Abstract
In the face of looming threats from multi-drug resistant microorganisms, there is a growing need for technologies that will enable rapid identification and drug susceptibility profiling of these pathogens in health care settings. In particular, recent progress in microfluidics and nucleic acid amplification is pushing the boundaries of timescale for diagnosing bacterial infections. With a diverse range of techniques and parallel developments in the field of analytical chemistry, an integrative perspective is needed to understand the significance of these developments. This review examines the scope of new developments in assay technologies grouped by key enabling domains of research. First, we examine recent development in nucleic acid amplification assays for rapid identification and drug susceptibility testing in bacterial infections. Next, we examine advances in microfluidics that facilitate acceleration of diagnostic assays via integration and scale. Lastly, recent developments in biosensor technologies are reviewed. We conclude this review with perspectives on the use of emerging concepts to develop paradigm-changing assays.
Keywords: microfluidics, molecular diagnostics, polymerase chain reaction, PCR, antibiotic susceptibility testing, high-resolution melting analysis, point-of-care
1. INTRODUCTION
In the era of precision medicine, the ability to guide the right drug selection for the indicated patients at the right time remains an elusive goal in clinical bacteriology. General principles for growth-based bacteriology techniques have fundamentally persisted over the past century. Observation of the morphological and biochemical features of isolated pathogens after prolonged cultivation remains the standard approach for identification, with antibiotics as the primary mode of intervention being tested to monitor drug susceptibility. Although laboratory automation has made great strides in improving standardization, throughput, cost, and efficiency, the lengthy processing time over days continues to bottleneck informed therapeutic decision making. For critical systemic infections, such as sepsis, in which every hour delay in effective treatment drastically increases mortality (1), time is of the essence. Managing high-risk infections in acute care settings without definitive diagnosis warrants the use of empiric broad-spectrum rather than etiologically targeted treatment regimens. Antibiotic over- and misuses not only lead to toxic iatrogenic complications (e.g., Clostridium difficile colitis) but drive the emergence and spread of multi-drug resistant organisms. Consequently, clinicians have fewer treatment options, particularly in the most dire patients. Accurate identification of etiologic agents is essential for establishing diagnosis, selecting antibiotics, initiating infection control measures as needed, and providing clues to underlying sources of infections to guide additional treatment modalities. In addition, antibiotic susceptibility testing (AST) to determine drug-resistant phenotypes is critical to ensure effectiveness of the antibiotic being administered. Timely pathogen identification and AST to enable early clinical decision making and effective antibiotic stewardship are integral parts of the overall strategy to improve the clinical outcome of infectious diseases and curtail the spread of antibiotic resistance.
A diverse range of analytical techniques has been developed to solve this important problem. In particular, burgeoning interest and research activities in nucleic acid amplification assays, microfluidics, and biosensors are not only pushing the boundaries of timescale for diagnosing bacterial infections but also showing promise for eventual clinical use. The potential of these analytical techniques, coupled with the yet-to-be-fulfilled goal in clinical applications, will undoubtedly fuel continued research and developments.
In an effort to establish an integrative perspective of recent progresses and provide guidance for future research in this space, we review and examine the scope of new developments in assay technologies grouped by related topics. We begin by surveying current technologies used for pathogen identification and AST, which helps establish an appreciation for the relevance of the emerging techniques. Next, we examine recent development in nucleic acid–based assays for rapid identification and AST of bacterial infections. We discuss recent developments in rapid nucleic acid amplification technologies (NAATs) and NAAT-based techniques incorporating phenotypic assays to overcome the shortcomings of genetic markers. We then examine advances in microfluidics that facilitate acceleration of diagnostic assays via integration and scale. Various types of emerging techniques for assay process integration are discussed, in addition to scale-based techniques for isolating and analyzing single pathogens. Lastly, recent developments in biosensor technologies are reviewed.
2. THE CURRENT LANDSCAPE OF COMMERCIALLY AVAILABLEIDENTIFICATION AND ANTIBIOTIC SUSCEPTIBILITY TESTING TECHNOLOGIES
Observation of current technologies leads to three broad categories of assays for pathogen identification and AST profiling, ranked by their proximity to the specimen source and required assay time. The first category involves assay techniques that require culture-positive specimens and downstream bacteria identification and AST that are both time consuming. The historical gold standard, for example, consists of a two-step culture process (2, 3). For bloodstream infections, a blood specimen is cultured for up to 120 h (median time to positivity is 15 h), with continuous monitoring for growth. Positive blood culture is subsequently evaluated using Gram stain microscopy and subcultured to generate isolates for identification and AST with an additional 24–48 h period. Automated instruments may substitute various aspects of this workflow to minimize labor in high-volume facilities. In samples with sufficient titer for direct measurement, such as urinary tract infection and bacterial meningitis, the presence of pathogens is screened directly from specimen via Gram stain microscopy and cultured on selective media for identification and subsequent AST. Genomics-based techniques such as 16S sequencing on isolates provide detailed phylogenetic information, although the time and resources expended on this method of identification make it less suitable for rapid diagnostic applications.
The next category involves assay techniques developed to enhance the speed and accuracy of pathogen identification from culture-positive specimens. The most significant development involves matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry, developed and marketed by diagnostic instrument manufacturers such as Bruker (4) and bioMérieux (5). This method enables protein-based identification of a broad range of bacteria and fungi directly from cultured samples on the order of minutes. Another development is fluorescence in situ hybridization (FISH), which utilizes fluorescent probes specific to the pathogen of interest to directly stain and monitor its presence via microscopy or luminometer. AccuProbe (Hologic)andQuickFISH/PNAFISH(OpGen)aresomeofthemorecommonlyknowncommercially available probes for this application (6).The integrated identification/AST assay instrument fromAccelerateDiagnosticsalsorepresentsthiscategory.Afteragelelectro-filtrationstep,theAccelerate Pheno system uses FISH for species identification and automated time-lapse microscopy on individual bacterial cells for phenotypic AST with minimum inhibitory concentration (MIC) reporting directly from positive blood cultures. Recently, panel-based approaches to pathogen identification and genotypic resistance testing gave rise to multiplexed polymerase chain reaction (PCR) assays targeting specific pathogens with limited resistant markers in culture-positive samples. Platforms for blood culture testing such as the Verigene Gram-Positive/Gram-Negative Blood Culture Tests (Luminex) and BD Max (Becton Dickinson) represent this category.
The last category of delivery represents assay technologies that provide identification and/or AST results directly from patient specimens without the initial overnight culture step. Current technologiesinthiscategoryareprimarilyrepresentedbytwotypes:sample-to-answerPCRassays and lateral-flow devices. Sample-to-answer PCR assays include the FilmArray platform (BioFire Diagnostics), a closed and fully automated system capable of detecting syndromic panels of bacteria and viruses directly from uncultured patient specimens by combining DNA extraction, nested multiplex PCR, and post-PCR melting curve analysis (7). The most recent iteration of these devices on the market is embodied by the T2MR platform (T2 Biosystems), where assay sensitivity is boosted by the combined use of direct PCR, allowing direct analysis of whole blood specimens with pathogen detection within hours of sample collection (8). Meanwhile, lateral-flow tests are typically based on immunoprecipitation and are employed in outpatient settings for point-of-care (POC) screening.
Although optimizations can be made at various steps of the clinical workflow, rapid pathogen diagnostics is only achieved when results are delivered directly from patient specimens without the initial culture step. This is especially relevant in the context of bloodstream infections (Table 1), where rapid diagnosis and treatment can substantially improve patient outcome (1). However, the current generation of assays leaves much to be desired in terms of functionality. For example, most assays currently available for rapid identification or AST require cultured pathogen isolates as inputs. Furthermore, the utility of current assays is limited to identifying one or a defined panel of pathogens. The lack of provisions for comprehensive AST is a major gap that continues to limit current platforms as adjuncts to culture-based testing. Several emerging technologies seek to achieve comprehensive identification and AST directly from clinical specimens by leveraging concepts that we discuss in subsequent sections.
Table 1.
Summary of commercially available rapid identification/antibiotic susceptibility testing technologies for bloodstream infections. Adapted from Edmiston et al. (130)
| Technology | Pathogen identification | Detection of antibiotic resistance | Antibiotic susceptibility testing with MIC | Turnaround time/time to result from patient blood | Turnaround time/time to result from positive blood culture |
|---|---|---|---|---|---|
| Singleplex PCR | Yes | No | No | NA | 1–3 h |
| Multiplex PCR | Yes | Genotypic only | No | NA | 1–2 h |
| NAAT/microarray | Yes | Genotypic only | No | NA | 2.5 h |
| MALDI-TOF | Yes | No | No | NA | 24 h |
| PNA-FISH | Yes | No | No | NA | 1 h |
| T2MR assay | Yes | No | No | 3–5 h | NA |
Abbreviations: MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight; MIC, minimum inhibitory concentration; NA, not applicable; NAAT, nucleic acid amplification technology; PCR, polymerase chain reaction; PNA-FISH, peptide nucleic acid–fluorescence in situ hybridization.
3. ACCELERATING ASSAYS VIA NUCLEIC ACID AMPLIFICATION
Analytical techniques for pathogen identification and AST based on NAATs encompass a broad range. In particular, a wide range of techniques has been developed for achieving rapid pathogen identification. Toward achieving rapid, NAAT-based AST, while the detection of genotypic markers for drug resistance remains a focus, recent progresses have shifted to a pheno-molecular-based approach, in which NAAT is employed as the readout for accelerating phenotypic-based AST. Meanwhile, there has also been increased research activity and progress in measuring the response biomarkers (e.g., RNA) of the host (i.e., patient) as a means of providing complementary information and further accelerating diagnosis in the early stages of disease.
3.1. Pathogen Identification
Employing NAATs for achieving rapid pathogen identification has today become turnkey. Indeed, a wide range of techniques such as PCR, isothermal amplification, high-resolution melting analysis, digital PCR, next generation and third generation sequencing, and additional emerging amplification strategies have been developed. We highlight herein representative studies within each technique to illustrate the state of the art of each technique.
3.1.1. PCR-based NAAT.
PCR is considered one of the most promising and reliable NAATs to advance microbial diagnostics by improving both speed and accuracy. PCR can detect bacterial DNA from nonculturable organisms—a common clinical scenario when patients received prior antibiotics. Recent commercial platforms involving limited multiplex pathogen identification can survey syndromic panels of different pathogen classes (e.g., bacterial, viral, fungal) in an effort to reduce antibiotic usage if evidence of nonbacterial infection is generated. Whereas some platforms with sample-to-answer capability allow direct detection from nonsterile (e.g., sputum, stool) or noncomplex sterile (e.g., urine, cerebrospinal fluid) specimens, detection of bloodstream infections still requires postculture samples due to low starting pathogen loads (<1–100 colony-forming units/mL). Most culture-independent PCR assays for bloodstream infections suffer suboptimal sensitivity, which may in part be explained by insufficient blood volume used for testing or inefficient removal of PCR inhibitors hampering DNA amplification. The T2MR system allows for direct PCR from whole blood samples. It involves the use of magnetic nanoparticles to allow near background-free sensing of post-PCR products. In clinical trials, T2MR demonstrated a sensitivity of 91.1% and specificity of 99.4%, with a time to result of 4–5 h for direct whole blood detection. Unfortunately, it still suffers from limited pathogen coverage (only five bacterial and Candida species in each).
Given the myriad of potential pathogens in bloodstream infections, an agnostic and catch-all detection strategy is ideally suited in this scenario. PCR followed by electrospray ionization mass spectrometry (PCR/ESI-MS)–based technology such as Abbott’s Plex-identification/IRIDICA (withdrawn from market) offers broad-spectrum detection capable of identifying approximately 780 microbial species that include >95% of eubacterial species associated with human infection, as well as four antibiotic-resistant markers (9–11). This technology combines PCR and downstream ESI-MS to amplify universal target genes [e.g., bacterial 16S ribosomal RNA (rRNA)] and discriminate amplicon sequence variants, allowing for identification of one or more species directly from whole blood samples within 8 h. A study analyzing 331 whole blood specimens from patients suspected to have bloodstream infections shows 91% sensitivity and 99% specificity with this assay (11). Recent platforms have also offered random-access features and automation that increase workflow efficiency. Additionally, improvement in PCR chemistries, such as rapid and extreme PCR, might also allow for accelerated PCR speed, resulting in shorter overall assay times (12, 13). One limitation common to PCR diagnostics is that the interpretation of positive results with negative culture remains challenging in terms of differentiating true infection from normal microbiota, transient colonizer, or sample contaminant. On the other hand, without exhaustive multiplex detection of pathogens, a negative result does not rule out infection. Together with the lack of reliable AST information, PCR-based diagnostics remains constrained as an adjunctive test to conventional cultures.
3.1.2. Isothermal NAAT.
Although PCR is the reference standard method for NAAT, its use is limited by the requirement for a thermocycler and downstream data analysis. In contrast, isothermal amplification techniques, which include rolling cycle amplification (RCA), strand-displacement amplification (SDA), helicase-dependent amplification (HDA), recombinase polymerase amplification (RPA), and loop-mediated amplification (LAMP), are mostly performed at one specific temperature and therefore do not require a thermocycling instrument. Among isothermal amplification strategies, LAMP has the most potential for NAAT. Studies showed increased sensitivity from regular PCR for detection of common bacterial pathogens such as Escherichia coli, Listeria monocytogenes, and Staphylococcus aureus (14–16). A LAMP study reported a 97% sensitivity and 100% specificity for identification of E. coli in 45 min (15). Taking advantage of magnesium pyrophosphate production as a byproduct of LAMP, measurement of results is usually performed by colorimetric detection using dyes such as SYBR Green or real-time optical instruments (17). Other detection methods include gel electrophoresis, immunoassays, electrochemical detection, and pH sensing (18). Integration of LAMP with microfluidic devices and downstream fluorescence capture allows for an even faster and more sensitive use of LAMP for nucleic acid detection (19). Despite its simplicity and sensitivity, LAMP still suffers from poor specificity due to contaminations and high false-positive rates. Future optimization of its chemistry is needed for improved accuracy and reproducibility.
3.1.3. High-resolution melting analysis.
High-resolution melting analysis (HRMA) is a simple, yet practical post-PCR amplification method that can create sequence-dependent melting curves completed through seamless closed-tube integration with quantitative PCR (qPCR) (20–22). For the past decade, this technique coupled with broad-range PCR has been used as a fingerprinting solution for large-scale bacterial identification and has demonstrated its clinical feasibility in the diagnosis of various infectious diseases (23–33). Aside from the commonly targeted 16S rRNA gene, amplicons generated from targeting the bacterial internal transcribed spacer (ITS) region were shown to create rich melting curve profiles that enhanced the breadth and analytical specificity of HRMA for species-level identification (34). Coupled with a machine-learned curve-classification algorithm, HRMA was reported to have 90% accuracy in identifying bacterial pathogens from culture-positive blood culture samples (34).The algorithm allowed for automated differentiation of bacterial species based on its unique melting curve profile when analyzed against an archived melting curve database (Figure 1a). Because the reference database can be incrementally updated and improved with the addition of more strains, the assay is easily expandable. Traditional bulk HRMA cannot resolve all species in a polymicrobial infection, as each nucleic acid sequence in the mixture will contribute to the ensembled, composite melting curve, which is impossible to decouple into individually contributing species. To overcome this limitation, digital PCR has been combined with digital HRMA to resolve heterogenous populations of target cells/DNA with absolute quantification (31, 35).
Figure 1.
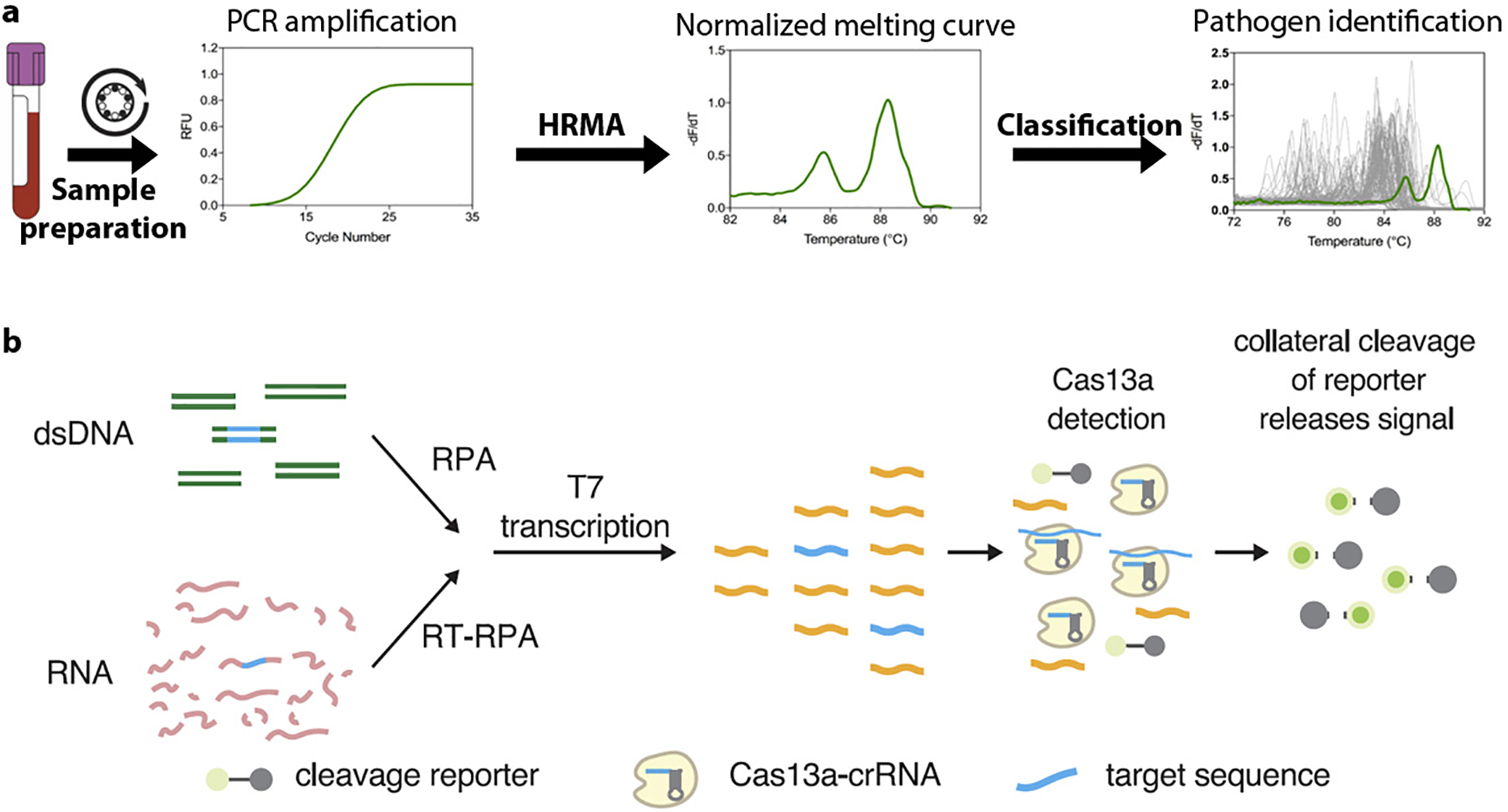
Emerging demonstrations of NAAT-based pathogen identification. (a) With HRMA, species were identified by their unique melting curves. After PCR amplification and HRMA, the raw melting curve of an unknown species was normalized and transformed into a derivative curve. An adaptive algorithm was then matched the unknown curve (green line) against an archived melting curve database (gray lines) to find the best fit. (b) Schematic of the SHERLOCK platform. The DNA or RNA target extracted from biological samples is amplified by RPA (RT-RPA or RPA, respectively). RPA products are detected in a reaction mixture containing T7 RNA polymerase, Cas13, a target-specific crRNA, and an RNA reporter that fluoresces when cleaved. Figure adapted with permission from Reference 48. Copyright 2017, AAAS. Abbreviations: crRNA, CRISPR RNA; HRMA, high-resolution melting analysis; NAAT, nucleic acid amplification technology; PCR, polymerase chain reaction; RFU, reflective fluorescence unit; RT, reverse transcription; RPA, recombinase polymerase amplification; SHERLOCK, specific high-sensitivity enzymatic reporter unlocking.
3.1.4. Digital PCR.
Conventional NAATs rely on bulk methods, in which the whole population of cells is analyzed based on genetic content. The detection sensitivity for such bulk methods is compromised, especially when analyzing challenging samples such as blood, by a high excess of background constituents, such as human DNA, bystander bacterial DNA, and PCR inhibitors (36). Reliable detection of targets becomes challenging in samples containing low concentrations of causative agents. The detection time becomes significantly longer as the target cell concentration in the sample decreases. Digital PCR (dPCR) can address these problems. dPCR platforms are capable of massive sample partitioning into minute PCR reaction volumes in which the effective target concentration in each reaction is drastically enhanced with diluted background/inhibitors, allowing for even higher detection sensitivity, resolution, speed, and precision without the need for standards. Studies involving dPCR for identification of foodborne pathogens showed higher sensitivity than regular PCR for lower numbers of pathogens with quantitative capability (37, 38). Moreover, pathogen diagnostics at the single-cell level can also offer valuable clinical information for guiding management by (a) resolving polymicrobial infections, (b) differentiating bystanders from true pathogens, and (c) correlating pathogen load with disease severity and treatment efficacy. There are currently no US Food and Drug Administration (FDA)–approved dPCR systems for diagnostic use in United States. Commercial dPCR systems for research use only include the QX200 Droplet Digital PCR System (Bio-Rad), Raindrop Digital PCR System (RainDance Technologies), qdPCR 37K IFC (Fluidigm), and QuantStudio 3D Digital PCR System (Life Technologies).
3.1.5. Next-generation and third-generation sequencing.
With the increasing affordability of next-generation sequencing (NGS) techniques, microbial sequencing has been utilized for pathogen identification with promising accuracy and time to result (39). One such application is metagenomics NGS, a method in which all of the nucleic acid (DNA and/or RNA) of a specimen is sequenced in parallel. In addition to yielding hypothesis-free, culture-independent pathogen detection directly from clinical specimens, NGS has the potential to enable epidemiological typing, pathogen evolution, detection of toxins and virulence, and host immune response to the offending pathogen. Although the work is still in progress, it has shown that data obtained through NGS can also be utilized to infer or predict phenotypic resistance. But as we discuss below, the correlation between genotypic data and clinical phenotype is not always true. Bulky and expensive sequencing instruments pose big challenges for NGS to be used as a POC test, but the recent invention of a portable MinION nanopore sequencing device may enable the use of sequencing for future clinical adoption. This portable device measures the changes in electrical current as a single-stranded DNA passes through the nanopore and uses the signal to determine the nucleotide sequence of the DNA strand (40). The sequence data are analyzed in real time, enabling results in the shortest time possible. An early study analyzing the MinION nanopore sequencing showed that bacterial species and strain information could be generated within 1 h of sequencing time (after DNA extraction and library preparation), and the results on initial and complete drug resistance profiles were available within 2 h and 12 h, respectively (41). Because fragments of genomic DNA from pathogens that cause infection at various locations within the body can be found in purified plasma cell-free DNA (cfDNA), metagenomic sequencing has been utilized for noninvasive detection of pathogen-derived cell-free plasma DNA in infected patients (42–44). Horiba et al. (45) reported that bacteria detected by NGS in plasma/serum were identical to the dominant bacteria isolated in blood culture for 8 out of 12 patients with bloodstream infections. Despite increasing reports of NGS successes, several hurdles still need to be addressed. These include differentiation of colonization and contaminants from infection, method standardization, as well as data storage, protection, analysis, and interpretation.
3.1.6. Other emerging nucleic acid–based technologies.
A number of innovative methods have also been developed for pathogen identification, with the aim of improving on overall speed, accuracy, and integration into clinical workflows. Using direct whole blood samples, researchers reported duplex DNA-invading γ-modified peptide nucleic acids (γPNAs) for rapid identification of bacterial and fungal pathogens. This HelixBind technology allows for the interaction between amplicon and γ-PNA followed by the formation of stable hybrid structures when perfect binding is accomplished. These structures then immobilize the target amplicon onto magnetic beads, enabling optical detection via chemiluminescence, while mismatched γ-PNAs are excluded. This platform provides species-level information in under 2.5 h and shows >95% sensitivity and >90% overall correlation to blood culture findings through the analysis of 61 clinical specimens (46).
A nonamplification method called universal microbial diagnostics exposes microbial genomic DNA to a collection of DNA probes to identify the pathogens present (47). The probes are randomly generated nucleotides that freely hybridize to different spots and different extents on various bacterial genomes. By measuring the degree to which the sample hybridizes with the collection of random probes, the technique can detect the presence and estimate the concentrations of the various bacteria in the sample. Owing to this random DNA probe design and structure, the test is universal, inexpensive, rapid, and phylogenetically informative: It can classify novel mutants with their closest known relatives (47).
One recent technology is the modification of CRISPR-based systems for highly specific and sensitive detection of RNA and DNA. The SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) platform combines the collateral effect of the Cas system with isothermal amplification for nucleic acid detection. The procedure starts with amplification by RPA, T7 RNA polymerase transcription of amplified DNA to RNA and, finally, the detection of target RNA by Cas13a collateral RNA cleavage-mediated release of reporter signal for fluorescence detection (Figure 1b) (48). By targeting the 16S rRNA gene V3 region where conserved flanking sequences allow universal RPA primers to be used across bacterial species with variable internal sequences, the method provided successful species differentiation and detection of low quantities of pathogen RNA/DNA with single base mismatch specificity (48–51). The reagents can also be lyophilized for long-term storage and portability.
3.2. Rapid Antibiotic Susceptibility Testing
Although NAAT offers substantial advantages in speed and sensitivity compared to other techniques, AST remains a challenge. Currently available techniques focus on the detection of genotypic markers for drug resistance, such as methicillin-resistant genes for S. aureus and vancomycin-resistant genes for enterococci (Table 2). NAAT-based detection of genotypic markers can yield results within a few hours, compared to phenotypic assays using standard culture-based approaches such as broth microdilution or the gradient plate method. A genotypic approach to testing drug resistance is appropriate when there is a direct causal relationship between the gene and the resistance phenotype and where the resistance gene panel is involved in a single or a few mechanisms of action for a particular drug. An example is quinolone resistance in Neisseria gonorrhoeae, which can be largely explained by a mutation in the DNA gyrase gene gyrA (52). Unfortunately, these constitute only a handful of potential cases for antibiotic resistance and do not provide a viable strategy for evaluating the vast majority of phenotypic drug resistance where multiple metabolic pathways are involved (53). Furthermore, treatment guidelines are not only determined by the presence of resistance genes but also by the level of resistance, which requires determining the MIC of drugs. Due to these factors, phenotypic assays continue to be utilized as the gold standard for AST despite a lengthy assay time.
Table 2.
List of NAAT platforms with genotypic resistance detection that are approved by the US Food and Drug Administration (FDA). Adapted from Bard & Lee (53) with permission from Elsevier
| Assay | Company | Specimen type | Method | Resistance gene | Run time |
|---|---|---|---|---|---|
| Xpert MRSA/SA Gen 3 | Cepheid | Blood culture | RT-PCR | Methicillin (mecA, mecC) | ~1 h |
| Xpert MTB/RIF | Cepheid | Sputum | RT-PCR | rpoB (Mycobacterium tuberculosis) | < 2 h |
| Xpert MRSA/SA SSTI | Cepheid | Swab | RT-PCR | mecA (Staphylococcus aureus) | ~1 h |
| BD Max StaphSR | Becton Dickinson | Blood culture | RT-PCR | mecA (Staphylococcus aureus) | ~1.5 h |
| Verigene Gram-Positive Blood Culture | Luminex | Blood culture | Microarray |
mecA (Staphylococcus aureus) vanA, vanB (Enterococcus spp.) |
2.5 h |
| Verigene Gram-Negative Blood Culture | Luminex | Blood culture | Microarray | CTX-M, IMI, VIM, KPC, NDM, OXA (Enterobacteriaceae) | 2.5 h |
| FilmArray Blood Culture identification | BioFire Diagnostics | Blood culture | Nested PCR |
mecA (Staphylococcus aureus) vanA, vanB (Enterococcus spp.) KPC (Enterobacteriaceae) |
1 h |
Abbreviations: NAAT, nucleic acid amplification technology; PCR, polymerase chain reaction.
Although NAAT is primarily a method for genotypic resistance detection, recent developments show that it can be used to make phenotypic observations for AST. The concept of phenol-molecular AST that couples culture and PCR for AST was introduced in the early 2000s (54, 55). The high sensitivity of PCR for detecting genomic content as a surrogate for bacterial growth greatly shortens the timescale of AST to hours as opposed to days needed in conventional culture-based methods. In addition to time saving, the use of PCR to perform phenotypic AST also confers an additional layer of specificity compared to conventional AST. Since the initial studies, several innovations led to rapid phenotypic AST and MIC determination using NAAT. A proof-of-concept study by Waldeisen et al. (56) demonstrated the use of quantitative PCR as a tool for generating antibiogram from E. coli spiked in blood with as short as 8 h of culture. In this scheme, bacterial growth was performed under AST conditions similar to broth microdilution and was measured using the PCR threshold cycle number (Cq).Susceptibility was measured by a large gap in Cq compared to a no-drug control, while resistance would be indicated by a minimal gap in Cq compared to the control. Similarly, using 16S PCR, Beuving et al. (57) detected the growth of a variety of bacteria species from positive blood bottle cultures within 9 h. Moreover, species-specific primers and probes can also be used to monitor growth of a specific target in the presence of other organisms. To this end, Chen et al. (58) recently demonstrated a gonorrhea-specific assay capable of measuring growth that is specific to N. gonorrhoeae from a complex sample matrix containing endogenous vaginal flora. In this work, the authors also demonstrated the use of direct PCR to further shorten the time to result by abridging the traditional nucleic acid isolation step. To demonstrate the potential utility to guide antibiotic selection for individualized treatment, the direct PCR assay was applied to test seven N. gonorrhoeae strains against three antibiotic agents, penicillin, tetracycline, and ciprofloxacin, with <4 h of drug exposure.
Broad amplification of multiple species of interest using universal PCR primers in a single sample can also be differentiated using a postamplification analysis technique, pushing this approach closer to the ultimate goal of integrated pathogen identification and AST. To illustrate this capability, a recent method proposed by Athamanolap et al. (59) implemented 16S PCR and HRMA to perform integrated bacterial identification and antibiotic susceptibility testing (Figure 2a). Integrated pathogen identification and AST of samples containing multiple bacterial species (polymicrobial samples) were also demonstrated when the assay was performed in a digital PCR-HRM format (59). More recently, Andini et al. (60) demonstrated the use of PCR-HRMA of the ITS region between the 16S and 23S for broad bacterial detection and AST directly from whole blood. The entire assay process, including sample pre-enrichment, antibiotic incubation, bacterial identification, and AST, could be completed in as few as 8 h.
Figure 2.
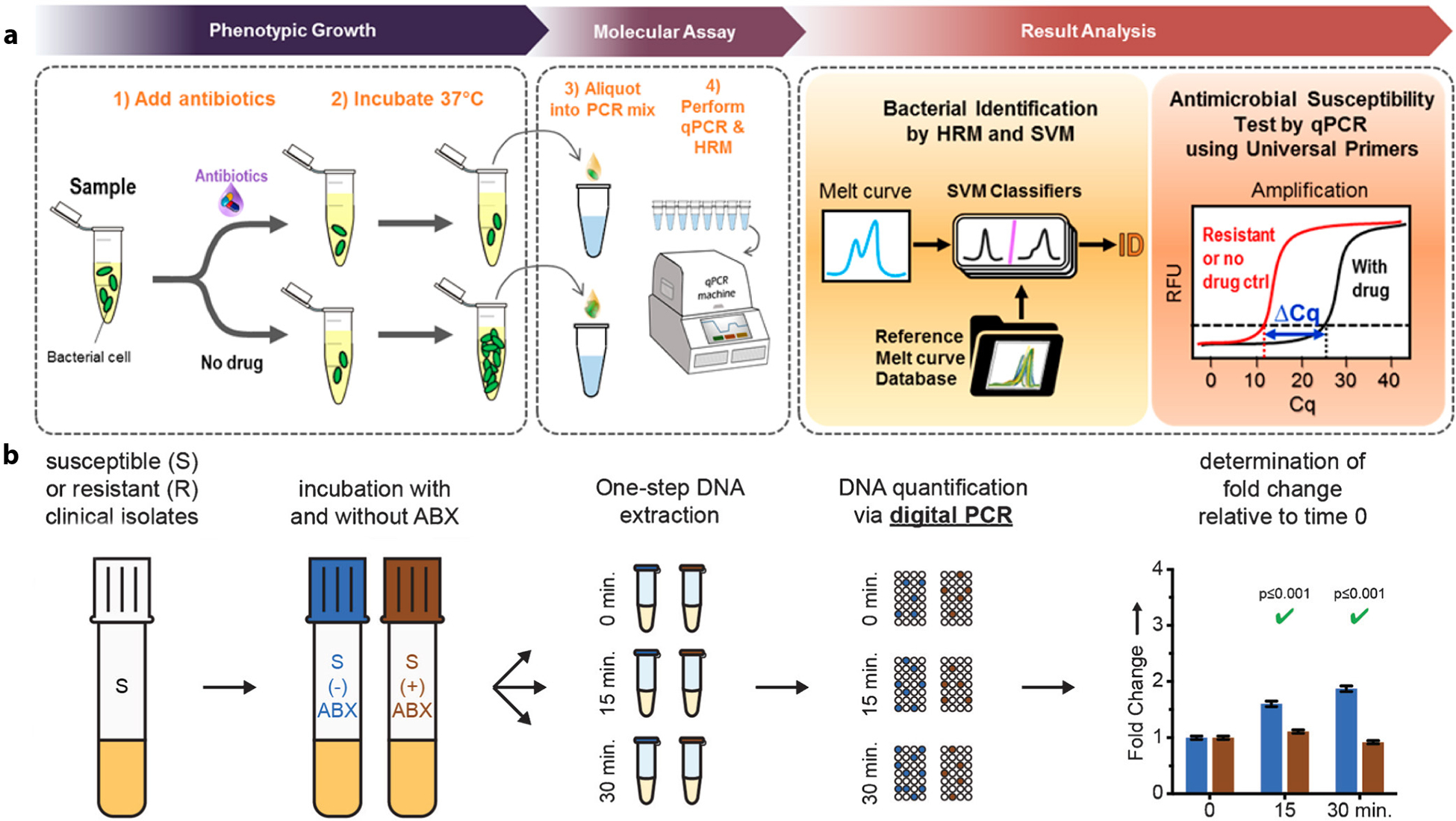
Emerging demonstrations of NAAT-based, pheno-molecular AST. Pheno-molecular AST uses NAAT to measure bacterial growth under phenotypic AST and takes advantage of the speed and sensitivity of NAAT to achieve rapid and robust AST. Typical assays start with the brief incubation of a bacteria sample with and without antibiotics. AST can then be determined by measuring the differences of DNA quantity between reactions with and without antibiotics, via either quantitative PCR or digital PCR, as bacteria that do not grow in the presence of the antibiotic (i.e., susceptible) would have less bacterial DNA than the no-antibiotic control. (a) The pheno-molecular AST concept has been coupled with broad-based, real-time PCR and HRM. In this case, unknown bacteria in the sample can be identified by matching the newly generated melt curve to the database of previously built melt curves via a machine-learning algorithm. AST is determined via a measurable ΔCq between the antibiotic sample and the no-antibiotic control. Panel adapted with permission from Reference 59. Copyright 2017, American Chemical Society. (b) The pheno-molecular AST concept has also been coupled with digital PCR. Following the incubation of samples with and without ABX for different time courses, antibiotic susceptibility of the pathogen tested is determined via digital PCR based on the fold change relative to time 0. Panel adapted with permission from Reference 61. Copyright 2016, John Wiley & Sons. Abbreviations: ABX, antibiotics; AST, antibiotic susceptibility testing; Cq, cycle number; HRM, high-resolution melting; NAAT, nucleic acid amplification technology; PCR, polymerase chain reaction; qPCR, quantitative PCR; RFU, relative fluorescence unit; SVM, support vector machine.
In pheno-molecular AST, antibiotic susceptibility is determined by differentiating the bacterial DNA or RNA quantity between drug-treated samples and no-drug controls (Figure 2a). The required incubation (antibiotic exposure) time for definite assessment of antibiotic susceptibility ranges from <1 h for pathogens of rapid replication rates such as E. coli to >10 h for fastidious pathogens (54, 56–60). Although the minimum incubation time is mainly dependent on the pathogen–drug combination, it can be reduced by using digital methods with high-quantification resolution capable of detecting a smaller change in bacterial nucleic acids. The scheme of digital AST was initially demonstrated by Schoepp et al. (61) who performed digital quantification of DNA replication using droplet digital PCR (the QX200 system by Bio-Rad). This enabled determination of antibiotic susceptibility of clinical isolates (E. coli) from urinary tract infections after 15 min of antibiotic exposure (Figure 2b). A subsequent study employing ultrafast digital LAMP in place of dPCR to perform AST further shortened the assay workflow to allow the completion of AST of E. coli directly from clinical urine samples within 30 min (62).
The assay time for NAAT-based phenotypic AST to deliver robust antibiogram results currently still needs to be confined to several hours due to the variability of bacterial replication rates in different pathogen–antibiotic combinations (56, 58). It is anticipated that further acceleration of this approach will take place by adopting the more abundant RNA markers in place of genomic DNA. Although the metabolic response of bacterial pathogens to antibiotic exposure may be complex, recent studies using RNA sequencing have helped to develop genome-scale metabolic models to better correlate phenotypic changes with RNA expression levels (63). Such models may be utilized to develop assays that can rapidly detect transient changes in RNA expression levels attributed to drug exposure. Furthermore, NAAT-specific optimizations to accelerate assay time, including extreme PCR (64) and direct PCR (65), could also be employed to further improve the time to result.
3.3. Host Response Markers
Biomarkers often serve as proxies for host response to a disease, informing disease progression and severity. Easily measurable host analytes such as procalcitonin are sensitive markers for inflammation and infections but have limited disease specificity, making them poor diagnostic tools. The development of microarray and omics technologies allows for recognition of the changes in nucleic acids of the host to inform not only disease pathology but also disease type. Such nucleic acid biomarkers provide the missing information that pathogen diagnostics alone cannot currently determine, for example, infection versus colonization, while they concurrently deliver prognostic and diagnostic information (66).
3.3.1. Nucleic acid biomarkers as rapid diagnostic tools.
Nucleic acid detection of host response is mainly based on transcriptomic studies and looks at changes in RNA level as an early measure of gene expression, which includes different RNA molecules such as mRNA and noncoding RNA. The pattern of genes that is expressed and their level of expression differ between cells according to conditions, defining the physiological state of each cell (67). Measuring the amount of mRNA enables the interpretation of a patient’s response to external stimuli, such as infection, and furthers our understanding of the molecular regulatory mechanisms that underlie the disease. Recent studies have reported microRNAs (miRNAs) as promising markers for disease diagnostics. These small noncoding RNAs are involved in posttranslational regulation and have been shown to feature distinct expression signatures in various infectious diseases (68).They are also ideal targets for diagnostics due to their resistance to boiling, freeze-thaw cycles, and decay (69, 70).
There are various methods for RNA quantification. The most preferred method is real-time quantitative reverse-transcription PCR (qRT-PCR), which is ideal when a set of identified targets is to be tested. It is a common and simple technique that is suitable for validation studies. However, the prerequisite to detect numerous target mRNAs might be a challenge for conventional qRT-PCR with limited multiplexing capability. Microarrays and RNA sequencing (RNA-seq) allow for whole transcriptomic analysis. Currently available arrays and microfluidic systems, such as those offered by NanoString and Fluidigm, are able to quantitate multiple mRNAs rapidly and in high-multiplex format. The ability of RNA-seq for unbiased capture and novel marker discovery triumphs over microarrays with their limited probes, although it comes with longer time, higher cost, and more elaborate data analysis. The set of markers for different conditions established through RNA-seq is commonly validated using qRT-PCR.
3.3.2. Biomarker signatures for infectious diseases.
The information based on host response could lead to early and improved diagnostics to better tailor therapy and improve outcome. Existing studies focused on the fundamental diagnostic questions, for example, infectious inflammation/SIRS (sepsis) versus noninfectious SIRS and bacterial versus viral, which would help clinicians better determine the course of therapy. A 42-gene classifier (SeptiCyte) was reported to differentiate sepsis from noninfectious SIRS in humans (71). The SeptiCyte classifier could detect sepsis 86–92% of the time. A recent analysis of publicly available data resulted in an 11-gene signature (Sepsis MetaScore) that was able to differentiate infection versus sterile inflammation with a mean ROC AUC of 0.87 and 0.83 in nine discovery and nine validation cohorts, respectively (72).
Distinct gene expression patterns between viral and bacterial infections were identified in pediatric patients with respiratory infections and febrile illnesses (73, 74). A study in an adult population of hospitalized patients with bacterial or viral lower respiratory tract infections discovered a 10-gene classifier that discriminated viral and bacterial etiologies with 91% accuracy in the training set and 96% accuracy in two validation cohorts compared to procalcitonin, which had 38% sensitivity and 91% specificity in the same population (75). Another bacterial/viral metascore based on a 7-gene signature capable of robust discrimination of bacterial and viral infections was identified and validated in 30 independent validation cohorts of more than 2,400 whole blood and peripheral blood mononuclear cell (PBMC) samples from patients with bacterial or viral infections with an AUC of 0.93 (76).
Owing to the systemic nature of a typical sample matrix (blood), detection of the host response is less prone to sampling error compared to detection of a rare pathogen. Without the need for cell enrichment, sample preparation should also be simpler and faster than pathogen detection. The use of RNA, however, requires the cautious handling of samples to maintain RNA integrity and purity, which can be labor intensive. Different study designs using different cell types ranging from whole blood to specific immune cells generated various signatures for similar questions. Therefore, there have to be further validations of reported host markers from the most informative immune cell type on large cohorts to finalize the minimum set of classifiers. Not until these classifiers are validated and normalized across different populations could they be of use clinically. Current transcriptomic markers can only differentiate down to pathogen classes. More work is necessary to establish possible host transcriptomics markers that can classify to a specific pathogen. Aside from transcriptomics, the advancing research in epigenomics might identify other classifier for early host response markers that allows for a faster and more sensitive test. With the development of simultaneous host and pathogen sequencing, it may also be possible to generate comprehensive information rapidly in a single assay. For the technology to be compatible in a clinical setting, it must have suitable platforms capable of multiplexed quantitative gene expression analysis. Many emerging techniques and materials have been employed for the multiplex detection of nucleic acids (RNA), such as electrochemical detection, microfluidic-based lab-on-a-chip devices, and nanomaterials, including the MinION nanopore sequencing device (67). Coupled with the appropriate pathogen diagnostic ideally on the same platform, host response markers can provide complementary information and further accelerate diagnosis in the early stage of disease when an extremely low pathogen titer may be present (76).
4. ACCELERATING ASSAYS VIA MICROFLUIDICS
Microfluidics encompasses a broad set of technologies that are used to manipulate fluids on a small scale. Due to the ubiquity of fluids in bioassays for pathogen identification and AST, microfluidic concepts have been studied extensively as the means to enhance assay performance in speed, resolution, and accuracy. A recent review on this topic highlights many of the emerging microfluidic technologies for rapid pathogen identification and AST (77). Microfluidics has the opportunity to accelerate bacterial identification and AST in two ways. First, fluidic manipulation at a small scale allowsminiaturizationandintegrationofsampleprocessingstepsintoasinglemodule, exemplified by the sample-to-answer feature found in commercial POC diagnostic platforms. This allows the end user to replace discrete processing steps for nucleic acid purification, assay preparation, and pathogen detection with a single step, resulting in a shorter time to result. A second opportunity is found in the inherent advantages of scale drive enhancement. This advantage is often realized through discretizing bulk biochemical assays into a large number of small volumes (femtoliters to nanoliters), which offers ready means for high-sensitivity and rapid pathogen detection.
4.1. Integration of Microfluidics-Driven Assays
Integration of assay protocols into a disposable cartridge has been the focus of the field of micro-total analysis systems since its inception (78). This is especially important for rapid diagnostic applications, where sample preparation is typically required to bridge the gap between a NAAT and patient specimen. Substantial progress has been made in the last two decades by both industry and academia. A recent review article on this topic discusses the range of commercially available integrated assay systems (79). Commercialized assays utilizing fluidic manifold-based actuation, such as the BioFire FilmArray and Cepheid GeneXpert platforms, as well as newer platforms based on the electrowetting-on-dielectric method, such as the GenMark ePlex platform (80), have already delivered sample-to-answer times within or approaching 1 h (7, 80, 81).Newer technologies in this domain seek to refine sample preparation steps to further accelerate and miniaturize assays for pathogen detection (Figure 3).
Figure 3.
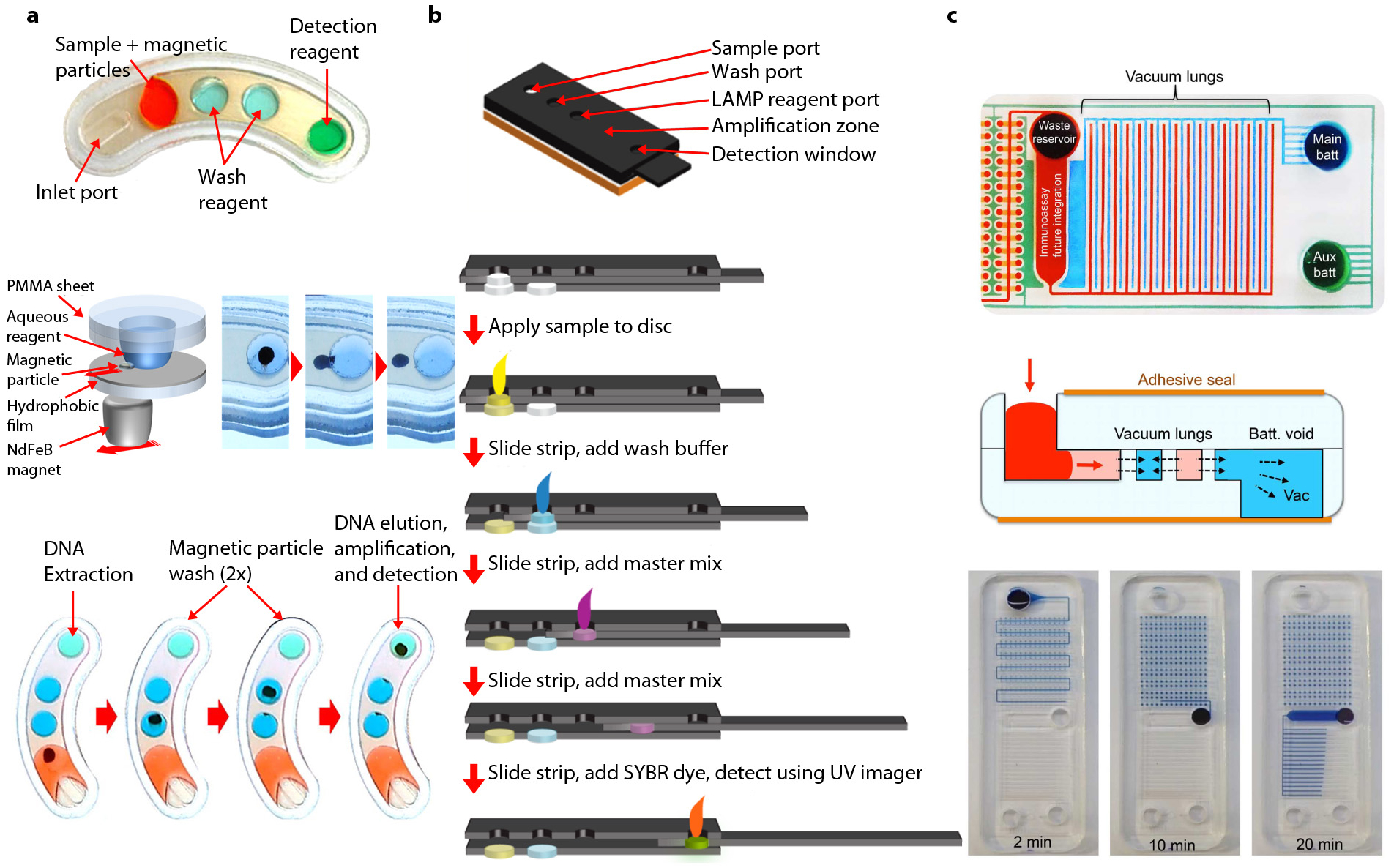
Emerging technologies for microfluidic assay integration. (a) Example of droplet magnetofluidic assay integration. (Top) A magnetofluidic assay cartridge with aqueous reagents for DNA extraction and amplification. (Middle) Overview of droplet magnetofluidic manipulation. Left panel shows a schematic of aqueous droplet anchored by a hydrophilic poly(methyl methacrylate) (PMMA) substrate. Magnetic particles are actuated on a hydrophobically coated surface via a rare-earth neodymium (NdFeB) magnet. Magnetic particles can be extracted from droplets via translational motion of the magnet. (Bottom) Overview of cartridge operation. DNA extraction, particle washing, elution, and amplification are all achieved on a single cartridge, with each process linked via translocation of magnetic particles between each reagent droplet. Panel adapted with permission form Reference 82 under the terms of the Creative Commons Attribution 4.0 International License, http://creativecommons.org/licenses/by/4.0. (b) Example of a paper analytical device–based integration. (Top) Three-dimensional rendering of an analytical device for loop-mediated amplification (LAMP) assay. (Bottom) Overview of device operation. Each reagent for DNA extraction, washing, and amplification is loaded sequentially into a moving paper matrix at each step of the assay. Panel adapted with permission form Reference 92. Copyright 2015, American Chemical Society. (c) Example of vacuum-driven integration. (Top) Image of a vacuum-driven device with interdigitating vacuum batteries highlighted in blue and green. (Middle) Principle of operation. The vacuum pulls air out of fluidic channels through gas-permeable walls, allowing fluids to be pulled through via negative pressure. (Bottom) Equipment-free loading and automatic sample compartmentalization. Panels adapted with permission from Reference 100 under the terms of the Creative Commons Attribution 4.0 International License, http://creativecommons.org/licenses/by/4.0.
A recent technique involves the use of magnetic particles for droplet magnetofluidics (Figure 3a) (82, 83). This method utilizes mechanical actuation of magnetic particles to integrate biochemical processes without fluidic actuation. This technology was developed as an outcome of substantial proof-of-concept work in the early 2010s (84–86) and was followed by recent platforms demonstrating the integration of solid-phase extraction chemistry for a variety of molecular diagnostic applications (82, 87–91). A key feature of this technology is that fluidic actuation is not driven by pressure sources, but by magnetic actuators, which are used to steer the motion of magnetic particles across reagents. With the help of mechanical lysis and chemical lysis reagents, nucleic acid extraction from clinical samples was demonstrated within a 10-min time frame (82, 87).Because an entire cartridge is a passive scaffold for reagent storage, fabrication of such devices is much simpler than for other integrated devices. These features make this technology an attractive option for POC applications. In the work demonstrated by Shin et al. (82), a NAAT assay cartridge capable of detecting chlamydia from clinical specimens was produced for under US$2 using a simple lamination-based fabrication method.
Paper analytical devices (PADs) represent another significant area of microfluidic integration (Figure 3b). A seminal review article on this topic captures many of the technology’s defining features (92). Briefly, PADs utilize a cellulose-based matrix to passively transport analytes across various zones. A key premise of this approach is that such devices can be manufactured and distributed at a low cost, making them ideal for global health applications. While previous iterations of PADs have been limited in scope and sensitivity due to their association with either chemical or molecular binding-based assays, recent development in this field has demonstrated PADs with NAAT capabilities (93–96). Isothermal assays such as LAMP (95, 97), HDA (98), RCA (99), and RPA (94) have been reported, with RPA showing the most rapid time-to-result at 20 min (94). While earlier studies could only utilize paper as a substrate for the hybridization-based visualization of products (99) or as a medium for dry reagent storage (94),a recent study by Rodriguez et al. (95) demonstrated a higher level of integration by performing a sample preparation that included lysis and filtration in addition to in-matrix amplification and visualization of nucleic acid targets.
Recent development in flow-based microfluidic assay integration has generated a new class of devices utilizing vacuum reservoirs to drive fluids without the need for external actuation. The vacuum-driven approach utilizes a vacuum reservoir in the form of a gas-permeable polymer base to drive fluidic actuation from the inlet toward reaction chambers. Although these devices operate in an analogous manner to PADs, their ability to perform reactions in the liquid phase without the need for a fiber matrix to perform fluidic actuation helps to retain the performance of the base assay chemistry. Furthermore, microfluidic features allow these devices to leverage concepts such as sample digitization (Figure 3c) (100) and sedimentation-based filtration (101) to expand the functionality of the device to a greater extent than a PAD-based design allows. Exemplified by a decade of proof-of-concept work in this field, the vacuum reservoir approach to fluidic actuation provides an attractive alternative to disposable, instrument-free assay devices for pathogen identification. As of now, efforts to commercialize this technology are being led by university-based start-ups, including Diassess and mFluiDx (100, 101).
4.2. Scale-Driven Enhancement of Antibiotic Susceptibility Testing
While microfluidics can be used to accelerate assays via process integration, it also enables measurement of phenomena that are difficult to decipher in an ensemble measurement. Confining individual bacterial cells in a small reaction volume leads to a reduced background and locally concentrated analytes or biomarkers released form the cell, effectively shortening the time to obtain an observable signal for bacterial identification and AST (102–104). A recent review of droplet microfluidics illustrates the enhancement effect of reduced reaction volume on traditional bulk assays (105).
Surrogates for phenotypic effects for AST include (a) cell viability as measured by fluorescent indicators, such as resazurin and membrane-impermeable nucleic acid binding dyes and (b) growth as measured by cell size and count. Techniques based on cell viability indicators have seen substantial development as early as 1997, aided by the availability of the flow cytometer as a commercial tool for scale reduction and parallelization (106–108). Earlier work by Jepras et al. (108) demonstrated the use of flow cytometry and the membrane potential probe bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] to identify the effects of azithromycin, cefuroxime, and ciprofloxacin on E. coli within 2 h of incubation. Subsequent studies have expanded on this approach by using microfluidics-enabled compartmentalization to reduce the reaction volume several orders of magnitude smaller than can be achieved using flow cytometry. Boedicker et al. (102) demonstrated single-cell susceptibility testing through confinement of individual cells with antibiotics in nanoliter plugs that flow from a microfluidic device into an attached Teflon tubing, where incubation took place. This system is capable of analyzing the MIC of cefoxitin for S. aureus within 7 h by measuring the fluorescent viability indicator. A recent study by Kaushik et al. (109) demonstrated the use of a droplet microfluidic device that integrates droplet generation and incubation and in-line fluorescence detection to perform single-cell AST (Figure 4a). Measuring the growth of a single bacterium in a small droplet (20 pL) facilitates the rapid assessment of antibiotic resistance within 1 h using the resazurin assay (109). In droplet microfluidics–based AST, the sample can be processed continuously and, thus, its volume is not limited by the footprint of the device. This unique feature renders droplet microfluidics suitable for direct analysis of clinical samples of low bacterial load without the need for sample preculture.
Figure 4.
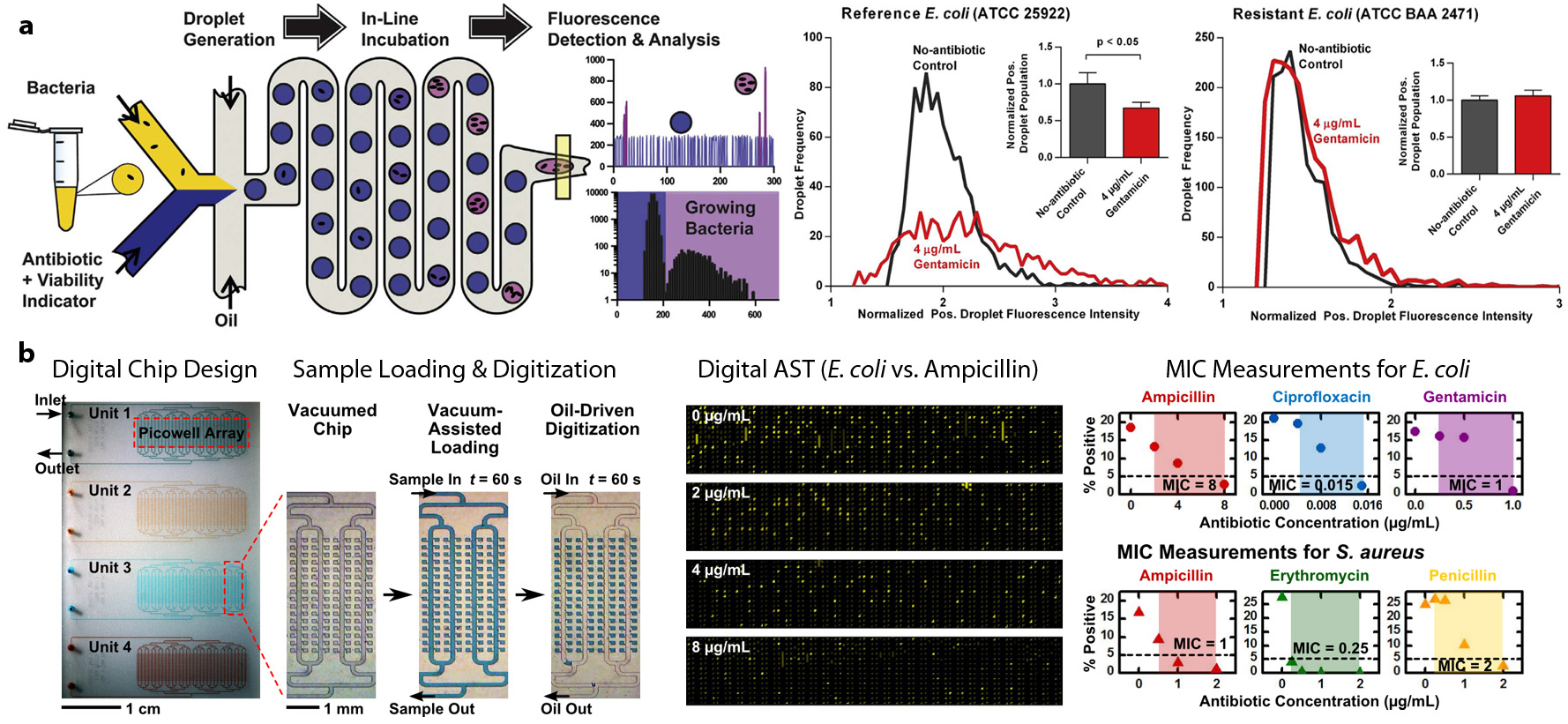
Emerging microfluidic devices for scale-driven enhancement of antibiotic susceptibility testing (AST). (a) A microfluidic device with fully integrated droplet generation, incubation, and in-line fluorescence detection is developed to perform single-cell bacterial growth detection and AST. Scaling the reaction chambers in the form of 20-pL droplets within this device enabled detection of single-cell Escherichia coli growth and its susceptibility/resistance to gentamicin in as little as 1 h. Panel adapted with permission from Reference 109. Copyright 2017, Elsevier. (b) A microfluidic digital array device for antimicrobial susceptibility testing. The microfluidic chip adopts a modular and scalable design for testing multiple antibiotic conditions in the same chip. Bacteria can be reliably digitized in 250-pL chambers via vacuum-assisted loading and oil-driven digitization. MICs (minimum inhibitory concentrations) for E. coli and Staphylococcus aureus against various antibiotics were measured using the digital chip. Panel adapted with permission from Reference 111. Copyright 2016, Chemical & Biological Microsystems Society.
Although techniques based on flow cytometry and droplet encapsulation can achieve sample digitization, they also require the use of specialized instruments that may limit a broader use of scale-enhanced AST. Some of these challenges can be mitigated using self-digitizing microfluidic arrays. Utilizing fixed-dimension chambers to compartmentalize samples obviates the need for complex fluidics and in-line optical detection, which can substantially simplify the assay workflow. Early work by Weibull et al. (110) demonstrated the use of microfabricated nanowells to parallelize broth microdilutions, demonstrating MIC determination from uropathogenic E. coli isolates within 4 h. Parallelization in this example helped to accommodate a broad range of different antibiotics and concentrations on a single device. Hsieh et al. (111, 112) developed a microfluidic digital array platform for single-cell growth detection and AST (Figure 4b). They employed vacuum-assisted sample loading and oil-driven sample digitization to stochastically confine single bacteria in picoliter chambers. Employing a resazurin-based bacterial growth assay, they were able to demonstrate accurate digital AST for E.coli and S.aureus against several antibiotics in 3 h (111). The microfluidic digital array–based resazurin assay was also demonstrated as a facile approach for rapid and precise quantification of viable bacteria that is critical to many microbiological applications (112). Another study by Avesar et al. (113) utilized a self-digitizing array device to compartmentalize bacterial suspension into 8-nL reaction chambers for a resazurin-based growth assay. Although this device did not achieve single-cell isolation, it was able to perform growth-based AST from bacterial suspensions within 5.5 h of incubation.
An alternative approach to AST is to measure growth directly via single-cell imaging. While single bacterial cells are challenging to resolve in free solution due to their size and motion, various techniques can be used to confine bacterial growth within boundaries that can be consistently imaged and quantified. Earlier work in this field utilized a digital microscopy system to obtain time-lapse image stacks from flow cells containing bacterial cells in AST incubation; the method was capable of performing AST of methicillin-resistant S. aureus within 3 h (114). Another pioneering study by Choi et al. (115) utilized an agarose matrix to immobilize bacterial cells prior to AST, where morphological changes were monitored to determine antibiotic susceptibility from bacterial cells within 4 h of incubation. Subsequent research by Malmberg et al. (116) combined the concept of direct cell imaging with microfluidics-based gradient generation to obtain an antibiogram from S. aureus culture as early as 2 h from positive blood cultures. Although these studies demonstrated substantial improvement in speed and convenience over conventional AST, bacterial growth detection at a single-cell resolution had yet to be achieved. To address this gap, a recent article by Baltekin et al.(117) described a method of performing broth microdilution in microchannels (Figure 5a) to evaluate ciprofloxacin resistance from clinical isolates of E. coli within 30 min. Geometric confinement of bacteria in channels enabled clear visualization of growth as the dividing bacteria stack along the length of the microchannel (Figure 5b). Due to the ability of microfluidics-assisted single-cell imaging to directly observe growth at the highest possible resolution, it is anticipated that this technique will continue to find relevance in rapid bacterial AST applications.
Figure 5.
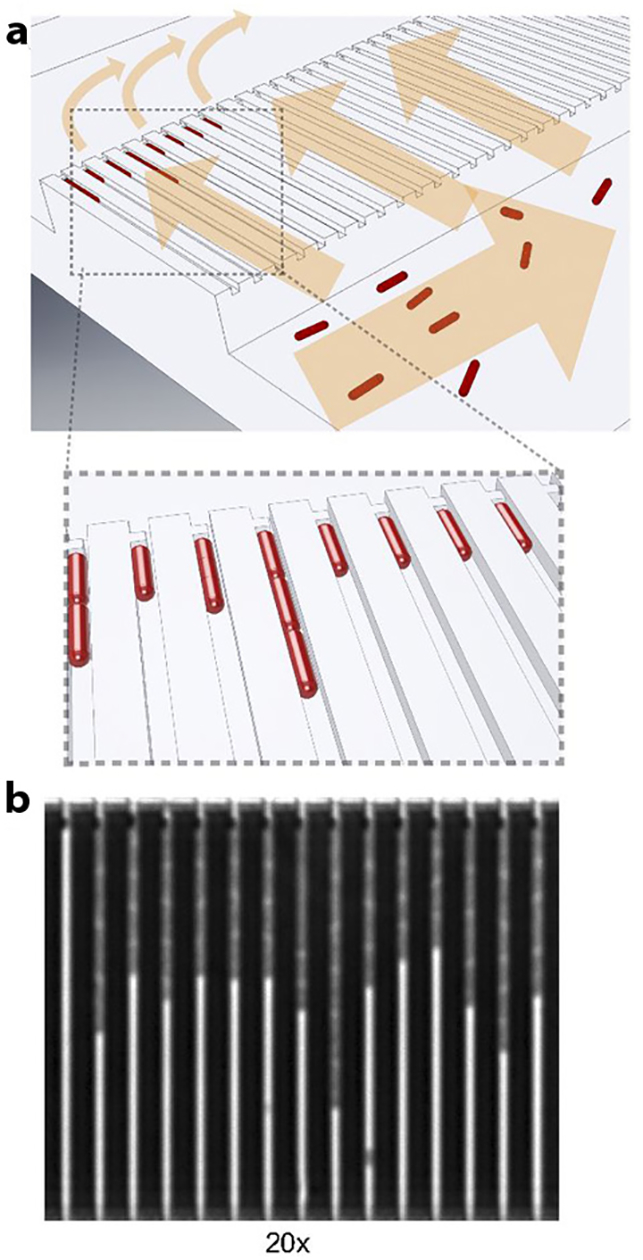
Rapid antibiotic susceptibility testing (AST) via microfluidics-enhanced single-cell imaging. (a) Cells are first seeded into the device via a large fluidic reservoir, which feeds the cells into microfluidic channels. The channels are connected to an outlet via a sieve, which keeps the cells retained in the channels. (b) Scanning electron microscope image reveals bacterial growth in the microfluidic channels. Panel adapted with permission from Reference 117. Copyright 2017, PNAS.
5. BIOSENSOR-BASED APPROACHES FOR RAPID PATHOGENIDENTIFICATION AND ANTIBIOTIC SUSCEPTIBILITY TESTING
In addition to advances in NAAT and microfluidics, recent developments in biosensors have also led to the discovery of new methods of performing rapid pathogen identification and AST. These techniques are able to generate a signal directly from the presence of bacterial cells without biochemical reactions, which saves a substantial amount of assay time from sample preparation and signal amplification. Many publications have discussed sensory elements based on immunological or nucleic acids–based binding of analytes, and some have found utility in various commercial assay platforms incorporating a biosensing element for multiplexing and specificity. However, technical challenges such as restrictive analyte binding conditions and assay sensitivity have often limited the utility of these elements for direct detection of analytes without a preamplification step. Here, we highlight several recent techniques that have demonstrated the potential to accelerate pathogen identification and AST beyond what current assays have been able to deliver.
Impedance-based biosensing is a rapidly expanding field for bioanalytical systems, with applications ranging from virus particle detection to genome sequencing (118, 119). The principle of this approach relies on the change in the electrical impedance as a pathogen crosses the sensing area between two electrodes. Sensing is achieved when the crossing either obstructs the electrical connection in an electrolyte solution (as in Coulter counters) or generates a new electrical connection by short circuiting the electrodes (as in toggle switches). Although the design and construction of the sensing elements present technical challenges, having the ability to directly observe the physical presence of an analyte without secondary probes or assay reagents allows for the rapid, unperturbed measurement of samples. In a recent example, Ali et al.(120) demonstrated the use of silver nanowires to substantially enhance the sensitivity of contact-based impedance sensing. Using the transient response of the sensing element, the authors could differentiate between three species of bacterial pathogens within 8 min of exposure to the sample. This was substantially faster than immunoaffinity-based biosensors that were reported to take 84 min (121).
Another promising approach for AST utilizes mechanical sensing that measures the change in the mechanical property of a microcantilever upon its interaction with bacterial cells. Several microcantilever biosensors for pathogen detection have been developed (122, 123). The sensing principles rely on the binding of target bacteria on the cantilever surface conjugated with pathogen-specific receptors and subsequently translating the binding event into mechanical signals as either a cantilever deflection (absorption stress) or a shift in resonance frequency (mass). Though highly sensitive, cantilever-based biosensors are often limited by the need to perform the measurements in an air environment and not in a liquid medium that otherwise leads to a poor performance due to liquid dampening (124). To rectify this limitation, Longo et al. (125) presented a biosensor that exploits the low-frequency fluctuations of an atomic force microscope (AFM) cantilever (<1 kHz) to characterize the activity of bacteria (Figure 6a). In this study, the movement of bacteria led to an increase in the amplitude of the fluctuations of the sensor that varies as a function of the medium present in the measurement chamber. Such an effect has facilitated the characterization of bacterial metabolism to determine their susceptibility to drugs, before the bacteria replicate (125). An interesting variation of this approach is the use of alternative methods of isolating bacteria at the tip of the cantilever. Etayash et al. (126) developed a microcantilever with an embedded microchannel chemically functionalized with receptors to capture the bacteria passing through the channel (Figure 6b). This sensor device incorporated a multimodal detection approach to measure changes in resonance frequency, cantilever deflection, and infrared absorption as a result of bacteria absorption inside the cantilever. The observation of a distinct nanomechanical response when the bacteria (e.g., E. coli) were exposed to antibiotics has facilitated the development of a biosensor device for detection of bacteria and their drug susceptibility (126). The development of a cantilever biosensor with an embedded microchannel was originally demonstrated by Burg et al. (127). In this earlier study, the use of suspended microchannel resonators with a low resonator mass (100 ng) enabled mass detection at a single-cell resolution. More recent developments of this technology toward applications in infectious diagnostics have demonstrated the feasibility of rapid AST by mass measurement (128). Considering that the growth measurement is already being performed in real time at a single-cell resolution, it is worth noting that the time frame observed here may represent the physical limit of AST when bacterial division is used as the main phenotypic indicator for evaluating antibiotic susceptibility.
Figure 6.
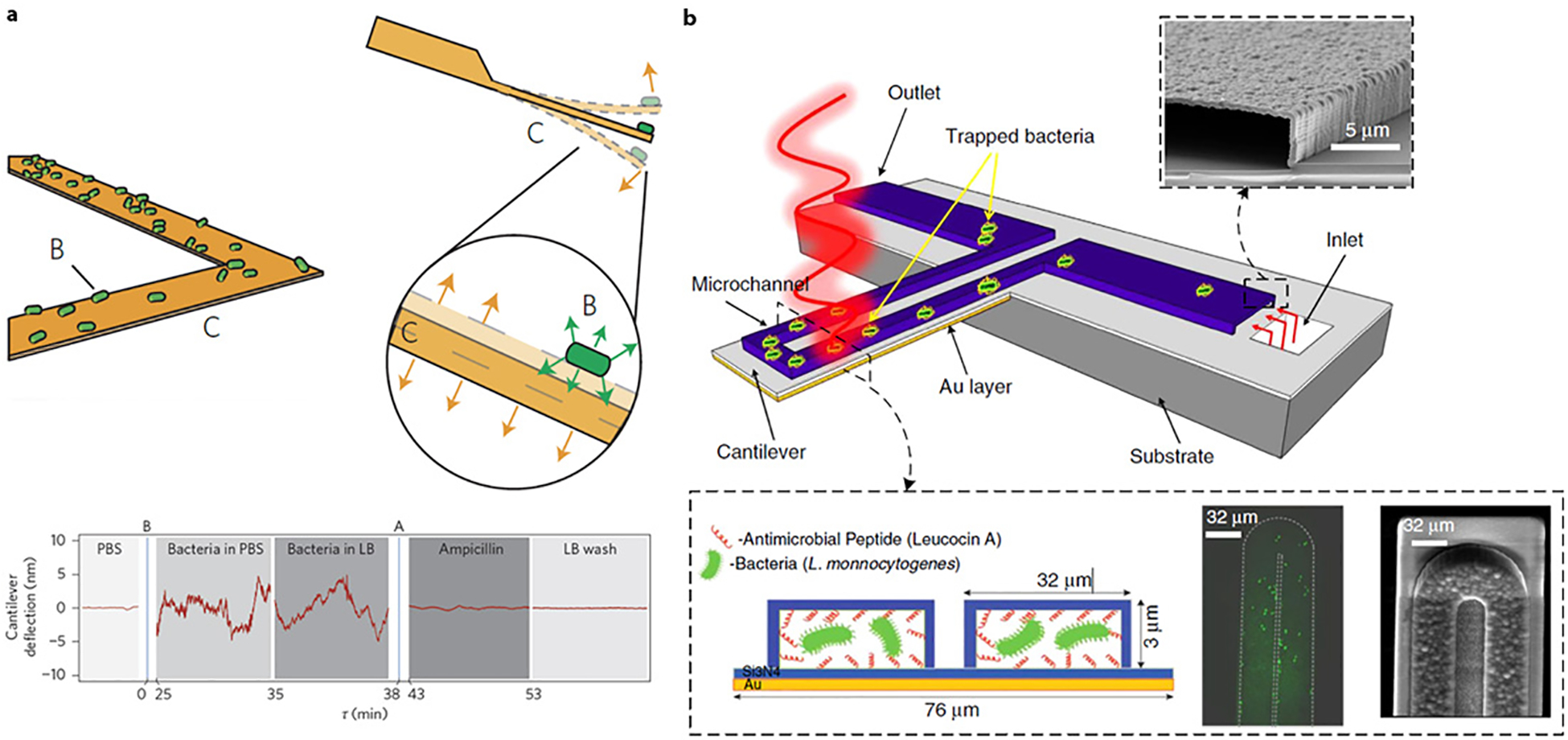
(a) Cantilever biosensor for detecting bacterial susceptibility to antibiotics. (Top) The cantilever is conjugated with a bacteria-specific receptor to capture the target bacteria. The attachment of bacteria to the cantilever leads to a change in cantilever fluctuation. (Bottom) Deflection of the cantilever for the antibiotic susceptibility testing (AST) experiments involving Escherichia coli. Panel adapted with permission form Reference 125. Copyright 2013, Springer Nature. (b) Microfluidic cantilever for detecting bacteria and their antibiotic susceptibility. (Top) The microfluidic channel filled with bacteria supported on a silicon substrate and irradiated with a specific wavelength of tunable infrared light. (Bottom) The inner surface of the cantilever’s microchannel was functionalized with a bacteria-targeted receptor. Shown on the right are fluorescent and scanning electron microscope images from the top side of the microchannel. Panel adapted with permission from Reference 126 under the terms of the Creative Commons Attribution 4.0 International License, http://creativecommons.org/licenses/by/4.0. Abbreviations: LB, lysogeny broth; PBS, phosphate buffered saline.
6. CONCLUSION AND PERSPECTIVES
Our survey of recent progress in analytical techniques for pathogen identification and AST reveals several key ideas. First is the prominence of NAAT in recently commercialized assays and its potential for further acceleration. While previous generations of NAAT were limited in speed and workflow by sample preparation steps, recent advances in technologies such as direct PCR and microfluidic integration suggest that these gaps may likely become narrower in the near term. Increasing compatibility of NAAT with phenotypic assays can become a significant innovation as the workflow shifts away from traditional culture-based microbiology toward a molecular assay-based one.
Although pathogen identification has benefited from the emergence of NAAT-based techniques, similar advancement has yet to be observed for AST. We found that most technologies pursuing the acceleration of AST are still in the nascent stages, and substantial follow-up work is expected to bridge the gap between proof-of-concept and a platform that is ready for clinical use. Both single-cell microfluidics and biosensor-enabled signal transduction that we discussed seem to push the theoretical limits for AST based on the measurement of bacterial growth. For these techniques to accelerate AST in a clinically meaningful way, our understanding of antibiotic resistance and standards will also need to expand from the current metrics based on end-point viability to those based on tolerance profiles at the level of an individual bacterium. Some efforts are already underway to elucidate bacterial physiology at this scale (129).
Our discussion has focused on recent developments in analytical techniques, with emphasis on time-saving measures from an engineering perspective. As such, biomarker discovery has not been a major topic in our discussion. However, it should be noted that new biomarkers can play a transformative role in how diagnostic practices are shaped. In the way that the development of PCR aided the discovery of genetic biomarkers and transformed diagnostic practices for pathogen detection, new analytical techniques such as NGS can result in the discovery of new biomarkers for pathogen identification and antibiotic resistance. Some of the scale-based microfluidic tools and biosensors discussed in this review are methods that can be used to study pathogens at unprecedented spatial and temporal resolutions. It is hoped that they can be leveraged for the discovery of new biomarkers for faster and more comprehensive pathogen identification and AST.
It is anticipated that the next generation of diagnostic tests will not depend on a single technique, but rather a combination of the techniques discussed in this review. Our findings show that many of the concepts in biosensor technologies overlap with microfluidics, NAAT, and NGS, and each have strengths to complement the advantages of others. This crossover effect is already present in the current generation of diagnostic platforms, where microfluidic assay integration and biosensor-based detection are coupled with NAAT-based assay chemistry. We might expect the migration of current bulk assays into a digital format to lead the change, where the enhancement in resolution can help foster a demand for new biosensors or assays to yield additional information from each reaction.
ACKNOWLEDGMENTS
The authors would like to acknowledge the US National Institutes of Health (grants R01AI117032,R01AI137272,R01AI138978,and U54EB007958) for funding. The authors would also like to thank Aniruddha Kaushik for the discussion and input when preparing this review.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, et al. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med 34:1589–96 [DOI] [PubMed] [Google Scholar]
- 2.Lagier JC, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. 2015. Current and past strategies for bacterial culture in clinical microbiology. Clin. Microbiol. Rev 28:208–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis 49:1749–55 [DOI] [PubMed] [Google Scholar]
- 4.Patel R 2015. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem 61:100–11 [DOI] [PubMed] [Google Scholar]
- 5.Murray PR. 2012. What is new in clinical microbiology-microbial identification by MALDI-TOF mass spectrometry: a paper from the 2011William Beaumont Hospital Symposium on molecular pathology. J. Mol. Diagn 14:419–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez RM, Bauerle ER, Fang FC, Butler-Wu SM. 2014. Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J. Clin. Microbiol 52:2521–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leber AL, Everhart K, Daly JA,Hopper A,Harrington A, et al. 2018. Multicenter evaluation of BioFire FilmArray Respiratory Panel 2 for detection of viruses and bacteria in nasopharyngeal swab samples. J. Clin. Microbiol 56 10.1128/JCM.01945-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zacharioudakis IM, Zervou FN, Mylonakis E. 2018. T2 magnetic resonance assay: overview of available data and clinical implications. J. Fungi 4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, et al. 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev. Mol. Diagn 10:399–415 [DOI] [PubMed] [Google Scholar]
- 10.Metzgar D, Frinder M, Lovari R, Toleno D, Massire C, et al. 2013. Broad-spectrum biosensor capable of detecting and identifying diverse bacterial and Candida species in blood. J. Clin. Microbiol 51:2670–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzgar D, Frinder MW, Rothman RE, Peterson S, Carroll KC, et al. 2016. The IRIDICA BAC BSI assay: rapid, sensitive and culture-independent identification of bacteria and Candida in blood. PLOS ONE 11:e0158186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrar JS, Wittwer CT. 2015. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin. Chem 61:145–53 [DOI] [PubMed] [Google Scholar]
- 13.Wittwer CT, Garling DJ. 1991. Rapid cycle DNA amplification: time and temperature optimization. Biotechniques 10:76–83 [PubMed] [Google Scholar]
- 14.Misawa Y, Yoshida A, Saito R, Yoshida H, Okuzumi K, et al. 2007. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J. Infect. Chemother 13:134–40 [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Li Y, Wang L, You L, Xu Z, et al. 2010. Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. Mol. Biol. Rep 37:2183–88 [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Li Y, Chu J, Xu Z, Zhong Q. 2012. Development and application of a simple loop-mediated isothermal amplification method on rapid detection of Listeria monocytogenes strains. Mol. Biol. Rep 39:445–49 [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Lowe SB, Gooding JJ. 2014. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron 61:491–99 [DOI] [PubMed] [Google Scholar]
- 18.Safavieh M, Kanakasabapathy MK, Tarlan F, Ahmed MU, Zourob M, et al. 2016. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomater. Sci. Eng 2:278–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rane TD, Chen LB, Zec HC, Wang TH. 2015. Microfluidic continuous flow digital loop-mediated isothermal amplification (LAMP). Lab Chip 15:776–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery JL, Sanford LN, Wittwer CT. 2010. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev. Mol. Diagn 10:219–40 [DOI] [PubMed] [Google Scholar]
- 21.Wittwer CT. 2009. High-resolution DNA melting analysis: advancements and limitations. Hum. Mutat 30:857–59 [DOI] [PubMed] [Google Scholar]
- 22.Wittwer CT, Reed GH, Kent JO. 2007. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8:597–608 [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Ramachandran P, Rothman R, Hsieh YH, Hardick A, et al. 2009. Rapid identification of biothreat and other clinically relevant bacterial species by use of universal PCR coupled with high-resolution melting analysis. J. Clin. Microbiol 47:2252–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothman R, Ramachandran P, Yang S, Hardick A, Won H, et al. 2010. Use of quantitative broad-based polymerase chain reaction for detection and identification of common bacterial pathogens in cerebrospinal fluid. Acad. Emerg. Med 17:741–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Won H, Rothman R, Ramachandran P, Hsieh YH, Kecojevic A, et al. 2010. Rapid identification of bacterial pathogens in positive blood culture bottles by use of a broad-based PCR assay coupled with high-resolution melt analysis. J. Clin. Microbiol 48:3410–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardick J, Won H, Jeng K, Hsieh YH, Gaydos CA, et al. 2012. Identification of bacterial pathogens in ascitic fluids from patients with suspected spontaneous bacterial peritonitis by use of broad-range PCR (16S PCR) coupled with high-resolution melt analysis. J. Clin. Microbiol 50:2428–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Park S, Yang S, Wang TH. 2010. An all-in-one microfluidic device for parallel DNA extraction and gene analysis. Biomed. Microdevices 12:1043–49 [DOI] [PubMed] [Google Scholar]
- 28.Jeng K, Yang S, Won H, Gaydos CA, Hsieh YH, et al. 2012. Application of a 16S rRNA PCR-high-resolution melt analysis assay for rapid detection of Salmonella bacteremia. J. Clin. Microbiol 50:1122–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeng K, Gaydos CA, Blyn LB, Yang S, Won H, et al. 2012. Comparative analysis of two broad-range PCR assays for pathogen detection in positive-blood-culture bottles: PCR-high-resolution melting analysis versus PCR-mass spectrometry. J. Clin. Microbiol 50:3287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masek BJ, Hardick J, Won H, Yang S, Hsieh YH, et al. 2014. Sensitive detection and serovar differentiation of typhoidal and nontyphoidal Salmonella enterica species using 16S rRNA gene PCR coupled with high-resolution melt analysis. J. Mol. Diagn 16:261–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraley SI, Hardick J, Masek BJ, Athamanolap P, Rothman RE, et al. 2013. Universal digital high-resolution melt: a novel approach to broad-based profiling of heterogeneous biological samples. Nucleic Acids Res. 41:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Athamanolap P, Parekh V, Fraley SI, Agarwal V, Shin DJ, et al. 2014. Trainable high resolution melt curve machine learning classifier for large-scale reliable genotyping of sequence variants. PLOS ONE 9:e109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraley SI, Athamanolap P, Masek BJ, Hardick J, Carroll KC, et al. 2016. Nested machine learning facilitates increased sequence content for large-scale automated high resolution melt genotyping. Sci. Rep 6:19218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andini N, Wang B, Athamanolap P, Hardick J, Masek BJ, et al. 2017. Microbial typing by machine learned DNA melt signatures. Sci. Rep 7:42097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Keefe CM, Pisanic TR, Zec HC, Overman MJ, Herman JG, Wang TH. 2018. Facile profiling of molecular heterogeneity by microfluidic digital melt. Sci. Adv 4:eaat6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikkari S, McLaughlin IJ, Bi W, Dodge DE, Relman DA. 2001. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol 39:1956–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cremonesi P, Cortimiglia C, Picozzi C, Minozzi G, Malvisi M, et al. 2016. Development of a droplet digital polymerase chain reaction for rapid and simultaneous identification of common foodborne pathogens in soft cheese. Front. Microbiol 7:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang M, Koh I, Lee SJ, Cheong JH, Kim P. 2017. Droplet-based microtumor model to assess cell-ECM interactions and drug resistance of gastric cancer cells. Sci. Rep 7:41541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, et al. 2017. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol 243:16–24 [DOI] [PubMed] [Google Scholar]
- 40.Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, et al. 2012. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. PNAS 109:6217–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao MD, Ganesamoorthy D, Elliott AG, Zhang H, Cooper MA, Coin LJ. 2016. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinION™ sequencing. Gigascience 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abril MK, Barnett AS, Wegermann K, Fountain E, Strand A, et al. 2016. Diagnosis of Capnocytophaga canimorsus sepsis by whole-genome next-generation sequencing. Open Forum Infect. Dis 3:ofw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong DK, Blauwkamp TA, Kertesz M, Bercovici S, Truong C, Banaei N. 2018. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis 92:210–13 [DOI] [PubMed] [Google Scholar]
- 44.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, et al. 2015. Noninvasive monitoring of infection and rejection after lung transplantation. PNAS 112:13336–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horiba K, Kawada JI, Okuno Y, Tetsuka N, Suzuki T, et al. 2018. Comprehensive detection of pathogens in immunocompromised children with bloodstream infections by next-generation sequencing. Sci. Rep 8:3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nölling J, Rapireddy S, Amburg JI, Crawford EM, Prakash RA, et al. 2016. Duplex DNA-invading γ-modified peptide nucleic acids enable rapid identification of bloodstream infections in whole blood. MBio 7:e00345–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aghazadeh A, Lin AY, Sheikh MA, Chen AL, Atkins LM, et al. 2016. Universal microbial diagnostics using random DNA probes. Sci. Adv 2:e1600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, et al. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, et al. 2018. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360:436–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, et al. 2016. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538:270–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360:439–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbiol. Rev 27:587–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bard JD, Lee F. 2018. Why can’t we just use PCR? The role of genotypic versus phenotypic testing for antimicrobial resistance testing. Clin. Microbiol. Newsl 40:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rolain JM, Mallet MN, Fournier PE, Raoult D. 2004. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother 54:538–41 [DOI] [PubMed] [Google Scholar]
- 55.Hunfeld KP, Bittner T, Rodel R, Brade V, Cinatl J. 2004. New real-time PCR-based method for in vitro susceptibility testing of Anaplasma phagocytophilum against antimicrobial agents. Int. J. Antimicrob. Agents 23:563–71 [DOI] [PubMed] [Google Scholar]
- 56.Waldeisen JR, Wang T, Mitra D, Lee LP. 2011. A real-time PCR antibiogram for drug-resistant sepsis. PLOS ONE 6:e28528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PF. 2011. Antibiotic susceptibility testing of grown blood cultures by combining culture and real-time polymerase chain reaction is rapid and effective. PLOS ONE 6:e27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Shin DJ, Zheng S, Melendez JH, Gaydos CA, Wang TH. 2018. Direct-qPCR assay for coupled identification and antimicrobial susceptibility testing of Neisseria gonorrhoeae. ACS Infect. Dis 4:1377–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Athamanolap P, Hsieh K, Chen L, Yang S, Wang TH. 2017. Integrated bacterial identification and antimicrobial susceptibility testing using PCR and high-resolution melt. Anal. Chem 89:11529–36 [DOI] [PubMed] [Google Scholar]
- 60.Andini N, Hu A, Zhou L, Cogill S, Wang TH, et al. 2018. A “culture” shift: broad bacterial detection, identification, and antimicrobial susceptibility testing directly from whole blood. Clin. Chem 10.1373/clinchem.2018.290189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoepp NG, Khorosheva EM, Schlappi TS, Curtis MS, Humphries RM, et al. 2016. Digital quantification of DNA replication and chromosome segregation enables determination of antimicrobial susceptibility after only 15 minutes of antibiotic exposure. Angew. Chem. Int. Ed 55:9557–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoepp NG, Schlappi TS, Curtis MS, Butkovich SS, Miller S, et al. 2017. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl.Med 9:eaal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jensen PA, Zhu Z, van Opijnen T. 2017. Antibiotics disrupt coordination between transcriptional and phenotypic stress responses in pathogenic bacteria. Cell Rep. 20:1705–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farrar JS, Wittwer CT. 2015. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin. Chem 61:145–53 [DOI] [PubMed] [Google Scholar]
- 65.Woodman ME. 2008. Direct PCR of intact bacteria (colony PCR). Curr. Protoc.Microbiol 9:A.3D.1–6 [DOI] [PubMed] [Google Scholar]
- 66.Holcomb ZE, Tsalik EL, Woods CW, McClain MT.2017. Host-based peripheral blood gene expression analysis for diagnosis of infectious diseases. J. Clin. Microbiol 55:360–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gliddon HD, Herberg JA, Levin M, Kaforou M. 2018. Genome-wide host RNA signatures of infectious diseases: discovery and clinical translation. Immunology 153:171–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, et al. 2017. Circulating microRNAs as potential biomarkers of infectious disease. Front. Immunol 8:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. PNAS 105:10513–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. 2010. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38:7248–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sutherland A, Thomas M, Brandon RA, Brandon RB, Lipman J, et al. 2011. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit. Care 15:R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sweeney TE, Shidham A, Wong HR, Khatri P. 2015. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci. Transl. Med 7:287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu X, Yu J, Crosby SD, Storch GA. 2013. Gene expression profiles in febrile children with defined viral and bacterial infection. PNAS 110:12792–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, et al. 2007. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109:2066–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. 2015. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis 212:213–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sweeney TE, Wong HR, Khatri P. 2016. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med 8:346ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Yang X, Zhao W. 2017. Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol. 22:585–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kovarik ML, Gach PC, Ornoff DM, Wang Y, Balowski J, et al. 2012. Micro total analysis systems for cell biology and biochemical assays. Anal. Chem 84:516–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maurer FP, Christner M, Hentschke M, Rohde H. 2017. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: implications for patient care and antimicrobial stewardship programs. Infect. Dis. Rep 9:6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Babady NE, England MR, Jurcic Smith KL, He T, Wijetunge DS, et al. 2018. Multicenter evaluation of the ePlex Respiratory Pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J. Clin. Microbiol 56 10.1128/JCM.01658-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, et al. 2017. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio 8 10.1128/MBIO.00812-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shin DJ, Athamanolap P, Chen L, Hardick J, Lewis M, et al. 2017. Mobile nucleic acid amplification testing (mobiNAAT) for Chlamydia trachomatis screening in hospital emergency department settings. Sci. Rep 7:4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shin DJ, Wang TH. 2014. Magnetic droplet manipulation platforms for nucleic acid detection at the point of care. Ann. Biomed. Eng 42:2289–302 [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Park S, Liu K, Tsuan J, Yang S, Wang TH. 2011. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip 11:398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Long Z, Shetty AM, Solomon MJ, Larson RG. 2009. Fundamentals of magnet-actuated droplet manipulation on an open hydrophobic surface. Lab Chip 9:1567–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Wang TH.2013. Full-range magnetic manipulation of droplets via surface energy traps enables complex bioassays. Adv. Mater 25:2903–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shin DJ, Trick AY, Hsieh YH, Thomas DL, Wang TH. 2018. Sample-to-answer droplet magnetofluidic platform for point-of-care hepatitis C viral load quantitation. Sci. Rep 8:9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiou CH, Shin DJ, Zhang Y, Wang TH. 2013. Topography-assisted electromagnetic platform for blood-to-PCR in a droplet. Biosens. Bioelectron 50:91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shin DJ, Zhang Y, Wang TH. 2014. A droplet microfluidic approach to single-stream nucleic acid isolation and mutation detection. Microfluid. Nanofluid 17:425–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stark A, Shin DJ, Pisanic T 2nd, Hsieh K, Wang TH. 2016. A parallelized microfluidic DNA bisulfite conversion module for streamlined methylation analysis. Biomed. Microdevices 18:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stark A, Shin DJ, Wang TH. 2018. A sample-to-answer droplet magnetofluidic assay platform for quantitative methylation-specific PCR. Biomed. Microdevices 20:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Connelly JT, Rolland JP, Whitesides GM.2015. “Paper machine” formolecular diagnostics. Anal. Chem 87:7595–601 [DOI] [PubMed] [Google Scholar]
- 93.Yang Y, Noviana E, Nguyen MP, Geiss BJ, Dandy DS, Henry CS. 2017. Paper-based microfluidic devices: emerging themes and applications. Anal. Chem 89:71–91 [DOI] [PubMed] [Google Scholar]
- 94.Magro L, Jacquelin B, Escadafal C, Garneret P, Kwasiborski A, et al. 2017. Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci. Rep 7:1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. 2016. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 16:753–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaur N, Toley BJ. 2018. Paper-based nucleic acid amplification tests for point-of-care diagnostics. Analyst 143:2213–34 [DOI] [PubMed] [Google Scholar]
- 97.Yang Z, Xu G, Reboud J, Ali SA, Kaur G, et al. 2018. Rapid veterinary diagnosis of bovine reproductive infectious diseases from semen using paper-origami DNA microfluidics. ACS Sens. 3:403–9 [DOI] [PubMed] [Google Scholar]
- 98.Shetty P, Ghosh D, Singh M, Tripathi A, Paul D. 2016. Rapid amplification of Mycobacterium tuberculosis DNA on a paper substrate. RSC Adv. 6:56205–12 [Google Scholar]
- 99.Ali MM, Aguirre SD, Xu Y, Filipe CD, Pelton R, Li Y. 2009. Detection of DNA using bioactive paper strips. Chem. Commun 43:6640–42 [DOI] [PubMed] [Google Scholar]
- 100.Yeh EC, Fu CC, Hu L, Thakur R, Feng J, Lee LP. 2017. Self-powered integrated microfluidic pointof-care low-cost enabling (SIMPLE) chip. Sci. Adv 3:e1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL, Ross BM, Park Y, et al. 2011. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS). Lab Chip 11:845–50 [DOI] [PubMed] [Google Scholar]
- 102.Boedicker JQ, Li L, Kline TR, Ismagilov RF. 2008. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 8:1265–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rane TD, Zec HC, Puleo C, Lee AP, Wang TH. 2012. Droplet microfluidics for amplification-free genetic detection of single cells. Lab Chip 12:3341–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang DK, Ali MM, Zhang KX, Huang SS, Peterson E, et al. 2014. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat. Commun 5:5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaushik AM, Hsieh K, Wang TH. 2018. Droplet microfluidics for high-sensitivity and high-throughput detection and screening of disease biomarkers. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol 10:e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Álvarez-Barrientos A, Arroyo J, Cantón R, Nombela C, Sánchez-Pérez M. 2000. Applications of flow cytometry to clinical microbiology. Clin. Microbiol. Rev 13:167–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Belkum A, Dunne WM.2013. Next-generation antimicrobial susceptibility testing. J. Clin. Microbiol 51:2018–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jepras RI, Paul FE, Pearson SC, Wilkinson MJ. 1997. Rapid assessment of antibiotic effects on Escherichia coli by bis-(1,3-dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob. Agents Ch 41:2001–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaushik AM, Hsieh K, Chen L, Shin DJ, Liao JC, Wang TH. 2017. Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosens. Bioelectron 97:260–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weibull E, Antypas H, Kjall P, Brauner A, Andersson-Svahn H, Richter-Dahlfors A. 2014. Bacterial nanoscale cultures for phenotypic multiplexed antibiotic susceptibility testing. J. Clin. Microbiol 52:3310–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsieh K, Zec HC, Chen L, Kaushik A, Wang TH. 2016. Rapid, accurate, and general single-cell antibiotic susceptibility test in digital bacteria picoarray. In The 20th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2016, pp. 174–75. Dublin, Ireland, October 9–13 [Google Scholar]
- 112.Hsieh K, Zec HC, Chen L, Kaushik AM, Mach KE, et al. 2018. Simple and precise counting of viable bacteria by resazurin-amplified picoarray detection. Anal. Chem 90:9449–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Avesar J, Rosenfeld D, Truman-Rosentsvit M, Ben-Arye T, Geffen Y, et al. 2017. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. PNAS 114:E5787–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fredborg M, Rosenvinge FS, Spillum E, Kroghsbo S,Wang M, Sondergaard TE. 2015. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur J. Clin. Microbiol. Infect. Dis 34:2385–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choi J, Yoo J, Lee M, Kim EG, Lee JS, et al. 2014. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med 6:267ra174. [DOI] [PubMed] [Google Scholar]
- 116.Malmberg C, Yuen P, Spaak J, Cars O, Tangden T, Lagerback P. 2016. A novel microfluidic assay for rapid phenotypic antibiotic susceptibility testing of bacteria detected in clinical blood cultures. PLOS ONE 11:e0167356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baltekin O, Boucharin A, Tano E, Andersson DI, Elf J. 2017. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. PNAS 114:9170–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, et al. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vogel R, Willmott G, Kozak D, Roberts GS, Anderson W, et al. 2011. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal. Chem 83:3499–506 [DOI] [PubMed] [Google Scholar]
- 120.Ali S, Hassan A, Hassan G, Eun CH, Bae J, et al. 2018. Disposable all-printed electronic biosensor for instantaneous detection and classification of pathogens. Sci. Rep 8:5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi X, Kadiyala U, VanEpps JS, Yau ST. 2018. Culture-free bacterial detection and identification from blood with rapid, phenotypic, antibiotic susceptibility testing. Sci. Rep 8:3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mader A, Gruber K, Castelli R, Hermann BA, Seeberger PH, et al. 2012. Discrimination of Escherichia coli strains using glycan cantilever array sensors. Nano Lett. 12:420–23 [DOI] [PubMed] [Google Scholar]
- 123.Wang JH, Morton MJ, Elliott CT, Karoonuthaisiri N, Segatori L, Biswal SL. 2014. Rapid detection of pathogenic bacteria and screening of phage-derived peptides using microcantilevers. Anal. Chem 86:1671–78 [DOI] [PubMed] [Google Scholar]
- 124.Longo G, Kasas S. 2014. Effects of antibacterial agents and drugs monitored by atomic force microscopy. Wires Nanomed. Nanobiotechnol 6:230–44 [DOI] [PubMed] [Google Scholar]
- 125.Longo G, Alonso-Sarduy L, Rio LM, Bizzini A, Trampuz A, et al. 2013. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol 8:522–26 [DOI] [PubMed] [Google Scholar]
- 126.Etayash H, Khan MF, Kaur K, Thundat T. 2016. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat. Commun 7:12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, et al. 2007Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446:1066–69 [DOI] [PubMed] [Google Scholar]
- 128.Schneider BK, Harris P, et al. 2016. Rapid antimicrobial susceptibility tests by mass measurement on a 96-well plate. Paper presented at the ASM Microbe Conference, Boston, June 16–20 [Google Scholar]
- 129.Meylan S, Andrews IW, Collins JJ. 2018. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172:1228–38 [DOI] [PubMed] [Google Scholar]
- 130.Edmiston CE, Garcia R, Barnden M, DeBaun B, Johnson HB. 2018. Rapid diagnostics for bloodstream infections: a primer for infection preventionists. Am. J. Infect. Control 46:1060–68 [DOI] [PubMed] [Google Scholar]


