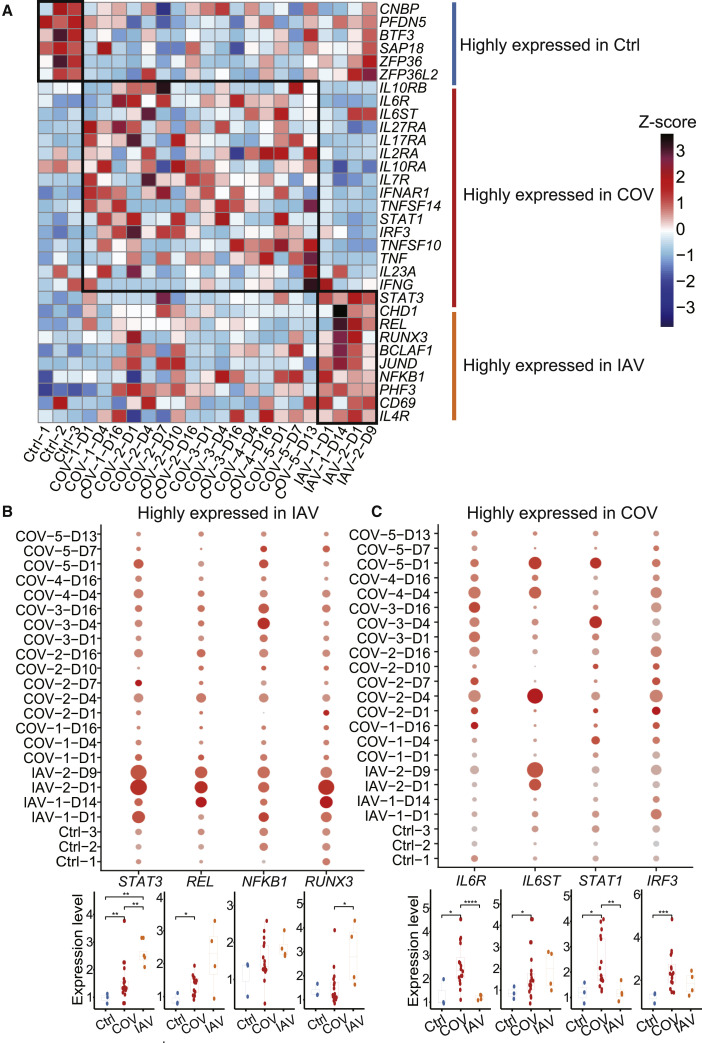
Next, we sought to identify the immune molecular signatures associated with COVID-19 and IAV infection. Thus, we compared the expression of cytokines, cytokine receptors, and transcription factors in T cell subsets, NK cells, and DCs among COVID-19 patients, IAV patients, and healthy controls (Figures 4 A and S4A). As the gene clustering pattern suggested, upregulated genes in COVID-19 patients mostly encode proinflammatory cytokines, cytokine receptors, and IFN-responsive transcription factors, while in IAV patients, proinflammatory transcription factors and virus-interacting host factors seem to be highly expressed.
Figure 4.
Hallmarks of COVID-19 Compared to IAV (Revealed by Single-Cell Analysis of Cytokines, Cytokine Receptors, and Transcription Factors)
(A) The relative expression level (Z score) of key cytokines, cytokine receptors, and transcription factors among COVID-19 patients, IAV patients, and healthy controls in the activated CD4+ T cells population.
(B) The expression level of four representative genes highly expressed in activated CD4+ T cells of IAV patients. Upper panel: the color of each dot in the dot plot indicates expression level of the gene; dot size represents the fraction of cells expressing the gene in activated CD4+ T cells population. Lower panel: difference in gene expression among samples from COVID-19 patients (COV) (n = 16), IAV patients (IAV) (n = 4), and healthy donors (Ctrl) (n = 3). Each dot in the boxplot represents the average expression level of a gene in the activated CD4+ T cells population in one sample. Student’s t test was applied to test the significance of the difference. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Similar to (B), showing four representative genes highly expressed in the activated CD4+ T cells of COVID-19 patients.
See also Figure S4.
Multiple key transcription factors involved in the host immune response, including STAT3, NFKB1, and REL, were upregulated in various cell types from IAV patients (Figures 4A and S4A). NFKB1 and REL encode active subunits of NF-κB heterodimer, one of the hallmark transcription factors activated by IAV infection (Ludwig and Planz, 2008). Activation of NF-κB plays a key role in regulating the proinflammatory innate immune response and adaptive immune response. Previous work showed that IL-6 and IL-10, known activators of STAT3 signaling, are highly increased in severe IAV patients (Yu et al., 2011). Expression of STAT3 was elevated in IAV patients compared to COVID-19 patients and healthy controls and seemed to correlate with time post admission. In addition, RUNX3 expression was upregulated in activated CD4+ T cells of IAV patients compared to the other groups (Figure 4B). It has been suggested that RUNX3 induction is a key step for CD4+ T cells to acquire cytotoxic activity, whereas another study showed that RUNX3 induced by IAV infection through the NF-κB pathway promoted apoptosis in airway epithelial cells (Gan et al., 2015). The role of RUNX3 in the T-cell-mediated immune response remains to be determined. Several pro-viral host factors such as CHD1, BCLAF1, and PHF3 that contribute to viral infection, replication, and immune evasion were also upregulated in IAV patients (Figures 4A and S4A) (Shapira et al., 2009). To see how response of these genes changes over time, we divided the disease processes of IAV patients into two stages (Table S3). We did not find any gene exhibiting statistically significant time-dependent regulation, although some them had trends toward increased or decreased expression (Figures S4B and S4C).
For COVID-19 patients, STAT1, a major transcription factor activated in response to interferon, was upregulated in activated CD4+ T cells, cytotoxic CD8+ T cells, naive T cells, and DCs (Figures 4A and S4A). Several proinflammatory factors, including TNF and TNFSF14, were elevated in activated CD4+ T cells, cytotoxic CD8+ T cells, MAIT cells, and NK cells (Figures S4A and S4E), suggesting enhanced effector function and memory cell development (Desai et al., 2018). Expression of IL6 was not detectable in PBMCs of any patients or healthy controls. We measured plasma concentrations of IL-6 during hospitalization of these COVID-19 patients and after leaving the hospital (Figure S4D). Plasma IL-6 were above the normal range (0.0–7.0 pg/mL) (Chen et al., 2020) in two out of five patients when admitted to the hospital, dropped back to normal during hospitalization, and remained stable after recovery. There was a rising phase of IL-6 in most COVID-19 patients, indicating an active inflammatory response, which is also observed in IAV patients (Yu et al., 2011). The patient with severe symptoms had a much higher level of IL-6 when hospitalized, and it took longer to return to normal plasma IL-6 levels, which corresponded to disease severity.
IL-6 exerts its function by binding to IL-6R. The IL-6-IL-6R complex then binds to GP130 (IL-6 receptor subunit beta [IL6ST]), a common signal-transducing chain shared by several related cytokines (Mihara et al., 2012). Expression of IL6R was elevated in activated CD4+ T cells, naive T cells, and DCs of COVID-19 patients compared to IAV patients and healthy controls (Figures 4C and S4A). Meanwhile, expression of IL6ST was upregulated in various cell types from COVID-19 patients and IAV patients compared to healthy controls. It has been reported that IL-6R is often shed from the membrane of activated T cells, whereupon it binds the soluble form of IL-6 and acts in trans upon cells expressing IL6ST. Such IL-6 trans-signaling via IL6ST may contribute significantly to the proinflammatory properties of IL-6 (Wolf et al., 2014), emphasizing the potential importance of IL6ST upregulation observed in the current study.
Based on clinical observations that plasma concentrations of inflammatory cytokines, such as IFN-α2, IL-7, IL-17, and IL-10 were higher in COVID-19 patients than in healthy adults (Huang et al., 2020), we compared expression levels of their corresponding receptors in activated CD4+ T cells, cytotoxic CD8+ T cells, MAIT cells, and NK cells across COVID-19 patients, IAV patients, and healthy controls. (Figure S4E). Upregulation of IFNAR1, the α chain of the interferon α/β receptor, was significant in these cell types in COVID-19 patients compared to healthy controls. Notably, along with IFNAR1, receptor subunits for IL-7, IL-17, and IL-27 were substantially elevated in activated CD4+ T cells of COVID-19 patients compared to other groups, indicating that CD4+ T cells might be involved in major inflammatory response to cytokines.
Finally, we examined temporal changes in their expression (Figures S4B and S4C). Although some IFN-responsive transcription factor genes, such as STAT1 and IRF3, tend to have decreased expression levels over time, none of these patterns were statistically significant due to the limited number of patients. Future studies with more patients and more time points will address this question.
Discussion
Emerging and re-emerging viruses pose a continuous threat to human health (Gao, 2018). The new SARS-CoV-2 virus infection causes severe pulmonary disease and complications with significant morbidities and mortalities. Currently, there is no optimal treatment or effective drug for this fatal lung disease. Our current understanding of the host immune response to SARS-CoV-2 infection is limited, making it difficult to design urgently needed novel therapeutics. Here, we performed scRNA-seq on PBMCs from COVID-19 patients. Overall, the infection of SARS-CoV-2 has little impact on the composition of immune cells in PBMCs. Among all the immune cell clusters, the percentage of plasma cells was increased significantly among all five COVID-19 patients compared with healthy controls. This increase in plasma cells could produce multiple protective neutralizing antibodies. These B-cell-derived antibodies generated in response to SARS-CoV-2 are critical for preventing death from acute respiratory tract infections and providing continued protection from future infection-induced illness and/or death. In addition, blood plasma containing protective neutralizing antibodies from recovered patients is currently being encouraged for use to treat severely affected patients.
IFNs can be induced by viral infection to exert antiviral functions and balance virus control and immune pathology during this process. In SARS-CoV-2-infected ICU patients, plasma IFNγ and TNF-α levels were higher (Huang et al., 2020). IFNα2 and IFNγ have also been strongly associated with lung injury in COVID-19 (Liu et al., 2020). In addition, IFNγ and granulocyte-macrophage colony-stimulating factor (GM-CSF) coexpressing Th1 cells were only found in intensive care unit (ICU) patients infected by SARS-CoV-2 compared to healthy controls (Zhou et al., 2020c). Consistently, “IFN-I response” was enriched in different PBMC subsets of COVID-19 patients in our study. Furthermore, expression of ISGs, including ISG15, IFI44L, MX1, and XAF1, was significantly upregulated in these patients compared to three healthy controls, indicating that strong antiviral functions by IFNs may be triggered (Perng and Lenschow, 2018). Moreover, severe COVID-19 patients showed a stronger response to IFNs and virus infection compared to mild patients and healthy ones. This indicates that the intensity of the interferon response may both indicate the severity of COVID-19 disease and distinguish COVID-19 patients from healthy individuals. Additionally, the dynamics of the IFN response suggest the individual differences and indicate that the timing of IFN therapy against SARS-CoV-2 infection is critical.
According to our results, upregulated XAF1 expression may be involved in increased T cell apoptosis in COVID-19 patients, cooperating with other genes, including IRF1, TP53, BCL2L11, and CASP3 (Jeong et al., 2018; Zou et al., 2012). Consistently, TP53 expression was increased in COVID-19 patients in another COVID-19 study (Xiong et al., 2020). In addition to XAF1-induced apoptosis, the extrinsic pathway of apoptosis, including TNF-α/TNFR1 and Fas/FasL path (Elmore, 2007), were also found to be involved in different cell subtypes in COVID-19 patients. Moreover, plasma TNF-α was reported to be increased in severe cases of COVID-19 (Chen et al., 2020; Huang et al., 2020), and TNF expression was upregulated in PBMCs of COVID-19 patients in our study, demonstrating a correlation between increased TNF-α secretion and TNF-α-induced apoptosis in COVID-19 patients (Rath and Aggarwal, 1999). XAF1 can be induced by TNF-α as well as IFN and functions as an alternative pathway for TNF-α-induced apoptosis (Straszewski-Chavez et al., 2007). XAF1 was also reported to collaborate with TNFSF10 (TRAIL) to promote Dengue virus-induced apoptosis (Zhang et al., 2019). Taken together, we found that the upregulated genes relevant to XAF1, TNF, and Fas pathways may lead to increased T cell apoptosis in COVID-19 patients.
We analyzed the expression of cytokines, cytokines receptors, and transcription factors that are essential in the immune responses to viral infection and pinpointed gene expression patterns in COVID-19 patients that differ from IAV patients. The upregulation of cytokine receptors was in accordance with increased serum cytokine levels, which might enhance the cytokine-mediated inflammatory responses. The fact that expression of most cytokines was not detected in PBMCs might indicate that serum cytokines largely arise from the infection site, i.e., the lower respiratory tract for COVID-19. We found that plasma concentrations of IL-6 are above normal levels in the majority of COVID-19 patients, consistent with other studies (Chen et al., 2020). Elevated IL-6 was also reported in IAV patients (Yu et al., 2011), and it is an immune signature in patients with acute respiratory stress syndrome (Wang et al., 2020c), which is associated with mortality caused by cytokine release syndrome. Currently, treatments blocking IL-6 or IL-6R have been approved for patients with pneumonia and elevated IL-6], and a small-sized study has demonstrated the efficacy of Tocilizumab, a monoclonal antibody against IL-6R, in alleviating clinical symptoms (Xu et al., 2020b). Our data show that expression of IL6R and IL6ST was upregulated in patients with COVID-19, and we suggest that increased expression of IL6R and IL6ST might synergize with elevated IL-6 to induce a strong inflammatory response, indicating that patients might benefit from IL-6 or IL-6R antagonist treatment.
Limitations of the Study
There are important limitations to the interpretation of this study. First, because only a limited number of patients were examined in our study, especially for IAV (two patients), the differences we identified between COVID-19 and IAV during infection need to be further validated by larger clinical trials and/or by further studies. Second, our study focuses on the single-cell transcriptome of PBMCs in blood. If we can combine our data with data from lesion sites such as the lung, our analysis will be more systematic and thus the conclusions would be more comprehensive.
Together, our study visualized the dynamic landscape of immune responses during the disease process in COVID-19 patients compared to IAV patients at the single-cell transcriptome level. These results not only represent the immune molecular and cellular signatures during the clinical process of COVID-19 but also suggest avenues to both diagnostic biomarkers and therapeutic targets, urgently needed for this novel disease.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| COVID-19 patients PBMCs | Fifth Medical Center of PLA General Hospital | N/A |
| IAV patients PBMCs | Beijing Ditan Hospital, Capital Medical University | N/A |
| Healthy control PBMCs | Fifth Medical Center of PLA General Hospital | N/A |
| Critical Commercial Assays | ||
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat#52904 |
| Novel Coronavirus (2019-nCoV) Nucleic Acid Detection kit | BioGerm | Cat#ZC-HX-201-2 |
| DNBelab C Series Single-Cell Library Prep Set | MGI | Cat#1000021082 |
| Qubit™ ssDNA Assay Kit | Thermo Fisher Scientific | Cat#Q10212 |
| Magnetic bead purification | Miltenyi Biotech | Cat#130-090-101 |
| Deposited Data | ||
| Data files for single-cell RNA sequencing (raw data) | This paper | CNP0001102 |
| Data files for single-cell RNA sequencing (processed data) | This paper | CNP0001102 |
| Software and Algorithms | ||
| Cell Ranger Single-Cell Software Suite | 10xgenomics | versions 2.1.0 http://10xgenomics.com/ |
| STAR software(v2.5.3) | Alex Dobin (dobin@cshl.edu), Cold Spring Harbor; Dobin et al., 2013 | N/A |
| Bowtie2 | Ben Langmead, Johns Hopkins University; Langmead and Salzberg, 2012 | N/A |
| Seurat (v3.1) R toolkit | Satija Lab | https://github.com/satijalab/seurat |
| Mfuzz | Kumar and Futschik, 2007 | http://www.bioconductor.org/packages/release/bioc/html/Mfuzz.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, William Jun Liu (liujun@ivdc.chinacdc.cn).
Materials Availability
All unique reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
Raw and processed data are available on CNGB Nucleotide Sequence Archive (CNSA:https://db.cngb.org/cnsa) with accession number CNP0001102. The codes supporting the current study are available from the corresponding authors on request.
Experimental Model and Subject Details
Five patients with COVID-19 were recruited in the Fifth Medical Center of PLA General Hospital from January to February 2020. Patients were divided into mild and severe groups according to the Fifth Revised Trial Version of the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidance. Influenza virus infection, respiratory syncytial virus infection and adenovirus infection were excluded in all patients with COVID-19. Two patients with IAV were recruited in the Beijing Ditan Hospital, Capital Medical University from January to February 2018. Two patients with IAV were diagnosed as having severe infection. The clinical characteristics of these patients are provided in Tables S1 and S2.
Method Details
Ethics statement
The study was approved by the Ethics Committees of the Fifth Medical Center of PLA General Hospital and Beijing Ditan Hospital, Capital Medical University. Written informed consent was obtained from all participants (including one from a teenager’s parents).
Clinical information
Throat swab and blood samples were collected from patients at various time-points after hospitalization. Sample collection, processing, and laboratory testing were conducted complied with WHO guidance. Viral RNA was extracted from throat swabs using the QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer’s instructions. SARS-CoV-2-infected patients were confirmed SARS-CoV-2 Nucleic Acid Detection Kit (BioGerm).
All clinical information including demographic data, medical history, symptoms, signs, and laboratory data were collected from patient medical records. The laboratory data includes blood routine, lymphocyte subsets, infection-related biomarkers, inflammatory cytokines. The total number of leukocytes, percentage of neutrophils and percentage of lymphocytes in peripheral blood were counted using hemocytometer. The number and percentage of lymphocyte subsets were analyzed using the FACSCanto flow cytometer for COVID-19 patients on admission. C-reactive protein and Lactic acid were detected by the Beckman automatic biochemical analyzer. Interleukin 6 (IL-6) was detected using the ROCHE Elecsys IL-6 assay.
Peripheral blood mononuclear cell (PBMC) collection
PBMCs were isolated from EDTA anticoagulated venous blood of COVID-19 patients, IAV patients or healthy donors using a FicollR Paque Plus (Sigma Aldrich) solution according to standard density gradient centrifugation methods. Cells were harvested and counted via Cellaca MX high-throughput cell counter (Nexcelom Bioscience). The PBMCs were resuspended in 90% fetal bovine serum (FBS, HyClone), 10% DMSO freezing media and frozen using a Nalgene® Mr. Frosty Cryo 1°C Freezing Container (Thermo Fisher Scientific) in a −80°C freezer for 24 h before being transferred to liquid nitrogen for long-term storage.
Single-cell suspension preparation
Frozen vials of PBMCs were rapidly thawed in a 37°C water bath for ∼2 min, and the vials were removed when a tiny ice crystal was left. Thawed PBMCs were quenched with 4 mL 37°C pre-warmed 1X phosphate-buffered saline (PBS, Thermo Fisher Scientific) and supplemented with 10% FBS. Cells were centrifuged at 500 x g for 10 min at room temperature each time. The supernatant was removed, and the cell pellet was resuspended in 3 mL 1X PBS containing 0.04% bovine serum albumin (BSA, Sangon Biotech), passed through a 40 μm cell strainer (Falcon), and then centrifuged. Dead cells were removed by magnetic bead purification (Miltenyi Biotech) according to the manufacturer’s protocol before scRNA-seq was performed. Cells were resuspended with cell resuspension buffer at a viable cell concentration of 1,000 cells/μL.
Single-cell RNA-seq with DNBelab C4 system
The DNBelab C Series Single-Cell Library Prep Set (MGI) was utilized as previously described (Liu et al., 2019) for single-cell RNA-seq library preparation. In brief, the single-cell suspensions were converted to barcoded scRNA-seq libraries through steps including droplet encapsulation, emulsion breakage, mRNA captured bead collection, reverse transcription, cDNA amplification and purification. Indexed sequencing libraries were constructed according to the manufacturer’s protocol. The sequencing libraries were quantified by Qubit ssDNA Assay Kit (Thermo Fisher Scientific). The sequencing libraries were sequenced by the DIPSEQ T1 sequencer at China National GeneBank (CNGB). The read structure was paired-end with Read 1, covering 30 bases inclusive of 10-bp cell barcode 1, 10-bp cell barcode 2 and 10-bp unique molecular identifier (UMI), and Read 2 containing 100 bases of transcript sequence, and 10-bp sample index.
Quantification and Statistical Analysis
Single-cell RNA-seq data processing (Alignment, Barcode Assignment, and UMI Counting)
The raw FASTQ files were transformed using custom Perl scripts into Cell Ranger-specific FASTQ files. These FASTQ files were then processed individually using a modified version of Cell Ranger count pipeline, which made use of the STAR software (v2.5.3) (Dobin et al., 2013) to align cDNA reads to the GRCh38.p5 human reference genome. Aligned reads were then filtered for valid cell barcodes and UMIs to generate gene-cell matrices for downstream analysis.
Detection of SARS-CoV-2 transcript
The genome of SARS-CoV-2 was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) with the accession number GenBank:NC_045512.2. The raw sequencing reads were then aligned to the genome by STAR (v2.5.3) (Dobin et al., 2013) and Bowtie2 (Langmead and Salzberg, 2012) using a modified annotation GTF file.
Unsupervised clustering
Cell clustering was performed by Seurat (v3.1) R toolkit (Butler et al., 2018) (https://github.com/satijalab/seurat). Genes expressed in less than 3 cells were filtered out and cells with less than 300 or more than 6000 genes detected were excluded. In order to deal with the batch effect, the “NormalizeData” and “FindVariableGene” functions were performed respectively for each sample (n = 23). After that, these 23 batches were integrated together using “FindIntergrationAnchors” and “IntegrateData” function with dims parameter set to 20. Then, the integrated dataset was scaled and PCA was calculated. The first 20 PCs were used to construct a SNN network and a graph-based clustering approach, louvain algorithm, was applied to identify cell clusters with the resolution set to 1. Finally, UMAP was applied to visualize the clustering result in 2D space. To further regress out the influence of red blood cells, we excluded this cluster of cells as well as 10 hemoglobin genes in the expression matrix (HBA1,HBA2, HBB, HBG1, HBG2, HBQ1, HBD, HBM, HBE1 and HBZ), and performed clustering again using the same method described above.
Cluster marker analysis and cell type annotation
To annotate each cluster to a specific immune cell type, we selected some classic markers for immune cells and used violin plots (Figure 1D) and UMAP feature plots (Figure S1) to annotate cell types. The following genes were used for cell type annotation: CD3G, CD4, CD8A, CD8B (T cells); KLRF1, XCL1 (NKs); MS4A1 (B cells); CD27 (Memory B cells) IGHG1, MZB1 (Plasma cells); CD68 (Monocytes); CD1C, LYZ (DCs); MKI67, TOP2A (Cycling cells); GZMA (Cytotoxic CD8+ T cells / NKs); CCR7, SELL (Naive T cells); PPBP (Megakaryocytes); CD34 (Stem cells).
Differential expressed genes (DEGs) analysis and GO enrichment
We performed DEG analysis using the “FindMarkers” function in Seurat R package. The cell populations we compared were input as ident.1 and ident.2, respectively. The fold changes of the mean expression level of genes between the selected cell populations were calculated. To find the function of upregulated genes (adjusted p < 0.01, Foldchange > 2), we used the function compareCluster (fun = "enrichGO,” pvalueCutoff = 0.1, pAdjustMethod = "BH,” OrgDb = "org.Hs.eg.db,ontBP”) of ChIPseeker R package (v.1.22.1) (Yu et al., 2015).
Identifying time dependent transcriptional program in COVID-19 patients
We used Mfuzz (Kumar and Futschik, 2007) to identify time dependent transcriptional program in COVID-19 patients. The samples from COVID-19 patients were grouped according to the disease progression stage (Table S3). First, the average expression of each gene was calculated for each stage. Next, we added 0.000001 for every gene to avoid 0 in the expression matrix which was not acceptable for Mfuzz. Then, the “filter.std(min.std=0),” “standardize()” and “mestimate()” functions were performed for preprocessing according to the tutorial. After that, we clustered the genes into 6 different expression programs (Figure 3D). For the GO analysis of the 6 programs, we first excluded genes with the maximum expression less than 1. Only GO terms with adjusted P value < 0.05 were shown in the figure (Figure 3E).
Boxplot
All of the boxplots in this paper were performed using “ggboxplot()” function in ggpubr R package. Each point represents for one sample. The sample number of each group was as follow: Healthy donors (Ctrl) (n = 3), COVID-19 patients (COV) (n = 16), IAV patients (IAV) (n = 4). The horizontal line within each box represents the median, and the top and bottom of each box indicate the 75th and 25th percentile. Student’s t test was applied to test the significance of the difference using “stat_compare_means()” function.
Quantification and Statistical Analysis
Statistical analysis was performed using R (version 3.6.1). Wilcoxon rank-sum test and Student’s t test were used in this study. ∗ p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Acknowledgments
We thank Yuxuan Liu (BGI) and Bo Li (BGI) for assisting the scRNA-seq library preparation. We also thank Dr. Andres Keller (Saarland University, Germany) for helpful discussion and evaluation of the DNBelab C4 technology. The study was supported by CAMS Research Units of Adaptive Evolution and Control of Emerging Viruses (2018RU009), National Mega-projects for Infectious Diseases (2020ZX10001016-005-001), and Beijing New Star Plan of Science and Technology (Z181100006218080). W.J.L. is supported by the Excellent Young Scientist Program of the National Natural Science Foundation of China (81822040) and the National Youth Talent Support Program. The study was supported by the National Natural Science Foundation of China (81971564); the Science, Technology and Innovation Commission of Shenzhen Municipality (JSGG20180508152912700); and Shenzhen Key Laboratory of Single-Cell Omics (ZDSYS20190902093613831). Editing support was received from Life Sciences Editors.
Author Contributions
W.J.L., L.L, G.F.G., and F.-S.W. designed and supervised the study. W.J.L., L.L., F.-S.W., and G.F.G. conceived the project. C.B., P.Y., Y.Z., R.S., N.Z., G.L., L.Y., J.Y., X. Wang., Y.L., F.M., P.M., S.Z., Y.S., Z.X., and G.D. collected clinical samples and analyzed the clinical and treatment data. Z.W., C.L., Y. Yu, M.W., Y. Yuan, Q.D., Y.W., G.H., Y.Z., B.Y., and J.Z. performed the experiments. L.Z., Z.Z., Q.G., Q.X., X. Wu, H.-X.S., Y.Z., L.L., and W.J.L. analyzed the data. P.L., Y.B., Y.S., G.W., and Z.C. contributed to fruitful discussions and key ideas. L.Z., Q.G., Q.X., H.-X.S., Z.L., L.L, Y.Z., W.J.L, F.-S.W., and G.F.G. wrote the manuscript. J.W., X.X., L.L, Y.Z., W.J.L., and G.F.G. participated in the manuscript editing and discussion.
Declaration of Interests
Employees of BGI have stock holdings in BGI.
Published: July 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.07.009.
Supplemental Information
References
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P., Tahiliani V., Hutchinson T.E., Dastmalchi F., Stanfield J., Abboud G., Thomas P.G., Ware C.F., Song J., Croft M., Salek-Ardakani S. The TNF superfamily molecule LIGHT promotes the generation of circulating and lung-resident memory CD8 T cells following an acute respiratory virus infection. J. Immunol. 2018;200:2894–2904. doi: 10.4049/jimmunol.1701499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H., Hao Q., Idell S., Tang H. Transcription factor Runx3 is induced by influenza A virus and double-strand RNA and mediates airway epithelial cell apoptosis. Sci. Rep. 2015;5:17916. doi: 10.1038/srep17916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.F. From “A”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Guo J., Zou Z., Liu J., Cao B., Zhang S., Li H., Wang W., Sheng M., Liu S. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat. Commun. 2014;5:3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.I., Kim J.W., Ko K.P., Ryu B.K., Lee M.G., Kim H.J., Chi S.G. XAF1 forms a positive feedback loop with IRF-1 to drive apoptotic stress response and suppress tumorigenesis. Cell Death Dis. 2018;9:806. doi: 10.1038/s41419-018-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Futschik M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu T., Fan F., Liu Y., Wu L., Junkin M., Wang Z., Yu Y., Wang W., Wei W. A portable and cost-effective microfluidic system for massively parallel single cell transcriptome profiling. bioRxiv. 2019 doi: 10.1101/818450. [DOI] [Google Scholar]
- Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020;7:1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., Gao G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S., Planz O. Influenza viruses and the NF-kappaB signaling pathway - towards a novel concept of antiviral therapy. Biol. Chem. 2008;389:1307–1312. doi: 10.1515/BC.2008.148. [DOI] [PubMed] [Google Scholar]
- Mihara M., Hashizume M., Yoshida H., Suzuki M., Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. (Lond.) 2012;122:143–159. doi: 10.1042/CS20110340. [DOI] [PubMed] [Google Scholar]
- Ochiai K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., Sciammas R. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y.-C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju S., Kometani K., Kurosaki T., Shaw A.S., Egawa T. The adaptor molecule CD2AP in CD4 T cells modulates differentiation of follicular helper T cells during chronic LCMV infection. PLoS Pathog. 2018;14:e1007053. doi: 10.1371/journal.ppat.1007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath P.C., Aggarwal B.B. TNF-induced signaling in apoptosis. J. Clin. Immunol. 1999;19:350–364. doi: 10.1023/a:1020546615229. [DOI] [PubMed] [Google Scholar]
- Rolfes M.A., Foppa I.M., Garg S., Flannery B., Brammer L., Singleton J.A., Burns E., Jernigan D., Olsen S.J., Bresee J., Reed C. Annual estimates of the burden of seasonal influenza in the United States: A tool for strengthening influenza surveillance and preparedness. Influenza Other Respir. Viruses. 2018;12:132–137. doi: 10.1111/irv.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straszewski-Chavez S.L., Visintin I.P., Karassina N., Los G., Liston P., Halaban R., Fadiel A., Mor G. XAF1 mediates tumor necrosis factor-alpha-induced apoptosis and X-linked inhibitor of apoptosis cleavage by acting through the mitochondrial pathway. J. Biol. Chem. 2007;282:13059–13072. doi: 10.1074/jbc.M609038200. [DOI] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical Features of 69 Cases with Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J., Rose-John S., Garbers C. Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine. 2014;70:11–20. doi: 10.1016/j.cyto.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang X., Zhao B., Wang J., Zhu Z., Teng Z., Shao J., Shen J., Gao Y., Yuan Z., Wu F. Intensive cytokine induction in pandemic H1N1 influenza virus infection accompanied by robust production of IL-10 and IL-6. PLoS ONE. 2011;6:e28680. doi: 10.1371/journal.pone.0028680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhang F., Chen D., Yang W., Duan S., Chen Y. Combined effects of XAF1 and TRAIL on the apoptosis of lung adenocarcinoma cells. Exp. Ther. Med. 2019;17:4663–4669. doi: 10.3892/etm.2019.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Zhang H., Liu K., Gao G.F., Liu W.J. Human T-cell immunity against the emerging and re-emerging viruses. Sci. China Life Sci. 2017;60:1307–1316. doi: 10.1007/s11427-017-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B., Chim C.S., Pang R., Zeng H., Dai Y., Zhang R., Lam C.S., Tan V.P., Hung I.F., Lan H.Y., Wong B.C. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol. Carcinog. 2012;51:422–432. doi: 10.1002/mc.20807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data are available on CNGB Nucleotide Sequence Archive (CNSA:https://db.cngb.org/cnsa) with accession number CNP0001102. The codes supporting the current study are available from the corresponding authors on request.