SUMMARY
Understanding how flowers form is an important problem in plant biology, as human food supply depends on flower and seed production. Flower development also provides an excellent model for understanding how cell division, expansion and differentiation are coordinated during organogenesis. In the model plant Arabidopsis thaliana, floral organogenesis requires AINTEGUMENTA (ANT) and AINTEGUMENTA‐LIKE 6 (AIL6)/PLETHORA 3 (PLT3), two members of the Arabidopsis AINTEGUMENTA‐LIKE/PLETHORA (AIL/PLT) transcription factor family. Together, ANT and AIL6/PLT3 regulate aspects of floral organogenesis, including floral organ initiation, growth, identity specification and patterning. Previously, we used RNA‐Seq to identify thousands of genes with disrupted expression in ant ail6 mutant flowers, indicating that ANT and AIL6/PLT3 influence a vast transcriptional network. The immediate downstream targets of ANT and AIL6/PLT3 in flowers are unknown, however. To identify direct targets of ANT regulation, we performed an RNA‐Seq time‐course experiment in which we induced ANT activity in transgenic plants bearing an ANT‐glucocorticoid receptor fusion construct. In addition, we performed a ChIP‐Seq experiment that identified ANT binding sites in developing flowers. These experiments identified 200 potential ANT target genes based on their proximity to ANT binding sites and differential expression in response to ANT. These 200 candidate target genes were involved in functions such as polarity specification, floral organ development, meristem development and auxin signaling. In addition, we identified several genes associated with lateral organ growth that may mediate the role of ANT in organ size control. These results reveal new features of the ANT transcriptional network by linking ANT to previously unknown regulatory targets.
Keywords: Arabidopsis thaliana, flower development, AINTEGUMENTA (ANT), AINTEGUMENTA‐LIKE (AIL)/PLETHORA (PLT), RNA‐Seq, ChIP‐Seq, organ polarity, organ growth, auxin signaling
Significance Statement
Flower development has been extensively studied, but our knowledge of gene regulatory networks generating floral organs with characteristic forms remains incomplete. This study used a highly targeted genomic approach to identify 200 genes likely to be direct targets of regulation by AINTEGUMENTA (ANT), a transcription factor regulating flower patterning and growth. Our set of identified target genes include those with known roles in polarity specification, floral organ development, lateral organ growth and auxin signaling.
INTRODUCTION
Flowers supply fruits, seeds and grains to the human diet, and are a subject of fascination for their beauty and morphological diversity. Molecular genetic studies in Arabidopsis have identified many regulatory factors that control the initiation and subsequent development of flowers. Flower primordia arise in the periphery of the inflorescence meristem at sites of auxin maxima, during the reproductive phase of the plant life cycle (Benkova et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005). At these sites, AUXIN RESPONSE FACTOR 5/MONOPTEROS (ARF5/MP) upregulates the expression of LEAFY (LFY) and two AINTEGUMENTA‐LIKE/PLETHORA (AIL/PLT) genes, AINTEGUMENTA (ANT) and AINTEGUMENTA‐LIKE 6 (AIL6)/PLETHORA 3 (PLT3), to specify these primordia as flowers and promote their outgrowth (Yamaguch et al., 2013). Within flower primordia, floral organ primordia are initiated at defined positions within whorls and subsequently adopt one of four fates according to the ABCE model (reviewed by Krizek and Fletcher, 2005). Class‐A and ‐E gene activities in whorl 1 specify sepal identity, class‐A, ‐B and ‐E gene activities in whorl 2 specify petal identity, class‐B, ‐C and ‐E gene activities in whorl 3 specify stamen identity, and class‐C and ‐E gene activities in whorl 4 specify carpel identity.
Most class‐A, ‐B, ‐C and ‐E floral organ identity genes encode MADS‐domain transcription factors that act in higher order protein complexes to regulate gene expression (Smaczniak et al., 2012). Genomic studies on these transcription factors have begun to reveal the gene regulatory networks that control the development of each floral organ type (Kaufmann et al., 2009; Kaufmann et al., 2010; Wuest et al., 2012; O’Maoiléidigh et al., 2013). These studies indicate that floral organ identity proteins regulate a large number of genes throughout floral organ development, with distinct genes being regulated at different stages of organ development (as reviewed by Stewart et al., 2016; Yan et al., 2016). Direct regulatory targets include other transcription factors as well as genes involved in plant hormone signaling pathways and developmental processes, such as pattern formation and morphogenesis. Furthermore, the floral organ identity proteins appear to repress the expression of genes that specify a leaf developmental program (O’Maoiléidigh et al., 2013).
Genetic studies have uncovered multiple roles for ANT and AIL6/PLT3 during the initiation and subsequent development of floral organ primordia. ANT and AIL6/PLT3 promote the initiation of floral organ primordia at defined positions within the floral meristem, and also prevent premature differentiation of both primordial and floral meristem cells (Krizek, 2009; Krizek and Eaddy, 2012). ANT and AIL6/PLT3 are required for the proper expression of the class‐B and ‐C floral organ identity genes APETALA 3 (AP3) and AGAMOUS (AG), respectively, and consequently for the elaboration of petal and stamen fates (Krizek, 2009). ANT and AIL6/PLT3 also regulate the growth of developing floral organs, thus contributing to morphogenesis and the attainment of correct organ size (Elliott et al., 1996; Klucher et al., 1996; Krizek, 1999; Mizukami and Fischer, 2000). Despite their well‐established importance in many facets of flower development, we currently know very little about the downstream target genes that are activated or repressed by ANT and AIL6/PLT3. Without this knowledge, we lack understanding of the biological processes regulated by these transcription factors that contribute to the sculpting of each floral organ type.
A genomic study comparing the transcriptomes of the wild type and the ant ail6 double mutants identified thousands of differentially expressed (DE) genes, consistent with the numerous roles of ANT and AIL6/PLT3 in floral organogenesis and the severe phenotypic consequences of losing ANT and AIL6/PLT3 functions (Krizek et al., 2016). This experiment suggested that ANT and AIL6/PLT3 functions are intimately coupled within a vast transcriptional network regulating floral organogenesis. To distinguish between short‐term and longer‐term consequences of loss of ANT and AIL6/PLT3 activities, we have adopted more focused genomic studies. We performed two complementary experiments: RNA‐Seq analysis of floral buds at 2, 4 and 8 h after the induction of ANT activity, and chromatin immunoprecipitation followed by sequencing (ChIP‐Seq) analysis of stage‐6/7 flowers. Together, these studies identified 200 genes that are both differentially expressed after ANT induction and are bound by ANT and are thus likely to be direct targets of ANT regulation. Our experiments suggest that ANT controls floral organogenesis through the direct regulation of growth and patterning genes as well as auxin responses.
RESULTS
RNA‐Seq identifies genes differentially expressed after the activation of ANT‐GR
To identify direct targets of ANT regulation, we created a line of transgenic plants containing an inducible form of ANT in which the ligand‐binding domain of the glucocorticoid receptor (GR) was fused to the coding region of ANT. In this system, the GR domain blocks migration of the ANT‐GR fusion partner into the nucleus, rendering it incapable of activating or repressing the expression of target genes. Applying the steroid dexamethasone (dex) causes a conformational change in GR that permits entry of ANT‐GR into the nucleus, where it can bind to target promoters and regulate gene expression. Applying dex to 35S:ANT‐GR inflorescences led to the production of larger flowers and caused male sterility, similar to the phenotypes observed in 35S:ANT inflorescences (Figure S1) (Krizek, 1999; Yamaguchi et al., 2013). This established that the ANT‐GR line contained inducible ANT activity that could be triggered by the application of dex.
We performed a time‐course experiment using the 35S:ANT‐GR line, in which floral buds were collected 2, 4 and 8 h following treatment with dex or treatment with solvent only (mock) as a negative control. Whole inflorescences corresponding to floral buds at stages 1–12 were collected as four matched pairs of treatment and control samples and processed for RNA sequencing. Following sequencing and alignment to the Arabidopsis reference genome, each sample yielded between 13 and 29 million aligned fragments, which were then analyzed for differential expression. Using a false discovery rate (FDR) of 0.05, we identified 1195 genes that were DE at one or more time points (Appendix S1). More DE genes were present at 4 h (746) and 8 h (609) after treatment, compared with 2 h (324) after treatment (Figure 1a). The log2 fold change (logFC) values of the DE genes ranged from 3.15 to −1.80. There were 106 genes that were DE at all three time points. Approximately equal numbers of upregulated (836) and downregulated (843) genes were identified.
Figure 1.
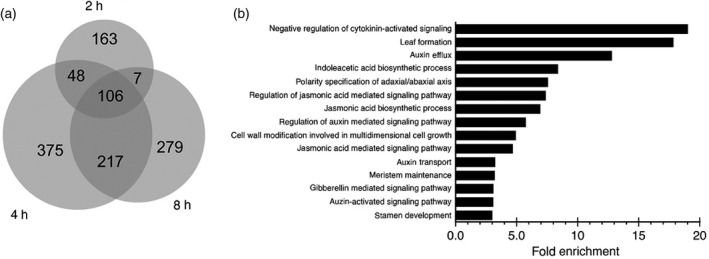
Genes differentially expressed (DE) in mock‐ and dex‐treated 35S:ANT‐GR inflorescences.
(a) Venn diagram showing the overlap between DE genes identified at 2, 4 and 8 h after treatment. The Venn diagram was created with biovenn (Hulsen et al., 2008).
(b) Biological process gene ontology (GO) terms enriched in DE genes.
To visualize gene expression changes over time, we developed an interactive r shiny app (called show gene expression) that plots individual gene expression data over the time course of the experiment (https://bitbucket.org/krizeklab) (Figure S2a–d). The app is available in the inducible‐ant‐rna‐seq repository (/DifferentialExpression/ShowGeneExpression). After launching the shiny app in the rstudio application, users can generate plots showing the expression of a gene of interest after entering its gene identifier. Gene expression can be visualized as: ‘Sample RPKM’, with individual treatment and time points selected; ‘Group RPKM’, which averages the four replicates for each sample; or ‘Expression over time’, in which the data can be visualized with lines, points or arrows that indicate up‐ or downregulation between the control and treated samples. The ‘gene info’ tab provides a link to The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org) for further information about the gene. We used this app to investigate the expression of candidate target genes and explore their functions.
Differentially expressed genes were enriched in terms associated with development and hormone physiology
We used enrichment analysis to identify gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) functional annotation categories over‐represented among the 1195 DE genes. We further investigated individual gene functions using TAIR for members of these categories.
Many DE genes had functions related to lateral organ development, consistent with the role of ANT as a regulator of organogenesis. These included: leaf formation (GO:0010338); polarity specification of adaxial/abaxial axis (GO:0009944); and stamen development (GO:0048443) (Figure 1b). Seven genes that regulate lateral organ polarity were differentially expressed (Table S1). These genes included: three YABBY genes – CRABS CLAW (CRC), YAB3 and YAB5; one class‐III HD‐ZIP gene – PHABULOSA (PHB); one KANADI gene – KAN2; ASYMMMETRIC LEAVES 1 (AS1); and BLADE ON PETIOLE 1 (BOP1). Thus, both adaxial (PHB, AS1 and BOP1) and abaxial (CRC, YAB3, YAB5 and KAN2) genes were identified as potential targets of ANT regulation. The identification of genes regulating adaxial/abaxial axis specification is consistent with previous genetic work suggesting that ANT contributes to organ polarity (Nole‐Wilson and Krizek, 2006). Leaf development GO category genes include several genes regulating organ growth, including KLUH (KLU/CYP78A5), GROWTH REGULATING FACTORS (GRF3, GRF6 and GRF8) and ANGUSTIFOLIA 3/GRF1‐INTERACTING FACTOR 1 (AN3/GIF1).
Several genes with known roles in floral organ development were DE. These included floral organ identity genes APETALA 1 (AP1), APETALA 2 (AP2) and SEPALLATA 3 (SEP3), as well as genes that function in later aspects of stamen development [SQUAMOSA PROMOTER‐BINDING PROTEIN‐LIKE 8 (SPL8), EXCESS MICROSPOROCYTES 1 (EMS1)] and carpel development [SPATULA (SPT), CRC, STRUBBELIG (SUB)] (Table S1).
In addition to genes involved in lateral organ development, seven DE genes involved in meristem maintenance (GO:0010073) were identified: CORYNE (CRN), BARELY ANY MERISTEM 3 (BAM3), CLAVATA3 INSENSITIVE RECEPTOR KINASE 4 (CIK4), MINI ZINC FINGER 2 (MIF2) and FANTASTIC FOUR 1, 2 and 3 (FAF1, FAF2 and FAF3) (Table S1). This suggests that ANT has a role in meristem maintenance, in addition to its role in lateral organ development, consistent with genetic work showing the importance of ANT and AIL6/PLT3 in preventing the premature differentiation of floral meristem cells (Krizek and Eaddy, 2012).
Many hormone‐related functions were enriched in the DE genes (Appendix S2), suggesting that ANT function is closely linked with multiple aspects of hormone physiology. These GO terms included: negative regulation of the cytokinin‐activated signaling pathway (GO:0080037); regulation of the jasmonic acid‐mediated signaling pathway (GO:2000022); regulation of the auxin‐mediated signaling pathway (GO:0010928); regulation of the jasmonic acid‐mediated signaling pathway (GO:0009867); regulation of the gibberellin‐mediated signaling pathway (GO:0010476); regulation of the auxin‐activated signaling pathway (GO:0009734); hormone biosynthesis – indoleacetic acid biosynthetic process (GO:000009684) and jasmonic acid biosynthetic process (GO:0009695); and hormone transport – auxin efflux (GO:0010315) and auxin transport (GO:0060918) (Figure 1b). Additional analysis using the KEGG pathway framework also found links with cytokinin, auxin, and gibberellin hormone signaling pathways (Figure S3). DE genes associated with the synthesis/metabolism, transport, signaling and response for cytokinin, auxin, gibberellin, abscisic acid and jasmonic acid (JA) are shown in Table S2. These results suggest that ANT plays roles in the metabolism of auxin and JA (and perhaps cytokinin and gibberellin) while also influencing signaling pathways downstream of cytokinin, auxin, gibberellin, abscisic acid and JA. Our earlier RNA‐Seq investigation of ant ail6 double mutants inflorescences previously linked ANT function with auxin biosynthesis and JA signaling, but not with cytokinin or gibberellin signaling (Krizek et al., 2016).
ChIP‐Seq identifies ANT genomic binding sites in stage‐6/7 flowers
To identify genome‐wide ANT binding sites, we performed chromatin immunoprecipitation in combination with next‐generation sequencing (ChIP‐Seq). For these studies, we used a transgenic line in which an ANT‐VENUS gene fusion was expressed under the control of the ANT promoter in the ant mutant background. Fusion of ANT with VENUS, a rapidly folding variant of YELLOW FLUORESCENT PROTEIN (YFP), allows the immunoprecipitation of DNA bound to ANT by a commercially available antibody against GREEN FLUORESCENT PROTEIN (GFP). The transgene fully rescued the ant‐4 mutant phenotype, as assayed by petal measurements and floral organ counts, indicating that the ANT‐VENUS fusion protein has full ANT activity (Tables S3, S4). The ANT:ANT‐VENUS ant‐4 line was crossed into a genetic background in which flower development can be synchronized (i.e. AP1:AP1‐GR ap1 cal) (O’Maoiléidigh et al., 2015), allowing us to collect large numbers of flowers of the same developmental stage. Inflorescences from two biological replicates of AP1:AP1‐GR ap1 cal and AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant‐4 were harvested 5 days after dex treatment, when they are composed of stage‐6/7 flowers.
In our ChIP‐Seq experiment, we identified 1113 ANT binding peaks in stage‐6/7 flowers using visual analytics and the integrated genome browser (igb). These peaks were associated with 1081 unique genes (Appendix S3). Almost half of the peaks (48%) are present upstream of the gene near the transcriptional start site (TSS), with the remaining peaks overlapping the start of transcription (18%), sitting within the gene (15%), overlapping the end of transcription (5%), sitting downstream of the gene (14%) or encompassing the gene (1%) (Figure 2a,b).
Figure 2.
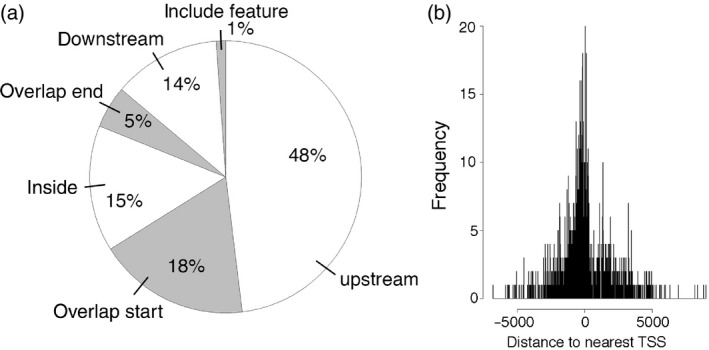
Position of ANT ChIP‐Seq peaks relative to the closest gene.
(a) Pie chart showing the position of ANT ChIP‐Seq binding peaks relative to the closest gene. Almost half of the peaks are upstream of the closest gene (48%). The remaining peaks either overlap with the start of the gene (18.0%), are within the gene (15%), overlap with the end of the gene (5%), are downstream of the gene (14%) or overlap the entire gene (1%).
(b) Position of ANT binding peak relative to the transcriptional start site (TSS) of the closest gene.
Genes associated with ANT binding sites include regulators of polarity specification, floral organ development and meristem development
To gain insight into the set of genes associated with ANT ChIP‐Seq peaks, we performed a GO enrichment analysis (Appendix S4; Figure 3). Several of the identified GO terms were the same or similar to those identified in the ANT‐GR RNA‐Seq experiment. Within the biological process GO category, a number of developmental terms were identified that relate to adaxial/abaxial polarity, floral organ development and meristem development, including the following: polarity specification of adaxial/abaxial axis (GO:0009944); floral organ formation (GO:0048449); meristem determinacy (GO:0010022); meristem initiation (GO:0010014); plant ovule development (GO:0048481); stamen development (GO:0048443); and regulation of flower development (GO:0009909) (Figure 3a). Other over‐represented developmental GO biological process terms include: stomatal complex morphogenesis (GO:0010103); cell fate specification (GO:0001708); and leaf development (GO:0048366) (Figure 3a). Two hormone‐related over‐represented GO biological process terms are the response to gibberellin (GO:0009739) and the auxin‐activated signaling pathway (GO:0009734), which were also identified in the RNA‐Seq experiment (Figure 3a); however, several other hormone‐related GO terms, such as those related to cytokinin and JA signaling identified in the RNA‐Seq experiment, were not enriched in the ChIP‐Seq experiment.
Figure 3.
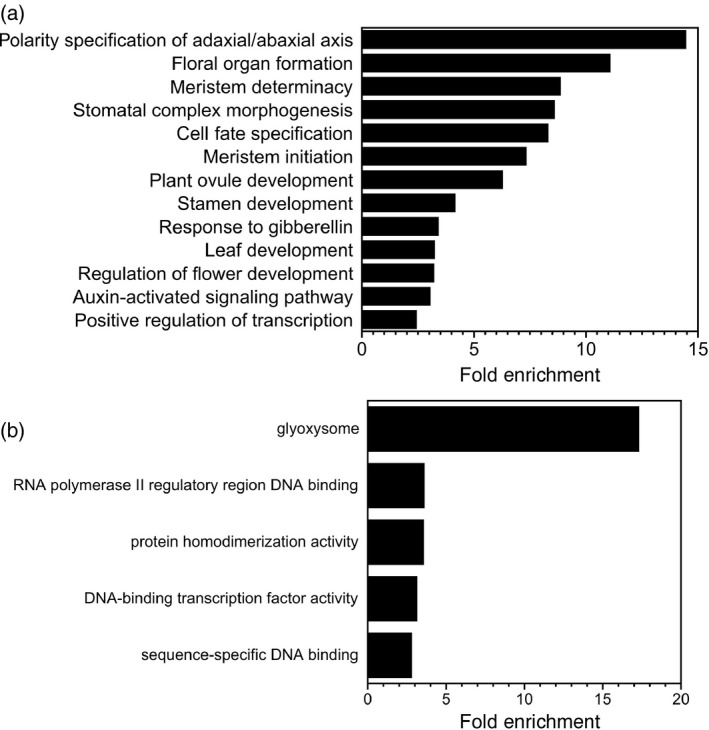
Gene ontology (GO) enrichment analyses on genes associated with ANT ChIP‐Seq binding peaks.
(a) Biological process GO terms enriched in genes associated with ANT binding peaks.
(b) Molecular function and cellular component GO terms enriched in genes associated with ANT binding peaks.
The biological process GO term ‘positive regulation of transcription, DNA‐templated’ (GO:0045893) was also over‐represented, which suggests that ANT regulates the expression of other transcription factors (Figure 3a). This is also supported by the identification of the following enriched molecular function GO terms: RNA polymerase II regulatory region DNA binding (GO:0001012); DNA‐binding transcription factor activity (GO:0003700); and sequence‐specific DNA binding (GO:0043565) (Figure 3b). Another enriched molecular function GO term was protein homodimerization activity (GO:0042803) (Figure 3b).
The enriched cellular component GO term glyoxysome (GO:0009514) was also identified (Figure 3b). This category includes two enzymes, isocitrate lyase (ICL) and malate synthase (MS), specific to the glyoxylate cycle that occurs in glyoxysomes.
Two hundred DE genes bound by ANT are likely to be direct targets of ANT regulation
The most likely direct targets of ANT regulation are genes that are both bound by ANT and are differentially expressed in response to changes in ANT activity. There were 200 genes shared between the set of RNA‐Seq DE genes and the set of ChIP‐Seq bound genes (Figure 4a; Appendix S5). Although the total set of DE genes (1195) consists of slightly less upregulated genes (523) than downregulated genes (672), a much larger number of upregulated genes (154) are associated with ANT binding peaks as compared with downregulated genes (46).
Figure 4.
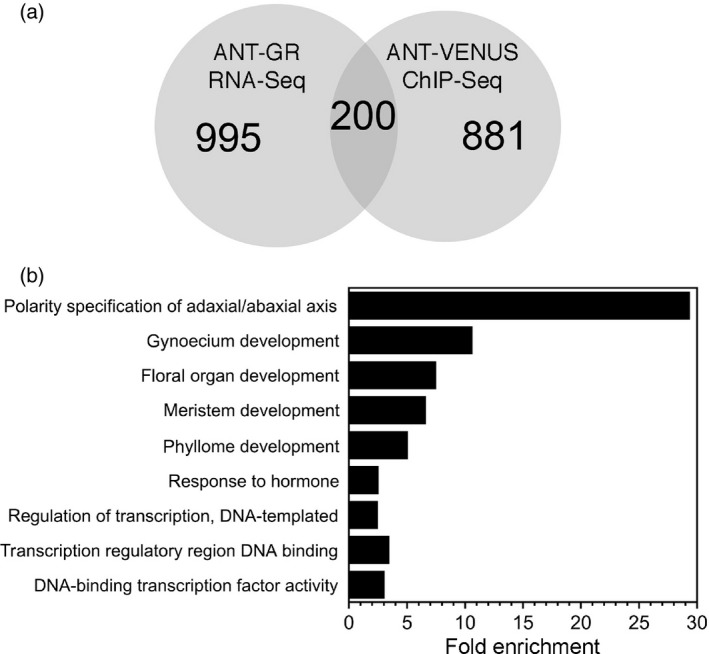
Differentially expressed (DE) genes that are bound by ANT.
(a) Venn diagram showing the overlap between the DE genes identified and those bound by ANT. The Venn diagram was created with biovenn (Hulsen et al., 2008).
(b) Biological process and molecular function GO terms enriched in genes that are both DE and bound by ANT.
Gene ontology (GO) term enrichment analysis on this set of 200 genes identified several GO terms similar to, or the same as, those identified for the entire set of ANT ChIP‐Seq associated genes (Appendix S6; Figure 4). In particular, three terms were enriched in both sets: polarity specification of adaxial/abaxial axis (GO:0009944); regulation of transcription, DNA‐templated (GO:0006355); and DNA‐binding transcription factor activity (GO:0003700) (Figure 4). Five polarity genes were identified: PHB, BOP1, AS1, KAN2 and YAB3 (Table 1). Other enriched GO terms were the following: gynoecium development (GO:0048467); floral organ development (GO:0048437); meristem development (GO:0048507); phyllome development (GO:0048827); and response to hormone (GO:0009725). Floral organ development genes include those involved in specifying floral organ identity (AP1, AP2 and SEP3), regulating cellular differentiation (EMS1 and SPL8) and controlling morphogenesis (SUB and SPT) (Table 1). Nine auxin genes were part of the hormone responses category, including those involved in biosynthesis (TAA1), signaling (AFB2, ARF6, ARF11, ARF18, IAA3/SHY2, IAA27 and PAP2) and responses (SAUR50 and SAUR14) (Table 2).
Table 1.
Developmental genes that are differentially expressed after ANT‐GR activation and are bound by ANT
| AGI locus code | Gene | log2 fold change (h) | ChIP‐Seq |
|---|---|---|---|
| Polarity specification | |||
| AT2G34710 | PHB | 0.256 (4); 0.346 (8) | Upstream; overlap start |
| AT3G57130 | BOP1 | −0.449 (4) | Upstream |
| AT2G37630 | AS1 | −0.235 (4); −0.175 (8) | Upstream |
| AT1G32240 | KAN2 | 0.227 (4); 0.255 (8) | Inside |
| AT4G00180 | YAB3 | 0.269 (2); 0.262 (4); 0.374 (8) | Upstream |
| Floral organ development | |||
| AT1G69120 | AP1 | −0.144 (4) | Upstream; overlap start |
| AT4G36920 | AP2 | 0.22 (2) | Upstream |
| AT1G24260 | SEP3 | 0.175 (8) | Inside |
| AT1G02065 | SPL8 | −0.341 (2); −0.589 (4); −0.534 (8) | Overlap start |
| AT5G07280 | EMS1 | −0.14 (4) | Upstream |
| AT1G11130 | SUB | 0.154 (4); 0.165 (8) | Overlap start |
| AT4G36930 | SPT | 0.269 (2); 0.297 (4); 0.418 (8) | Upstream |
| Growth genes | |||
| AT1G13710 | KLU/CYP78A5 | −0.281 (2); −0.272 (4) | Upstream |
| AT4G24150 | GRF8 | 0.317 (8) | Inside |
| AT5G28640 | AN3/GIF1 | 0.244 (2); 0.25 (4); 0.403 (8) | Overlap start, inside |
| AT4G03210 | XTH9 | 0.315 (2); 0.351 (4); 0.405 (8) | Upstream |
| AT2G47780 | SRP2 | 0.482 (4); 0.718 (8) | Overlap start |
| AT2G32710 | KRP4 | 0.293 (2); 0.299 (4); 0.331 (8) | Overlap end |
Table 2.
Hormone genes that are differentially expressed after ANT‐GR activation and are bound by ANT
| AGI locus code | Gene | log2 fold change (h) | ChIP‐Seq |
|---|---|---|---|
| Auxin | |||
| AT1G70560 | TAA1/WEI8 | 0.297 (4); 0.444 (8) | Inside |
| AT3G26810 | AFB2 | 0.185 (8) | Upstream |
| AT1G30330 | ARF6 | 0.132 (4) | Upstream |
| AT2G46530 | ARF11 | 0.376 (8) | Upstream |
| AT3G61830 | ARF18 | 0.198 (8) | Overlap end |
| AT1G04240 | SHY2/IAA3 | 0.285 (8) | Overlap start |
| AT4G29080 | PAP2/IAA27 | 0.14 (4) | Upstream |
| AT4G34760 | SAUR50 | 0.346 (8) | Upstream |
| AT4G38840 | SAUR14 | −0.478 (4) | Overlap start |
| Gibberellin | |||
| AT1G14920 | GAI | 0.133 (4); 0.181 (8) | Upstream |
| AT2G01570 | RGA | 0.246 (2); 0.259 (4); 0.334 (8) | Upstream |
| Brassinosteroid | |||
| AT1G78700 | BEH4 | 0.664 (2); 0.734 (4); 0.918 (8) | Inside |
| Abscisic acid | |||
| AT2G40330 | PYL6 | 0.552 (2) | Overlap start |
| Ethylene | |||
| AT1G15360 | WIN1/SHN1 | −0.212 (8) | Overlap start |
To confirm our RNA‐Seq and ChIP‐Seq results, we selected eight genes from this set of 200. The selected genes were not previously known to be regulated by ANT and were associated with several different biological processes. The eight genes included: two polarity genes, KAN2 and PHB; two hormone signaling genes, BES1/BZR1 HOMOLOG 4 (BEH4) and REPRESSOR OF GA (RGA); the reproductive organ development gene SPL8; and three genes that regulate lateral organ growth, AN3/GIF1, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9 (XTH9) and SMALL RUBBER PARTICLE PROTEIN2 (SRP2) (Table 1).
We examined the expression of these eight genes in an independent batch of mock‐ and dex‐treated 35S:ANT‐GR inflorescences. To determine whether the activation of these target genes required novel protein synthesis, we included inflorescences treated with the protein synthesis inhibitor cycloheximide (chx). Inflorescences were collected 4 h after the respective treatment. Similar changes in gene expression in inflorescences treated with dex + chx versus chx alone, as compared with those treated with dex versus mock, support the direct regulation of the gene by ANT. Seven of the eight DE genes showed expression changes independent of protein synthesis in 35S:ANT‐GR inflorescences (Figure 5). The lone exception was SRP2, which showed similar expression levels in chx and dex + chx samples (Figure 5). Thus, SRP2 may not be a direct target of ANT regulation, although it is possible that the use of whole inflorescences in the quantitative reverse transcription PCR (RT‐qPCR) experiment obscured developmental stage‐specific regulation of this gene by ANT in stage‐6/7 flowers.
Figure 5.
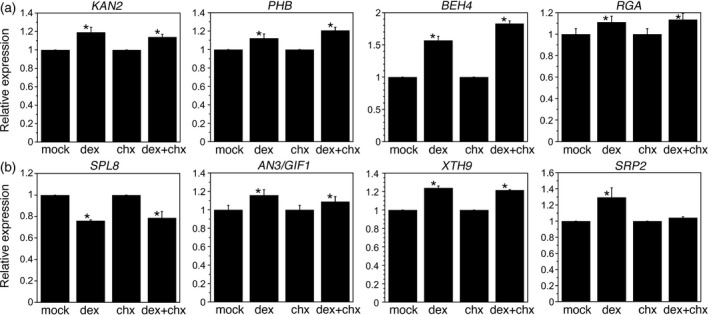
Several differentially expressed (DE) genes tested show protein synthesis‐independent gene expression changes after induction of ANT activity in 35S:ANT‐GR. Graphs show the relative expression of KAN2, PHB, BEH4, RGA, SPL8, AN3/GIF1, XTH9 and SRP2 after mock, dexamethasone (dex), cycloheximide (chx) and dex + chx treatments. Relative expression refers to mRNA levels in dex samples compared with mock samples and mRNA levels in dex + chx samples compared with chx samples. *The dex samples statistically differ from mock samples and the dex + chx samples statistically differ from chx samples, as determined by Students t‐test (P < 0.05). mRNA levels were measured 4 h after treatment. Graphs show means ± SDs of two biological replicates.
The ChIP‐qPCR experiments performed on these eight genes with an independent batch of ChIP DNA gave results that are similar to those seen by ChIP‐Seq (Figures 6 and S4). ANT binds to regions upstream or overlapping the 5' untranslated region (5'‐UTR) (PHB, RGA, SPL8, AN3/GIF1, XTH9 and SRP2) or within the gene body (KAN2 and BEH4) in stage‐6/7 flowers. No enrichment was observed in AP1:AP1‐GR ap1 cal inflorescences lacking ANT:ANT‐VENUS. ANT binds to genomic regions of both upregulated (KAN2, PHB, BEH4, RGA, AN3/GIF1, XTH9 and SRP2) and a downregulated (SPL8) gene.
Figure 6.
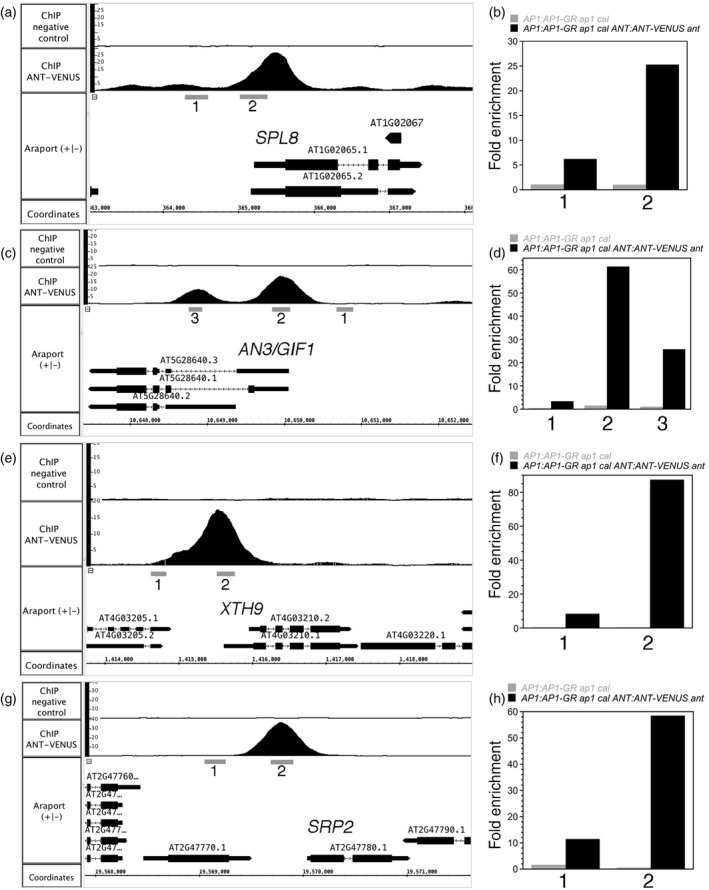
ChIP‐qPCR confirms that ANT binds to genomic regions upstream or within genes associated with stamen development (SPL8) and growth (AN3/GIF1, XTH9, SRP2). ChIP‐Seq coverage graphs for SPL8 (a), AN3/GIF1 (b), XTH9 (e), and SRP2 (g). Numbers below the gene indicate the regions tested for ANT binding by ChIP‐qPCR. ChIP‐qPCR data for SPL8 (b), AN3/GIF1 (d), XTH9 (f) and SRP2 (h). Grey bars show results from AP1:AP1‐GR ap1 cal and black bars show results from AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant. Numbers on the x axis correspond to the genomic regions indicated in the ChIP‐Seq coverage graphs.
DNA sequence motif analyses of ChIP‐Seq peaks
In previous work using SELEX, we identified the in vitro DNA binding motif of ANT (Nole‐Wilson and Krizek, 2000). To determine whether ANT binding peaks contain DNA sequences with similarity to this motif, we mapped putative ANT binding sites on a genome‐wide level using the fimo program from the meme software suite (Grant et al., 2011). We then compared the position of the fimo‐predicted sites to both the ChIP‐Seq peaks and to a set of randomized peaks of the same size. About 66.7% of the ChIP‐Seq peaks and 43.8% of the randomized peaks overlapped with a fimo site. Thus, ChIP‐Seq peaks were more likely than randomized peaks to contain a fimo‐predicted ANT binding site; however, nearly a third of the ChIP‐Seq peaks did not contain a fimo‐predicted ANT binding site.
We used meme‐chip from the meme suite to perform novel motif discovery (Machanick and Bailey, 2011). Our analysis used the DAP‐Seq database for motif discovery, which includes several AIL/PLT binding sites but not that of ANT (O’Malley et al., 2016). MEME‐ChIP identified seven motifs with an e‐value of 1.00E–10 or lower (Table 3). Two of these motifs, MEME‐1 (HNNNHGGCACRNWTH) and MEME‐3 (RCACRRWWHYCRAKG), were similar to the PLT1 and AIL6/PLT3 DAP‐Seq binding motifs, respectively (Figure 7a; Table 3). The PLT1 and AIL6/PLT3 binding sites consisted of a fairly long sequence with several conserved residues near each end of the site and fewer conserved nucleotides in the center. The ANT SELEX‐determined in vitro binding motif is similar to both of these sites (Figure 7a) (Nole‐Wilson and Krizek, 2000). The MEME‐1 motif had similarity to the first conserved part of the AIL/PLT sites, whereas the MEME‐3 motif had similarity to both conserved parts of AIL/PLT sites (Figure 7a). Thus, the identification of MEME‐1 and MEME‐3 motifs in ANT binding peaks suggests that the in vivo binding specificity of ANT resembles that determined in vitro, but with reduced conservation at several positions within the motif. Interestingly, in some MEME‐1 sites, a second motif was identified at a conserved distance from the MEME‐1 motif and resembled the second half of the AIL/PLT binding sites (Figure S5).
Table 3.
meme‐chip analysis of ANT ChIP‐Seq peaks
| Motif | Motif ID | Width | Sites | e‐value | Most similar motif |
|---|---|---|---|---|---|
| HNNNHGGCACRNWTH | MEME‐1 | 15 | 267 | 5.80E‐251 | PLT1 (AP2/ERF) |
| YTYTBTCTYTYTYTY | MEME‐2 | 15 | 400 | 2.00E‐169 | BPC5 (BBR/BPC) |
| RCACRRWWHYCRAKG | MEME‐3 | 15 | 263 | 7.30E‐99 | AIL6/PLT3 (AP2/ERF) |
| CGWGSC | DREME‐1 | 6 | 221 | 8.70E‐19 | BPE/bHLH31 (bHLH) |
| ARAGARAR | DREME‐2 | 8 | 348 | 5.30E‐16 | BPC1 (BBR/BPC) |
| AAARGHRGARARARAAADARAAVAAMAAA | MEME‐4 | 29 | 64 | 2.60E‐15 | VRN1 (ABI3/VP1) |
| GYRRRTSCCACGTG | MEME‐5 | 14 | 39 | 8.30E‐11 | PIF7 (bHLH) |
Figure 7.
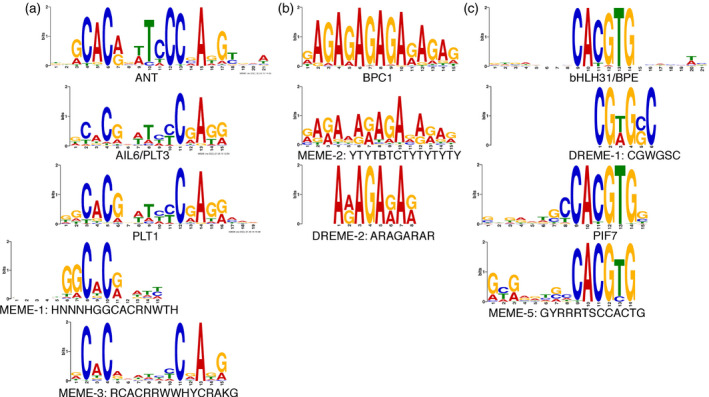
meme‐chip identifies sequences with similarity to AIL/PLT sites as well as other transcription factors.
(a) Sequence logos representing the DNA binding specificities of ANT, AIL6/PLT3, PLT1 and two motifs (MEME‐1 and MEME‐3) identified within ANT ChIP‐Seq binding peaks.
(b) Sequence logos representing the DNA binding specificity of BPC1 and two related motifs (MEME‐2 and DREME‐2) identified within ANT ChIP‐Seq binding peaks.
(c) Two bHLH binding sites from bHLH31/BPE and PIF7 and two related motifs (DREME‐1 and MEME‐5) identified within ANT ChIP‐Seq binding peaks.
meme‐chip identified several other enriched motifs in ANT binding peaks. These had similarity to the binding sites of other transcription factors. Two such motifs, MEME‐2 (YTYTBTCTYTYTYTY) and DREME‐2 (ARAGARAR), resembled the binding sites of BASIC PENTACYSTEINE (BPC) transcription factors (Figure 7b; Table 3). These transcription factors bind GA repeat sequences present in many plant promoters. Recently, BPC proteins were shown to bind to Polycomb response elements (PREs) and interact with components of Polycomb repressive complex 2 (PRC2) to mediate the silencing of gene expression by PRC2 (Xiao et al., 2017). Two additional motifs, MEME‐5 (GYRRRTSCCACGTG) and DREME‐1 (CGWGSC), resembled the binding sites of basic helix–loop–helix (bHLH) transcription factors (Figure 7c; Table 3). In particular, the identified motifs most closely match the DAP‐Seq binding sites of BIG PETAL (BPE)/bHLH31 and PHYTOCHROME INTERACTING FACTOR 7 (PIF7).
The ANT ChIP‐Seq gene set exhibited limited overlap with those from AP1, JAG and PLT2
To investigate where ANT acts within the hierarchy of known flower development regulators, we compared the set of genes bound by ANT to genes bound by other transcription factors involved in flower development, as determined by ChIP‐chip or ChIP‐Seq experiments. We compared ANT with the floral meristem identity protein LFY, the floral organ identity proteins AP1, AP3, PI, AG and SEP3, the growth regulator JAGGED (JAG) and AUXIN RESPONSE FACTOR 3/ETTIN (ARF3/ETT), which regulates gynoecium development (Winter et al., 2011; Wuest et al., 2012; O’Maoiléidigh et al., 2013; Pajoro et al., 2014; Schiessl et al., 2014; Simonini et al., 2017). Fisher’s exact tests were performed using the r package geneoverlap (Shen and Sinai, 2019). geneoverlap also calculates the Jaccard index to assess the overlap of two gene lists. A Jaccard index of 0 indicates no similarity between the gene lists whereas a value of 1 indicates that the lists are identical. Comparisons between the gene set of ANT with gene sets from LFY, AP3, PI, AG, SEP3 and ETT each gave a Jaccard index of 0.1, whereas the comparison between ANT and JAG and that between ANT and AP1 gave a Jaccard index of 0.2 (Figure S6). This degree of overlap is much less than that observed among the floral organ identity proteins AP3, PI and AG, which exhibit Jaccard indices of 0.4 or 0.5 (Figure S6). JAG also showed a Jaccard index of 0.2 with the floral organ identity proteins SEP3, AP1 and PI.
We also compared the set of ANT ChIP‐Seq bound genes with genes bound by two other AIL/PLT transcription factors: PLT2, which specifies stem cell identity in the root and regulates shoot phyllotaxy; and BABYBOOM (BBM), which promotes somatic embryogenesis (Boutilier et al., 2002; Aida et al., 2004; Prasad et al., 2011). The PLT2 ChIP‐Seq experiment used roots and the BBM ChIP‐Seq experiment used somatic embryos (BBM) (Horstman et al., 2015; Santuari et al., 2016). The comparison with PLT2 gave a Jaccard index of 0.2, whereas the comparison with BBM gave a Jaccard index of 0.1. It is interesting that ANT showed more overlap with the related PLT2 transcription factor, which primarily functions in roots, compared with most non‐related transcription factors that regulate floral organ identity.
ANT may repress SPL8 to promote petal growth
One of the 200 likely direct targets of ANT regulation is SPL8, a gene that acts in micro‐ and megasporogenesis (Unte et al., 2003). spl8 mutants produce slightly thinner flowers than the wild type and have stamens with smaller anthers and shorter filaments (Unte et al., 2003). SPL8 mRNA levels are reduced in dex‐treated 35S:ANT‐GR inflorescences, and ANT binds to its 5'‐UTR (Figures 5 and 6). This suggests that ANT can directly repress SPL8 expression. To investigate a possible genetic interaction between ANT and SPL8, we generated ant‐4 spl8‐1 double mutants. ant‐4 spl8‐1 flowers exhibit a partial suppression of the petal growth defects of ant‐4 (Figure 8a). Petal width and petal area are larger in ant‐4 spl8‐1 flowers, compared with ant‐4 flowers (Figure 8b). This finding suggests that SPL8 acts as a repressor of petal growth in the ant‐4 background and that one means by which ANT promotes petal growth is through the downregulation of SPL8. A role in petal development has not previously been noted for SPL8, although the gene is expressed in the margins of petals in stage‐8 flowers (Unte et al., 2003). The molecular mechanism by which SPL8 may repress petal growth in ant mutants is not clear, as spl8‐1 mutants have smaller petals than those in wild‐type flowers (Figure S7).
Figure 8.
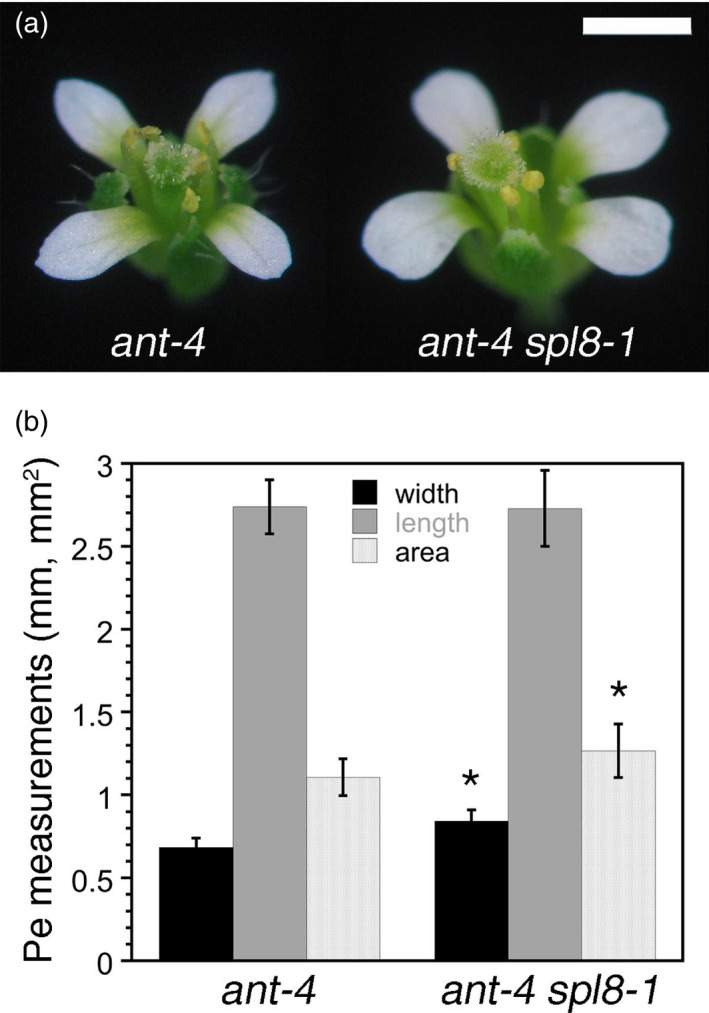
Mutations in SPL8 partially rescue the petal size defects of ant flowers.
(a) ant‐4 flower (left) and ant‐4 spl8‐1 flower (right).
(b) Petal width, length and area in ant‐4 and ant‐4 spl8‐1 flowers. *P < 0.05 (Student's t‐test). Scale bar: 1 mm.
DISCUSSION
ANT is a key regulator of floral organ growth that acts in a redundant manner with AIL6/PLT3 to regulate the initiation, identity and patterning of floral organs (Elliott et al., 1996; Klucher et al., 1996; Krizek, 1999; Mizukami and Fischer, 2000; Krizek, 2009). Using transcriptional profiling and genome‐wide mapping of ANT binding sites, we identified 200 genes that both responded to changes in ANT activity and were bound by ANT. This set of genes included auxin signaling genes as well as genes that specify floral organ identity, establish polarity, regulate growth and promote cell differentiation. Thus, our work begins to reveal the molecular means by which ANT regulates growth and patterning during floral organogenesis and identifies candidate direct target genes in the larger transcriptional network surrounding ANT.
Our studies reveal that ANT can act directly as both a transcriptional activator and a transcriptional repressor. The set of 200 likely direct targets of ANT regulation include both upregulated and downregulated genes. More upregulated genes (154) are associated with ANT binding peaks compared with downregulated genes (46), suggesting that ANT primarily acts as a transcriptional activator. Although our work suggests that ANT binds to DNA sequences in vivo with a similar sequence specificity as that in vitro not all ANT binding peaks contain an AIL/PLT‐like site, as defined by the MEME‐1 and MEME‐3 motifs determined with meme. Thus, ANT may bind directly to other DNA sequences and/or be recruited to different DNA sites via interaction with other transcription factors. The identification of DNA binding motifs of BPC and bHLH transcription factors within ANT peaks suggests that ANT may be recruited to some DNA sites via interaction with other classes of transcription factors.
A comparison of the set of genes bound by ANT with those bound by other floral regulators did not reveal strong overlap with any other gene set, as measured by the Jaccard index. This is consistent with ANT having functions that are distinct from transcription factors specifying floral meristem or floral organ identity. One limitation of these comparisons is the different tissues used in these studies. The LFY, JAG and ETT experiments were performed with whole inflorescences, whereas the SEP3, AP1, AP3, PI and AG experiments used stage‐5 flowers. Among the floral regulators tested, the highest overlap of ANT was seen with AP1 and JAG. JAG is a regulator of floral organ shape, which suggests that ANT and JAG may regulate some common target genes mediating floral organ growth. We also observed higher overlap with the root stem cell regulator PLT2 despite the different tissues used in the ChIP‐Seq experiments (stage‐6/7 flowers versus seedlings). The similar DNA binding specificities of ANT and PLT2 may result in the regulation of some common targets despite the distinct morphologies and functions of roots and flowers.
ANT directly regulates genes involved in auxin signaling
Genomic studies in the root have shown that AIL/PLT transcription factors directly regulate genes involved in auxin biosynthesis and transport. Our studies here imply that ANT directly regulates auxin signaling in developing flowers (Santuari et al., 2016; Zúñiga‐Mayo et al., 2019). Although our RNA‐Seq experiment suggested that ANT could regulate other hormone signaling pathways, including cytokinin, gibberellin and JA, the DE genes in these categories were not bound by ANT and are thus likely to be indirect targets of ANT regulation. It is also possible that these genes may be bound by ANT at a different stage of flower development, however, as the RNA‐Seq experiment used floral buds of stage 1–12, whereas the ChIP‐Seq experiment used stage‐6/7 buds.
Auxin is linked to many aspects of flower development, including floral organ initiation, primordium growth, stamen filament elongation, pollen maturation and gynoecium patterning, several of which overlap with ANT function (Cheng et al., 2006; Marsch‐Martînez and de Folter, 2016). Patterning of distinct tissues and cell types within the gynoecium appears to involve complex transcriptional networks and the precise distribution of hormones, particularly auxin and cytokinin (Zúñiga‐Mayo et al., 2019). Within the gynoecium, ANT acts redundantly with several other genes including REV to promote the development of the carpel marginal meristem, a meristematic region within the medial domain of the ovary that gives rise to multiple tissues, including the placentae, ovules and transmitting tract (Liu et al., 2000; Nole‐Wilson and Krizek, 2006; Azhakanandam et al., 2008; Krizek, 2009). In ant rev double mutants, medial domain development is disrupted and there is partial loss of the carpel marginal meristem (Nole‐Wilson et al., 2010). These defects are associated with reduced expression of the auxin biosynthetic enzyme TAA1 in stage‐7 gynoecium (Nole‐Wilson et al., 2010). We found that ANT induction activated TAA1 expression and that ANT bound to TAA1. These results suggest that ANT may be a direct regulator of TAA1 in this tissue. Mutation of the ANT binding site within the TAA1 gene would help to reveal whether ANT is required for the expression of this gene within the gynoecium.
ANT directly regulates genes that specify floral organ identity
Three of the 200 likely direct targets of ANT regulation were floral organ identity genes: the class‐A genes AP1 and AP2 and the class‐E gene SEP3. AP1 was downregulated after ANT induction, whereas AP2 and SEP3 were upregulated (Table 1). Expression of AP1, AP2 and SEP3 is initiated early in flower development (stage 1 for AP1 and AP2 and stage 2 for SEP3) and maintained in developing flowers (Mandel et al., 1992; Jofuku et al., 1994; Mandel and Yanofsky, 1998). Thus, the binding of ANT to the regulatory regions of these genes in stage‐6/7 flowers may reflect a role in maintaining (AP2 and SEP3) or limiting (AP1) expression in later stages of flower development.
The combined activities of class‐A and ‐E genes specify sepal identity, which is not disrupted in ant single mutants or ant ail6 double mutants; however, in combination with class‐B genes, class‐A and ‐E genes also contribute to the specification of petal identity in the second whorl. Petals are not present in ant ail6 flowers and expression of the class‐B gene AP3 is reduced (Krizek, 2009; Krizek et al., 2016). Although we detected binding of ANT to the regulatory regions of AP3 in stage‐6/7 flowers (Appendix S3), we failed to observe statistically significant differential expression of AP3 following induction of ANT activity. Failure to detect a significant difference between treatment and control samples in any time point was not a result of low AP3 expression, as AP3 was highly expressed in these samples. Instead, this negative result may have resulted from the timing of the floral stage examined, the bulk nature of the tissue collected or because ANT requires some other unknown factor to alter AP3 expression.
ANT directly regulates genes acting later in floral organogenesis that control growth and differentiation
We found that ANT directly regulates genes that act in the elaboration of organ size and shape and the differentiation of distinct cell types. We showed that ANT binds to two genes involved in stamen development: EXCESS MICROSPOROCYTES1 (EMS1) and SPL8. EMS1 encodes a leucine‐rich repeat receptor kinase that is required for tapetal cell differentiation (Zhao et al., 2002; Huang et al., 2016) and SPL8 encodes an SPB‐box transcription factor that is required for the normal development of anther sporogenic tissue (Unte et al., 2003; Xing et al., 2010). Both EMS1 and SPL8 were downregulated after induction of ANT activity, consistent with a role for ANT in inhibiting differentiation in early stages of floral organogenesis (Krizek and Eaddy, 2012).
Several known regulators of lateral organ growth were identified in the set of 200 DE genes bound by ANT. These were KLU, GRF8, AN3/GIF1, XTH9 and SRP2, which act as growth‐promoting genes, and KIP‐RELATED PROTEIN 4 (KRP4), which acts as a growth repressor (Table 1) (Hyodo et al., 2003; Kim et al., 2003; Kim and Kende, 2004; Anastasiou et al., 2007; Bemis and Torii, 2007; Kim et al., 2016). GRF8, AN3/GIF1, XTH9 and SRP2 were upregulated after ANT induction, suggesting that they mediate the role of ANT in promoting organ growth. Furthermore, AN3/GIF1 and XTH9 have largely overlapping expression patterns with ANT, which is consistent with ANT acting as an activator of these genes (Figure S8) (Hyodo et al., 2003).
The identification of members of the GRF/GIF pathway (GRF8 and AN3/GIF) as potential targets of ANT regulation is interesting, as GRFs and their miRNA regulator miR396 are key regulators of growth in many plant tissues (reviewed by Liebsch and Palatnik, 2020). In the root, GRF/GIF complexes repress AIL/PLT expression in transit‐amplifying cells to promote the proliferation of these cells, whereas PLT1 and PLT2 activate miR396 within the root stem cell niche to repress GRF expression and maintain stem cell identity (Rodriguez et al., 2015). The miR396‐GRF/GIF module also controls floral organ growth and meristematic competence within reproductive organs, but a connection with AIL/PLT function has not been described (Lee et al., 2014; Lee et al., 2018). ant an3 double mutants exhibit more severe defects in leaf growth than either single mutant, but there was no enhancement of the carpel marginal meristem defects within the gynoecium (Lee et al., 2014). Extensive genetic redundancies in the AIL/PLT, GRF and GIF gene families may complicate the interpretation of these results (Lee et al., 2014). Future studies will need to address whether AIL/PLT might directly regulate GRF/GIF expression or act indirectly through miR396 in the carpel marginal meristem, and whether AIL/PLT genes are targets of GRF/GIF regulation.
ANT may promote organ growth through the direct regulation of polarity genes
Seven genes associated with lateral organ polarity were identified in our RNA‐Seq experiment, of which five genes (PHB, BOP1, AS1, KAN2 and YAB3) were also next to an ANT binding site (Table 1). The DE genes included both upregulated and downregulated genes. We found ANT binding sites near or within four other genes that were not detected as DE. These included the adaxial genes REVOLUTA (REV) and BOP2 and the abaxial genes KAN2 and FILAMENTOUS FLOWER (FIL). The ANT binding peaks upstream of FIL and YAB3 overlapped an ANT binding site defined in vitro (Nole‐Wilson and Krizek, 2006).
Previous genetic work has suggested a role for ANT in lateral organ polarity. Although ant single mutants do no show defects in organ polarity, mutations in ANT combined with mutations in FIL produce plants with smaller leaves that exhibit polarity defects on both the adaxial and the abaxial surfaces of leaves (Nole‐Wilson and Krizek, 2006). ant fil yab3 triple mutants exhibit even more severe defects in leaf polarity and growth, and YAB3 and FIL expression is reduced in ant ail6 double mutants (Nole‐Wilson and Krizek, 2006; Krizek et al., 2016). Polarity defects were also observed in ant fil floral organs, and these defects were associated with reduced expression of PHB (Nole‐Wilson and Krizek, 2006). Together, these data suggest that ANT regulates organ polarity through the regulation of both adaxial‐ and abaxial‐specifying genes. As the juxtaposition of adaxial and abaxial cell types is required for outgrowth of the leaf lamina, our work suggests that one mechanism by which ANT controls lateral organ growth is through the direct regulation of polarity genes to establish distinct adaxial and abaxial domains within developing lateral organ primordia (Yamaguchi et al., 2012).
Overall, the work described here reveals that ANT can directly regulate the expression of target genes involved in various aspects of flower development, including floral organ identity, polarity, growth and cellular differentiation. Furthermore, our findings connect ANT function with several hormone pathways that may provide positional information for growth and patterning events during flower development.
EXPERIMENTAL PROCEDURES
Plant materials, growth conditions, genotyping and treatments
35S:ANT‐GR plants were grown on a soil mixture of Fafard 4P:perlite:vermiculite (8:1:1) in 16‐h days at a light intensity of approximately 160 µmol m−2 s−1 at 20°C. ANT:ANT‐VENUS ant‐4 AP1:AP1‐GR ap1 cal inflorescences were grown on a soil mixture of Fafard 4P:perlite:vermiculite (8:1:1) in 24‐h days at a light intensity of approximately 160 µmol m−2 s−1 at 20°C. ant‐4 and spl8‐1 were grown on a soil mixture of Fafard 4P:perlite:vermiculite (8:1:1) in 16 h days at a light intensity of approximately 160 µmol m−2 s−1 at 22°C. ant‐4 spl8‐1 double mutants were identified by genotyping for ant‐4 and spl8‐1, as described previously (Unte et al., 2003; Krizek, 2009). 35S:ANT‐GR plants for RNA‐Seq and RT‐qPCR were treated by pipetting a mock (0.1% ethanol + 0.015% Silwet), dex (10 µm dexamethasone + 0.015% Silwet), chx (10 µm cycloheximide + 0.015% Silwet + 0.1% ethanol) or dex + chx (10 µm dexamethasone + 10 µm cycloheximide + 0.015% Silwet) solution onto the inflorescences. AP1:AP1‐GR ap1 cal and AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant‐4 plants for ChIP‐Seq and ChIP‐qPCR were treated by pipetting a dex (10 µm dexamethasone + 0.015% Silwet) solution onto the inflorescences.
RNA‐Seq
35S:ANT‐GR inflorescences containing unopened floral buds (flowers stages 1–12) were collected in four batches at each time point (2, 4 and 8 h after treatment) consisting of two flats per batch, where dex was applied to one flat and a mock treatment was applied to the other flat. RNA was extracted from inflorescences using Trizol following the manufacturer’s instructions with cleanup and DNase treatment on a RNeasy column (Qiagen, https://www.qiagen.com). Sequencing libraries were prepared from four biological replicates using TruSeq Stranded mRNA sample preparation kit (Illumina, https://www.illumina.com) and sequenced on the Illumina HiSeq 2500 producing 100 base single‐end reads. Sequence reads were aligned to the reference A. thaliana genome (version TAIR9, released June 2009) using tophat and bowtie 2. Reads per gene were counted using featurecounts. Read counts were analyzed using edger. Differentially expressed genes were identified using an additive linear model with adjustment for batch (flat) effects. Source code for differential expression analysis is available in the project ‘git’ repository https://bitbucket.org/krizeklab. GO analyses were performed with amigo 2 (http://amigo.geneontology.org/amigo).
RT‐qPCR
RNA was isolated as described above for RNA‐Seq. First‐strand complementary DNA (cDNA) synthesis was performed using Quanta qScript cDNA SuperMix (Quanta BioSciences, https://www.quantabio.com), following the manufacturer’s instructions. qPCR was performed on a BioRad CFX96 using PerfeCTa sybr Green FastMix for iQ (Quanta BioSciences) and primers listed in Table S5. Data analyses were carried out as described previously (Krizek and Eaddy, 2012). Two biological replicates were analyzed for each experiment.
ChIP‐Seq and ChIP‐qPCR
Chromatin immunoprecipitation was performed similarly to that described by Yamaguchi et al. (2014), with the same buffers and solutions. Approximately 600 mg of inflorescence tissue consisting of stage‐6/7 flowers from AP1:AP1‐GR ap1 cal and ANT:ANT‐VENUS ant‐4 AP1:AP1‐GR ap1 cal plants was collected 5 days after dex treatment into a 2‐ml tube filled with 1.5 ml of cold 1× PBS on ice. The PBS was then removed and replaced with 10 ml of 22°C 1% methanol‐free formaldehyde (ThermoFisher Scientific, https://www.thermofisher.com) in 1× PBS and 0.015% Silwet L‐77 for 15 mins at room temperature. During this time, the tissue was vacuum infiltrated three times for 2 min each time. The fixative was removed and the cross‐linking was stopped with the addition of 10 ml of 0.125 m glycine, and then incubated for 5 min. During this time, the tissue was vacuum infiltrated once for 2 min. The tissue was rinsed three times with 10 ml of cold 1× PBS on ice, dried briefly on paper towels, frozen in liquid nitrogen and then stored at −80°C. The tissue was ground in liquid nitrogen and 2.5 ml of nuclei extraction buffer with protease inhibitors and β‐mercaptoethanol was added. The samples were filtered twice through Miracloth and centrifuged at 10 000 g for 5 min at 4°C. The pellet was resuspended in 107 µl of nuclei lysis buffer and left on ice for 30 min with occasional stirring with a pipet tip. An 893‐µl volume of ChIP dilution buffer without Triton X‐100 was added to bring the volume to 1 ml. The sample was loaded into a milliTUBE 1‐ml AFA Fiber tube (Covaris, https://covaris.com) and chromatin shearing was performed with a Covaris M220 Focused ultra‐sonicator (14 cycles of 75% peak power, 5 duty factor, 200 cycles/burst at 7°C). After sonication, 200 µl of ChIP dilution buffer with Triton X‐100 and 53 µl of 22% Triton X‐100 was added to each sample. The samples were centrifuged twice at 12 000 g for 10 min at 4°C. The sample was pre‐cleared by adding 50 µl of Dynabeads‐Protein A and incubating for 2 h at 4°C on a tube rotator. The sample was removed using a magnetic stand and transferred into a 1.5‐ml low adhesion tube. A 12.5‐µl sample was removed as the Input sample. A 50‐μl volume of GFP (A6455; Invitrogen, now ThermoFisher Scientific) coated Dynabeads was added to each sample and incubated for 4 h at 4°C.The samples were washed twice (5 min each at 4°C) with the following four cold‐wash buffers: low salt wash buffer, high salt wash buffer, 250 mm LiCl buffer, 0.5× Tris EDTA. Immunoprecipitated DNA was eluted from the Dynabeads by the addition of 50 µl of nuclei lysis buffer and a 30‐min incubation at 65°C on an Eppendorf ThermoMixer (Eppendorf, https://eppendorf.com). The elution was repeated a second time and the samples combined. Cross‐links were reversed by the addition of 6 µl of 5 m NaCl to the ChIP samples and an overnight incubation at 65°C. An 87.5‐µl volume of nuclei lysis buffer and 6 µl of 5 m NaCl was added to the input samples followed by overnight incubation at 65°C. The input and ChIP DNA was purified using a Qiagen PCR purification kit. Primers for ChIP‐qPCR are listed in Table S5. Fold enrichment was determined relative to a negative control, the transposon TA3.
Sequencing libraries were prepared from two biological replicates of input and ChIP DNA for stage‐6/7 flowers for both AP1:AP1‐GR ap1 cal and AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant‐4 using Accel‐NGS 2S DNA library kit (Swift Biosciences, https://swiftbiosci.com). The libraries were quantitated using the NEBNext Library Quant Kit for Illumina (New England Biolabs, https://international.neb.com) and sequenced on an Illumina HiSeq 2500 producing 150 base paired‐end reads. Sequence reads were aligned to the reference A. thaliana genome (version TAIR9, released June 2009) using bowtie 2. Examination of the coverage graphs revealed high reproducibility between the two ChIP‐Seq replicates. In addition, the input samples closely resembled the control untagged AP1:AP1‐GR ap1 cal samples. ANT binding peaks were identified using a visual analytics approach within the integrated genome browser (igb) (Freese et al., 2016). Specifically, coverage graphs were generated for the combined data from the two replicates. A difference coverage graph was generated by subtracting coverage graphs of the untagged sample (AP1:AP1‐GR ap1 cal) from the coverage graphs for the tagged sample (AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant‐4). Peaks were defined using the thresholding feature. A thresholding value of 5000 identified 90 peaks, whereas a thresholding value of 1000 identified 11 133 peaks. Further analyses were performed using the 1113 peaks identified with a threshold value of 1000. For each peak identified, chippeakanno was used to identify the gene with the closest transcription start site (TSS) (Zhu et al., 2010). geneoverlap was used to compare gene lists for different ChIP‐Seq data sets. GO analyses were performed with amigo 2 (http://amigo.geneontology.org/amigo). Source code for bioinformatic analyses is available in the project ‘git’ repository https://bitbucket.org/krizeklab.
Motif analysis with the MEME Suite
Genomic locations of putative ANT binding sites were determined using the fimo tool of meme suite (Grant et al., 2011). The in vitro defined ANT binding motif was used as a position‐specific prior (Nole‐Wilson and Krizek, 2000). Putative ANT binding sites were identified using a P value of 0.001 or lower. Novel motif discovery was performed with meme‐chip.
Petal measurements
Petal width, length and area were measured as described previously (Krizek, 2015).
ACCESSION NUMBERS
RNA‐Seq sequences are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA539947. ChIP‐Seq sequences are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA593434. Version‐controlled source code used to process and analyze data is available from https://bitbucket.org/krizeklab. Sequence alignments and coverage graphs are available for interactive visualization within igb (Nicol et al., 2009). To view the data in igb, readers may download and install the software from https://bioviz.org. Once installed, data sets from the study can be opened within igb by selecting the latest A. thaliana genome and then choosing RNA‐Seq and ChIP‐Seq folders within the Available Data Sets section of the Data Access Panel.
AUTHOR CONTRIBUTIONS
BAK and AEL designed the research. BAK carried out the experiments. BAK, ICB, YYH, NF and AEL performed the data analyses. BAK wrote the article. All authors edited the article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
Figure S2. Interactive r shiny app tool to display gene expression data in control (C) and treated (T) 35S:ANT‐GR samples. Control samples correspond to mock‐treated 35S:ANT‐GR inflorescences whereas treated samples correspond to dex‐treated 35S:ANT‐GR inflorescences. A. Sample RPKM for AT1G48660 that shows gene expression values for each of four biological replicates. B. Group RPKM for AT1G48660 which show average RPKM for the four replicates. C. Expression over time for AT1G48660. D. Gene info links for AT1G48660.
Figure S3. Hormone signaling pathways associated with changes in ANT activity. Blue indicates genes that are downregulated after induction of ANT activity. Orange indicates genes that are upregulated after induction of ANT activity. Mixed colored rectangles indicate classes in which some genes were downregulated whereas others were upregulated. The circles above and below the rectangle represent the number of downregulated (blue) and upregulated (orange) genes for such classes. Abbreviations: TFs, transcription factors.
Figure S4. ChIP‐qPCR confirms that ANT binds to genomic regions upstream or within genes associated with polarity specification (KAN2, PHB) and hormone signaling (BEH4, RGA). ChIP‐Seq coverage graphs for KAN2 (A), PHB (B), BEH4 (E), and RGA (G). Numbers below the gene indicate the regions tested for ANT binding by ChIP‐qPCR. ChIP‐qPCR data for KAN2 (B), PHB (D), BEH4 (F) and RGA (H). Grey bars show results from AP1:AP1‐GR ap1 cal and black bars show results from AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant. Numbers on the x‐axis correspond to the genomic regions indicated in the ChIP‐Seq coverage graphs.
Figure S5. Secondary motif identified in some MEME‐1 sites. Sequence logos for the DAP‐Seq site of PLT1 (top), the MEME‐1 motif (middle) and the secondary motif identified in some MEME‐1 sites (bottom).
Figure S6. Pairwise comparison heat map displaying the Jaccard index degree of overlap among whole‐genome ChIP data sets of floral regulators and AIL/PLT transcription factors. The ANT data set shows the highest level of overlap with those of AP1, JAG and PLT2. This degree of overlap is less than that observed among the floral organ identity proteins AP3, PI and AG. The heat map was created with heatmapper (heatmapper.ca) (Sabicki et al., 2016).
Figure S7. spl8‐1 flowers are smaller than wild‐type flowers. A. Col flower (left) and spl8‐1 flower (right). B. Graph showing petal width, length and area for Col and spl8‐1 flowers. *P < 0.05 (Student's t‐test). Scale bar: 1 mm.
Figure S8. AN3/GIF1 and XTH9 expression within developing flowers overlaps with ANT expression. AN3/GIF1 mRNA expression in the inflorescence meristem (A), stage‐2 flower (A), stage‐4 flower (B), stage‐6 flower (C), stage‐8 flower (D) and in the developing carpel (E). XTH9 mRNA expression in the inflorescence meristem (F), stage‐2 flower (F), stage‐4 flower (G), stage‐7 flower (H), stage‐8 flower (I) and in the developing carpel (J). All pictures taken at the same magnification. Abbreviations: IM, inflorescence meristem; st 2, stage‐2 flowers; st 4, stage‐4 flowers; st 6, stage‐6 flowers; st 7, stage‐7 flowers; and st 8, stage‐8 flowers. Scale bar: 50 µm. All pictures were taken at the same magnification.
Table S1. Developmental genes differentially expressed after ANT‐GR activation.
Table S2. Hormone genes differentially expressed after ANT‐GR activation.
Table S3. Petal area, length and width in Ler, ant‐4 and ANT:ANT‐VENUS ant‐4 flowers.
Table S4. Floral organ counts in Ler, ant‐4 and ANT:ANT‐VENUS ant‐4 flowers at positions 1–30 on the inflorescence.
Table S5. Primers used in this study.
Appendix S1. Genes differentially expressed in 35S:ANT‐GR inflorescences after dex treatment.
Appendix S2. Over‐represented gene ontology (GO) terms for 35S:ANT‐GR DE genes.
Appendix S3. Genes associated with ANT ChIP‐Seq peaks.
Appendix S4. Over‐represented gene ontology (GO) terms for genes associated with ANT ChIP‐Seq peaks.
Appendix S5. Genes DE in 35S:ANT‐GR and bound by ANT.
Appendix S6. Over‐represented gene ontology (GO) terms for genes DE in 35S:ANT‐GR and bound by ANT.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Science Foundation (NSF) grant IOS 1354452. The igb software was supported by National Institutes of Health grants R01‐GM103463 and R01‐GM121927. Data hosting is provided by the SciDas project, funded by NSF award 1659300 to principle investigator Frank (Alex) Feltus. We thank Roger Deal for helpful suggestions for ChIP experiments and ChIP‐Seq library preparation.
REFERENCES
- Aida, M. , Beis, D. , Heidstra, R. , Willemsen, V. , Blilou, I. , Galinha, C. , Nussaume, L. , Noh, Y.‐S. , Amasino, R. and Scheres, B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell, 119, 109–120. [DOI] [PubMed] [Google Scholar]
- Anastasiou, E. , Kenz, S. , Gerstung, M. , MacLean, D. , Timmer, J. , Fleck, C. and Lenhard, M. (2007) Control of plant organ size by KLUH/CYP78A5‐dependent intercellular signaling. Dev. Cell, 13, 843–856. [DOI] [PubMed] [Google Scholar]
- Azhakanandam, S. , Nole‐Wilson, S. , Bao, F. and Franks, R.G. (2008) SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 146, 1165–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis, S.M. and Torii, K.U. (2007) Autonomy of cell proliferation and developmental programs during Arabidopsis aboveground organ morphogenesis. Dev. Biol. 304, 367–381. [DOI] [PubMed] [Google Scholar]
- Benkova, E. , Michniewicz, M. , Sauer, M. , Teichmann, T. , Seifertova, D. , Jurgens, G. and Friml, J. (2003) Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell, 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Boutilier, K. , Offringa, R. , Sharma, V.K. et al . (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell, 14, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Dai, X. and Zhao, Y. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenase controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C. , Betzner, A.S. , Huttner, E. , Oakes, M.P. , Tucker, W.Q.J. , Gerentes, D. , Perez, P. and Smyth, D.R. (1996) AINTEGUMENTA, an APETALA2‐like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell, 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese, N.H. , Norris, D.C. and Loraine, A.E. (2016) Integrated genome browser: visual analytics platform for genomics. Bioinformatics, 32, 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, C.E. , Bailey, T.L. and Noble, W.S. (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics, 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G. , Ohno, C. , Das, P. , Sieber, P. , Reddy, G.V. , Long, J.A. and Meyerowitz, E.M. (2005) Patterns of auxin transport and gene expression during primodium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- Horstman, A. , Fukuoka, H. , Muiño, J.M. , Nitsch, L. , Guo, C. , Passarinho, P. , Sanchez‐Perez, G.F. , Immink, R. , Angenent, G.C. and Boutilier, K. (2015) AIL and HDG proteins act antagonistically to control cell proliferation. Development, 142, 454–464. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Zhang, T. , Linstroth, L. , Tillman, Z. , Otegui, M.S. , Owen, H.A. and Zhao, D. (2016) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 12, e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen, T. , de Vlieg, J. and Aikema, W. (2008) BioVenn‐a web application for the comparison and visualization of biological lists using area‐proportional Venn diagrams. BMC Genom. 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo, H. , Yamakawa, S. , Takeda, Y. , Tsuduki, M. , Yokota, A. , Nishitani, K. and Kohchi, T. (2003) Active gene expression of a xyloglucan endotransglucosylate/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana . Plant Mol. Biol. 52, 473–482. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D. , den Boer, B.G.W. , Van Montagu, M. and Okamuro, J.K. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2 . Plant Cell, 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, K. , Muino, J.M. , Jauregui, R. , Airoldi, C.A. , Smaczniak, C. , Krajewski, P. and Angenent, G.C. (2009) Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLOS Biol. 2009, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, K. , Wellmer, F. , Muino, J.M. et al . (2010) Orchestration of floral initiation by APETALA1. Science, 328, 85–89. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. and Kende, H. (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis . Proc. Natl Acad. Sci. USA, 101, 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H. , Choi, D. and Kende, H. (2003) The AtGRF family of putative transcription factos is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. [DOI] [PubMed] [Google Scholar]
- Kim, E.Y. , Park, K.Y. , Seo, Y.S. and Kim, W.T. (2016) Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170, 2494–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, K.M. , Chow, H. , Reiser, L. and Fischer, R.L. (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2 . Plant Cell, 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A. (1999) Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A. (2009) AINTEGUMENTA and AINTEGUMENTA‐LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 150, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A. (2015) AINTEGUMENTA‐LIKE genes have partly overlapping functions with AINTEGUMENTA but make distinct contributions to Arabidopsis thaliana flower development. J. Exp. Bot. 66, 4537–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A. and Eaddy, M. (2012) AINTEGUMENTA‐LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 78, 199–209. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A. and Fletcher, J.C. (2005) Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A. , Bequette, C.J. , Xu, K. , Blakley, I. , Fu, Z.Q. , Stratmann, J.W. and Loraine, A.E. (2016) RNA‐Seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA‐LIKE6 to cell wall remodeling and plant defense pathways. Plant Physiol. 171, 2069–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.H. , Wynn, A.N. , Franks, R.G. , Hwang, Y.‐S. , Lim, J. and Kim, J.H. (2014) The Arabidopsis thaliana GRF‐INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386, 12–24. [DOI] [PubMed] [Google Scholar]
- Lee, S.J. , Lee, B.H. , Jung, J.H. , Park, S.K. , Song, J.T. and Kim, J.H. (2018) GROWTH‐REGULATING FACTOR and GRF‐INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch, D. and Palatnik, J.F. (2020) MicroRNA miR396, GRF transcription factors and GIF co‐regulators: a conserved plant organ regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Franks, R.G. and Klink, V.P. (2000) Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA . Plant Cell, 12, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick, P. and Bailey, T.L. (2011) MEME‐ChIP: motif analysis of large DNA datasets. Bioinformatics, 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A. and Yanofsky, M.F. (1998) The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod. 11, 22–28. [Google Scholar]
- Mandel, M.A. , Gustafson‐Brown, C. , Savidge, B. and Yanofsky, M.F. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1 . Nature, 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Marsch‐Martînez, N. and de Folter, S. (2016) Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 29, 104–114. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y. and Fischer, R.L. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl Acad. Sci. USA, 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol, J.W. , Helt, G.A. , Blanchard, S.G.J. , Raja, A. and Loraine, A.E. (2009) The Integrated Genome Browser: free software for distribution and exploration of genome‐scale datasets. Bioinformatics, 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole‐Wilson, S. and Krizek, B.A. (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 28, 4076–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole‐Wilson, S. and Krizek, B.A. (2006) AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 141, 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole‐Wilson, S. , Azhakanandam, S. and Franks, R.G. (2010) Polar auxin transport together with AINTEGUMENTA and REVOLUTA coordinate early Arabidopsis gynoecium development. Dev. Biol. 346, 181–195. [DOI] [PubMed] [Google Scholar]
- O’Malley, R.C. , Huang, S.C. , Song, L. , Lewsey, M.G. , Bartlett, A. , Nery, J.R. , Galli, M. , Gallavotti, A. and Ecker, J.R. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Maoiléidigh, D.S. , Wuest, S.E. , Rae, L. et al . (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell, 25, 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Maoiléidigh, D.S. , Thomson, B. , Raganelli, A. , Wuest, S.E. , Ryan, P.T. , Kwasniewska, K. , Carles, C.C. , Graciet, E. and Wellmer, F. (2015) Gene network analysis of Arabidopsis thaliana flower development through dynamic gene perturbations. Plant J. 83, 344–358. [DOI] [PubMed] [Google Scholar]
- Pajoro, A. , Madrigal, P. , Muiño, J.M. et al . (2014) Dynamics of chromatin accessibility and gene regulation by MADS‐domain transcription factors in flower development. Genome Biol. 15, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, K. , Grigg, S.P. , Barkoulas, M. et al . (2011) Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 21, 1123–1128. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D. , Eva‐Rachele, P. , Stieger, P. , Mandel, T. , Baltensperger, K. , Bennett, M. , Traas, J. , Friml, J. and Kuhlemeier, C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature, 426, 255–260. [DOI] [PubMed] [Google Scholar]
- Rodriguez, R.E. , Ercoli, M.F. , Debernardi, J.M. , Breakfield, N.W. , Mecchia, M.A. , Sabatini, M. , Cools, T. , De Veylder, L. , Benfey, P.N. and Palatnik, J.F. (2015) MicroRNA miR396 regulates the switch between stem cells and transit‐amplifying cells in Arabidopsis roots. Plant Cell, 27, 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabicki, S. , Arndt, D. , Marcu, A. , Lian, Y. , Grant, J.R. , Maciejewski, A. and Wishart, D.S. (2016) Heatmapper: web‐enabled heat mapping for all. Nucleic Acids Res. 44, W147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari, L. , Sanchez‐Perez, G.F. , Luijten, M. et al . (2016) The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell, 28, 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl, K. , Muiño, J.M. and Sablowski, R. (2014) Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip‐related cell cycle inhibitors. Proc. Natl Acad. Sci. USA, 111, 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. and Sinai, M. (2019) GeneOverlap: Test and visualize gene overlaps. R package version 1220.
- Simonini, S. , Bencivenga, S. , Trick, M. and Ostergaard, L. (2017) Auxin‐induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell, 29, 1864–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak, C. , Immink, R.G.H. , Muino, J.M. et al . (2012) Characterization of MADS‐domain transcription factor complexes in Arabidopsis flower development. Proc. Natl Acad. Sci. USA, 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, D. , Graciet, E. and Wellmer, F. (2016) Molecular and regulatory mechanisms controlling floral organ development. FEBS J. 283, 1823–1830. [DOI] [PubMed] [Google Scholar]
- Unte, U.S. , Sorensen, A.‐M. , Pesaresi, P. , Gandikota, M. , Leister, D. , Saedler, H. and Huijser, P. (2003) SPL8, an SBP‐Box gene that affects pollen sac development in Arabidopsis. Plant Cell, 15, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, C.M. , Austin, R.S. , Blanvillain‐Baufumé, S. et al . (2011) LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell, 20, 430–443. [DOI] [PubMed] [Google Scholar]
- Wuest, S.E. , O’Maoiléidigh, D.S. , Rae, L. , Kwasniewska, K. , Raganelli, A. , Hanczaryk, K. , Lohan, A.J. , Loftus, B. , Graciet, E. and Wellmer, F. (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl Acad. Sci. USA, 109, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. , Jin, R. , Yu, X. et al . (2017) Cis and trans determinants of epigenetic silencing by Polycomb repressive comples 2 in Arabidopsis . Nat. Genet. 49, 1546–1552. [DOI] [PubMed] [Google Scholar]
- Xing, S. , Salinas, M. , Höhmann, S. , Berndtgen, R. and Huijser, P. (2010) miR156‐targeted and nontargeted SBP‐box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell, 22, 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T. , Nukazuka, A. and Tsukaya, H. (2012) Leaf adaxial‐abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53, 1180–1194. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, N. , Wu, M.‐F. , Winter, C.M. , Berns, M.C. , Nole‐Wilson, S. , Yamaguchi, A. , Coupland, G. , Krizek, B.A. and Wagner, D. (2013) A molecular framework for auxin‐mediated initiation of flower primordia. Dev. Cell, 24, 271–282. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, N. , Winter, C.M. , Wu, M.‐F. , Kwon, C.S. , William, D.A. and Wagner, D. (2014) Chromatin immunoprecipitation from Arabidopsis tissue. Arabidopsis Book, 2014(12), e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W. , Chen, D. and Kaufmann, K. (2016) Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 29, 154–162. [DOI] [PubMed] [Google Scholar]
- Zhao, D.Z. , Wang, G.F. , Speal, B. and Ma, H. (2002) The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine‐rich repeat receptor protein kinase that controls somatic and reproductive cell fares in the Arabidopsis anther. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.J. , Gazin, C. , Lawson, N.D. , Pagés, H. , Lin, S.M. , Lapointe, D.S. and Green, M.R. (2010) ChIPpeakAnno: a Bioconductor package to annotate ChIP‐seq and ChIP‐chip data. BMC Bioinformatics, 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga‐Mayo, V.M. , Gómez‐Felipe, A. , Herrera‐Ubaldo, H. and de Folter, S. (2019) Gynoecium development: networks in Arabidopsis and beyond. J. Exp. Bot. 70, 1447–1460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Interactive r shiny app tool to display gene expression data in control (C) and treated (T) 35S:ANT‐GR samples. Control samples correspond to mock‐treated 35S:ANT‐GR inflorescences whereas treated samples correspond to dex‐treated 35S:ANT‐GR inflorescences. A. Sample RPKM for AT1G48660 that shows gene expression values for each of four biological replicates. B. Group RPKM for AT1G48660 which show average RPKM for the four replicates. C. Expression over time for AT1G48660. D. Gene info links for AT1G48660.
Figure S3. Hormone signaling pathways associated with changes in ANT activity. Blue indicates genes that are downregulated after induction of ANT activity. Orange indicates genes that are upregulated after induction of ANT activity. Mixed colored rectangles indicate classes in which some genes were downregulated whereas others were upregulated. The circles above and below the rectangle represent the number of downregulated (blue) and upregulated (orange) genes for such classes. Abbreviations: TFs, transcription factors.
Figure S4. ChIP‐qPCR confirms that ANT binds to genomic regions upstream or within genes associated with polarity specification (KAN2, PHB) and hormone signaling (BEH4, RGA). ChIP‐Seq coverage graphs for KAN2 (A), PHB (B), BEH4 (E), and RGA (G). Numbers below the gene indicate the regions tested for ANT binding by ChIP‐qPCR. ChIP‐qPCR data for KAN2 (B), PHB (D), BEH4 (F) and RGA (H). Grey bars show results from AP1:AP1‐GR ap1 cal and black bars show results from AP1:AP1‐GR ap1 cal ANT:ANT‐VENUS ant. Numbers on the x‐axis correspond to the genomic regions indicated in the ChIP‐Seq coverage graphs.
Figure S5. Secondary motif identified in some MEME‐1 sites. Sequence logos for the DAP‐Seq site of PLT1 (top), the MEME‐1 motif (middle) and the secondary motif identified in some MEME‐1 sites (bottom).
Figure S6. Pairwise comparison heat map displaying the Jaccard index degree of overlap among whole‐genome ChIP data sets of floral regulators and AIL/PLT transcription factors. The ANT data set shows the highest level of overlap with those of AP1, JAG and PLT2. This degree of overlap is less than that observed among the floral organ identity proteins AP3, PI and AG. The heat map was created with heatmapper (heatmapper.ca) (Sabicki et al., 2016).
Figure S7. spl8‐1 flowers are smaller than wild‐type flowers. A. Col flower (left) and spl8‐1 flower (right). B. Graph showing petal width, length and area for Col and spl8‐1 flowers. *P < 0.05 (Student's t‐test). Scale bar: 1 mm.
Figure S8. AN3/GIF1 and XTH9 expression within developing flowers overlaps with ANT expression. AN3/GIF1 mRNA expression in the inflorescence meristem (A), stage‐2 flower (A), stage‐4 flower (B), stage‐6 flower (C), stage‐8 flower (D) and in the developing carpel (E). XTH9 mRNA expression in the inflorescence meristem (F), stage‐2 flower (F), stage‐4 flower (G), stage‐7 flower (H), stage‐8 flower (I) and in the developing carpel (J). All pictures taken at the same magnification. Abbreviations: IM, inflorescence meristem; st 2, stage‐2 flowers; st 4, stage‐4 flowers; st 6, stage‐6 flowers; st 7, stage‐7 flowers; and st 8, stage‐8 flowers. Scale bar: 50 µm. All pictures were taken at the same magnification.
Table S1. Developmental genes differentially expressed after ANT‐GR activation.
Table S2. Hormone genes differentially expressed after ANT‐GR activation.
Table S3. Petal area, length and width in Ler, ant‐4 and ANT:ANT‐VENUS ant‐4 flowers.
Table S4. Floral organ counts in Ler, ant‐4 and ANT:ANT‐VENUS ant‐4 flowers at positions 1–30 on the inflorescence.
Table S5. Primers used in this study.
Appendix S1. Genes differentially expressed in 35S:ANT‐GR inflorescences after dex treatment.
Appendix S2. Over‐represented gene ontology (GO) terms for 35S:ANT‐GR DE genes.
Appendix S3. Genes associated with ANT ChIP‐Seq peaks.
Appendix S4. Over‐represented gene ontology (GO) terms for genes associated with ANT ChIP‐Seq peaks.
Appendix S5. Genes DE in 35S:ANT‐GR and bound by ANT.
Appendix S6. Over‐represented gene ontology (GO) terms for genes DE in 35S:ANT‐GR and bound by ANT.
Supplementary Material


