Abstract
Objective
To compare the accuracies of quantitative computed tomography (CT) parameters and semiquantitative visual score in evaluating clinical classification of severity of coronavirus disease (COVID-19).
Materials and Methods
We retrospectively enrolled 187 patients with COVID-19 treated at Tongji Hospital of Tongji Medical College from February 15, 2020, to February 29, 2020. Demographic data, imaging characteristics, and clinical data were collected, and based on the clinical classification of severity, patients were divided into groups 1 (mild) and 2 (severe/critical). A semiquantitative visual score was used to estimate the lesion extent. A three-dimensional slicer was used to precisely quantify the volume and CT value of the lung and lesions. Correlation coefficients of the quantitative CT parameters, semiquantitative visual score, and clinical classification were calculated using Spearman's correlation. A receiver operating characteristic curve was used to compare the accuracies of quantitative and semi-quantitative methods.
Results
There were 59 patients in group 1 and 128 patients in group 2. The mean age and sex distribution of the two groups were not significantly different. The lesions were primarily located in the subpleural area. Compared to group 1, group 2 had larger values for all volume-dependent parameters (p < 0.001). The percentage of lesions had the strongest correlation with disease severity with a correlation coefficient of 0.495. In comparison, the correlation coefficient of semiquantitative score was 0.349. To classify the severity of COVID-19, area under the curve of the percentage of lesions was the highest (0.807; 95% confidence interval, 0.744–0.861: p < 0.001) and that of the quantitative CT parameters was significantly higher than that of the semiquantitative visual score (p = 0.001).
Conclusion
The classification accuracy of quantitative CT parameters was significantly superior to that of semiquantitative visual score in terms of evaluating the severity of COVID-19.
Keywords: COVID-19, Quantitative CT, Severity, Clinical classification
INTRODUCTION
A novel coronavirus, which was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the World Health Organization (1,2), has been isolated from lower respiratory tract specimens (3). The virus seems more infectious with regard to person-to-person transmission than the well-known 2003 severe acute respiratory syndrome coronavirus (SARS-CoV) (4). As of May 11, 2020, the global total number of confirmed cases of coronavirus disease (COVID-19) is 4006257 (88891 new cases), with 278892 deaths (4531 new cases), and the highest incidence has been in Europe, with 1731606 confirmed cases (23660 new cases) and 156603 deaths (1051 new cases) (5).
Similar to other types of coronaviral pneumonia, such as SARS and middle East respiratory syndrome (MERS), COVID-19 can also lead to acute respiratory distress syndrome and even death (6,7). Treatment options vary depending on the severity of the patient's condition (8), and patients with severe and critical cases of COVID-19 have a poor survival prognosis (9). It is clinically important to identify patients with severe COVID-19 early in the disease course.
Imaging plays an important role in the management of COVID-19 (10). In the detection of COVID-19, computed tomography (CT) is superior to chest radiography, and some small lesions are not visible on chest radiographs (10,11). Some studies have estimated the severity of COVID-19 based on CT characteristics (12,13) or semiquantitative visual CT scores (14,15). At present, there have been a few quantitative CT studies on COVID-19 (16). The aim of this study was to use an open-source software platform to compare the accuracy of the clinical classification of the severity of COVID-19 based on quantitative CT parameters and the semiquantitative visual score.
MATERIALS AND METHODS
The study protocol was approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. The need for informed consent was waived because of the retrospective nature of the study. The anonymous data were collected and analyzed to facilitate better clinical decision making and treatment.
Patient Selection and Clinical Classification of Severity
This study was a retrospective single-center study that included patients who were hospitalized because of the suspicion of a novel coronavirus infection and underwent both chest CT imaging and laboratory virus nucleic acid testing (reverse transcription polymerase chain reaction [RT-PCR] assay with throat swab samples) from February 15, 2020, to February 29, 2020. A total of 187 patients (mean age, 57.12 ± 15.57 years; 50.8% [95/187] men) were recruited for the analysis. Patients whose poor breath-holding ability led to poor image quality were excluded. The diagnosis of COVID-19 and clinical classification of severity were determined according to the new coronavirus pneumonia diagnosis and treatment plan (trial version 7) developed by the National Health Committee of the People's Republic of China (8). The patients were divided into two groups according to the clinical classification of severity: groups 1 (common) and 2 (severe and critical).
CT Scanning Protocol
All images were obtained using one of the three CT systems (uCT 780, United Imaging, Shanghai, China; Optima CT660, GE Healthcare, Milwaukee, WI, USA; Somatom Definition AS+, Siemens Healthineers, Erlangen, Germany) with patients placed in the supine position. The main scanning parameters were as follows: tube voltage, 120 kVp; automatic tube current modulation, 30–70 mAs; pitch, 0.99–1.22 mm; matrix, 512 × 512; slice thickness, 10 mm; and field of view, 350 × 350 mm. All images were then reconstructed with a slice thickness of 0.625–1.250 mm with the same increments.
Imaging Interpretation
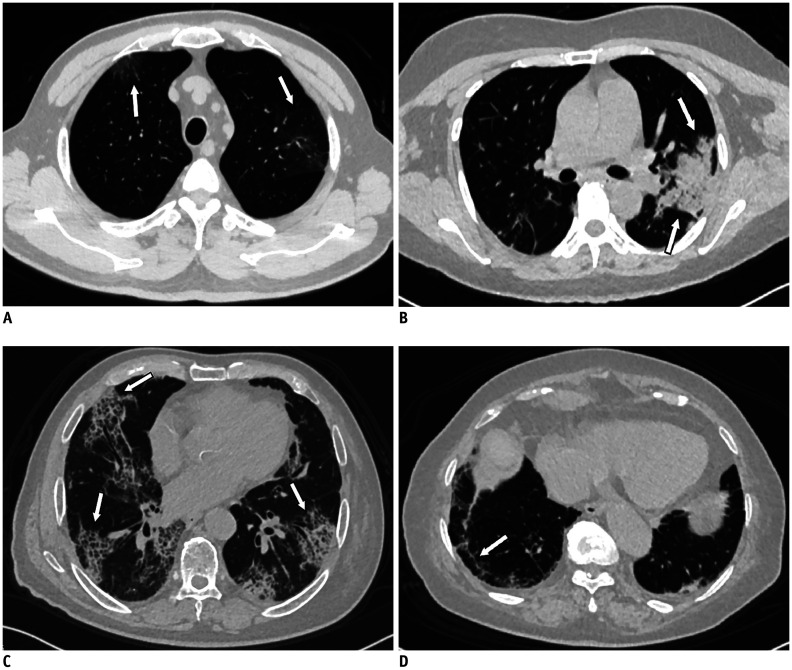
CT images were reviewed by two radiologists (with 15 and 9 years of experience in cardiothoracic imaging, respectively), who were blinded to the clinical classification results. As part of their initial training, the two radiologists analyzed 10 CT imaging studies and reached a consensus to calibrate the grading thresholds with the scoring systems. Subsequently, they reviewed all chest CT images and resolved discrepancies by discussion until they reached a consensus. The CT characteristics were described using the internationally standard nomenclature defined by the Fleischner Society glossary and peer-reviewed literature on viral pneumonia (17,18,19); the terms used were “ground-glass opacity (GGO),” “crazy-paving pattern,” and “consolidation” (Fig. 1A–C). In addition to the crazy-paving pattern, a common form of interstitial thickening is the fibrous cord (Fig. 1D).
Fig. 1. CT manifestations of coronavirus disease.
A. Chest CT shows GGO (arrows) under pleura in bilateral upper lobes (arrow). B. There is consolidation in left lower lobe extending towards pulmonary hilum (arrows). No air bronchogram is observed within lesion. C. Crazy-paving pattern is seen with symmetrical distribution in bilateral lungs (arrows). D. Localized fibrous cord is seen in subpleural area of right lower lobe (arrow). CT = computed tomography, GGO = ground-glass opacity
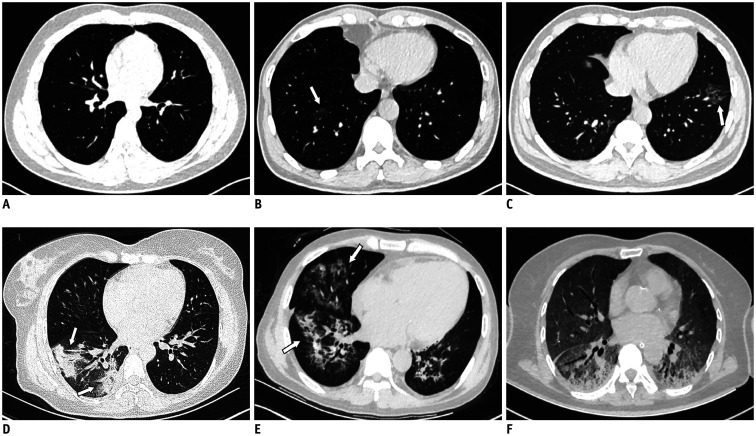
The readers assessed the presence, location, and density of pulmonary abnormalities on chest CT images and graded the abnormalities using the following semiquantitative visual scoring system (15,20,21): score 0, 0% involvement (Fig. 2A); score 1, less than 5% involvement (Fig. 2B); score 2, 5% to 25% involvement (Fig. 2C); score 3, 26% to 49% involvement (Fig. 2D); score 4, 50% to 75% involvement (Fig. 2E); and score 5, greater than 75% involvement (Fig. 2F). Each lobe was assigned a score of 0–5, with a possible total semiquantitative score of 0–25.
Fig. 2. 5-point scale for coronavirus disease based on CT images.
A. No lesion, score 0. B. Focal GGOs in right lower lobe (arrow) with less than 5% involvement, score 1. C. Patchy GGOs in left lower lobe (arrow) with 5% to 25% involvement, score 2. D. Wedge and strip consolidations in right lower lobe (arrows) with 26% to 49% involvement, score 3. E. GGOs combined with consolidation in right lower lobe (arrow) and middle lobes (arrow) with 50% to 75% involvement, score 4. F. Bilateral diffuse GGOs combined with consolidation with greater than 75% involvement, score 5.
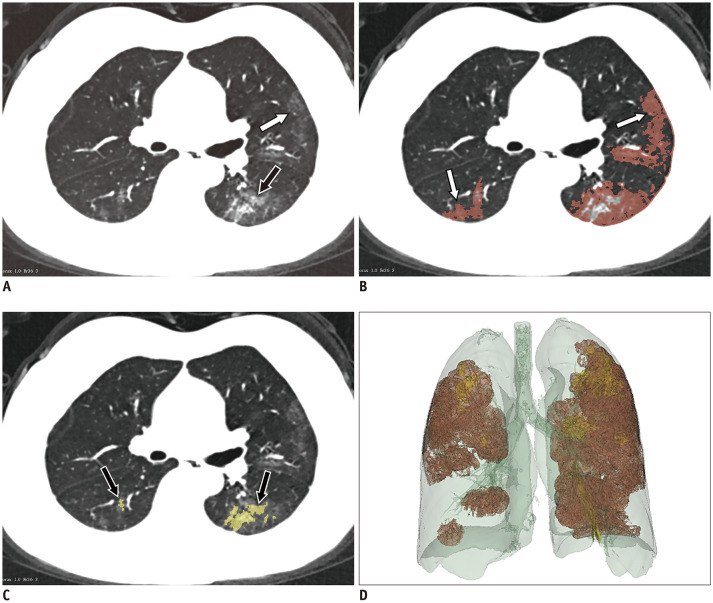
Quantitative CT parameters were assessed with the 3D Slicer software (version 4.10.2, https://www.slicer.org/), and image segmentation was executed by the two aforementioned radiologists. The pulmonary volume (Fig. 3D) and lesion volume were segmented, and lesion volume was divided into the volume of GGOs (Fig. 3A, B) and that of consolidation (Fig. 3A, C). Subsequently, the volume and mean CT value were calculated. The percentages of lesions, GGOs, and consolidation out of the total lung volume were calculated.
Fig. 3. GGOs and consolidation are segmented with 3D Slicer software on CT images.
A. GGO (white arrow) combined with consolidation (black arrow) in subpleural area. B, C. Manual delineation of scope of GGO (white arrows) and consolidation (black arrows) by threshold selection. D. Entire lesion fused with GGO and consolidation is shown in whole lung volume.
Data Analysis
The cutoff values for the quantitative CT parameters and semiquantitative visual score were calculated based on the maximum Youden index. Differences in all parameters between groups 1 and 2 were compared with the t test or chi-square test. Spearman's correlation was used to detect the correlations between the CT parameters and the clinical classification of severity. The receiver operating characteristic (ROC) curve analysis was used to select the method with superior accuracy in terms of determining the disease severity from among the quantitative CT parameters and semiquantitative visual score. Analyses were performed with the SPSS 25.0 statistical package (IBM Corp., Armonk, NY, USA) and MedCalc version 19.1.3 (MedCalc Software bvba, Mariakerke, Belgium). P values < 0.05 were considered statistically significant.
RESULTS
General Description and Clinical Classification
Five patients were excluded because of poor image quality. Fifty-nine patients (31.6%) were classified in group 1, while 128 (68.4%) were classified in group 2 on the day of the examination. The mean ages of the two groups were not significantly different; group 1 had a mean age of 55.4 ± 2.3 years, and group 2 had a mean age of 57.9 ± 1.3 years (p = 0.303). The sex distributions did not differ between the two groups (p = 0.993).
CT Characteristics and Semiquantitative Visual Score of COVID-19
Table 1 shows the comparison of the imaging characteristics, namely, interstitial thickening, air bronchogram, axial plane distribution, and unilateral or bilateral involvement, between groups 1 and 2. Among the 187 patients with COVID-19, 13 were negative on CT pulmonary imaging (Fig. 1), and all negative patients were in group 1. The lesions were primarily located in the subpleural area, with a possible extension towards the pulmonary hilum in large lesions.
Table 1. Conventional CT Parameters Characteristic of Coronavirus Disease.
| CT Parameters | Group 1 | Group 2 | χ2 | P |
|---|---|---|---|---|
| Imaging characteristic | 30.448 | < 0.001 | ||
| No lesion | 13 | 0 | ||
| Ground-glass opacities | 30 | 85 | ||
| Consolidation | 2 | 7 | ||
| Mixed pattern | 14 | 36 | ||
| Interstitial thickening | 8.284 | 0.016 | ||
| No interstitial thickening | 51 | 85 | ||
| Crazy-paving pattern | 4 | 25 | ||
| Interstitial fibrosis | 4 | 18 | ||
| Air-bronchogram | 1 | 5 | 0.636 | 0.425 |
| Axial plane distribution | 35.823 | < 0.001 | ||
| No lesion | 13 | 0 | ||
| Peripheral | 40 | 107 | ||
| Inner zone | 1 | 6 | ||
| Balance | 5 | 15 | ||
| Unilateral or bilateral involvement | 33.476 | < 0.001 | ||
| No lesion | 13 | 0 | ||
| Unilateral | 13 | 20 | ||
| Bilateral | 33 | 108 |
CT = computed tomography
The quantitative CT parameters showed significant differences in the volume (volumes of lesions, GGOs, and consolidation) and percentage of the lesions out of the total lung volume (percentages of lesions, GGOs, consolidation) between groups 1 and 2. Table 2 shows the comparison of the quantitative CT parameters. Compared to group 1, group 2 had larger values for the volume and percentage of the lesion out of the total lung volume (p < 0.001).
Table 2. Quantitative CT Parameters for Coronavirus Disease.
| Quantitative CT Parameters | Group 1 | Group 2 | t | P |
|---|---|---|---|---|
| Pulmonary volume (cm3) | 3912.4 ± 147.7 | 3662.2 ± 100.6 | 1.400 | 0.164 |
| Volume of ground-glass opacities (cm3) | 83.6 ± 18.5 | 298.5 ± 31.7 | -5.849 | < 0.001 |
| Volume of consolidation (cm3) | 28.1 ± 5.8 | 94.9 ± 11.7 | -5.366 | < 0.001 |
| Volume of lesions (cm3) | 108.7 ± 21.9 | 393.4 ± 36.6 | -6.683 | < 0.001 |
| Mean CT value of lung (HU) | -778 ± 10 | -755 ± 7 | -1.929 | 0.056 |
| Mean CT value of ground-glass opacities (HU) | -644 ± 26 | -655 ± 6 | 0.419 | 0.677 |
| Mean CT value of consolidation (HU) | -230 ± 10 | -223 ± 7 | -0.569 | 0.570 |
| Mean CT value of lesions (HU) | -561 ± 19 | -547 ± 15 | -0.580 | 0.563 |
| Percentage of ground-glass opacities (%) | 2.6 ± 0.8 | 9.9 ± 1.2 | -4.943 | < 0.001 |
| Percentage of consolidation (%) | 0.7 ± 0.2 | 3.2 ± 0.4 | -5.289 | < 0.001 |
| Percentage of lesions (%) | 3.3 ± 1.0 | 13.0 ± 1.4 | -5.622 | < 0.001 |
Table 3 shows the significant differences in the semiquantitative visual score for each lobe and that between groups 1 and 2 (p < 0.001).
Table 3. Semiquantitative Visual Score for Coronavirus Disease.
| Lobes | Group 1 | Group 2 | t | P |
|---|---|---|---|---|
| Right upper lobe | 0.92 ± 0.17 | 1.64 ± 0.14 | -3.363 | < 0.001 |
| Right middle lobe | 0.54 ± 0.15 | 1.31 ± 0.14 | -3.799 | < 0.001 |
| Right lower lobe | 1.54 ± 0.20 | 2.53 ± 0.14 | -4.097 | < 0.001 |
| Left upper lobe | 0.69 ± 0.15 | 1.60 ± 0.13 | -4.723 | < 0.001 |
| Left lower lobe | 1.31 ± 0.19 | 2.40 ± 0.16 | -4.500 | < 0.001 |
| Semiquantitative score | 4.49 ± 0.68 | 9.59 ± 0.58 | -4.919 | < 0.001 |
Correlation Analysis and ROC Analysis
Spearman's correlation analysis of the relationships between the imaging parameters (including the quantitative CT parameters and semiquantitative visual score) and the clinical classification of severity showed that the relationship was stronger for quantitative CT parameters than for the semiquantitative visual score (Table 4). In addition, the correlation of lesions and GGOs was stronger than that of consolidation in the comparison among the quantitative CT parameters.
Table 4. Spearman's Correlation Analysis of Imaging Data and Clinical Classification.
| Imaging Data | r | P |
|---|---|---|
| Volume of lesions | 0.489 | < 0.001 |
| Percentage of lesions | 0.495 | < 0.001 |
| Volume of ground-glass opacities | 0.464 | < 0.001 |
| Percentage of ground-glass opacities | 0.476 | < 0.001 |
| Volume of consolidation | 0.349 | < 0.001 |
| Percentage of consolidation | 0.358 | < 0.001 |
| Semiquantitative score | 0.349 | < 0.001 |
Table 5 shows the cutoff values, sensitivity, specificity, area under the curve (AUC) and 95% confidence interval when the chest CT images of all subjects were assessed with the two different systems: the quantitative CT parameters were more accurate than the semiquantitative visual score for determination of the severity of COVID-19. The results showed that the percentage of lesions had the highest AUC (0.807; 95% confidence interval, 0.744–0.861; p < 0.001) and specificity (93.2%; 95% confidence interval, 83.5–98.1%), and the percentage of GGOs had the highest sensitivity (93.0%; 95% confidence interval, 87.1–96.7%).
Table 5. AUC for Severity Classification of COVID-19 Using Quantitative CT Parameters and Semiquantitative Visual Score.
| Test Result Variable(s) | Cutoff | Sensitivity (%) | Specificity (%) | Youden's Index (%) | AUC | ||||
|---|---|---|---|---|---|---|---|---|---|
| Value | 95% CI | Value | 95% CI | Value | P | 95% CI | |||
| Volume of lesions (cm3) | 42.018 | 91.4 | 85.1–95.6 | 55.9 | 42.4–68.6 | 47.3 | 0.804 | < 0.001 | 0.739–0.858 |
| Percentage of lesions | 7.477 | 50.8 | 41.8–59.7 | 93.2 | 83.5–98.1 | 44.0 | 0.807 | < 0.001 | 0.744–0.861 |
| Volume of ground-glass opacities (cm3) | 28.780 | 92.2 | 86.1–96.2 | 54.2 | 40.8–67.3 | 46.4 | 0.788 | < 0.001 | 0.722–0.844 |
| Percentage of ground-glass opacities | 0.671 | 93.0 | 87.1–96.7 | 55.9 | 42.4–68.8 | 48.9 | 0.795 | < 0.001 | 0.730–0.851 |
| Semiquantitative score | 2.000 | 85.9 | 78.7–91.4 | 52.5 | 39.1–65.7 | 38.5 | 0.716 | < 0.001 | 0.646–0.780 |
AUC = area under curve, CI = confidence interval
We compared the AUCs between the quantitative CT parameters and the semiquantitative visual score (Fig. 4). Tables 5 and 6 show that the AUC of the quantitative CT parameters was significantly larger than that of the semiquantitative visual score (p < 0.05).
Fig. 4. ROC curves of quantitative CT parameters and semiquantitative visual score for identification of clinical classification of severity.
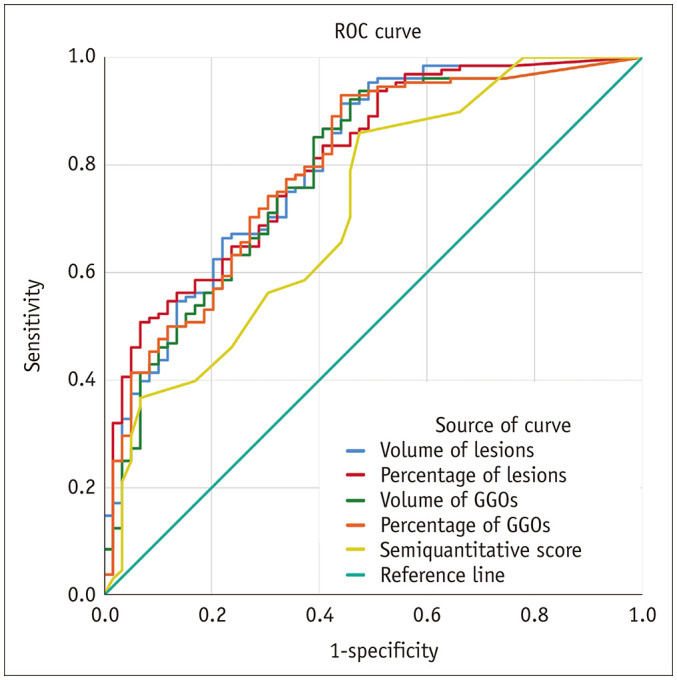
ROC = receiver operating characteristic
Table 6. Comparison of AUC between Quantitative CT Parameters and Semiquantitative Visual Score.
| Compared Parameter | Difference between AUCs | Standard Error | z | P |
|---|---|---|---|---|
| Volume of lesions vs. semiquantitative score | 0.087 | 0.0295 | 2.955 | 0.0031 |
| Percentage of lesions vs. semiquantitative score | 0.091 | 0.0285 | 3.198 | 0.0014 |
| Volume of ground-glass opacities vs. semiquantitative score | 0.072 | 0.0318 | 2.251 | 0.0244 |
| Percentage of ground-glass opacities vs. semiquantitative score | 0.079 | 0.0301 | 2.617 | 0.0089 |
DISCUSSION
The outbreak of COVID-19 has resulted in a global health emergency. COVID-19 shows radiological similarities to SARS and MERS pneumonia (22,23,24,25,26), with a predominance of bilateral GGOs and consolidative lesions in the peripheral lung (27,28).
Treatment strategies are determined by the disease severity (8), and CT is a reliable modality for determining the disease severity (29). Previous studies have assessed the severity of COVID-19 through the description of imaging features (12,13), semiquantitative visual scores (14,15), and deep learning algorithms (16). Deep learning algorithms vary across manufacturers, and the accuracy of automatic segmentation is lower than that of manual segmentation. The results are difficult to generalize. This study used an open-source software platform and confirmed that the correlation between quantitative CT parameters and the severity of COVID-19 was stronger than that between the semiquantitative visual score and the severity of COVID-19. Compared with the semiquantitative visual score, the quantitative CT parameters have superior accuracy.
The quantitative CT parameters of lesion involvement (volume and percentage) in COVID-19 patients were significantly associated with the clinical classification of severity, but the mean density of lesions was not; therefore, we can conclude that the clinical classification of the severity of COVID-19 is related to the degree of involvement but not to the degree of consolidation of pulmonary tissue. We obtained similar results in the correlation analysis, with the correlation coefficient of consolidation (volume and percentage) being significantly lower than those of GGOs and lesions.
In this study, the sensitivity and specificity of the quantitative CT parameters were higher than those of the semiquantitative visual score in terms of the degree of involvement. Therefore, we can conclude that the former has superior accuracy for the clinical classification of severity. The sensitivity of the GGO percentage was the highest, but the specificity was moderate, indicating that some patients had milder disease but larger GGOs; this supports the findings of some studies, which have shown that some patients have moderate CT characteristics but mild or no clinical symptoms at the time of admission and that disease progression occurs in these patients over the next few days (21,30). Therefore, the GGO percentage can not only be used to classify the clinical severity when the severity does not match the degree of involvement but also as a clinical indication for close observation. If the patient's condition changes, treatment should be upgraded immediately. Moreover, the specificity of the lesion percentage was the highest, but the sensitivity was moderate. One likely reason is that the lesion percentage more accurately reflects the degree of involvement of lesions and therefore, has a greater specificity. The maximum AUC of the precise quantitative CT parameters was 0.807, while that of the semiquantitative visual score was 0.716, with the difference being significant. Previous studies have shown that the AUC of the semiquantitative visual score is 0.918 (14) or 0.870 (15). However, we found that the proportions of patients in group 2 were significantly lower in those two studies than in this study (10.2%, 30.1%, and 68.4%, respectively). Therefore, it can be speculated that the higher proportion of patients in group 2 is the reason for the relatively lower AUC.
There are several limitations to this study. First, this was a single-center study, but our CT images were obtained with three CT scanners from different manufacturers, which reduced the bias that would be caused by using only one scanner. Furthermore, the patients in our study were inpatients, and most patients with mild COVID-19 are not hospitalized. Therefore, the proportion of patients in group 2 was higher than that in group 1. Consequently, the accuracy of classification could be further improved. Finally, this study only evaluated the clinical classification of the severity with CT images and did not analyze the influence of complications, such as hypertension and cardiovascular disease; however, neither the quantitative CT parameters nor the semiquantitative visual score analysis can be used to evaluate complications, so the effect on their comparison was limited.
In conclusion, the classification accuracy of quantitative CT parameters was significantly superior to that of semiquantitative visual score in terms of evaluating the severity of COVID-19. In particular, the clinical classification of the severity of COVID-19 significantly correlated with the quantitative volume-dependent parameters, and their accuracy was significantly superior to that of the semiquantitative visual score. The initial results for quantitative CT parameters in this study are encouraging, and these parameters may be seen as a viable alternative for predicting the severity of COVID-19.
Acknowledgments
The authors would like to acknowledge Yao Jing for his assistance in improving the English in this manuscript.
Footnotes
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Jiang X, Rayner S, Luo MH. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J Med Virol. 2020;92:476–478. doi: 10.1002/jmv.25708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorusso A, Calistri P, Petrini A, Savini G, Decaro N. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet Ital. 2020;56:5–10. doi: 10.12834/VetIt.2173.11599.1. [DOI] [PubMed] [Google Scholar]
- 3.Tan W, Zhao X, Ma X, Wang W, Niu P, Xu W, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019−2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9:469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Coronavirus disease 2019 (COVID-19). Situation report–112. WHO Web site; [Accessed May 12, 2020]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200511-covid-19-sitrep-112.pdf?sfvrsn=813f2669_2. Published May 11, 2020. [Google Scholar]
- 6.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Health Commission & National Administration of Traditional Chinese Medicine. Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7) Chin Med J. 2020;133:1087–1095. doi: 10.1097/CM9.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the fleischner society. Radiology. 2020 Apr 07; doi: 10.1148/radiol.2020201365. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi H, Qi X, Yoon SH, Park SJ, Lee KH, Kim JY, et al. Extension of coronavirus disease 2019 (COVID-19) on chest CT and implications for chest radiograph interpretation. Radiology: Cardiothoracic Imaging. 2020 Mar 30; doi: 10.1148/ryct.2020200107. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu KC, Xu P, Lv WF, Qiu XH, Yao JL, Gu JF, et al. CT manifestations of coronavirus disease-2019: a retrospective analysis of 73 cases by disease severity. Eur J Radiol. 2020 May 12; doi: 10.1016/j.ejrad.2020.108941. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am J Roentgenol. 2020;214:1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 14.Li K, Fang Y, Li W, Pan C, Qin P, Zhong Y, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur Radiol. 2020 Mar 25; doi: 10.1007/s00330-020-06817-6. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55:327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Han R, Ai T, Yu P, Kang H, Tao Q, et al. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiology: Cardiothoracic Imaging. 2020 Mar 30; doi: 10.1148/ryct.2020200075. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franquet T. Imaging of pulmonary viral pneumonia. Radiology. 2011;260:18–39. doi: 10.1148/radiol.11092149. [DOI] [PubMed] [Google Scholar]
- 18.Koo HJ, Lim S, Choe J, Choi SH, Sung H, Do KH. Radiographic and CT features of viral pneumonia. Radiographics. 2018;38:719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 19.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Muller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246:697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 20.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi WJ, Lee KN, Kang EJ, Lee H. Middle East respiratory syndrome-coronavirus infection: a case report of serial computed tomographic findings in a young male patient. Korean J Radiol. 2016;17:166–170. doi: 10.3348/kjr.2016.17.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ajlan AM, Ahyad RA, Jamjoom LG, Alharthy A, Madani TA. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. AJR Am J Roentgenol. 2014;203:782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 24.Wan YL, Tsay PK, Cheung YC, Chiang PC, Wang CH, Tsai YH, et al. A correlation between the severity of lung lesions on radiographs and clinical findings in patients with severe acute respiratory syndrome. Korean J Radiol. 2007;8:466–474. doi: 10.3348/kjr.2007.8.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong KT, Antonio GE, Hui DS, Lee N, Yuen EH, Wu A, et al. Thin-section CT of severe acute respiratory syndrome: evaluation of 73 patients exposed to or with the disease. Radiology. 2003;228:395–400. doi: 10.1148/radiol.2283030541. [DOI] [PubMed] [Google Scholar]
- 26.Das KM, Lee EY, Enani MA, AlJawder SE, Singh R, Bashir S, et al. CT correlation with outcomes in 15 patients with acute Middle East respiratory syndrome coronavirus. AJR Am J Roentgenol. 2015;204:736–742. doi: 10.2214/AJR.14.13671. [DOI] [PubMed] [Google Scholar]
- 27.Lee KS. Pneumonia associated with 2019 novel coronavirus: can computed tomographic findings help predict the prognosis of the disease? Korean J Radiol. 2020;21:257–258. doi: 10.3348/kjr.2020.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim KH, et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J Radiol. 2020;21:494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H. Outbreak of novel coronavirus (COVID-19): what is the role of radiologists? Eur Radiol. 2020 Feb 18; doi: 10.1007/s00330-020-06748-2. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei J, Li J, Li X, Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020 Jan 31; doi: 10.1148/radiol.2020200236. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]