Abstract
Purpose
Metastasis is a crucial cause of the high mortality in patients with lung cancer. Long non-coding RNAs (lncRNAs) are emerging as important players in the development and progression of human cancers. Here, we aimed to identify metastasis-associated lncRNA and to study its roles in the migration and invasion of lung cancer cells.
Materials and Methods
We screened differentially expressed lncRNAs between high- and low-metastatic lung cancer cell lines by using microarray and identified the target lncRNA TM4SF1-AS1. The effect of the TM4SF1-AS1 on the invasion and migration was evaluated through the wound healing experiment and transwell assay. The expression of related genes was assessed by RNA sequence and Western blotting.
Results
TM4SF1-AS1 was highly expressed in high metastatic lung cancer cell line, and it was also significantly up-regulated in lymph node metastatic lung cancer and was associated with lymph node metastasis. Overexpression of TM4SF1-AS1 promoted the migration and invasion of lung cancer cells. Overexpression of TM4SF1-AS1 decreased the expression of E-Cadherin and increased the expression of Vimentin, Snail and Twist, while knockdown of TM4SF1-AS1 exhibited the opposite trend. Furthermore, RNA sequence analysis revealed that some signaling pathways, including PI3K/AKT signaling pathway, were enriched upon TM4SF1-AS1 overexpression. Western blotting further confirmed that the PI3K/AKT signaling pathway was activated by TM4SF1-AS1.
Conclusion
This study illustrates that TM4SF1-AS1 promotes the migration and invasion of lung cancer cells by activating the PI3K/AKT signaling pathway. TM4SF1-AS1 might be a novel target of molecular treatment for lung cancer.
Keywords: lung cancer, long non-coding RNA, TM4SF1-AS1, metastasis, PI3K/AKT signaling pathway
Introduction
At present, lung cancer is the most commonly diagnosed cancer (11.6% of the total cases) and is the leading cause of cancer death (18.4% of the total cancer deaths) worldwide.1 In China, the situation is grim, too, with lung cancer being the most common and deadly malignancy.2 The main reason for the high mortality of lung cancer is its high invasion and metastasis, which is mediated by a multi-step process named as the metastatic cascade.3 In recent years, many factors and genes have been reported to contribute to this metastatic cascade of lung cancer.4–6 However, the invasion and metastasis of lung cancer is a multi-factor and multi-step process, which is controlled by a variety of internal and external factors and thus needs to be further explored.
LncRNAs are usually characterized by the size exceeding 200 nucleotides and apparent lack of detectable open reading frame. It is estimated that more than 90% of the mammalian genome is transcribed as non-coding RNAs.7 LncRNAs were previously considered as transcriptional noise. But recently, IncRNAs have been proved to be important regulators participating in the development of diseases, in particular cancer.8,9 These lncRNAs could regulate the biological behaviors of tumor cells in terms of genomic stability,10,11 transcription,12,13 metabolism14 and so on. Nevertheless, studies of lncRNAs in cancer are still at an early stage and need to be examined in more detail.
In this work, we screened the metastasis-associated lncRNAs in lung cancer cell lines and recognized the target lncRNA, transmembrane 4 superfamily 1-antisense1 (TM4SF1-AS1). We investigated the effect of TM4SF1-AS1 on the migration and invasion of lung cancer cells, and explored its possible molecular mechanism.
Materials and Methods
Patients and Clinical Samples
Forty-three human lung cancer tissues and adjacent non-tumor lung tissues were obtained from diagnosed lung cancer patients at the First Affiliated Hospital of Dalian Medical University (Dalian, China) in 2018. The samples were frozen in liquid nitrogen immediately after moving out of human body. Informed consent for tissue analysis was obtained before operation. The study was approved by the research ethics boards at the First Affiliated Hospital of Dalian Medical University.
Cell Culture
Human lung cancer cell lines (A549, NCI-H1299, NCI-H1975, NCI-H1650, H460-SM, 95D and 95C) and HEK293T were gotten from the American Type Culture Collection (ATCC). These cell lines were grown in RPMI-1640 medium (Gibco), while 95C, 95D and HEK293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco), supplemented with 10% FBS (Gibco) and double resistances of penicillin (100 units/mL)-streptomycin (100 μg/mL) (Gibco) at 37°C with 5% CO2. Cells were harvested during the logarithmic growth phase for subsequent experiments.
Array Hybridization and Microarray Analysis
Total RNA was extracted using TRIzol Reagent (Life technologies) following the manufacturer’s instructions. Total RNA was amplified, labeled and purified by Affymetrix WT PLUS Reagent Kit (Affymetrix) to obtain biotin-labeled cDNA according to the manufacturer’s instructions. Six arrays named Affymetrix Human Transcriptome Array 2.0 were used. More than six million probes were placed on the array, covering 44,699 coding genes and 22,829 non-coding genes. Array hybridization was performed and scanned by Affymetrix GeneChip Scanner3000. Raw data were then normalized by Expression Console. Fold change and T test statistical methods were used to screen the differential genes. The selection criteria were as follows: (1) Fold change ≤0.5 or ≥2; (2) p-value<0.05.
Quantitative Real-Time PCR (qPCR)
Total RNA was extracted by TRIzol Reagent (Life Technologies) and then qualified. Afterwards, first-strand complementary DNA (cDNA) was synthesized with a reverse transcriptase. The qPCR was accomplished with SYBR Green mix and the relative expression levels of genes were analyzed by the 2−ΔΔCt method. β-actin was served as an endogenous control. Specific primers are listed in Supplementary Table 1.
Stable Cell Line Construction
To establish stable cell lines with TM4SF1-AS1 overexpression or knockdown, vectors containing shRNA sequences against TM4SF1-AS1 were generated by cloning the shRNAs into pLKO.1, while TM4SF1-AS1 overexpression vectors were obtained by cloning the TM4SF1-AS1 into pLenti 6.3-IRES2-EGFP. HEK293T cells were then transfected with the indicated vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The viral supernatants were collected 48–72hrs after transfection. TM4SF1-AS1 shRNA lentiviruses were used to infect NCI-H1650 cells, while TM4SF1-AS1 overexpression lentiviruses infected NCI-H1299 cells. After 24hrs, cells were fed with 5 μg/mL puromycin-containing media to establish stable cell lines. The cell lines with stable overexpression or knockdown of TM4SF1-AS1were identified by using RT-qPCR. The shRNA sequences targeting TM4SF1-AS1 are shown in Supplementary Table 2.
Wound Healing Experiment
A total of 5×105 cells were seeded in six-well plates using a medium containing 10% FBS until confluence. The cell layer was scratched with a 200 μL pipette tip. Then, the cells were cultured in a serum-free medium. After 12 hrs and 24 hrs, the cells were rinsed with PBS and the wound closure was monitored under the microscope, respectively. The coverage of the scratched area at different positions was examined and the wound widths were measured by Image J software (National Institutes of Health, USA). Experiments were independently repeated at least three times.
Transwell Assay
The cell invasion and migration were assessed by using Transwell chambers (Costar). 1×105 cells in 500 μL serum-free medium were inoculated in the upper chamber coated with (invasion assay) or without (migration assay) Matrigel. Medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, the cells remaining on the upper membrane were removed with cotton wool. The invaded or migrated cells were fixed by formaldehyde and stained using 0.5% crystal violet. The number of cells was counted at least 6 unbiased (200×) fields. Experiments were independently repeated three times.
Western Blotting Analysis
Cells were lysed using RIPA protein extraction reagent (Beyotime) supplemented with a protease inhibitor cocktail (Roche) and phenylmethylsulfonyl fluoride (Roche). Identical quantities of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Sigma-Aldrich), followed by incubation with the primary antibodies. Equal loading amount of protein was confirmed by using β-actin. The membranes were then washed with PBST three times and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody. The Western blotting was visualized by using the ChemiDocTM XRS + (Bio-Rad).
RNA‐Sequence (RNA-Seq) and Analysis
The transcriptome sequencing profiles of TM4SF1-AS1 overexpression cells and control cells were carried out to detect the changes in the expression of downstream genes. Briefly, sequencing libraries were constructed by NEBNext Ultra RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s instructions. In order to select cDNA fragments of preferentially 250~300 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparation was sequenced on an Illumina Hiseq platform to produce 125 bp/150 bp paired-end reads. Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using Hisat2 (v2.0.5) and paired-end clean reads were aligned to the reference genome using Hisat2 (v2.0.5). The mapped reads of each sample were assembled by StringTie (v1.3.3b) (Mihaela Pertea.et al 2015) in a reference-based approach. FeatureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. And then FPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed. Correlations of significantly differentially expressed genes with cellular process, pathway and disease were analyzed by Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG) and DisGeNET.
Statistical Analysis
A statistical analysis was conducted using SPSS 21.0 or GraphPad Prism 7.0 software. Comparisons were determined using two-tailed Student’s t test or Mann–Whitney U. The association between TM4SF1-AS1 expression and various clinicopathological factors was evaluated using the χ2 test. All quantitative results are presented as mean ± SD. P value <0.05 was recognized to be statistically significant.
Results
Identification of Metastasis-Associated lncRNAs in Lung Cancer Cells
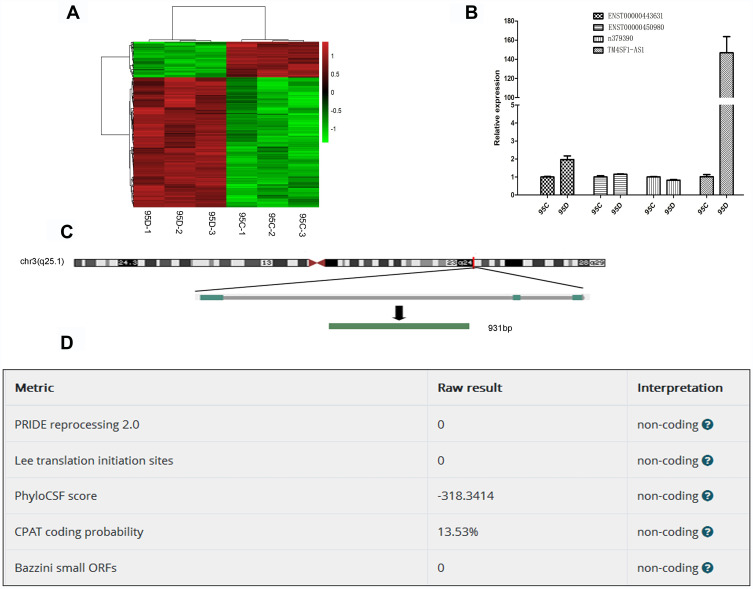
To find lncRNAs involved in lung cancer metastasis, we used microarray to detect the differentially expressed lncRNAs between high-metastatic human lung cancer cell line (95D) and paired low-metastatic human lung cancer cell line (95C). As shown in Figure 1A, 582 lncRNAs were significantly up-regulated, while 164 lncRNAs were remarkably down-regulated in 95D cell line, compared with 95C cell line. The top 10 up-regulated and down-regulated lncRNAs are listed in Supplementary Tables 3 and 4, respectively.
Figure 1.
Identification of metastasis-associated lncRNAs in lung cancer cell lines. (A) Heatmap of differentially expressed lncRNAs in 95C and 95D. Red color represents the high expression, and green color represents the low expression. 582 lncRNAs were significantly up-regulated, while 164 lncRNAs were remarkably down-regulated in 95D cell line, compared with 95C cell line. (B) Verification of differentially expressed lncRNAs by qPCR. (C) Genomic location of TM4SF1-AS1. (D) Confirmation of non-coding nature of TM4SF1-AS1 by using LNCipedia (https://lncipedia.org/).
According to fold change, abundance and sequence characteristics, four lncRNAs, TM4SF1-AS1, ENST00000443631, ENST00000450980 and n379390, were selected for subsequent verification. The expression of these four lncRNAs in 95C and 95D cell lines was examined by qPCR, and the results showed that among the four lncRNAs, TM4SF1-AS1 was significantly up-regulated in 95D cell line (Figure 1B), which was consistent with the microarray results. TM4SF1-AS1 was then selected as the target gene in the following studies. TM4SF1-AS1 is an antisense-lncRNA and is located on human chromosome 3 from 149378219 to 149386581 nucleotides (Figure 1C). The protein-coding potential of TM4SF1-AS1 is determined to be non-coding (Figure 1D).
TM4SF1-AS1 is Up-Regulated in Lymph Node Metastatic Lung Cancer and is Associated with Lymph Node Metastasis
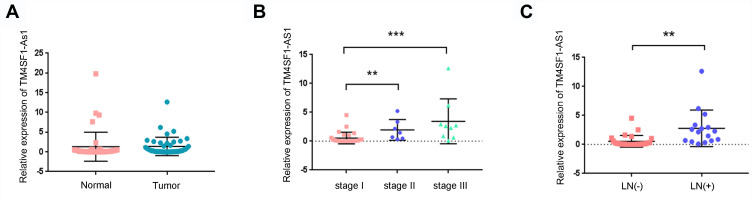
In order to examine the clinical significance of TM4SF1-AS1 in human lung cancer, we evaluated the expression of TM4SF1-AS1 in 43 paired human lung cancer specimens and adjacent non-tumor tissues using qPCR, and observed that the expression of TM4SF1-AS1 seldom showed a significant difference between the two groups (Figure 2A). However, TM4SF1-AS1 expression was dramatically up-regulated in lymphatic node-positive lung cancer compared with lymphatic node-negative tumor, as well as in TNM stage II and III cancers compared with TNM stage I tumor (Figure 2B and C). We then analyzed the relationship between TM4SF1-AS1 expression and clinicopathological features of lung cancer patients, and found that TM4SF1-AS1 expression was essentially correlated with differentiation, TNM stage and lymphatic node metastasis, independent of sex, age, histology or tumor size (Table 1).
Figure 2.
TM4SF1-AS1 expression in lung cancer tissues. (A) Expression of TM4SF1-AS1 in lung cancer tissues and adjacent non-tumor tissues was determined by qPCR. (B) TM4SF1-AS1 was up-regulated in patients with TNM stage II and III. (C) TM4SF1-AS1was up-regulated in patients with lymph node (LN)` metastasis. Statistical analysis was performed using Mann–Whitney U. **P < 0.01, ***P < 0.001.
Table 1.
Correlation Between TM4SF1-AS1 Expression and Clinicopathologic Characteristics in Lung Cancer Tissues
| Clinicopathologic Factor | Number of Cases | TM4SF1-AS1 | χ2 | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Age (years) | 1.149 | 0.2838 | |||
| ≤65 | 28 | 12 | 16 | ||
| >65 | 15 | 9 | 6 | ||
| Sex | 0.0244 | 0.8759 | |||
| Male | 22 | 11 | 11 | ||
| Female | 21 | 10 | 11 | ||
| Histology | 3.376 | 0.0661 | |||
| Squamous | 11 | 8 | 3 | ||
| Adenocarcinoma | 32 | 13 | 19 | ||
| Differentiation | 4.044 | 0.0443* | |||
| Well | 27 | 10 | 17 | ||
| Moderate + poor | 16 | 11 | 5 | ||
| Tumor size (cm) | 0.1961 | 0.6578 | |||
| ≤3 | 24 | 11 | 13 | ||
| >3 | 19 | 10 | 9 | ||
| Lymphatic node metastasis | 15.24 | <0.0001* | |||
| Negative | 27 | 7 | 20 | ||
| Positive | 16 | 14 | 2 | ||
| TNM stage | 15.26 | 0.0005* | |||
| I | 27 | 7 | 20 | ||
| II | 7 | 6 | 1 | ||
| III | 9 | 8 | 1 | ||
Notes: *Significant difference (P <0.05). The two-tailed Pearson χ2 test was employed to analyze the correlation of TM4SF1-AS1 expression with clinicopathologic factors.
TM4SF1-AS1 Promotes the Migration and Invasion of Lung Cancer Cells
For the sake of investigating the biological function of TM4SF1-AS1 in lung cancer metastasis, we first assessed the TM4SF1-AS1 expression in six lung cancer cell lines (Figure 3A). Our results revealed that NCI-H1299 cells exhibited low levels of TM4SF1-AS1. Therefore, NCI-H1299 cells were selected to stably express the TM4SF1-AS1. The successful overexpression of TM4SF1-AS1 in NCI-H1299 cell line was confirmed by qPCR (Figure 3B). Then, the results of wound healing and Transwell assays showed that the migration and invasion abilities of NCI-H1299 cells were obviously enhanced by TM4SF1-AS1 overexpression (Figure 3C and D). Next, we knocked down the TM4SF1-AS1 expression using short hairpin RNAs (shRNAs) in NCI-H1650 cells, which expressed relatively high levels of TM4SF1-AS1 (Figure 3E). As expected, two shRNAs against TM4SF1-AS1 (shTM4SF1-AS1-1 and shTM4SF1-AS1-2) reduced the migration and invasion abilities of NCI-H1650 cells compared to the negative control (shControl) (Figure 3F and G). Finally, Western blotting assays showed that TM4SF1-AS1 overexpression decreased the expression of E-Cadherin and increased the expression of Vimentin, Snail and Twist, while knockdown of TM4SF1-AS1 exhibited the opposite effects on the expression of these epithelial mesenchymal transition (EMT) markers (Figure 3H). Taken together, these results indicate that TM4SF1-AS1 enhances the migration and invasion of lung cancer cells.
Figure 3.
TM4SF1-AS1 enhances the migration and invasion of lung cancer cells. (A) RT-qPCR analysis of TM4SF1-AS1 expression in lung cancer cell lines (A549, NCI-H1299, NCI-H1975, NCI-H1650, H460-SM and 95D). (B) RT-qPCR analysis of TM4SF1-AS1 expression in NCI-H1299 cells stably overexpressing TM4SF1-AS1 or control vector. (C) Wound healing assay of NCI-H1299 cells overexpressing TM4SF1-AS1. Representative images (left) and quantification analysis (right) are shown (n = 3). (D) The invasion and migration of NCI-H1299 cells overexpressing TM4SF1-AS1 were evaluated by Transwell assay. Representative images (left) and quantification analysis (right) are shown (n = 3). (E) RT-qPCR analysis of TM4SF1-AS1 expression in NCI-H1650 cells knocking down TM4SF1-AS1. (F) Wound healing assay of NCI-H1650 cells with TM4SF1-AS1 knockdown. Representative images (left) and quantification analysis (right) are shown (n = 3). (G) The invasion and migration of NCI-H1650 cells with TM4SF1-AS1 knockdown were evaluated by Transwell assay. Representative images (left) and quantification analysis (right) are shown (n = 3). (H) Western blotting showing the expression of EMT markers in cell lines with TM4SF1-AS1 overexpression or knockdown. Statistical significance was assessed using two-tailed Student’s t test. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
TM4SF1-AS1 Alters Genome-Wide Gene Expression in Lung Cancer Cells
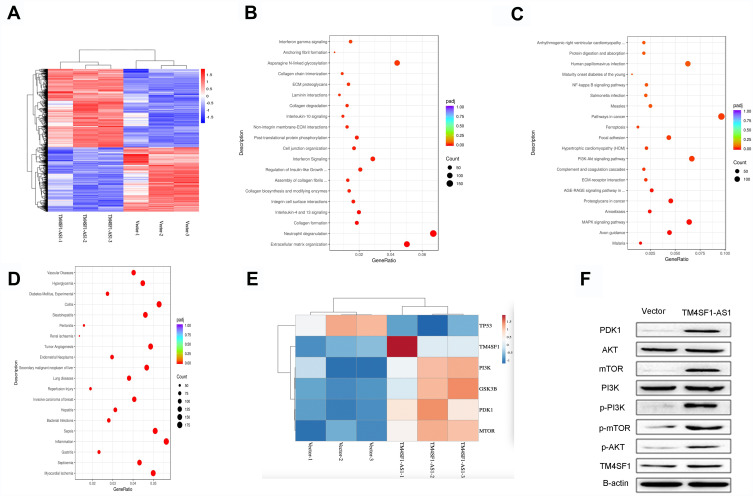
To explore the underlying mechanism of the enhanced migration and invasion of lung cancer cells by TM4SF1-AS1, we then carried out RNA-seq analysis to profile genome-wide gene expression in TM4SF1-AS1 overexpression and control cell lines. Four thousand two hundred and forty-four genes were differentially expressed in the two cell lines, with 2335 up-regulated genes and 1909 down-regulated genes upon TM4SF1-AS1 overexpression (Figure 4A). The Reactome analysis indicated that the differentially expressed genes were related to some biological behaviors, such as extracellular matrix organization, neutrophil degranulation, asparagine N-linked glycosylation, post-translational protein phosphorylation, cell junction organization and interferon signaling (Figure 4B). Furthermore, the KEGG analysis revealed that the most significantly altered pathways of differentially expressed genes included pathways in cancer, PI3K/AKT signaling pathway, MAPK signaling pathway, proteoglycans in cancer and focal adhesion (Figure 4C). Additionally, the analysis of the DisGeNET demonstrated that the differentially expressed genes were associated with several illnesses, such as tumor angiogenesis, secondary malignant neoplasm of liver, lung diseases, invasive carcinoma of breast and inflammation (Figure 4D).
Figure 4.
TM4SF1-AS1 activates PI3K/AKT signaling pathway. (A) Heatmap of differentially expressed genes in TM4SF1-AS1 overexpression and control cells. Red color represents the high expression, and blue color represents the low expression. 4244 genes were differentially expressed in the two cell lines, with 2335 up-regulated genes and 1909 down-regulated genes upon TM4SF1-AS1 overexpression. (B) Reactome analysis of the biological behaviors related to differentially expressed genes. (C) KEEG analysis of the enriched pathways of differentially expressed genes. (D) DisGeNET analysis of the illnesses related to differentially expressed genes. (E) A heatmap showing hierarchical clustering of five genes involved in the PI3K/AKT signaling pathway and the gene of TM4SF1. Red color represents the high expression, and blue color represents the low expression. (F) Western blotting showing the expression of PI3K/AKT signaling pathway markers and TM4SF1 in TM4SF1-AS1 overexpression and control cells.
TM4SF1-AS1 Regulates PI3K/AKT Signaling Pathway
Previous findings showed that TM4SF1, the host gene of TM4SF1-AS1, was associated with PI3K/AKT signaling pathway.15 Because TM4SF1 and many important factors of the PI3K/AKT signaling pathway were included in differentially expressed genes (Figure 4E), we aimed to investigate the effect of TM4SF1-AS1 on PI3K/AKT signaling pathway. To do it, we applied Western blotting to evaluate the expression of PDK1, AKT, mTOR, PI3K, p-PI3K, p-mTOR and p-AKT in TM4SF1-AS1 overexpression cells and control cells, respectively. As shown in Figure 4F, the expression levels of PDK1, mTOR, p-mTOR, p-PI3K and p-AKT were higher in TM4SF1-AS1 overexpression cells than those in control cells. Moreover, Western blotting showed that the expression of TM4SF1 was also increased in TM4SF1-AS1 overexpression cells (Figure 4F). In a word, our results suggest that TM4SF1-AS1 activates the PI3K/AKT signaling pathway in lung cancer cells.
Discussion
Lung cancer is one of the deadliest malignant tumors both in the world and in China.1,2 A better understanding of the underlying mechanisms and a search for potential biomarkers are urgently required to improve the survival rate of lung cancer patients. Recently, increasing evidences have indicated that the aberrant expression of lncRNA is involved in tumorigenesis and progression of lung cancer.16–18 In the present study, we screened the differentially expressed lncRNAs in high-(95D) and low-metastatic (95C) human lung cancer cell lines by microarray analysis and qPCR assay, and found that many lncRNAs were differentially expressed between the two cell lines and TM4SF1-AS1 was verified to be highly expressed in 95D cell line.
The aberrant expression of lncRNAs has been shown to be a key malignant phenotype in several human cancers, such as hepatocellular carcinoma,19 lung cancer20 and breast carcinoma.21 To confirm the high level of TM4SF1-AS1 in lung cancer, we examined the expression of TM4SF1-AS1 in lung cancer tissues. However, there was no significant difference between lung cancer specimens and adjacent non-tumor tissues. Interestingly, we found that TM4SF1-AS1 was overexpressed in lung cancer tissues with lymphatic metastasis or TNM stage of II and III, compared to lung cancer tissues without lymphatic metastasis or TNM stage I, respectively. It is possible that TM4SF1-AS1 is not related to tumorigenesis but to metastasis in lung cancer. Of course, due to the small number of cases of stage III, there are some limitations. In the future, we will further expand the sample size to conduct statistics. Furthermore, we observed that knockdown of TM4SF1-AS1 decreased the migration and invasion of NCI-H1650 cells, while overexpression of TM4SF1-AS1 increased the migration and invasion of NCI-H1299 cells. In sum, these findings demonstrate that TM4SF1-AS1 may be related to lung cancer metastasis.
It has been shown that EMT is closely related to the invasion and metastasis of tumor.22,23 The loss of E-cadherin is considered as a vital event in the EMT process, which is regulated by several transcription factors, including Snail, Slug and the Zeb family.24 Recently, many lncRNAs are also shown to play a crucial role in regulating the EMT process in cancer cells. Gao et al found that lncRNA NBR2 suppressed the progression of EMT in NSCLC by regulating the Notch1 pathway.25 lncRNA-CTS was reported to promote the TGF-β1-induced-EMT process and activate the SMAD/TGF pathway via miR-505 in cervical carcinoma cells.26 In this article, we found that TM4SF1-AS1 overexpression decreased the expression of E-cadherin and increased the expression of Vimentin, Snail and Twist simultaneously. Accordingly, it is speculated that TM4SF1-AS1 might prompt the invasion and metastasis of lung cancer cells by regulating the EMT.
The EMT process is influenced by many signaling pathways, such as TGF-β signaling pathways,27 Wnt Signaling pathways28 and PI3K/AKT signaling pathways.29 PI3K/AKT signaling pathway is involved in cell proliferation, differentiation, apoptosis and other cellular processes.30–32 Meanwhile, the PI3K/AKT signaling pathway takes part in tumor metastasis.33 Intriguingly, it has been reported that PI3K/AKT signaling pathway can be regulated by lncRNAs.34,35 TM4SF1-AS1 is located on chromosome 3q25.1 region and is the antisense RNA of TM4SF1. TM4SF1 is a small plasma membrane glycoprotein that belongs to the transmembrane 4 superfamily. TM4SF1 was identified as an oncogene and was shown to correlate with tumor cell metastasis.36,37 Meanwhile, TM4SF1 silencing significantly decreased the expressions of p-AKT, p-mTOR and p-P70.15
In order to further explore the mechanism of tumor metastasis regulated by TM4SF1-AS1, RNA-seq was performed in NCI-H1299 cells upon TM4SF1-AS1 overexpression. As expected, the results showed that TM4SF1-AS1 was involved in many biological processes and was connected with lots of diseases. In addition, TM4SF1-AS1 participated in many signaling pathways, for example PI3K/AKT signaling pathway and MAPK signaling pathway. Furthermore, the expression of TM4SF1 was higher in TM4SF1-AS1 overexpression cells than that in control cells. Collectively, these studies suggest that lncRNA TM4SF1-AS1 may promote the metastasis of lung cancer cells through the PI3K/AKT signaling pathway.
Conclusion
In summary, we identified TM4SF1-AS1 as a metastasis-associated lncRNA in lung cancer. TM4SF1-AS1 promotes the migration and invasion of lung cancer cells and activates the PI3K/AKT signaling pathway. Thus, TM4SF1-AS1 may serve as a potential biomarker for lung cancer metastasis, and targeting TM4SF1-AS1 may be a promising therapeutic strategy for lung cancer.
Acknowledgment
We thank Dr. Xuefeng Liu for the careful review of the manuscript.
Funding Statement
This work was supported by Liaoning Provincial Natural Science Foundation (No.20180551083) and Dalian medical scientific Research Foundation (No.1812015).
Abbreviations
lncRNAs, long non-coding RNAs; TM4SF1-AS1, transmembrane 4 superfamily 1-antisense1; cDNA, complementary DNA; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; KEGG, Kyoto Encyclopedia of Genes and Genomes; EMT, epithelial mesenchymal transition.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Langley RR, Fidler IJ. The seed and soil hypothesis revisited – the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Liu R, Zhu W, et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571(7763):127–131. doi: 10.1038/s41586-019-1340-y [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Zheng H, Zhan Y, et al. An emerging tumor invasion mechanism about the collective cell migration. Am J Transl Res. 2019;11(9):5301–5312. [PMC free article] [PubMed] [Google Scholar]
- 6.Altorki NK, Markowitz GJ, Gao D, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(316):1484–1488. doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- 8.Cho SW, Xu J, Sun R, et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173(6):1398–1412. doi: 10.1016/j.cell.2018.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1–2):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munschauer M, Nguyen CT, Sirokman K, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561(7721):132–136. doi: 10.1038/s41586-018-0453-z [DOI] [PubMed] [Google Scholar]
- 12.McHugh CA, Chen CK, Chow A, et al. TheXist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua JT, Ahmed M, Guo H, et al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. 2018;174(3):564–575. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Hu M, Zhou L, et al. Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2. Oncol Lett. 2019;18(3):2212–2219. doi: 10.3892/ol.2019.10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Xu Y, Xu J, et al. Role of TM4SF1 in regulating breast cancer cell migration and apoptosis through PI3K/AKT/mTOR pathway. Int J Clin Exp Pathol. 2015;8(8):9081–9088. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Gong J, Lu J, et al. Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am J Transl Res. 2019;11(9):6075–6083. [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X, Liu X, Zhang Z, Guan Y. lncRNA CCAT1 acts as a MicroRNA-218 sponge to increase gefitinib resistance in NSCLC by targeting HOXA1. Mol Ther Nucleic Acids. 2020;19:1266–1275. doi: 10.1016/j.omtn.2020.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davalos V, Esteller M. Disruption of long noncoding RNAs targets cancer hallmark pathways in lung tumorigenesis. Cancer Res. 2019;79(12):3028–3030. doi: 10.1158/0008-5472.CAN-19-0910 [DOI] [PubMed] [Google Scholar]
- 19.Lan T, Yuan K, Yan X, et al. lncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79(13):3220–3234. doi: 10.1158/0008-5472.CAN-18-4044 [DOI] [PubMed] [Google Scholar]
- 20.Zhu FY, Zhang SR, Wang LH, et al. LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur Rev Med Pharmacol Sci. 2019;23(19):8377–8390. doi: 10.26355/eurrev_201910_19149 [DOI] [PubMed] [Google Scholar]
- 21.Li P, Zhou B, Lv Y, et al. LncRNA HEIH regulates cell proliferation and apoptosis through miR-4458/SOCS1 axis in triple-negative breast cancer. Hum Cell. 2019;32(4):522–528. doi: 10.1007/s13577-019-00273-1 [DOI] [PubMed] [Google Scholar]
- 22.Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- 23.Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- 24.Loh CY, Chai JY, Tang TF, et al. The E-Cadherin and N-cadherin switch in epithelial-to-mesenchymal transition:signaling, therapeutic implications, and challenges. mCells. 2019;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao YP, Li Y, Li HJ, et al. LncRNA NBR2 inhibits EMT progression by regulating Notch1 pathway in NSCLC. Eur Rev Med Pharmacol Sci. 2019;23(18):7950–7958. doi: 10.26355/eurrev_201909_19011 [DOI] [PubMed] [Google Scholar]
- 26.Feng S, Liu W, Bai X, et al. LncRNA-CTS promotes metastasis and epithelial-to-mesenchymal transition through regulating miR-505/ZEB2 axis in cervical cancer. Cancer Lett. 2019;465:105–117. doi: 10.1016/j.canlet.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 27.Kaowinn S, Kim J, Lee J, et al. Cancer upregulated gene 2 induces epithelial-mesenchymal transition of human lung cancer cells via TGF-β signaling. Oncotarget. 2017;8(3):5092–5110. doi: 10.18632/oncotarget.13867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teeuwssen M, Fodde R. Wnt signaling in ovarian cancer stemness, EMT, and therapy resistance. J Clin Med. 2019;8(10):1658. doi: 10.3390/jcm8101658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu T, Zhao Y, Chen Y, et al. Down-regulated MAC30 expression inhibits breast cancer cell invasion and EMT by suppressing Wnt/β-catenin and PI3K/Akt signaling pathways. Int J Clin Exp Pathol. 2019;12(5):1888–1896. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Du P, Li Y, et al. TASP1 promotes gallbladder cancer cell proliferation and metastasis by up-regulating FAM49B via PI3K/AKT pathway. Int J Biol Sci. 2020;16(5):739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Jiang B, Zhu N, et al. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt pathway. Med Oncol. 2019;37(1):5. doi: 10.1007/s12032-019-1320-y [DOI] [PubMed] [Google Scholar]
- 32.Yan -Y-Y, Shi K-Y, Teng F, et al. A novel derivative of valepotriate inhibits the PI3K/AKT pathway and causes Noxa-dependent apoptosis in human pancreatic cancer cells. Acta Pharmacol Sin. 2020;41(6):835–842. doi: 10.1038/s41401-019-0354-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Han T, Bian Y, et al. Knockdown of SLCO4C1 inhibits cell proliferation and metastasis in endometrial cancer through inactivating the PI3K/Akt signaling pathway. Oncol Rep. 2020;43(3):919–929. doi: 10.3892/or.2020.7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Qian J, Li J, et al. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp Ther Med. 2019;18(1):435–442. doi: 10.3892/etm.2019.7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X, Wang Q, Tong L, et al. Long non-coding RNA FOXO1 inhibits lung cancer cell growth through down-regulating PI3K/AKT signaling pathway. Iran J Basic Med Sci. 2019;22(5):491–498. doi: 10.22038/ijbms.2019.31000.7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao C, Yao H, Liu H, Feng Y, Yang Z. TM4SF1 is a potential target for anti-invasion and metastasis in ovarian cancer. BMC Cancer. 2019;19(1):237. doi: 10.1186/s12885-019-5417-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan C, Liu N, Zheng D, Du J, Wang K. MicroRNA-206 inhibits metastasis of triple-negative breast cancer by targeting transmembrane 4 L6 family member 1. Cancer Manag Res. 2019;11:6755–6764. doi: 10.2147/CMAR.S199027 [DOI] [PMC free article] [PubMed] [Google Scholar]