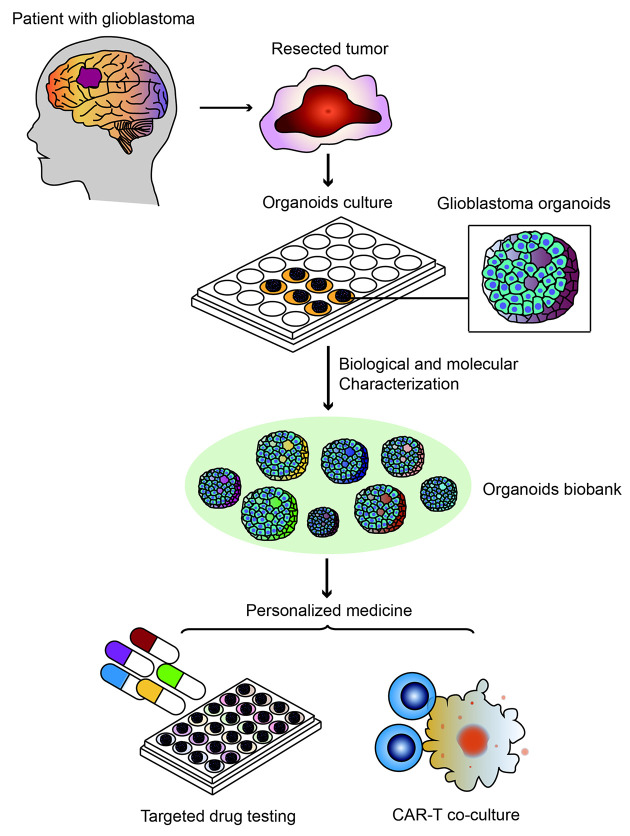
Recently, an original article titled “A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity” was published in Cell (1), a leading journal of scientific research worldwide. This paper reported that Fadi Jacob and colleagues established an organoids biobank with patient-derived glioblastoma organoids (GBOs), recapitulating the histological features, cellular diversity, gene expression and mutational profiles of their corresponding parental tumors. And functions of these organoids were confirmed both in vitro and in vivo xenograft models. In addition, by modeling targeted drug testing and chimeric antigen receptor T cell (CAR-T) immunotherapy, they demonstrated the application of GBOs in personalized medicine (1) (Figure 1). These discoveries provided an effective platform for basic and translational cancer research characterized by high degree of heterogeneity and contributed to the investigation of personalized medicine. Here in this commentary, we provide a concise explanation of this biobank and its impacts on clinical cancer therapy.
1. A GBOs biobank established by Fadi Jacob et al. used for recapitulating tumor heterogeneity and modeling personalized medicine. GBO, glioblastoma organoids; CAR-T, chimeric antigen receptor T cell.
Fadi Jacob et al. established an organoids biobank including 70 GBOs from different patients without dissociating resected tumor into single cells and with no added EGF/bFGF or extracellular matrix to avoid clonal selection, better preserving local cytoarchitecture and mimicking heterogeneity of original tumors (1). After biological and molecular characterization both in vitro and in vivo, these organoids were demonstrated to model targeted drug treatments and personalized CAR-T immunotherapy, providing important guidance for personalized medicine.
Introduction
Previous models are incapable of replicating tumor heterogeneity
Inter- and intra-tumoral heterogeneity has been demonstrated across a variety of carcinomas, including different genetic alterations such as single-nucleotide variants, small insertions or deletions (indels), somatic copy number alterations, fusion genes and structural variants, promoting clonal diversity, metastasis and chemoresistance and eventually leading to poor clinical outcomes (2). However, traditional two-dimensional (2D) cultured cell lines and patient-derived xenograft (PDX) models are incapable of replicating tumor heterogeneity of individual patients. For example, SPOP and FOXA1 mutations and CHD1 loss were not reported in 2D prostate cancer cell lines while they were detected in a new three-dimensional (3D) culture model, termed “organoids”, derived from patients with advanced prostate carcinoma (3). Therefore, organoids were gradually introduced in cancer research to mimic tumor heterogeneity and model personalized medicine.
Development of organoids technology in cancer research
Organoids are 3D culture systems replicating structures and functions of corresponding organ (4). The technology of organoids has developed rapidly in recent years. In 2009, Sato and colleagues cultured single Lgr5-positive intestinal stem cells in 3D medium artificially provided niche factors, such as R-spondin, EGF, noggin and Wnt. As a result, organotypic epithelial structures with proliferative crypt and differentiated villus compartments were generated (5). This protocol marked the starting point for organoids culture of multiple organs, including lung (6) , liver (7), colon (8), prostate (9), pancreas (10), stomach (11), esophagus (12), fallopian tube (13) and endometrium (14). Based on improved culture methodologies, organoids were introduced in the field of cancer research as well. In 2014, Dong Gao plated prostate cancer metastasis samples along with circulating tumor cells into prostate organoids culture and established organoids lines from six patients suffered from advanced prostate cancer (3). And from then on, organoids biobanks of prostate carcinoma (15), colon carcinoma (16), pancreatic carcinoma (17), gastric carcinoma (18), renal carcinoma (19), breast carcinoma (20), ovary carcinoma (21) and bladder carcinoma (22) were established, which could be expanded and cryopreserved for worldwide dissemination. The platform of tumor organoids provided a source for basic discoveries, including human cancer initiation, progression, invasion and metastasis (23), offering important guidance for clinical cancer therapy. In 2018, Georgios Vlachogiannis et al. demonstrated potential of patient-derived organoids to predict patient clinical outcomes, directly suggesting the application of tumor organoids in clinical therapy.
Current research of organoids in tumor heterogeneity
Although amount of tumor biobanks has been established, however, attention still should be paid on the ability of organoids in capturing tumor heterogeneity. For example, Oded Kopper and colleagues in Hans Clevers’ team generated an organoids platform for ovarian cancer (OC) mimicking intra- and inter-patient heterogeneity (21). They established 56 organoids lines from 32 patients, representing all main subtypes of OC and recapitulating histological and genomic features of pertinent lesions. Hence these organoids could be applied to test drug sensitivity commonly used in clinical treatment based on features of individual patients (21). Until now, not all the organoids biobanks have been demonstrated to preserve heterogeneity of individual patients and the degree to which organoids recapitulate key features of tumor heterogeneity of individual patients remains unclear in a variety of carcinomas.
Here, Fadi Jacob et al.provided firm proofs to show us an organoids biobank characterized by high degree of tumor heterogeneity in line with original tumors and demonstrated the application of this organoids platform in personalized medicine (1).
Perspectives
A new culture method preserving tumor heterogeneity
Writing in this paper, Fadi Jacob defined a new medium with no added EGF/bFGF or extracellular matrix to reduce further clonal selection or generation of specific cell populations, preserving the local cytoarchitecture and native cell-cell interactions of original tumors and promoting the maintenance of tumor heterogeneity (1). In fact, culture ingredients and culture methods have been adjusted several times to satisfy the growth of different organoids derived from a variety of carcinomas or different clinical stages of a certain tumor to better preserve tumor heterogeneity. For example, culture protocols of organoids derived from human and mouse ductal pancreatic cancer were provided by Sylvia F et al. without identifying heterogeneous patterns in detail (10). Based on this foundation, Takashi Seino established a pancreatic tumor organoids library including 3 functional subtypes with stem cell niche factor adjusted depending on Wnt and R-spondin (24). In addition, EGF removal was also reported to efficiently select KRAS mutant organoids while leaving wild-type KRAS organoids disappeared in pancreatic carcinoma, through which we could preserve more molecular features of original tumor (24).
Another important feature of organoids generation method in Fadi Jacob’s research is that they establish GBOs without mechanical or enzymatic dissociation of the resected tumor tissue into single cells. In fact,in vivotumors consist of cancer cells with different proliferation rates, including high-proliferated, low-proliferated and non-proliferated cells (1). Traditional organoids culture methods started from single cells were intended to undergo clonally selected process for high-proliferated cells in growth factor-rich media while reducing proportions of the relatively low-proliferated cells originally presented within parental tumors. However, these low-proliferated cells also play an important role in pathogenesis and chemoresistance and could not be ignored in clinical cancer therapy. Without resecting tumor tissues into a single cell, Fadi Jacob enabled growth, expansion and differentiation of most cancer cells. After modifying culture methods, tumor heterogeneity is well-maintained in GBOs biobank they established, demonstrated by morphology, genomic and transcriptomic analysis.
New proofs demonstrating recapitulation of tumor heterogeneity
In Fadi Jacob’ s research, they demonstrated the preservation of tumor heterogeneity using traditional methods such as bulk RNA sequencing (RNA-seq) and exome sequencing to investigate genomic and transcriptomic profiling of these cells, and more importantly, they showed new proofs about maintenance of cell-type heterogeneity (1). Previously, heterogeneity of many established tumor organoids was largely illustrated with similar expression profiles and genetic alterations as presented in original tumor tissues (3,24-26). In addition, using single-cell transcriptomic analysis, Fadi Jacob and colleagues identified cell-type heterogeneity and its molecular signatures in GBOs biobank both in short-time and long-time culture (1).
As a result, cells from both parental tumors and GBOs showed patient-specific clusters, while non-neoplastic cells such as macrophages/microglia, T cells and stromal cells clustered together at the single cell level. Moreover, macrophage/microglia cell types even exhibited similar expressions of immune-related genes in the corresponding tumors and GBOs, including cytokines such as TNF, IL1B and TGFB1, indicating the maintenance of certain features of tumor microenvironment within GBOs. Further, to prove whether this heterogeneity could be well-preserved with long-time organoids culture, they performed single-cell RNA-seq for GBOs both in short-time culture (8 weeks) and long-time culture (24 weeks) and found similar cell distributions as for GBOs and corresponding parental tumors (1).
Therefore, by analyzing the involved different cell types and their evolutions, Fadi Jacob provided new proofs for organoids to demonstrate the recapitulation of tumor heterogeneity. The analysis methods involved will be of great use in other organoids biobanks as well.
An important platform for personalized medicine
It is largely known that there are drawbacks in current PDX models for disease modeling and drug screening. For example, the efficiency of tumor formation is relatively low and the process of PDX establishment is time-consuming and money-consuming. In addition, there is tumor evolution in xenograft models. Therefore, tumor heterogeneity of individual patients is not well-preserved in PDX models and organoids models which could overcome these shortages are urgently needed (22). In this article, indeed, they proved advantages of organoids models over traditional PDX models with high efficiency of engraftment, and more importantly, these in vivo organoids showed robust infiltration and retention of key driver mutation expressions, suggesting a better platform for disease modeling.
The contribution for personalized medicine in Fadi Jacob’ s research is that they demonstrated the utility of GBOs to test personalized therapies both by correlating GBO mutational profiles with responses to specific drugs and by modeling CAR-T immunotherapy in a co-culture system (1), providing important guidance for clinical cancer therapy. In this article for drug response test, firstly, they explored targeted drug treatments for specific signaling pathways based on somatic mutations involved in therapeutic resistance. For example, MEK inhibitor trametinib significantly reduced Ki-67+ cells in NF-1 mutant GBOs but not in the EGFR-mutated or PI3K-mutated GBOs, suggesting the application of organoids in reflecting heterogeneous drug responses.
And a more salient and novel finding is that organoids could be used to model personalized CAR-T immunotherapy (1). The co-culture systems in organoids were reported in some carcinomas. For example, Takashi Seino cultured human pancreatic tumor organoids with cancer-associated fibroblasts (CAF) with the purpose of modifying tumor microenvironment and providing a Wnt niche for organoids to better mimic every stage of original tumor (24). However, co-culture systems for organoids, especially co-cultured with immune cells to investigate potential therapeutic targets have not been illustrated clearly until now and they still need to be explored further. In Fadi Jacob’ s research, they co-cultured GBOs with CAR-T cells and tested ability of CAR-T cells in targeting and killing EGFRvIII+ cells in organoids, which is important in immunotherapy but hard to directly manipulate in patients, demonstrating the utility of organoids in guiding personalized antigen-specific CAR-T treatment.
Taken together, these GBOs provided important guidance for clinical cancer treatment based on status of individual patients and showed new co-culture methods in personalized immunotherapy.
Limitations and future applications
While the organoids biobank they established largely resembled heterogeneity of corresponding tumors, however, significant impediments still need to be overcomed to improve the whole successive generation rate according to different mutations or tumor stage. It is reported that the vast majority (96.4%) of wide type isocitrate dehydrogenase 1 (IDH1-WT) tumors have formed plentiful organoids while there was only a reduced success rate (66.7%) in generating organoids with IDH1 mutant tumors. And organoids derived from clinical recurrent tumors also showed a low generation rate (1). To resolve this problem, one possible approach we considered is to modify ingredients in culture media such as changing niche factors or basement membrane (matrigel). Another possible approach is to introduce CAFs or immune cells to create a better tumor microenvironment for every subtype of original tumor. In general, methods to preserve individual tumor features and to develop a success rate in generating organoids still need to be explored further.
Despite limitations in this research, Fadi Jacob et al. have made a great breakthrough in capturing tumor heterogeneity using organoids biobank and exploring personalized therapeutic strategies. With the fast-developed culture methods of organoids in recent years, more biobanks replicating features of a variety of carcinomas or mimicking different clinical stages of original tumors will be established. And these biobanks will undoubtedly continue to be a source for cancer research allowing for worldwide dissemination. In addition, along with personalized CAR-T immunotherapy mentioned in this research, more guidance will be provided in clinical cancer therapy based on patient derived organoids, which will shed a light on personalized medicine in the near future as well.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Jacob F, Salinas RD, Zhang DY, et al A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e122. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turajlic S, Sottoriva A, Graham T, et al Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–16. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 3.Gao D, Vela I, Sboner A, et al Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossi G, Manfrin A, Lutolf MP Progress and potential in organoid research. Nat Rev Genet. 2018;19:671–87. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, Vries RG, Snippert HJ, et al Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche . Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Rock JR, Onaitis MW, Rawlins EL, et al Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–5. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huch M, Gehart H, van Boxtel R, et al Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung P, Sato T, Merlos-Suarez A, et al Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–7. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 9.Karthaus WR, Iaquinta PJ, Drost J, et al Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–75. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boj SF, Hwang CI, Baker LA, et al Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartfeld S, Bayram T, van de Wetering M, et al In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–36.e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWard AD, Cramer J, Lagasse E Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–11. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M, Hoffmann K, Brinkmann V, et al The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turco MY, Gardner L, Hughes J, et al Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. 2017;19:568–77. doi: 10.1038/ncb3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drost J, Karthaus WR, Gao D, et al Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347–58. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimokawa M, Ohta Y, Nishikori S, et al Visualization and targeting of LGR5+ human colon cancer stem cells . Nature. 2017;545:187–92. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Holtzinger A, Jagan I, et al Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21:1364–71. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Yuen ST, Xu J, et al Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 19.Batchelder CA, Martinez ML, Duru N, et al Three dimensional culture of human renal cell carcinoma organoids. PLoS One. 2015;10:e0136758. doi: 10.1371/journal.pone.0136758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs N, de Ligt J, Kopper O, et al A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–86. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Kopper O, de Witte CJ, Lõhmussaar K, et al An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25:838–49. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee SH, Hu W, Matulay JT, et al Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–28.e17. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuveson D, Clevers H Cancer modeling meets human organoid technology. Science. 2019;364:952–5. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 24.Seino T, Kawasaki S, Shimokawa M, et al Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–67.e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Broutier L, Mastrogiovanni G, Verstegen MM, et al Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan HHN, Siu HC, Law S, et al A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–97. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]