Abstract
Viruses are associated with several human diseases that infect a large number of individuals, hence directly affecting global health and economy. Owing to the lack of efficient vaccines, antiviral therapy and emerging resistance strains, many viruses are considered as a potential threat to public health. Therefore, researches have been developed to identify new drug candidates for future treatments. Among them, antiviral research based on natural molecules is a promising approach. Phospholipases A2 (PLA2s) isolated from snake venom have shown significant antiviral activity against some viruses such as Dengue virus, Human Immunodeficiency virus, Hepatitis C virus and Yellow fever virus, and have emerged as an attractive alternative strategy for the development of novel antiviral therapy. Thus, this review provides an overview of remarkable findings involving PLA2s from snake venom that possess antiviral activity, and discusses the mechanisms of action mediated by PLA2s against different stages of virus replication cycle. Additionally, molecular docking simulations were performed by interacting between phospholipids from Dengue virus envelope and PLA2s from Bothrops asper snake venom. Studies on snake venom PLA2s highlight the potential use of these proteins for the development of broad-spectrum antiviral drugs.
Keywords: Snake venom, Phospholipases A2, Virus, Antiviral drugs
1. Viral diseases: a public health problem
Viruses are associated to several endemic diseases, including Enterovirus [1], HPV (Human papillomavirus) [2], HIV (Human immunodeficiency virus) [3] and HSV (Herpes simplex virus) [4], as well as in outbreaks as Ebola virus, ZIKV (Zika virus), Influenza virus, YFV (Yellow Fever virus), DENV (Dengue virus) and, currently, the SARS-CoV-2 (Severe Acute Respiratory Coronavirus 2) [[5], [6], [7], [8], [9]]. Most of the reported outbreaks since 1980 were related to virus infections [10], which are still a global burden for public health and economy. In addition, due to their genetic diversity, viruses are able to infect a wide range of hosts that can result in host jumps after zoonotic contacts [11,12].
Pandemics caused by viruses are usually severe and can claim up to million lives, as shown during the pandemic of H1N1 in 1918 [13], H1N1 ‘swine flu’ in 2009 [14] and Coronavirus Disease 2019 (COVID-19) [15], that infected 4,993,470 people and caused 327,738 deaths until May 22, 2020 in worldwide according to World Health Organization. Furthermore, the global incidence of dengue has grown dramatically in recent decades. It is estimated that 100 to 400 million cases of dengue occur annually worldwide [16].
Viral infection depends on the successful replication into the host cells [17,18]. In general, the replicative cycle starts by the viral particle attaching to specific receptors in the surface of host cells that triggers the viral entry by endocytosis (non-enveloped or enveloped virus), membrane fusion (enveloped virus) and direct penetration [[19], [20], [21]]. After internalization, the capsid is released into the cytoplasm, allowing viral genome uncoating [22], which is replicated to produce copies of the genome and translated to viral proteins. In the endoplasmic reticulum (ER) and Golgi complex, the viral structure is assembled, matured and then forwarded to host cell membrane, where the progeny of virus particles is released [23].
Currently, specific antiviral drugs and vaccines are not sufficient to control emerging and reemerging viral diseases [24,25]. Thus, the discovery of novel antiviral drugs is mandatory. In general, antiviral therapy is the only approach to specifically treat viral infections, abrogating viral replicative cycle [26]. However, due to the high genetic variability, viruses can rapidly acquire resistance to antiviral treatment, especially RNA viruses [[27], [28], [29]]. Furthermore, antiviral therapy and the prolonged treatment can cause several adverse effects, including gastrointestinal effects, fatigue, headache, neuropathy and liver toxicity [[30], [31], [32]]. In addition, there are no antivirals to all diseases and the only course is supportive therapy and, thereby, numerous innovative drugs have been developed from natural prototypes such as aspirin (anti-inflammatory) and morphine (analgesic) [33].
In this way, a diversity of compounds isolated from natural sources has set grounds for further advances in drug development against various diseases [34]. Among them, many drugs based on snake venoms were approved by the FDA or are involved in preclinical or clinical trials for a variety of therapeutic applications [[35], [36], [37], [38], [39]]. The development of snake venom-derived drugs gained a significant improve since the discovery of bradykinin-potentiating peptides (BPP) isolated from the Brazilian arrowhead viper (Bothrops jararaca) venom, which allowed the development of captopril, an inhibitor of the angiotensin-converting enzyme that is widely used against hypertensive process [40,41]. Besides that, other snake venom-derived drugs have been found in clinical use, such as tirofiban and eptifibatide (antiplatelet agents) [42,43], batroxobin, moojenin and vivostat (anticoagulant agents) [[44], [45], [46], [47]]. Other drugs which comprise molecules from snake venom as scaffolds are also being explored in preclinical studies [48].
Due to this therapeutic potential, snake venom toxins have been widely explored for the discovery of new bioactive compounds and stand out as an alternative source for therapeutics for a variety of diseases, including life-threatening viral illnesses [[49], [50], [51], [52]].
2. Phospholipases A2 from snake venom
Phospholipases (EC 3.1.) family is widely distributed in nature and includes hydrolase enzymes, which are essential for phospholipid metabolism and for the regulation of membrane lipids, membrane composition, signaling, digestion, and inflammation [53]. These proteins are classified into four major families (A, B, C and D) based on the site cleaved in the phospholipid molecule [54,55].
Among phospholipases family, the Phospholipases A2 (PLA2s) are the most studied group [53,54]. These enzymes hydrolyze 2-acyl ester bond to 2-sn phospholipids, releasing free fatty acid and lysophospholipids [56,57]. The free fatty acids (arachidonic acid) can be converted into eicosanoids (prostaglandins, thromboxanes, prostacyclins and leukotrienes), which are associated to a range of physiological and pathological effects, such as inflammation and platelet activation. In addition, the lysophospholipids are also related to a variety of physiological roles in cell signaling [53,58].
PLA2s are classified into six groups: cytosolic (cPLA2), Ca(2+)-independent (iPLA2), platelet-activating factor acetylhydrolase (PAF-AH), lysosomal PLA2 (LyPLA2), adipose specific PLA2 (AdPLA2) and secretory PLA2 (sPLA2) [53]. In addition, the sPLA2s are divided into the following groups: IA, IB, IIA, IIB, IIC, IID, IIE, IIF, III, V, IX, X, XIA, XIB, XII, XIII and XIV [53,54]. PLA2s from snake venom belong to the group of secreted type of enzymes (sPLA2s) and can be classified into the structural group IB (in Elapidae snake venoms), which exhibits homology to the mammalian pancreatic juice PLA2, and also into the group IIA (in Viperidae snake venoms), that is homologous to the mammalian ‘inflammatory’ PLA2 [59]. Although the PLA2s family is more frequent in snake venom, recent proteome studies have demonstrated that Phospholipases B (PLB) can also be found in snake venom [[60], [61], [62]].
sPLA2s are proteins with molecular mass of about 14 kDa, pH optimum at 7 and share a conserved catalytic mechanism based on a His/Asp dyad using Ca2+ as an essential cofactor for the catalytic activity. Group II of sPLA2s presents an extended C-terminal segment (5–7 amino acids) [63,64] and is subdivided into two main subgroups, depending on the amino acid residue at position 49 in the protein primary structure. Aspartate (Asp49 or D49) sPLA2s are enzymatically active, while lysine (Lys49 or K49) sPLA2s present no enzymatic activity [[65], [66], [67]]. However, there are further variants, as the serine (Ser49), asparagine (Asn49) or arginine (Arg49) [[68], [69], [70]].
Lys49 PLA2s are devoid of catalytic activity due to their inability to bind Ca2+, a key cofactor for PLA2 activity. Although the lack of enzymatic activity, the Lys49 PLA2 homologues have shown to display toxicity, especially myotoxicity [67]. The toxicity of Lys49 proteins can be related to a cluster of cationic and hydrophobic/aromatic amino acid residues located at the C-terminal region of these toxins [71,72].
Therefore, the cytotoxicity of sPLA2 is probably mediated by the interaction between the C-terminal region and the plasma membrane [73,74]. Moreover, the PLA2 effects can be mediated through the integrins and other receptors, such as vascular endothelial growth factor receptor-2 (VEGFR-2), and M–Type receptors [[75], [76], [77]]. Recently, it was demonstrated that the cell surface nucleolin interacts with and internalizes the PLA2-like Bothrops asper myotoxin-II, which is responsible to mediate its toxic activity [78].
sPLA2s from snake venom can act on cell membranes of specific tissues inducing several pharmacological actions such as myotoxicity, neurotoxicity, cardiotoxicity, platelet aggregation activation or inhibition, hypotension, edema among others [57,79]. In this scenario, these proteins have emerged as a potential therapeutic model, since numerous studies have focused on their microbicidal [80], antitumor [[81], [82], [83]], antiangiogenesis [84], antiparasitic [[85], [86], [87]] and antiviral activities [51].
The development of efficient antiviral therapies has become a global health emergency. In this sense, several researches have demonstrated the antiviral activity of sPLA2s from snake venom against human viruses, including DENV, YFV, HCV and others [[88], [89], [90], [91], [92]]. Hence, the present review aimed to summarize the sPLA2s from snake venom that were previously described to possess antiviral activity, highlighting the mechanisms of action of sPLA2s against different stages of virus replication cycle (Table 1 ).
Table 1.
sPLA2s from snake venom with antiviral effects.
| Source/species | Protein name | EC50/used dosage | Virus | Proposed action mechanism (inhibition) | Reference |
|---|---|---|---|---|---|
| Bothrops asper | Mt–I | 50 μg/mL | DENV-1, 2, 3 | Entry (virucidal activity) |
[92] |
| 50 μg/mL | YFV | ||||
| 1.5 ng/mL (EC50) | DENV-2 | ||||
| Mt–II | 50 μg/mL | DENV-1, 2, 3 | |||
| 50 μg/mL | YFV | ||||
| 2768 ng/mL (EC50) | DENV-2 | ||||
| Bothrops jararacussu | BthTX-I | 4.8 ng/μL (EC50) | DENV-2 | Entry (virucidal activity) |
[88] |
| 7.063 ng/μL (EC50) | YFV | ||||
| 57.3 ng/μL (EC50) | DENV-2 | Entry (interfering in adsorption) |
|||
| 25.0 ng/μL (EC50) | YFV | ||||
| 69.0 ng/μL (EC50) | DENV-2 | Entry (interfering in internalization) |
|||
| 23.4 ng/μL (EC50) | YFV | ||||
| Bothrops leucurus | BlK-PLA2 | 20 μg/mL | DENV-1, 2 3 | Replication (interfering in host cell components) |
[91] |
| BlD-PLA2 | 20 μg/mL | ||||
| Crotalus durissus terrificus | Crotoxin | 0.001 ng/μL (EC50) | DENV-2 | Entry (virucidal activity) |
[[88], [89], [90],97] |
| 0.00045 ng/μL (EC50) | YFV | ||||
| 0.0046 ng/μL (EC50) | ROCV | ||||
| 0.0036 ng/μL (EC50) | MAYV | ||||
| 0.0054 ng/μL (EC50) | OROV | ||||
| – | HIV-1,2 | ||||
| 10 μg/mL | HCV | ||||
| 0.018 ng/μL (EC50) | DENV-2 | Entry (interfering in adsorption) |
|||
| 0.0365 ng/μL (EC50) | YFV | ||||
| 34.4 ng/μL (EC50) | DENV-2 | Entry (interfering in internalization) |
|||
| 13.7 ng/μL (EC50) | YFV | ||||
| 0.05 ng/μL (EC50) | DENV-2 | Replication (interfering in host cell components) |
|||
| 0.04 ng/μL (EC50) | YFV | ||||
| 10 μg/mL | HCV | Release | |||
| PLA2-CB (subunit of crotoxin) | 0.00003 ng/μL (EC50) | DENV-2 | Entry (virucidal activity) |
[[88], [89], [90]] | |
| 0.0037 ng/μL (EC50) | YFV | ||||
| 0.021 ng/μL (EC50) | ROCV | ||||
| 0.066 ng/μL (EC50) | MAYV | ||||
| 0.0067 ng/μL (EC50) | OROV | ||||
| 10 μg/mL | HCV | ||||
| 0.044 ng/μL (EC50) | DENV-2 | Entry (interfering in adsorption) |
|||
| 0.01647 ng/μL (EC50) | YFV | ||||
| 17.2 ng/μL (EC50) | DENV-2 | Entry (interfering in internalization) |
|||
| 3.3 ng/μL (EC50) | YFV | ||||
| 10 μg/mL | HCV | ||||
| 10 μg/mL | HCV | Entry (interfering in host cell components) |
|||
| 0.06 ng/μL (EC50) | DENV-2 | Replication (interfering in host cell components) |
|||
| 0.26 ng/μL (EC50) | YFV | ||||
| 6.08 μg/mL (EC50) | HCV | ||||
| PLA2-IC | 0.0137 ng/μL (EC50) | DENV-2 | Entry (virucidal activity) |
[88] | |
| 0.0054 ng/μL (EC50) | YFV | ||||
| 0.133 ng/μL (EC50) | DENV-2 | Entry (interfering in adsorption) |
|||
| 0.268 ng/μL (EC50) | YFV | ||||
| 21.6 ng/μL (EC50) | DENV-2 | Entry (interfering in internalization) | |||
| 0.775 ng/μL (EC50) | DENV-2 | Replication (interfering in host cell components) |
|||
| 1.30 ng/μL (EC50) | YFV | ||||
| Naja mossambica mossambica | CM-II-sPLA2 | 0.036 ng/mL (EC50) | HCV | Entry (virucidal activity) |
[114] |
| 0.031 ng/mL (EC50) | DENV | ||||
| 1.34 ng/mL (EC50) | JEV | ||||
| 10,000 ng/mL (EC50) | MERS-CoV | ||||
| >10,000 ng/mL (EC50) | SINV | ||||
| >10,000 ng/mL (EC50) | FLUAV | ||||
| >10,000 ng/mL (EC50) | SeV | ||||
| 2300 ng/mL (EC50) | VSNJV | ||||
| 5.4 ng/mL (EC50) | HIV-1 | ||||
| >10,000 ng/mL (EC50) | HSV-1 | ||||
| >10,000 ng/mL (EC50) | CV-B3 | ||||
| >10,000 ng/mL (EC50) | EMCV | ||||
| NmmCMIII | 0.4 nM (EC50) | HIV-1 isolates | Entry (interfering in host cell components) |
[107] | |
| Naja nigricollis | Nigexine | 0.4 nM (EC50) | HIV-1 isolates | ||
| Oxyuranus scutellatus | Taipoxin | 0.8 nM (EC50) | HIV-1 isolates |
CV-B3 (Coxsackievirus B3; Picornaviridae); DENV (Dengue virus); EMCV (Encephalomyocarditis virus; Picornaviridae); FLUAV (Influenza A virus); HCV (Hepatitis C virus); HIV (Human immunodeficiency virus); HSV (Herpes simplex virus); JEV (Japanese encephalitis virus); MAYV (Mayaro virus); MERS-CoV (Middle East respiratory syndrome coronavirus); OROV (Oropouche virus); ROCV (Rocio virus); SeV (Sendai virus); SINV (Sindbis virus); VSNJV (Vesicular stomatitis New Jersey virus); YFV (Yellow Fever virus).
3. sPLA2s from snake venom with antiviral effects
3.1. Crotoxin, PLA2-CB (basic chain of crotoxin) and PLA2-IC from Crotalus durissus terrificus venom
The venom of Crotalus durissus terrificus (C. d. terrificus), a South American rattlesnake, is composed by a large number of molecules with biological activities, such as crotoxin, crotamin, PLA2 “inter-cro” (PLA2-IC), convulxin and gyroxin [93,94]. Crotoxin, which comprehends more than a half of the dry weight of C. d. terrificus venom, is a heterodimeric compound composed by the PLA2-CB (a basic phospholipase component) and crotapotin (an acidic nontoxic catalytically inactive protein) [95,96]. Villarrubia and coworkers [97] reported that crotoxin has anti-HIV (HIV-1, 2) effect by a direct interaction with Gag p24 glycoprotein on the viral surface, which appears to abrogate the HIV anchoring to host cell.
Furthermore, Muller and colleagues [88] working with diverse sPLA2s isolated from C. d. terrificus venom explored different approaches to unveil the potent antiviral activity mediated by crotoxin, PLA2-CB and PLA2-IC against DENV-2 and YFV (enveloped virus). The authors demonstrated that all investigated sPLA2s promoted a significant inhibition of DENV-2 and YFV entry into VERO E6 cells by a direct action on the viral particles (virucidal activity), and by interfering in the adsorption and internalization steps (early stages of the viral replication cycle) [88]. Besides that, cell pretreatment with three sPLA2s was able to protect host cell against flaviviruses infection after 7 days by the reduction in the number of plaque formation. Interestingly, sPLA2s treatment after viral infection promoted an enhancement of load viral, indicating that antiviral effect occurs in the early stages of viral infection [88]. In addition, the researchers gained insights into the role of catalytic sites of the tested sPLA2s, proposing the use of a sPLA2 without catalytic activity (BthTX-I) isolated from Bothrops jararacussu [98].
BthTX-I revealed antiviral activity against YFV and DENV-2 in the virucidal, adsorption and internalization assays. Interestingly, as shown to other catalytically-active sPLA2s at 100 ng/μL, BthTX-I at the same concentration was also able to inhibit YFV entry by virucidal activity (100%), interfering in adsorption (77%) and internalization (78%) [88]. Although BthTX-I showed antiviral activity, the effective concentration 50% (EC50) values obtained for this toxin were extremely higher when compared to the catalytically-active sPLA2s. For example, for the half-maximum virucidal activity against YFV, this toxin required 7.063 ng/μL, while crotoxin, PLA2-CB and PLA2-IC demanded 0.00045, 0.0037 and 0.0054 ng/μL, respectively. In a similar way against DENV-2, BthTX-I acted at 4.8 ng/μL, in contrast to the crotoxin, PLA2-CB and PLA2-IC that required 0.001, 0.00003 and 0.0137 ng/μL, respectively. As shown, the huge differences of EC50 values between BthTX-I and enzymatically active proteins reflect that the enzymatic activity is an important factor for the antiviral activity of sPLA2s [88].
In a further study, PLA2-CB and crotoxin inhibited virus entry by virucidal activity against other enveloped viruses, such as Rocio virus (ROCV; Flaviviridae family), Oropouche virus (OROV; Bunyaviridae family), and Mayaro virus (MAYV; Togaviridae family). However, these compounds did not show virucidal effect against Coxsackie B5 virus (CV-B5; Picornaviridae family; non-enveloped virus), hence suggesting that the possible antiviral action occurs upon the lipid bilayer viral envelope [89]. To corroborate these findings, it was demonstrated that preincubating DENV-2 with PLA2-CB or crotoxin resulted in an increase of exposure and degradation of viral RNA [89]. Also, Russo and collaborators [99] expressed two recombinant PLA2-CB isoforms through a prokaryotic system and noted that both rPLA2-CB1 and rPLA2-CB2 maintained the viral inhibitory activity against CHIKV, DENV-2, YFV and ZIKV when compared to the native sPLA2-CB. Additionally, Muller and colleagues [88,89] suggested that the mechanism of action of PLA2-CB isolated from C. t. terrificus against DENV can occur through an interaction with components on the host cell surface or mainly due to the glycerophospholipid cleavage on the virus envelope, destabilizing viral E proteins and resulting in the viral envelope disruption and RNA viral exposure before the infection of host cells.
In order to gain insights into the antiviral mechanism of sPLA2s obtained from C. t. terrificus, Shimizu and colleagues [90] showed that PLA2-CB inhibited HCVcc JFH-1 virus strain entry and replication in Huh 7.5 cells, and crotoxin blocked virus entry and release, suggesting that these proteins possess multiple antiviral effects against HCV. Moreover, the authors also reported that PLA2-CB significantly decrease the levels of lipid droplets, which are essential for the HCV replication complex, and reduced the levels of HCV NS5A protein due to the replication inhibition, evidencing that besides the action on virus entry, PLA2-CB is able to disrupt HCV replication probably by an interference in lipid metabolism of host cell [90,100,101].
3.2. BlK-PLA2 and BlD-PLA2 from Bothrops leucurus venom
Both BlK-PLA2 (Lys49 sPLA2s) and BlD-PLA2 (Asp49 sPLA2s) are two basic sPLA2s isolated from Bothrops leucurus venom, a pit viper (white-tailed-jararaca) commonly found in the northeast of Brazil [102]. Cecilio and coworkers [91] showed that the pretreatment of LLC-MK2 cells (Rhesus Monkey Kidney Epithelial cells) with each isoform of Bl-PLA2 followed by viral infection was able to inhibit DENV infectivity (serotypes 1, 2 and 3), measured by qRT-PCR quantification of the DENV viral load in the cell supernatants after virus infection. On the other hand, Bl-PLA2s treatment after viral entry was not capable of inhibiting viral replication, then suggesting that the antiviral effect occurs upon components on the surface of the host cell membrane. The authors did not assess the potential virucidal mechanism of Bl-PLA2s against DENV. However, they suggested that the possible mechanism of action of Bl-PLA2s does not depend exclusively on their catalytic site. The Lys49-BlK-PLA2 treatment was able to interfere in the viral load, indicating that the functional effect mediated by Bl-PLA2s also may occur due to the presence of pharmacological domains on the enzyme surface that would allow the interaction with host cell proteins, as well as the enzymatic activity [91]. The authors hypothesized that the DENV RNA level reduction is mediated by the intracellular action of Bl-PLA2s due to the higher penetrability capacity of basic sPLA2s, in comparison to neutral and acidic enzymes [91,103].
3.3. Mt-I and Mt-II from Bothrops asper venom
Bothrops asper is a viperid specie found in Central America and its venom contains significant concentrations of acid and basic sPLA2 enzymes [104]. The B. asper venom has both the basic enzymatically-active sPLA2 (Mt-I) and the catalytically-inactive sPLA2-like protein (Mt-II) [74].
Brenes and collaborators [92] investigated the antiviral potential triggered by both Mt-I and Mt-II isoforms isolated from B. asper venom. The authors showed that these sPLA2s at concentration of 50 μg/mL completely blocked virus entry by a virucidal action against members of Flaviviridae family, such as DENV and YFV, while exhibited moderate to negligible effects against other enveloped viruses (HSV-1, HSV-2, Influenza A H3N2 and Vesicular stomatitis VSV) or non-enveloped viruses (Sabin Poliovirus 1, 2 and 3). Interestingly, for the half-maximum virucidal activity against DENV-2, Mt-I required 1.5 ng/mL, while Mt-II acted at 2768 ng/mL, revealing that Mt-I is extremely more potent than Mt-II [92]. Investigating the role of the enzymatic activity in the inhibitory effect upon DENV-2, it was promoted the inactivation of the catalytic activity of Mt-I with p-bromophenacyl bromide (pBPB). The data showed that the chemical inactivation of Mt-I resulted in a reduction of the virucidal potency, indicating the relevant role of the enzymatic action against viral infection [92]. Even without enzymatic activity, the C-terminal region of Mt-II, which encompasses the amino acid residues 115–129, is responsible for the membrane-permeabilizing effect caused in many cellular types [67], as well as its bactericidal activity [105]. Notwithstanding that, the authors demonstrated that, even at high concentrations, the synthetic peptide “p115” corresponding to the C- terminal region of Mt-II (amino acid residues 115–129) did not inhibit DENV-2 [92]. Thus, the authors speculate that the weak virucidal effect of Mt-II may be intrinsic or more possible related to a trace contamination with Mt-I, where the total chromatographic separation for these toxins is hardly achieved [92].
In addition, it was suggested that Mt-I acts by a direct virucidal mechanism that depends on its enzymatic activity, which may hydrolyze viral envelope phospholipids and disrupt the viral envelope of flaviviruses leading to the impairment of the infection. Also, the mode of action of Mt-I and Mt-II is not related to an effect on host cell, since cell treatment after infection did not interfere in viral replication [92]. Furthermore, in a pretreatment assay, it was demonstrated a partial reduction of viral plaques, that may be explained by a slight cytotoxic action of Mt-I on cells [92]. Finally, the higher antiviral activity of Mt-I against Flaviviridae viruses in comparison to other enveloped virus families may be related to the specific structural organization, physicochemical composition, curvature and fluidity of viral envelope from flaviviruses, which may positively affect the catalytic activity of Mt-I against this family [106].
3.4. Taipoxin (Oxyuranus scutellatus), nigexine (Naja nigricollis) and NmmCMIII (Naja mossambica mossambica)
In a previous study, Fenard and colleagues [107] demonstrated anti-HIV-1 effects of different sPLA2s from snake venom, such as taipoxin (Oxyuranus scutellatus venom), NmmCMIII (Naja mossambica mossambica venom) [108,109] and nigexine (Naja nigricollis venom) [110]. Investigating the possible mode of action of some of these sPLA2s, it was observed that despite their enzymatic activity, NmmCMIII and taipoxin did not show virucidal effects against HIV-1, but promoted an efficient inhibition of HIV-1 entry by preventing the intracellular release of HIV-1 Gap p24 proteins from the viral capsid [107].
The blockage of HIV entry appears to not depend exclusively on sPLA2s catalytically active, which was confirmed through two manners: i) the use of inhibitors of sPLA2s activity, such as phenacyl bromide, aristolochic acid or oleoyloxyethylphosphocholine, that were not able to interfere in the blockage of virus entry mediated by sPLA2s; ii) the use of cleavage products of sPLA2s, such as arachidonic acid, lysophosphatidylethanolamine, lysophosphatidic acid, oleoyl-lysophosphatidylcholine and palmitoyl-lysophosphatidylcholine, which were also not able to inhibit virus entry [107]. In addition, competition binding assays between sPLA2s and host cells showed extremely low dissociation constant (K) values for NmmCMIII, taipoxin and nigexine, suggesting that the inhibition of HIV-1 entry triggered by sPLA2s is more probably linked to sPLA2s binding membrane receptors of host cells than their enzymatic activity [107,111].
3.5. CM-II-sPLA2 from Naja mossambica mossambica venom
CM-II-sPLA2 is a secreted PLA2 isoform isolated from Naja mossambica mossambica venom [112,113]. Recently, Chen and coworkers [114] reported that this sPLA2 possesses a potent dose-dependent virucidal activity that impairs the entry of enveloped viruses from budding through the endoplasmic reticulum, such as HCV, DENV and JEV (Japanese encephalitis virus) belonging to the Flaviviridae family. In contrast, CM-II-sPLA2 demonstrated a low antiviral activity against other enveloped viruses by: i) budding through the plasma membrane, as observed for SINV (Sindbis virus; Togaviridae), SeV (Sendai virus; Paramyxoviridae), FLUAV (Influenza A virus; Orthomyxoviridae), VSNJV (Vesicular stomatitis New Jersey virus; Rhabdoviridae) and HIV-1 (Retroviridae); ii) budding through the trans-Golgi network, as seen for HSV- 1 (Herpes simplex virus type 1; Herpesviridae); iii) budding through the ER-Golgi intermediate compartment, as for MERS-CoV (Middle East respiratory syndrome coronavirus; Coronaviridae). Additionally, the slight effect was also observed against non-enveloped viruses, such as EMCV (Encephalomyocarditis virus; Picornaviridae) and CV-B3 (Coxsackievirus B3; Picornaviridae) [114].
The disruption of viral envelope by CM-II-sPLA2 appears to be directly related to its enzymatic activity, which was confirmed by the use of manoalide (a specific sPLA2 inhibitor) that inhibited the virucidal activity of CM-II-sPLA2 against HCV and DENV [114]. Moreover, the selectivity of CM-II-sPLA2 for virus buds through endoplasmic reticulum may be related to the differences in the phospholipid contents and physicochemical characteristics (thickness and sturdiness) that can differ among the different routes of viral budding, which would enhance the sensitivity to CM-II-sPLA2 mediated by hydrolysis against HCV, DENV and JEV [[114], [115], [116], [117]].
4. Proposed antiviral mechanism of sPLA2s from snake venom
Findings from the current literature about the antiviral activity of toxins (Table 1) are heterogeneous, since authors developed a variety of assays/models using different sPLA2s and viruses. The virucidal model corresponds to the strategy in which the toxins act directly on the virus particles before infecting the cell monolayer; in the pre-infection model, the uninfected monolayers are previously treated with toxins before viral infection; and in the post-infection model, cell monolayers are adsorbed with the virus followed by toxin treatment. In this sense, many studies raised the following questions: in which stages of virus replication are the sPLA2s able to interfere? Does the antiviral action of sPLA2s depend on their catalytic activity? Based on this, we summarize a possible model of antiviral action mediated by sPLA2s from snake venom.
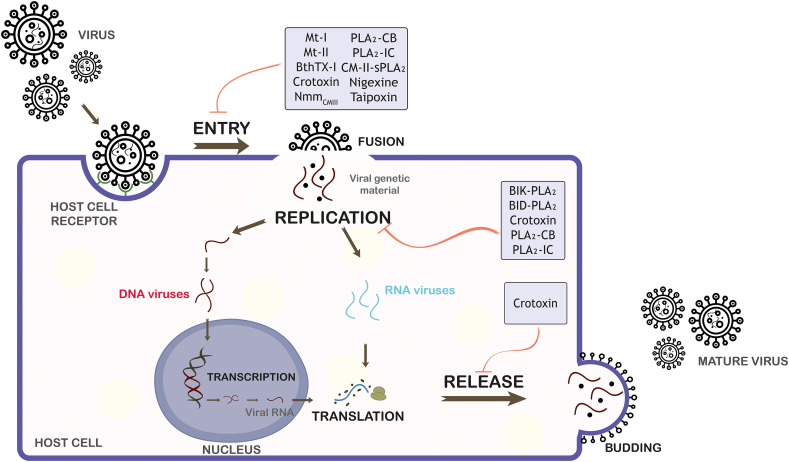
As discussed above, sPLA2s have demonstrated to be potent antiviral inhibitors by interfering in different stages of virus replicative cycle as entry steps, replication and release (Fig. 1 ). Current studies have reported that the antiviral action of sPLA2s on steps of viral cycle can occur through a direct action upon viral particle and/or by an interaction with virus or host cell components.
Fig. 1.
Schematic representation of the mechanisms of action of sPLA2s from snake venom on viral replicative cycle. sPLA2s, which possess antiviral activity, are indicated in early and/or late stages of the viral life cycle: entry, replication and release. Mt-I and Mt-II (Bothrops asper), BthTX-I (Bothrops jararacussu), crotoxin, PLA2-CB and PLA2-IC (Crotalus durissus terrificus), CM-II-sPLA2 and NmmCMIII (Naja mossambica mossambica), nigexine (Naja nigricollis), taipoxin (Oxyuranus scutellatus), BlK-PLA2 and BlD-PLA2 (Bothrops leucurus) are demonstrated.
Regarding the virucidal activity, studies have shown that both catalytically-active sPLA2s (crotoxin, PLA2-IC, PLA2-CB, Mt-I and CM-II-sPLA2) and catalytically-inactive sPLA2s (Mt-II and BthTX-I) indicated virucidal activity preferentially against enveloped viruses, such as DENV (serotypes 1, 2 and 3), YFV, ROCV, OROV, MAYV, HCV, JEV, MERS-CoV, SINV, FLUAV, SeV, VSNJV, HIV-1 and HSV-1 [[88], [89], [90],92,114].
It is proposed that the potent virucidal activity of sPLA2s against enveloped viruses is likely associated with the ability that catalytically-active sPLA2s have to cleave glycerophospholipids in the virus lipid envelope, and it is reasonable to propose that sPLA2s also present domains that are capable to interact with viral envelope components, which could lead to viral envelope disruption, hence resulting in exposure of the viral content (viral inactivation) and compromising the early stages of viral replication. Additionally, Muller and colleagues [88,89], through a steric and electrostatic analysis of the interaction of PLA2-CB with the DENV envelope lipid bilayer, showed that PLA2-CB probably accesses the DENV lipid bilayer through the pores found on each of the twenty 3-fold vertices in the E protein shell on the DENV surface, which would allow the glycerophospholipid cleavage on the virus envelope and destabilization of the E proteins. Interestingly, it has been demonstrated that the structural organization and lipid composition of viral envelope may influence the antiviral efficiency of some sPLA2s, suggesting that the virucidal mechanism mediated by sPLA2s is specific [92].
Independent studies have revealed that sPLA2s such as crotoxin, PLA2-IC, PLA2-CB, Bl-PLA2 and BthTX-I are also able to dramatically impact the entry, replication and release of viruses by targeting host cell components [[88], [89], [90], [91]]. To gain insights into these viral cycle stages, it was demonstrated that NmmCMIII, taipoxin and nigexine prevented the intracellular release of HIV-1 Gap p24 proteins from the viral capsid (inhibition virus entry) by a direct binding to membrane receptors of host cells [107]. In addition, PLA2-CB was able to disrupt HCV replication probably by an interference in lipid metabolism of host cell [90].
It was demonstrated through the use of both specific sPLA2 inhibitors and the catalytically-inactive sPLA2s that the antiviral effect of the major tested catalytically-active sPLA2s, such as crotoxin, PLA2-IC, PLA2-CB, BlD-PLA2, Mt-I and CM-II-sPLA2 is significantly higher when presented their functional catalytic site to sPLA2s with no enzymatic activity (BthTX-I, BlK-PLA2 and Mt-II) [88,91,92,114].
In order to corroborate with the data from the current literature, we performed docking simulations between the sPLA2s from Bothrops asper venom and three phospholipids found in the DENV envelope, which are 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC), 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE) and 1-Palmitoyl-2-oleoylphosphatidylserine (POPS) [118]. The molecular docking simulations were done by using the 3D crystal structure of Mt-I (PDB: 5TFV) and Mt-II (PDB ID 4YV5) retrieved from Protein Database (https://www.rcsb.org/).
We simulated the interaction in the enzymatic site of sPLA2s with palmitoyl phospholipids (head or complete structure) using AutoDock Vina software [119]. The predicted affinity between sPLA2s and palmitoyl phospholipids (head or complete) was similar (Table 2 ). Concerning to the interaction with the phospholipids head, Mt-I showed a higher affinity to POPC, POPE and POPS (−5.1, −4.7 and −5.0 kcal/mol, respectively) related to Mt-II (Table 2). When we simulated the docking with the complete palmitoyl phospholipid molecule, Mt-I also has a higher affinity than Mt-II (Table 2).
Table 2.
Docking simulations between Mt-I and Mt-II with palmitoyl phospholipids (head or complete structure) from DENV envelope.
| PLA2s affinity (kcal/mol) | Phospholipids |
|||||
|---|---|---|---|---|---|---|
| POPC |
POPE |
POPS |
||||
| Head | Complete | Head | Complete | Head | Complete | |
| Mt-I | −5.1 | −5.4 | −4.7 | −5.1 | −5.0 | −5.4 |
| Mt-II | −4.3 | −4.5 | −4.3 | −4.7 | −4.3 | −4.8 |
POPC - 1-palmitoyl-2-oleoylphosphatidylcholine; POPE - 1-palmitoyl-2-oleoylphosphatidylethanolamine; POPS - 1-palmitoyl-2-oleoylphosphatidylserine.
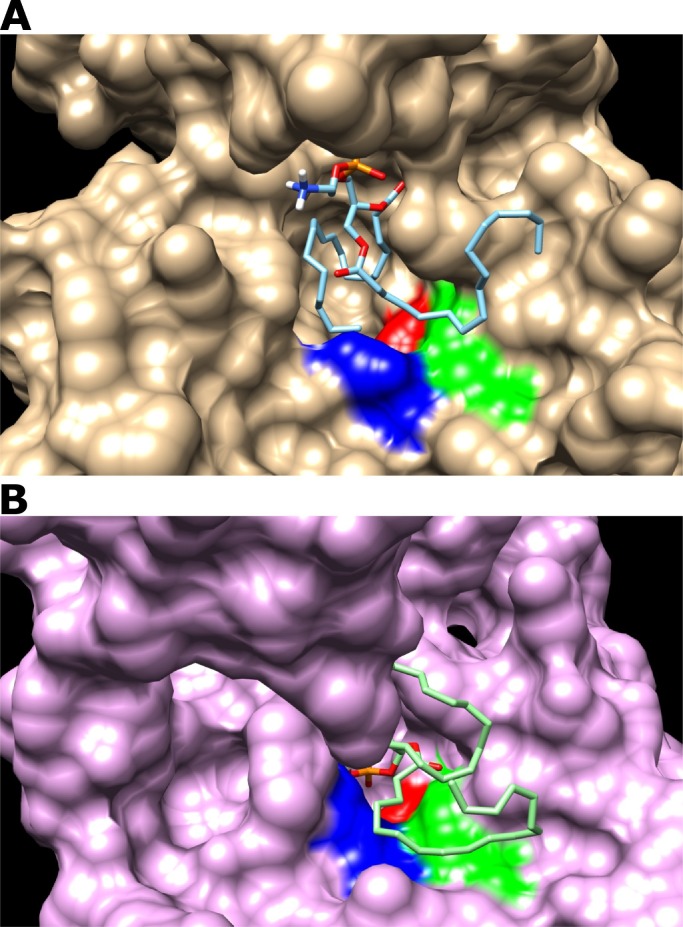
Although we do not observe a strong difference in the affinity between Mt-I and Mt-II for palmitoyl phospholipids, it is possible to note structural variation in the enzymatic site of these two toxins. Compared to Mt-II, the enzymatic site of Mt-I (Fig. 2A) is more suitable due to a smaller aspartic acid radical group. The van der Waals radii volume of aspartic acid is 91 and hence it is more prominent, while the lysine has a volume of 135, and this results in less space in enzymatic site entrance in Mt-II (Fig. 2B). This difference could create an enzymatic site more restricted to palmitoyl phospholipid entrance/binding and be partially responsible for the absence of enzymatic activity in Mt-II [120]. In addition, the enzymatic activity of Mt-I can be attributed to highly conserved catalytic site formed by the amino acid residues His48, Asp49, Tyr52 and Asp99. Asp49 coordinates the hydrolysis reaction of phospholipids together with the residues of the Ca2+ binding loop, essential in the catalytic activity of PLA2s. The substitution of lysine residue at the same position affects the ability of this protein to bind to Ca2+, resulting in the absence of catalytic activity [92].
Fig. 2.
Docking simulations between sPLA2s and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC). Mt-I (A) and Mt-II (B) are showed as surface and the enzymatic site is colored. The amino acids from the enzymatic site are H48 in red, D49 (Mt-I) or K49 (Mt-II) in blue, and Y52 in green. The POPC is showed as wire structure. The aspartic acid has a smaller volume than lysine, which may result in a less open entrance in Mt-II (panel B) compared with Mt-I (panel A). H: histidine; D: aspartate; K: lysine; Y: tyrosine. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Despite the stronger antiviral activity is associated with the enzymatic activity, the antiviral mechanism of sPLA2s does not depend exclusively on their catalytic site, since Lys49 sPLA2s and inhibited catalytically-active sPLA2s were also able to show antiviral effects, suggesting that sPLA2s may possess different mechanisms of action. However, additional studies with different Lys49 from snake venom are required to better characterize the antiviral potential of this protein class.
Functional and structural studies have described that the activity of Lys49 PLA2s from snake venom toward cell membranes in myotoxic mechanism involves an allosteric transition, and the participation of two independent interaction sites with the target membrane [67,72,[121], [122], [123]]. The action of Lys49 PLA2s is related to a cluster of cationic and hydrophobic/aromatic amino acid residues located at the C-terminal region of this toxin. These two conserved regions in most Lys49-PLA2s are designed by “cationic membrane-docking site” (MDoS), which are formed by the strictly conserved C-terminal residues (Lys115 and Arg118), eventually aided by other cationic and exposed residues such as Lys20, Lys80, Lys122 and Lys127; and the “hydrophobic membrane-disruption site” (MDiS) formed by residues of Leu121 and Phe125. The key step for protein activation is the binding of a fatty acid at the hydrophobic channel, which leads to allosteric transition and structure stabilization exposing MDoS and MDiS to the membrane, following by the insertion of the MDiS region from both monomers into the target membrane. This penetration disrupts the lipid bilayer, causing alterations in the membrane permeability, highlighted by a prominent influx of ions (i.e., Ca2+ and Na+), and eventually, irreversible intracellular alterations and cell death [123].
According to myotoxic mechanism of Lys49 PLA2s from viperid snake venoms, it is proposed that the fatty acids which are important to protein activation may come from membrane phospholipid hydrolysis by catalytic PLA2s (Asp49), highlighting the synergism between Asp49 PLA2s and Lys49 PLA2 in snake envenomation [124]. In this way, the antiviral effects of the Lys49 PLA2s from snake venom, showed in this review, may be associated to fatty acids from the catalytic activity of cytosolic PLA2 (cPLA2) from virus lipid envelope, once it was demonstrated that enzymatic activity of the cPLA2 is required for replication of various virus [[125], [126], [127]]. Muller and colleagues [126] showed that the pharmacological inhibition of a cellular phospholipase, cPLA2, using a specific small-molecule inhibitor, drastically reduces coronavirus RNA synthesis and, as a consequence, protein accumulation and the production of infectious virus progeny. In addition, cPLA2 activity was shown to be critically involved in the production of infectious progeny of HCV and DENV [128].
5. Concluding remarks: sPLA2s as a possible useful tool for the development of antiviral compounds
The present review highlighted that PLA2s from snake venom have become valuable as pharmacological tools and/or therapeutic approaches due to their extremely high specificity and potent activity against microbial infection. Regarding to antiviral properties, we highlighted the following remarks: (i) the antiviral effects of sPLA2s can be mediated by either a dependent or independent catalytic mechanism; (ii) sPLA2s-antiviral effects are more evident against enveloped virus; (iii) sPLA2s promoted the blockage of viral entry into host cells by the direct action on the viral particle, resulting in glycerophospholipids cleavage and destabilization of viral envelope proteins; (iv) the structural organization, physicochemical composition, and the curvature and fluidity of viral envelope may influence in the antiviral efficiency of some sPLA2s; (v) sPLA2s promoted the blockage of entry, replication and release of virus probably by the interference on the host cell components.
The structure and function of sPLA2s from snake venom have been widely explored. Homology studies with sPLA2s have demonstrated highly conserved regions in these proteins capable of disrupting the integrity of membranes and provoking many pharmacological effects. Despite extensive studies on sPLA2s in over decades, there is few of them focusing on mechanistic aspects of the antiviral activities and to date are limited to in vitro and in silico models. It is important to note that these sPLA2s showed damage effects in vivo, such as myotoxicity and inflammation. Thus, further in vivo studies for attesting antiviral effects of sPLA2s need to be addressed to investigate their safety, toxicity and pharmacokinetics. Taken together, new structural and functional studies with sPLA2s are essential to discover new relevant motifs responsible for the antiviral activities that would allow the future use of these proteins or peptides for the design of antiviral drugs, capable of ensuring more stability and targeting the specific site of action.
Acknowledgements
The authors thank Federal University of Bahia (UFBA), Federal University of Uberlândia (UFU) and the Brazilian funding agencies, CNPq, CAPES, FAPEMIG and PPSUS for providing financial support to this study: PPSUS (SUS0037/2018), FAPEMIG (CBB-APQ-01637-15, CBB-APQ-01401-17, APQ-00587-14 and APQ-03385-18), CAPES (Coordination for the Improvement of Higher Education Personnel – Code 001), Royal Society – Newton Advanced Fellowship (grant reference NA 150195) and National Institute of Science and Technology in Theranostics and Nanobiotechnology INCT-TeraNano-CNPq/CAPES/FAPEMIG, grant numbers CNPq-465669/2014-0.
References
- 1.Lugo D., Krogstad P. Enteroviruses in the early 21st century: new manifestations and challenges. Curr. Opin. Pediatr. 2016;28(1):107–113. doi: 10.1097/MOP.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braaten K.P., Laufer M.R. Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev. Obstet. Gynecol. 2008;1(1):2–10. [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd P.J., Garnett G.P., Hallett T.B. Examining the promise of HIV elimination by ‘test and treat’ in hyperendemic settings. AIDS (London, England) 2010;24(5):729–735. doi: 10.1097/QAD.0b013e32833433fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koelle D.M., Corey L. Herpes simplex: insights on pathogenesis and possible vaccines. Annu. Rev. Med. 2008;59:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- 5.Carroll M.W., Matthews D.A., Hiscox J.A., Elmore M.J., Pollakis G., Rambaut A., Hewson R., Garcia-Dorival I., Bore J.A., Koundouno R., Abdellati S., Afrough B., Aiyepada J., Akhilomen P., Asogun D., Atkinson B., Badusche M., Bah A., Bate S., Baumann J., Becker D., Becker-Ziaja B., Bocquin A., Borremans B., Bosworth A., Boettcher J.P., Cannas A., Carletti F., Castilletti C., Clark S., Colavita F., Diederich S., Donatus A., Duraffour S., Ehichioya D., Ellerbrok H., Fernandez-Garcia M.D., Fizet A., Fleischmann E., Gryseels S., Hermelink A., Hinzmann J., Hopf-Guevara U., Ighodalo Y., Jameson L., Kelterbaum A., Kis Z., Kloth S., Kohl C., Korva M., Kraus A., Kuisma E., Kurth A., Liedigk B., Logue C.H., Ludtke A., Maes P., McCowen J., Mely S., Mertens M., Meschi S., Meyer B., Michel J., Molkenthin P., Munoz-Fontela C., Muth D., Newman E.N., Ngabo D., Oestereich L., Okosun J., Olokor T., Omiunu R., Omomoh E., Pallasch E., Palyi B., Portmann J., Pottage T., Pratt C., Priesnitz S., Quartu S., Rappe J., Repits J., Richter M., Rudolf M., Sachse A., Schmidt K.M., Schudt G., Strecker T., Thom R., Thomas S., Tobin E., Tolley H., Trautner J., Vermoesen T., Vitoriano I., Wagner M., Wolff S., Yue C., Capobianchi M.R., Kretschmer B., Hall Y., Kenny J.G., Rickett N.Y., Dudas G., Coltart C.E., Kerber R., Steer D., Wright C., Senyah F., Keita S., Drury P., Diallo B., de Clerck H., Van Herp M., Sprecher A., Traore A., Diakite M., Konde M.K., Koivogui L., Magassouba N., Avsic-Zupanc T., Nitsche A., Strasser M., Ippolito G., Becker S., Stoecker K., Gabriel M., Raoul H., Di Caro A., Wolfel R., Formenty P., Gunther S. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature. 2015;524(7563):97–101. doi: 10.1038/nature14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heukelbach J., Alencar C.H., Kelvin A.A., de Oliveira W.K., de Goes Cavalcanti L. Pamplona. Zika virus outbreak in Brazil. Journal of infection in developing countries. 2016;10(2):116–120. doi: 10.3855/jidc.8217. [DOI] [PubMed] [Google Scholar]
- 7.Neumann G., Noda T., Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins N.D., Barrett A.D. Live attenuated yellow fever 17D vaccine: a legacy vaccine still controlling outbreaks in modern day. Curr. Infect. Dis. Rep. 2017;19(3):14. doi: 10.1007/s11908-017-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith K.F., Goldberg M., Rosenthal S., Carlson L., Chen J., Chen C., Ramachandran S. Global rise in human infectious disease outbreaks. J. R. Soc. Interface. 2014;11(101) doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulliam J.R. Viral host jumps: moving toward a predictive framework. EcoHealth. 2008;5(1):80–91. doi: 10.1007/s10393-007-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K., Wang W., Eden J.S., Shen J.J., Liu L., Holmes E.C., Zhang Y.Z. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556(7700):197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 13.Palese P. Influenza: old and new threats. Nat. Med. 2004;10(12 Suppl):S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 14.Krammer F. Emerging influenza viruses and the prospect of a universal influenza virus vaccine. Biotechnol. J. 2015;10(5):690–701. doi: 10.1002/biot.201400393. [DOI] [PubMed] [Google Scholar]
- 15.Qin L., Sun Q., Wang Y., Wu K.F., Chen M., Shia B.C., Wu S.Y. vol. 17. 2020. Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index. (7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R., Simmons C.P., Scott T.W., Farrar J.J., Hay S.I. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haig D.M. Subversion and piracy: DNA viruses and immune evasion. Res. Vet. Sci. 2001;70(3):205–219. doi: 10.1053/rvsc.2001.0462. [DOI] [PubMed] [Google Scholar]
- 18.Ryu W.-S. Antivir. Ther. 2017:367–381. [Google Scholar]
- 19.Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorley J.A., McKeating J.A., Rappoport J.Z. Mechanisms of viral entry: sneaking in the front door. Protoplasma. 2010;244(1–4):15–24. doi: 10.1007/s00709-010-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aleksandrowicz P., Marzi A., Biedenkopf N., Beimforde N., Becker S., Hoenen T., Feldmann H., Schnittler H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011;204(Suppl. 3):S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marintcheva B. 2018. Introduction to Viral Structure, Diversity and Biology; pp. 1–26. (Parts of this chapter were originally published in Marintcheva B. A box of paradoxes: the fascinating world of viruses. Bridgew Rev 2013;32(2):25–8. http://vc.bridgew.edu/br_rev/vol32/iss2/8 and are reproduced here with the permission of the editor) [Google Scholar]
- 23.Rodenhuis-Zybert I.A., Wilschut J., Smit J.M. Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010;67(16):2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann S.H., McElrath M.J., Lewis D.J., Del Giudice G. Challenges and responses in human vaccine development. Curr. Opin. Immunol. 2014;28:18–26. doi: 10.1016/j.coi.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Maslow J.N. The cost and challenge of vaccine development for emerging and emergent infectious diseases. Lancet Glob. Health. 2018;6(12):e1266–e1267. doi: 10.1016/S2214-109X(18)30418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammadi Pour P., Fakhri S., Asgary S., Farzaei M.H., Echeverria J. The signaling pathways, and therapeutic targets of antiviral agents: focusing on the antiviral approaches and clinical perspectives of anthocyanins in the management of viral diseases. Front. Pharmacol. 2019;10:1207. doi: 10.3389/fphar.2019.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D.K., Chung R.T. Overview of direct-acting antiviral drugs and drug resistance of hepatitis C virus. Methods in molecular biology (Clifton, N.J.) 2019;1911:3–32. doi: 10.1007/978-1-4939-8976-8_1. [DOI] [PubMed] [Google Scholar]
- 29.Takashita E. Influenza polymerase inhibitors: mechanisms of action and resistance. Cold Spring Harbor perspectives in medicine. 2020 doi: 10.1101/cshperspect.a038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carr A., Cooper D.A. Adverse effects of antiretroviral therapy. Lancet (London, England) 2000;356(9239):1423–1430. doi: 10.1016/S0140-6736(00)02854-3. [DOI] [PubMed] [Google Scholar]
- 31.Lai K.K., Gang D.L., Zawacki J.K., Cooley T.P. Fulminant hepatic failure associated with 2′,3′-dideoxyinosine (ddI) Ann. Intern. Med. 1991;115(4):283–284. doi: 10.7326/0003-4819-115-4-283. [DOI] [PubMed] [Google Scholar]
- 32.Montessori V., Press N., Harris M., Akagi L., Montaner J.S. Adverse effects of antiretroviral therapy for HIV infection. Can. Med. Assoc. J. 2004;170(2):229–238. [PMC free article] [PubMed] [Google Scholar]
- 33.Dias D.A., Urban S., Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2(2):303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brahmachari G. 2011. Natural Products in Drug Discovery: Impacts and Opportunities — An Assessment; pp. 1–199. [Google Scholar]
- 35.Mohamed Abd El-Aziz T., Soares A. Garcia, Stockand J.D. Snake venoms in drug discovery: valuable therapeutic tools for life saving. Toxins. 2019;11(10) doi: 10.3390/toxins11100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoni R. The use of snake venom-derived compounds for new functional diagnostic test kits in the field of haemostasis. Pathophysiol. Haemost. Thromb. 2005;34(4–5):234–240. doi: 10.1159/000092430. [DOI] [PubMed] [Google Scholar]
- 37.Millard M., Odde S., Neamati N. Integrin targeted therapeutics. Theranostics. 2011;1:154–188. doi: 10.7150/thno/v01p0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh C.Y., Kini R.M. From snake venom toxins to therapeutics—cardiovascular examples. Toxicon. 2012;59(4):497–506. doi: 10.1016/j.toxicon.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Calderon L.A., Sobrinho J.C., Zaqueo K.D., de Moura A.A., Grabner A.N., Mazzi M.V., Marcussi S., Nomizo A. vol. 2014. 2014. Antitumoral Activity of Snake Venom Proteins: New Trends in Cancer Therapy; p. 203639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ondetti M.A., Williams N.J., Sabo E.F., Pluscec J., Weaver E.R., Kocy O. Angiotensin-converting enzyme inhibitors from the venom of Bothrops jararaca. Isolation, elucidation of structure, and synthesis. Biochemistry. 1971;10(22):4033–4039. doi: 10.1021/bi00798a004. [DOI] [PubMed] [Google Scholar]
- 41.Cushman D.W., Ondetti M.A. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension (Dallas, Tex.: 1979) 1991;17(4):589–592. doi: 10.1161/01.hyp.17.4.589. [DOI] [PubMed] [Google Scholar]
- 42.Scarborough R.M., Rose J.W., Hsu M.A., Phillips D.R., Fried V.A., Campbell A.M., Nannizzi L., Charo I.F. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m. barbouri. J. Biol. Chem. 1991;266(15):9359–9362. [PubMed] [Google Scholar]
- 43.Gan Z.R., Gould R.J., Jacobs J.W., Friedman P.A., Polokoff M.A. Echistatin. A potent platelet aggregation inhibitor from the venom of the viper, Echis carinatus. J. Biol. Chem. 1988;263(36):19827–19832. [PubMed] [Google Scholar]
- 44.Funk C., Gmur J., Herold R., Straub P.W. Reptilase-R—a new reagent in blood coagulation. Br. J. Haematol. 1971;21(1):43–52. doi: 10.1111/j.1365-2141.1971.tb03415.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang D.S., Hanamoto M., Fang F., Ohba M., Ishii M., Kimura F., Higaki E., Senga H. Defibrinogenating effect of batroxobin (Defibrase) in rats and inhibition of migration of human vascular smooth muscle cells by the plasma of batroxobin-treated rats in vitro. Atherosclerosis. 2001;156(1):73–80. doi: 10.1016/s0021-9150(00)00628-6. [DOI] [PubMed] [Google Scholar]
- 46.de Morais N.C., Neves Mamede C.C., Fonseca K.C., de Queiroz M.R., Gomes-Filho S.A., Santos-Filho N.A., Bordon Kde C., Beletti M.E., Sampaio S.V., Arantes E.C., de Oliveira F. Isolation and characterization of moojenin, an acid-active, anticoagulant metalloproteinase from Bothrops moojeni venom. Toxicon. 2012;60(7):1251–1258. doi: 10.1016/j.toxicon.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Graziano F., Maugeri R., Basile L., Meccio F., Iacopino D.G. Aulogous fibrin sealant (Vivostat((R))) in the neurosurgical practice: part II: vertebro-spinal procedures. Surg. Neurol. Int. 2016;7(Suppl. 3):S77–S82. doi: 10.4103/2152-7806.174894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waheed H., Moin S.F., Choudhary M.I. Snake venom: from deadly toxins to life-saving therapeutics. Curr. Med. Chem. 2017;24(17):1874–1891. doi: 10.2174/0929867324666170605091546. [DOI] [PubMed] [Google Scholar]
- 49.Bailey P., Wilce J. Venom as a source of useful biologically active molecules. Emergency medicine (Fremantle, W.A.) 2001;13(1):28–36. doi: 10.1046/j.1442-2026.2001.00174.x. [DOI] [PubMed] [Google Scholar]
- 50.Soares A.M., Zuliani J.P. Toxins of animal venoms and inhibitors: molecular and biotechnological tools useful to human and animal health. Curr. Top. Med. Chem. 2019;19(21):1868–1871. doi: 10.2174/156802661921191024114842. [DOI] [PubMed] [Google Scholar]
- 51.da Mata E.C., Mourao C.B., Rangel M., Schwartz E.F. Antiviral activity of animal venom peptides and related compounds. The journal of venomous animals and toxins including tropical diseases. 2017;23:3. doi: 10.1186/s40409-016-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meenakshisundaram R., Sweni S., Thirumalaikolundusubramanian P. Hypothesis of snake and insect venoms against human immunodeficiency virus: a review. AIDS Res. Ther. 2009;6:25. doi: 10.1186/1742-6405-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burke J.E., Dennis E.A. Phospholipase A2 biochemistry. Cardiovasc. Drugs Ther. 2009;23(1):49–59. doi: 10.1007/s10557-008-6132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filkin S.Y., Lipkin A.V., Fedorov A.N. Phospholipase superfamily: structure, functions, and biotechnological applications. Biochemistry. Biokhimiia. 2020;85(Suppl. 1):S177–s195. doi: 10.1134/S0006297920140096. [DOI] [PubMed] [Google Scholar]
- 55.Aloulou A., Ali Y.B., Bezzine S., Gargouri Y., Gelb M.H. Phospholipases: an overview. Methods in molecular biology (Clifton, N.J.) 2012;861:63–85. doi: 10.1007/978-1-61779-600-5_4. [DOI] [PubMed] [Google Scholar]
- 56.Arni R.K., Ward R.J. Phospholipase A2—a structural review. Toxicon. 1996;34(8):827–841. doi: 10.1016/0041-0101(96)00036-0. [DOI] [PubMed] [Google Scholar]
- 57.Kini R.M. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42(8):827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Austin S.C., Funk C.D. Insight into prostaglandin, leukotriene, and other eicosanoid functions using mice with targeted gene disruptions. Prostaglandins & other lipid mediators. 1999;58(5–6):231–252. doi: 10.1016/s0090-6980(99)00041-6. [DOI] [PubMed] [Google Scholar]
- 59.Gutierrez J.M., Lomonte B. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon. 2013;62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 60.Boldrini-Franca J., Cologna C.T., Pucca M.B., Bordon K.C., Amorim F.G., Anjolette F.A., Cordeiro F.A., Wiezel G.A., Cerni F.A., Pinheiro-Junior E.L., Shibao P.Y., Ferreira I.G., de Oliveira I.S., Cardoso I.A., Arantes E.C. Minor snake venom proteins: structure, function and potential applications. Biochim. Biophys. Acta, Gen. Subj. 2017;1861(4):824–838. doi: 10.1016/j.bbagen.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 61.Nicolau C.A., Carvalho P.C., Junqueira-de-Azevedo I.L., Teixeira-Ferreira A., Junqueira M., Perales J., Neves-Ferreira A.G., Valente R.H. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca. J. Proteome. 2017;151:214–231. doi: 10.1016/j.jprot.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 62.Mendez R., Bonilla F., Sasa M., Dwyer Q., Fernandez J., Lomonte B. Proteomic profiling, functional characterization, and immunoneutralization of the venom of Porthidium porrasi, a pitviper endemic to Costa Rica. Acta Trop. 2019;193:113–123. doi: 10.1016/j.actatropica.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 63.Schaloske R.H., Dennis E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 2006;1761(11):1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 64.Six D.A., Dennis E.A. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta. 2000;1488(1–2):1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 65.Maraganore J.M., Merutka G., Cho W., Welches W., Kezdy F.J., Heinrikson R.L. A new class of phospholipases A2 with lysine in place of aspartate 49. Functional consequences for calcium and substrate binding. J. Biol. Chem. 1984;259(22):13839–13843. [PubMed] [Google Scholar]
- 66.Angulo Y., Lomonte B. Biochemistry and toxicology of toxins purified from the venom of the snake Bothrops asper. Toxicon. 2009;54(7):949–957. doi: 10.1016/j.toxicon.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Lomonte B., Rangel J. Snake venom Lys49 myotoxins: from phospholipases A(2) to non-enzymatic membrane disruptors. Toxicon. 2012;60(4):520–530. doi: 10.1016/j.toxicon.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 68.Wei J.F., Wei X.L., Chen Q.Y., Huang T., Qiao L.Y., Wang W.Y., Xiong Y.L., He S.H. N49 phospholipase A2, a unique subgroup of snake venom group II phospholipase A2. Biochim. Biophys. Acta. 2006;1760(3):462–471. doi: 10.1016/j.bbagen.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 69.Polgar J., Magnenat E.M., Peitsch M.C., Wells T.N., Clemetson K.J. Asp-49 is not an absolute prerequisite for the enzymic activity of low-M(r) phospholipases A2: purification, characterization and computer modelling of an enzymically active Ser-49 phospholipase A2, ecarpholin S, from the venom of Echis carinatus sochureki (saw-scaled viper) The Biochemical journal. 1996;319(Pt 3):961–968. doi: 10.1042/bj3190961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ownby C.L., Selistre de Araujo H.S., White S.P., Fletcher J.E. Lysine 49 phospholipase A2 proteins. Toxicon. 1999;37(3):411–445. doi: 10.1016/s0041-0101(98)00188-3. [DOI] [PubMed] [Google Scholar]
- 71.Kini R.M., Evans H.J. A common cytolytic region in myotoxins, hemolysins, cardiotoxins and antibacterial peptides. Int. J. Pept. Protein Res. 1989;34(4):277–286. doi: 10.1111/j.1399-3011.1989.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 72.Nunez C.E., Angulo Y., Lomonte B. Identification of the myotoxic site of the Lys49 phospholipase A(2) from Agkistrodon piscivorus piscivorus snake venom: synthetic C-terminal peptides from Lys49, but not from Asp49 myotoxins, exert membrane-damaging activities. Toxicon. 2001;39(10):1587–1594. doi: 10.1016/s0041-0101(01)00141-6. [DOI] [PubMed] [Google Scholar]
- 73.Ambrosio A.L., Nonato M.C., de Araujo H.S., Arni R., Ward R.J., Ownby C.L., de Souza D.H., Garratt R.C. A molecular mechanism for Lys49-phospholipase A2 activity based on ligand-induced conformational change. J. Biol. Chem. 2005;280(8):7326–7335. doi: 10.1074/jbc.M410588200. [DOI] [PubMed] [Google Scholar]
- 74.Lomonte B., Angulo Y., Moreno E. Synthetic peptides derived from the C-terminal region of Lys49 phospholipase A2 homologues from viperidae snake venoms: biomimetic activities and potential applications. Curr. Pharm. Des. 2010;16(28):3224–3230. doi: 10.2174/138161210793292456. [DOI] [PubMed] [Google Scholar]
- 75.Lambeau G., Ancian P., Nicolas J.P., Beiboer S.H., Moinier D., Verheij H., Lazdunski M. Structural elements of secretory phospholipases A2 involved in the binding to M-type receptors. J. Biol. Chem. 1995;270(10):5534–5540. doi: 10.1074/jbc.270.10.5534. [DOI] [PubMed] [Google Scholar]
- 76.Akalu A., Cretu A., Brooks P.C. Targeting integrins for the control of tumour angiogenesis. Expert Opin. Investig. Drugs. 2005;14(12):1475–1486. doi: 10.1517/13543784.14.12.1475. [DOI] [PubMed] [Google Scholar]
- 77.Jimenez-Charris E., Lopes D.S., Gimenes S.N.C., Teixeira S.C., Montealegre-Sanchez L., Solano-Redondo L., Fierro-Perez L., Rodrigues Avila V.M. Antitumor potential of Pllans-II, an acidic Asp49-PLA2 from Porthidium lansbergii lansbergii snake venom on human cervical carcinoma HeLa cells. Int. J. Biol. Macromol. 2019;122:1053–1061. doi: 10.1016/j.ijbiomac.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 78.Massimino M.L., Simonato M., Spolaore B., Franchin C., Arrigoni G., Marin O., Monturiol-Gross L., Fernandez J., Lomonte B., Tonello F. Cell surface nucleolin interacts with and internalizes Bothrops asper Lys49 phospholipase A2 and mediates its toxic activity. Sci. Rep. 2018;8(1):10619. doi: 10.1038/s41598-018-28846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kini R.M., Evans H.J. A model to explain the pharmacological effects of snake venom phospholipases A2. Toxicon. 1989;27(6):613–635. doi: 10.1016/0041-0101(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 80.Diniz-Sousa R., Caldeira C.A.S., Kayano A.M., Paloschi M.V., Pimenta D.C., Simoes-Silva R., Ferreira A.S., Zanchi F.B., Matos N.B., Grabner F.P., Calderon L.A., Zuliani J.P., Soares A.M. Identification of the molecular determinants of the antibacterial activity of LmutTX, a Lys49 phospholipase A2 homologue isolated from Lachesis muta muta snake venom (Linnaeus, 1766) Basic Clin. Pharmacol. Toxicol. 2018;122(4):413–423. doi: 10.1111/bcpt.12921. [DOI] [PubMed] [Google Scholar]
- 81.Prinholato da Silva C., Costa T.R., Paiva R.M., Cintra A.C., Menaldo D.L., Antunes L.M., Sampaio S.V. Antitumor potential of the myotoxin BthTX-I from Bothrops jararacussu snake venom: evaluation of cell cycle alterations and death mechanisms induced in tumor cell lines. The journal of venomous animals and toxins including tropical diseases. 2015;21:44. doi: 10.1186/s40409-015-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Azevedo F.V., Lopes D.S., Cirilo Gimenes S.N., Ache D.C., Vecchi L., Alves P.T., Guimaraes Dde O., Rodrigues R.S., Goulart L.R., Rodrigues Vde M., Yoneyama K.A. Human breast cancer cell death induced by BnSP-6, a Lys-49 PLA(2) homologue from Bothrops pauloensis venom. Int. J. Biol. Macromol. 2016;82:671–677. doi: 10.1016/j.ijbiomac.2015.10.080. [DOI] [PubMed] [Google Scholar]
- 83.de Vasconcelos Azevedo F.V.P., Zoia M.A.P., Lopes D.S., Gimenes S.N., Vecchi L., Alves P.T., Rodrigues R.S., Silva A.C.A., Yoneyama K.A.G., Goulart L.R., de Melo Rodrigues V. Antitumor and antimetastatic effects of PLA2-BthTX-II from Bothrops jararacussu venom on human breast cancer cells. Int. J. Biol. Macromol. 2019;135:261–273. doi: 10.1016/j.ijbiomac.2019.05.164. [DOI] [PubMed] [Google Scholar]
- 84.Bazaa A., Luis J., Srairi-Abid N., Kallech-Ziri O., Kessentini-Zouari R., Defilles C., Lissitzky J.C., El Ayeb M., Marrakchi N. MVL-PLA2, a phospholipase A2 from Macrovipera lebetina transmediterranea venom, inhibits tumor cells adhesion and migration. Matrix Biol. 2009;28(4):188–193. doi: 10.1016/j.matbio.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 85.D.C. Nunes, M.M. Figueira, D.S. Lopes, D.L. De Souza, L.F. Izidoro, E.A. Ferro, M.A. Souza, R.S. Rodrigues, V.M. Rodrigues, K.A. Yoneyama, BnSP-7 toxin, a basic phospholipase A2 from Bothrops pauloensis snake venom, interferes with proliferation, ultrastructure and infectivity of Leishmania (Leishmania) amazonensis, Parasitology 140(7) (2013) 844–54. [DOI] [PubMed]
- 86.Borges I.P., Castanheira L.E., Barbosa B.F., de Souza D.L., Silva R.J. da, Mineo J.R., Tudini K.A., Rodrigues R.S., Ferro E.A., de Melo Rodrigues V. Anti-parasitic effect on Toxoplasma gondii induced by BnSP-7, a Lys49-phospholipase A2 homologue from Bothrops pauloensis venom. Toxicon. 2016;119:84–91. doi: 10.1016/j.toxicon.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 87.Rodrigues J.P., Vasconcelos Azevedo F.V.P., Zoia M.A.P., Maia L.P., Correia L.I.V., Costa-Cruz J.M., de Melo Rodrigues V., Goulart L.R. The anthelmintic effect on Strongyloides venezuelensis induced by BnSP- 6, a Lys49-phospholipase A2 homologue from Bothrops pauloensis venom. Curr. Top. Med. Chem. 2019;19(22):2032–2040. doi: 10.2174/1568026619666190723152520. [DOI] [PubMed] [Google Scholar]
- 88.Muller V.D., Russo R.R., Cintra A.C., Sartim M.A., Alves-Paiva Rde M., Figueiredo L.T., Sampaio S.V., Aquino V.H. Crotoxin and phospholipases A(2) from Crotalus durissus terrificus showed antiviral activity against dengue and yellow fever viruses. Toxicon. 2012;59(4):507–515. doi: 10.1016/j.toxicon.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 89.Muller V.D., Soares R.O., dos Santos N.N., Jr., Trabuco A.C., Cintra A.C., Figueiredo L.T., Caliri A., Sampaio S.V., Aquino V.H. Phospholipase A2 isolated from the venom of Crotalus durissus terrificus inactivates dengue virus and other enveloped viruses by disrupting the viral envelope. PLoS One. 2014;9(11):e112351. doi: 10.1371/journal.pone.0112351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shimizu J.F., Pereira C.M., Bittar C., Batista M.N., Campos G.R.F., Silva S. da, Cintra A.C.O., Zothner C., Harris M., Sampaio S.V., Aquino V.H., Rahal P., Jardim A.C.G. Multiple effects of toxins isolated from Crotalus durissus terrificus on the hepatitis C virus life cycle. PLoS One. 2017;12(11):e0187857. doi: 10.1371/journal.pone.0187857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cecilio A.B., Caldas S., Oliveira R.A., Santos A.S., Richardson M., Naumann G.B., Schneider F.S., Alvarenga V.G., Estevão-Costa M.I., Fuly A.L., Eble J.A., Sanchez E.F. Molecular characterization of Lys49 and Asp49 phospholipases A₂ from snake venom and their antiviral activities against dengue virus. Toxins. 2013;5(10):1780–1798. doi: 10.3390/toxins5101780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brenes H., Loría G.D., Lomonte B. Potent virucidal activity against Flaviviridae of a group IIA phospholipase A(2) isolated from the venom of Bothrops asper. Biologicals. 2020;63:48–52. doi: 10.1016/j.biologicals.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 93.Prado-Franceschi J., Brazil O.V. Convulxin, a new toxin from the venom of the South American rattlesnake Crotalus durissus terrificus. Toxicon. 1981;19(6):875–887. doi: 10.1016/0041-0101(81)90085-4. [DOI] [PubMed] [Google Scholar]
- 94.Alexander G., Grothusen J., Zepeda H., Schwartzman R.J. Gyroxin, a toxin from the venom of Crotalus durissus terrificus, is a thrombin-like enzyme. Toxicon. 1988;26(10):953–960. doi: 10.1016/0041-0101(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 95.Hendon R.A., Fraenkel-Conrat H. Biological roles of the two components of crotoxin. Proc. Natl. Acad. Sci. U. S. A. 1971;68(7):1560–1563. doi: 10.1073/pnas.68.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rubsamen K., Breithaupt H., Habermann E. Biochemistry and pharmacology of the crotoxin complex. I. Subfractionation and recombination of the crotoxin complex. Naunyn-Schmiedebergs Archiv fur Pharmakologie. 1971;270(3):274–288. doi: 10.1007/BF00997027. [DOI] [PubMed] [Google Scholar]
- 97.Villarrubia V.G., Costa L.A., Díez R.A. Fosfolipasas A2 segregadas (sPLA2):¿amigas o enemigas? ¿Actores de la resistencia antibacteriana y antivirus de la inmunodeficiencia humana? Med. Clin. 2004;123(19):749–757. doi: 10.1016/s0025-7753(04)74656-4. [DOI] [PubMed] [Google Scholar]
- 98.Cintra A.C., Marangoni S., Oliveira B., Giglio J.R. Bothropstoxin-I: amino acid sequence and function. J. Protein Chem. 1993;12(1):57–64. doi: 10.1007/BF01024915. [DOI] [PubMed] [Google Scholar]
- 99.Russo R.R., Junior N.N. Dos Santos, Cintra A.C.O., Figueiredo L.T.M., Sampaio S.V., Aquino V.H. vol. 164. 2019. Expression, Purification and Virucidal Activity of Two Recombinant Isoforms of Phospholipase A2 From Crotalus durissus terrificus Venom; pp. 1159–1171. (4) [DOI] [PubMed] [Google Scholar]
- 100.Bartenschlager R., Lohmann V., Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013;11(7):482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 101.Masaki T., Suzuki R., Murakami K., Aizaki H., Ishii K., Murayama A., Date T., Matsuura Y., Miyamura T., Wakita T., Suzuki T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 2008;82(16):7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Higuchi D.A., Barbosa C.M., Bincoletto C., Chagas J.R., Magalhaes A., Richardson M., Sanchez E.F., Pesquero J.B., Araujo R.C., Pesquero J.L. Purification and partial characterization of two phospholipases A2 from Bothrops leucurus (white-tailed-jararaca) snake venom. Biochimie. 2007;89(3):319–328. doi: 10.1016/j.biochi.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 103.Chijiwa T., Tokunaga E., Ikeda R., Terada K., Ogawa T., Oda-Ueda N., Hattori S., Nozaki M., Ohno M. Discovery of novel [Arg49]phospholipase A2 isozymes from Protobothrops elegans venom and regional evolution of Crotalinae snake venom phospholipase A2 isozymes in the southwestern islands of Japan and Taiwan. Toxicon. 2006;48(6):672–682. doi: 10.1016/j.toxicon.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 104.Alape-Giron A., Sanz L., Escolano J., Flores-Diaz M., Madrigal M., Sasa M., Calvete J.J. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J. Proteome Res. 2008;7(8):3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- 105.Paramo L., Lomonte B., Pizarro-Cerda J., Bengoechea J.A., Gorvel J.P., Moreno E. Bactericidal activity of Lys49 and Asp49 myotoxic phospholipases A2 from Bothrops asper snake venom—synthetic Lys49 myotoxin II-(115-129)-peptide identifies its bactericidal region. Eur. J. Biochem. 1998;253(2):452–461. doi: 10.1046/j.1432-1327.1998.2530452.x. [DOI] [PubMed] [Google Scholar]
- 106.Berg O.G., Gelb M.H., Tsai M.D., Jain M.K. Interfacial enzymology: the secreted phospholipase A(2)-paradigm. Chem. Rev. 2001;101(9):2613–2654. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- 107.Fenard D., Lambeau G., Valentin E., Lefebvre J.C., Lazdunski M., Doglio A. Secreted phospholipases A(2), a new class of HIV inhibitors that block virus entry into host cells. J. Clin. Invest. 1999;104(5):611–618. doi: 10.1172/JCI6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lambeau G., Barhanin J., Schweitz H., Qar J., Lazdunski M. Identification and properties of very high affinity brain membrane-binding sites for a neurotoxic phospholipase from the taipan venom. J. Biol. Chem. 1989;264(19):11503–11510. [PubMed] [Google Scholar]
- 109.Cupillard L., Mulherkar R., Gomez N., Kadam S., Valentin E., Lazdunski M., Lambeau G. Both group IB and group IIA secreted phospholipases A2 are natural ligands of the mouse 180-kDa M-type receptor. J. Biol. Chem. 1999;274(11):7043–7051. doi: 10.1074/jbc.274.11.7043. [DOI] [PubMed] [Google Scholar]
- 110.Chwetzoff S., Tsunasawa S., Sakiyama F., Menez A. Nigexine, a phospholipase A2 from cobra venom with cytotoxic properties not related to esterase activity. Purification, amino acid sequence, and biological properties. J. Biol. Chem. 1989;264(22):13289–13297. [PubMed] [Google Scholar]
- 111.Lambeau G., Lazdunski M. Receptors for a growing family of secreted phospholipases A2. Trends Pharmacol. Sci. 1999;20(4):162–170. doi: 10.1016/s0165-6147(99)01300-0. [DOI] [PubMed] [Google Scholar]
- 112.Joubert F.J. Naja mossambica mossambica venom. Purification, some properties and the amino acid sequences of three phospholipases A (CM-I, CM-II and CM-III) Biochim. Biophys. Acta. 1977;493(1):216–227. doi: 10.1016/0005-2795(77)90275-6. [DOI] [PubMed] [Google Scholar]
- 113.Ahmad T., Lawrence A.J. Purification and activation of phospholipase A2 isoforms from Naja mossambica mossambica (spitting cobra) venom. Toxicon. 1993;31(10):1279–1291. doi: 10.1016/0041-0101(93)90401-4. [DOI] [PubMed] [Google Scholar]
- 114.Chen M., Aoki-Utsubo C., Kameoka M. vol. 7. 2017. Broad-spectrum Antiviral Agents: Secreted Phospholipase A(2) Targets Viral Envelope Lipid Bilayers Derived From the Endoplasmic Reticulum Membrane; p. 15931. (1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vacklin H.P., Tiberg F., Fragneto G., Thomas R.K. Phospholipase A2 hydrolysis of supported phospholipid bilayers: a neutron reflectivity and ellipsometry study. Biochemistry. 2005;44(8):2811–2821. doi: 10.1021/bi047727a. [DOI] [PubMed] [Google Scholar]
- 116.van Meer G., de Kroon A.I. Lipid map of the mammalian cell. J. Cell Sci. 2011;124(Pt 1):5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 117.Monje-Galvan V., Klauda J.B. Modeling yeast organelle membranes and how lipid diversity influences bilayer properties. Biochemistry. 2015;54(45):6852–6861. doi: 10.1021/acs.biochem.5b00718. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Q., Hunke C., Yau Y.H., Seow V., Lee S., Tanner L.B., Guan X.L., Wenk M.R., Fibriansah G., Chew P.L., Kukkaro P., Biukovic G., Shi P.Y., Shochat S.G., Grüber G., Lok S.M. The stem region of premembrane protein plays an important role in the virus surface protein rearrangement during dengue maturation. J. Biol. Chem. 2012;287(48):40525–40534. doi: 10.1074/jbc.M112.384446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Higgs P.G. A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol. Direct. 2009;4:16. doi: 10.1186/1745-6150-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lomonte B., Moreno E., Tarkowski A., Hanson L.A., Maccarana M. Neutralizing interaction between heparins and myotoxin II, a lysine 49 phospholipase A2 from Bothrops asper snake venom. Identification of a heparin-binding and cytolytic toxin region by the use of synthetic peptides and molecular modeling. J. Biol. Chem. 1994;269(47):29867–29873. [PubMed] [Google Scholar]
- 122.dos Santos J.I., Soares A.M., Fontes M.R. Comparative structural studies on Lys49-phospholipases A(2) from Bothrops genus reveal their myotoxic site. J. Struct. Biol. 2009;167(2):106–116. doi: 10.1016/j.jsb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 123.Fernandes C.A., Borges R.J., Lomonte B., Fontes M.R. A structure-based proposal for a comprehensive myotoxic mechanism of phospholipase A2-like proteins from viperid snake venoms. Biochim. Biophys. Acta. 2014;1844(12):2265–2276. doi: 10.1016/j.bbapap.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 124.Mora-Obando D., Fernandez J., Montecucco C., Gutierrez J.M., Lomonte B. Synergism between basic Asp49 and Lys49 phospholipase A2 myotoxins of viperid snake venom in vitro and in vivo. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vijay R., Hua X., Meyerholz D.K., Miki Y., Yamamoto K., Gelb M., Murakami M., Perlman S. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J. Exp. Med. 2015;212(11):1851–1868. doi: 10.1084/jem.20150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Müller C., Hardt M., Schwudke D., Neuman B.W., Pleschka S., Ziebuhr J. Inhibition of cytosolic phospholipase A(2)α impairs an early step of coronavirus replication in cell culture. J. Virol. 2018;92(4) doi: 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Allal C., Buisson-Brenac C., Marion V., Claudel-Renard C., Faraut T., Dal Monte P., Streblow D., Record M., Davignon J.L. Human cytomegalovirus carries a cell-derived phospholipase A2 required for infectivity. J. Virol. 2004;78(14):7717–7726. doi: 10.1128/JVI.78.14.7717-7726.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Menzel N., Fischl W., Hueging K., Bankwitz D., Frentzen A., Haid S., Gentzsch J., Kaderali L., Bartenschlager R., Pietschmann T. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 2012;8(7) doi: 10.1371/journal.ppat.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]