Abstract
Background
Circular RNAs (circRNAs), a group of covalently closed non-coding RNAs, serve critical regulatory roles in many human cancers, including oral squamous cell carcinoma (OSCC). The purpose of this study was to investigate the functional role of circular RNA ITCH (circ-ITCH) in OSCC and the underlying mechanisms.
Methods
RT-qPCR analysis was applied to detect the expression levels of circ-ITCH in OSCC tissues and cell lines. MTT assay and flow cytometer analysis were used to evaluate the effects of circ-ITCH overexpression on the proliferation and apoptosis of OSCC cells. Bioinformatics analysis and dual-luciferase reporter assay were applied to determine the binding relation between circ-ITCH and miR-421 as well as PDCD4 mRNA and miR-421.
Results
Our results showed that circ-ITCH expression was remarkably decreased in OSCC tissues and cell lines. Low circ-ITCH expression was strongly associated with adverse clinicopathological characteristics of OSCC patients. Moreover, functional assays demonstrated that circ-ITCH overexpression significantly inhibited OSCC cell proliferation and induced cell apoptosis. Our data further uncovered that circ-ITCH could bind directly to miR-421 and block its repression on PDCD4 in OSCC. MiR-421 expression was significantly increased in OSCC tissues and was inversely correlated with circ-ITCH expression. Notably, miR-421 restoration blocked the tumor-suppressive role of circ-ITCH in OSCC cells.
Conclusion
In conclusion, our study reveals that circ-ITCH serves as a tumor suppressor in OSCC partly by regulating miR-421/PDCD4 axis.
Keywords: oral squamous cell carcinoma, circular RNA ITCH, miR-421, apoptosis, PDCD4
Introduction
Oral squamous cell carcinoma (OSCC), accounting for approximately 90% of all oral cancer cases, is a leading cause of cancer-related mortality worldwide.1 Despite recent advancements in cancer prevention strategies and therapeutic approaches, the prognostic outcomes of patients with OSCC remain dismal, with the 5-year survival rate of only 50%.2 Accordingly, it is worthy and important for us to elucidate the molecular mechanisms of OSCC and to identify novel therapeutic targets for OSCC patients.
Circular RNAs (circRNAs), a group of endogenous non-coding RNAs, are featured by their covalently closed loop structures without a 5ʹ cap or a 3ʹ Poly A tail. Compared with their linear counterparts, circRNAs are stable and conserved in eukaryotic cells.3 CircRNAs were previously regarded as by-products of splicing errors,4 but with the development of RNA sequencing technology, they have been confirmed to serve an important role in the initiation and progression of many human diseases, especially in tumors.5 To be specific, circular RNA itchy E3 ubiquitin-protein ligase (circ-ITCH) was demonstrated to function as a tumor suppressor in multiple human cancers, such as lung cancer,6 ovarian carcinoma7 and colorectal cancer.8 In the present study, we aimed to study the functional role of circ-ITCH in OSCC.
Patients and Methods
Patients and Tissue Samples
Paired OSCC tissues and adjacent normal tissues were collected from 103 cases of patients who underwent surgical resection at Hainan General Hospital (Haikou, China). All patients did not receive any therapy before surgery. All collected tissue samples were histologically characterized by pathologists, then immediately frozen in liquid nitrogen and stored at −80°C until further use. This study had received the approval from the Ethics Committee of Hainan General Hospital, and written informed consent was obtained from all participants.
Cell Culture and Transfection
The normal human oral keratinocytes (HOK) and five human OSCC cell lines (SCC6, SCC9, SCC25, HN4, and HN6), provided by American Type Cell Collection (Manassas, VA, USA), were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and antibiotics (100 μg/mL streptomycin and 100 U/mL penicillin; Sigma-Aldrich, St Louis, MO, USA) at 37°C in a humidified incubator with 5% CO2.
MiR-421 mimics (miR-421), negative mimics control (miR-NC), miR-421 inhibitor (anti-miR-421), negative inhibitor control (anti-miR-NC) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). To generate circ-ITCH-overexpressing plasmid, human circ-ITCH cDNA sequence was amplified and subcloned into the pcD-ciR vector (Geenseed Biotech Inc., Guangzhou, China). Cells grown at 70–80% confluence were transfected with the vectors or oligonucleotides using Lipofectamine 2000 (Invitrogen). After 48 h, the cells were collected for further experiments.
RT-qPCR Analysis
Total RNA was extracted with Trizol reagent (Beyotime, Shanghai, China). The complementary DNA (cDNA) was synthesized with 1 μg RNA as template using the PrimeScript™ RT reagent kit (TaKaRa, Dalian, China). Thereafter, qPCR analysis was performed using the SYBR Premix Ex Taq II kit (TaKaRa) on the LightCycler 480 System II (Roche Diagnostics, Rotkreuz, Switzerland). The 2−ΔΔCt method was used to calculate the relative gene expression.9 GAPDH or U6 was used as the endogenous reference.
MTT Assay
Cell proliferation was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells (5×103 cells/well) were seeded into 96-well plates. At each time point, 10 μL MTT solution (5 mg/mL; Sigma-Aldrich) was added to each well, followed by incubation for additional 4 h at 37°C. After discarding the supernatant, 150 μL DMSO (Sigma-Aldrich) was added to dissolve formazan, and the absorbance of each well was read at 570 nm on a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Cell Apoptosis Analysis
Cell apoptosis was evaluated using Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). Cells were harvested, resuspended in 1× binding buffer, and double-stained with 5 μL Annexin V-FITC and 5 μL PI. After incubation for 15 min at room temperature in the dark, the stained cells were analyzed using a flow cytometer (BD Biosciences) with the BD CellQuest™ Pro software.
Western Blot Analysis
Total protein was extracted with RIPA lysis buffer (Beyotime). About 30 μg of protein extracts were fractionated by SDS-PAGE gel, and then electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% skim milk for 2 h, the membranes were incubated overnight at 4°C with the primary antibodies, followed by incubation with the HRP-conjugated secondary antibody for 1 h at room temperature. The blots were visualized using the enhanced chemiluminescence reagents (Bio-Rad Laboratories, Hercules, CA, USA). GAPDH was used as the loading control.
Subcellular Fractionation
Cytoplasmic and nuclear RNA were isolated using PARIS™ Kit (Ambion, Austin, TX, USA). All isolated RNA was subjected to RT-qPCR analysis, normalizing to U6 (nucleus control) and GAPDH (cytoplasm control).
Dual-Luciferase Reporter Assay
The sequences of circ-ITCH or PDCD4 mRNA containing the miR-421 binding sites were amplified and subcloned into the psiCHECK-2 vector (Promega, Madison, WI, USA). The mutant construct was generated using the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen). HEK-293 cells were co-transfected with the reporter plasmid and miR-421 mimics or negative mimics control using Lipofectamine 2000; 48 h later, cells were lysed, and the luciferase activity was observed by the Dual-Luciferase Reporter Assay system (Promega).
Statistical Analysis
All statistical analyses were performed using Graphpad Prism (version 6.01) software (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS version 18.0 software (SPSS Inc., Chicago, IL, USA). Experimental data were recorded as the means ± standard deviation (SD) from at least three independent replicates. The differences between groups were analyzed using Student’s t-test or one-way analysis of variance followed by Tukey’s test. The correlation between the expression levels of circ-ITCH and miR-421 in OSCC tissues was analyzed by Pearson correlation analysis. P values of less than 0.05 were considered as statistically significance.
Results
circ-ITCH is Downregulated in OSCC
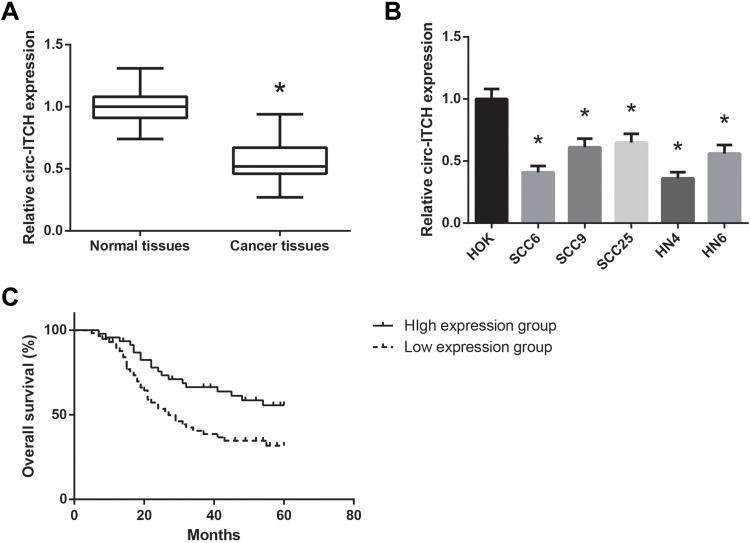
We first investigated the expression pattern of circ-ITCH in OSCC. Through RT-qPCR analysis, circ-ITCH was found to be significantly downregulated in OSCC tissues compared to adjacent normal tissues (Figure 1A). Besides, circ-ITCH expression was also significantly decreased in a panel of OSCC cell lines (SCC6, SCC9, SCC25, HN4, and HN6), compared with normal HOK cells (Figure 1B).
Figure 1.
circ-ITCH is downregulated in OSCC. (A) RT-qPCR analysis for the expression levels of circ-ITCH in OSCC tissues and adjacent normal tissues. (B) RT-qPCR analysis for the expression levels of circ-ITCH in OSCC cell lines and normal HOK cells. (C) Association of circ-ITCH expression with overall survival of OSCC patients. *P<0.05 vs. normal tissues or HOK cells.
According to the median circ-ITCH expression level, these patients were further allocated into low circ-ITCH expression group (N=57) and high circ-ITCH expression group (N=46), and we observed that low circ-ITCH expression was strongly related to lymph node metastasis (P=0.035) and advanced TNM stage (P=0.027) of OSCC patients (Table 1), but harbored no close association with other parameters. Kaplan–Meier survival analysis further indicated that patients with lower circ-ITCH expression in OSCC tissues had significantly shorter overall survival (P=0.010; Figure 1C).
Table 1.
Correlation Between Clinicopathological Characteristics and circ-ITCH Expression in 103 OSCC Patients
| Characteristics | Total Number (N=103) | circ-ITCH Expression | P value | |
|---|---|---|---|---|
| Low (N=57) | High (N=46) | |||
| Age (years) | 0.261 | |||
| ≥55 | 60 | 36 | 24 | |
| <55 | 43 | 21 | 22 | |
| Gender | 0.725 | |||
| Male | 63 | 34 | 29 | |
| Female | 40 | 23 | 17 | |
| Tumor differentiation | 0.234 | |||
| Well | 56 | 28 | 28 | |
| Moderate + Poor | 47 | 29 | 18 | |
| Lymph node metastasis | 0.035 | |||
| Yes | 36 | 25 | 11 | |
| No | 67 | 32 | 35 | |
| TNM stage | 0.027 | |||
| I–II | 64 | 30 | 34 | |
| III–IV | 39 | 27 | 12 | |
circ-ITCH Suppresses Proliferation but Induces Apoptosis in OSCC Cells
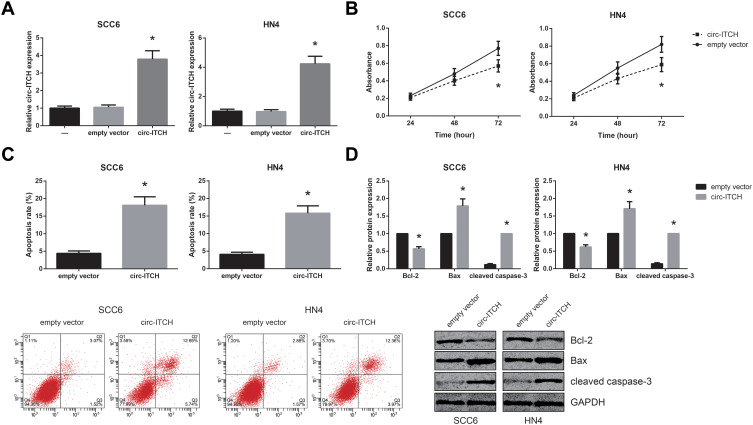
The functional roles of circ-ITCH in OSCC were further investigated by multiple experiments. SCC6 and HN4 cells were chosen for further analysis since they exhibited the lowest expression levels of circ-ITCH among the tested OSCC cell lines. As shown in Figure 2A, after transfection with circ-ITCH-overexpressing plasmid, circ-ITCH expression was remarkably increased in both SCC6 and HN4 cells. MTT assay demonstrated that overexpression of circ-ITCH significantly inhibited the proliferation of SCC6 and HN4 cells (Figure 2B). Flow cytometry analysis further revealed that the percentage of apoptotic cells were obviously increased when circ-ITCH was silenced in SCC6 and HN4 cells (Figure 2C). Furthermore, as shown in Figure 2D, circ-ITCH overexpression led to the increased levels of Bax and cleaved caspase-3 in SCC6 and HN4 cells, accompanied by the downregulation of Bcl-2.
Figure 2.
Circ-ITCH suppresses proliferation but induces apoptosis in OSCC cells. (A) RT-qPCR analysis for the expression levels of circ-ITCH in OSCC cells after transfection. (B) MTT assay for the proliferation of OSCC cells after transfection. (C) Flow cytometry analysis for the apoptosis of OSCC cells after transfection. (D) Western blot analysis for the expression levels of apoptosis-related proteins in OSCC cells after transfection. *P<0.05 vs. empty vector-transfected cells.
circ-ITCH Serves as a ceRNA for miR-421 in OSCC
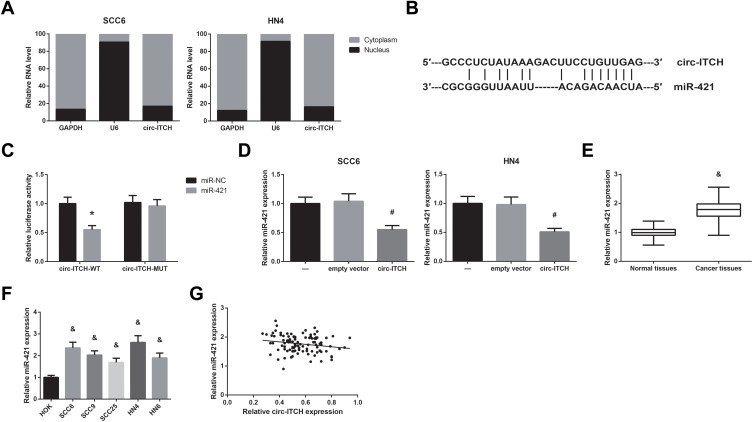
We further observed that circ-ITCH was mainly located in the cytoplasm of SCC6 and HN4 cells (Figure 3A), indicating its potential role as a ceRNA to interact with miRNAs, and through the Starbase online software (http://starbase.sysu.edu.cn/index.php), we identified the potential binding sites for miR-421 on circ-ITCH fragment (Figure 3B). Dual-luciferase reporter assay was then carried out to verify the prediction, and the results demonstrated that co-transfection of miR-421 mimics led to an about 50% decline of the luciferase activity of circ-ITCH-WT in HEK-293 cells, but this effect was abolished when the binding sites were mutated (Figure 3C). Besides, we also found that circ-ITCH overexpression significantly decreased miR-421 expression in SCC6 and HN4 cells (Figure 3D). MiR-421 expression was notably increased in the OSCC tissue samples compared with adjacent normal tissues (Figure 3E), and this expression pattern was further observed in a panel of OSCC cell lines (Figure 3F). In addition, a significant inverse correlation between circ-ITCH and miR-421 expression was confirmed in OSCC tissues (r=−0.199, P=0.044; Figure 3G).
Figure 3.
Circ-ITCH serves as a ceRNA for miR-421 in OSCC. (A) The subcellular distribution of circ-ITCH in OSCC cells. (B) The predicted binding site of miR-421 within circ-ITCH fragment. (C) Dual-luciferase reporter assay for the luciferase activity in HEK-293 cells. (D) RT-qPCR analysis for the expression levels of miR-421 in OSCC cells after transfection. (E) RT-qPCR analysis for the expression levels of miR-421 in OSCC tissues and adjacent normal tissues. (F) RT-qPCR analysis for the expression levels of miR-421 in OSCC cell lines and normal HOK cells. (G) Pearson correlation analysis for the correlation between the expression levels of circ-ITCH and miR-421 in OSCC tissues. *P<0.05 vs. miR-NC-transfected cells; #P<0.05 vs. empty vector-transfected cells; &P<0.05 vs. normal tissues or HOK cells.
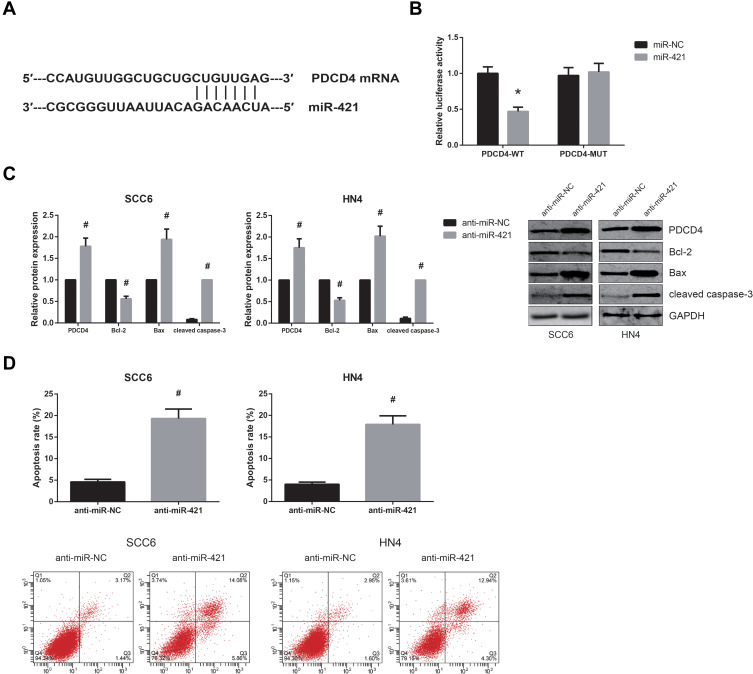
MiR-421 Directly Targets PDCD4 in OSCC
Based on TargetScan software (http://www.targetscan.org), PDCD4 was predicted as a direct target of miR-421 (Figure 4A), and dual-luciferase reporter assay further showed that co-transfection with miR-421 mimics markedly inhibited the luciferase activity of PDCD4-WT in HEK-293 cells (Figure 4B). What’s more, miR-421 inhibition significantly increased PDCD4 protein expression in SCC6 and HN4 cells (Figure 4C), accompanied by the decreased expression of Bcl-2 and the increased levels of Bax and cleaved caspase-3. MiR-421 inhibition also led to the enhanced apoptosis of SCC6 and HN4 cells (Figure 4D).
Figure 4.
MiR-421 directly targets PDCD4 in OSCC. (A) The predicted binding site of miR-421 within PDCD4 mRNA. (B) Dual-luciferase reporter assay for the luciferase activity in HEK-293 cells. (C) Western blot analysis for the expression levels of PDCD4 protein and other apoptosis-related proteins in OSCC cells after transfection. (D) Flow cytometry analysis for the apoptosis of OSCC cells after transfection. *P<0.05 vs. miR-NC-transfected cells; #P<0.05 vs. anti-miR-NC-transfected cells.
MiR-421 Blocks the Tumor Suppressive Role of circ-ITCH in OSCC
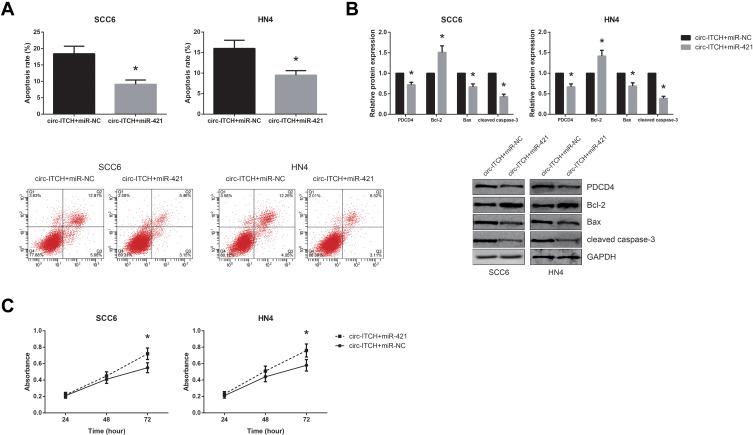
Rescue experiments were then performed to verify whether circ-ITCH exerts its role partly through miR-421. As shown in Figure 5A and B, the increased apoptosis rates of SCC6 and HN4 cells with circ-ITCH overexpression were notably diminished by co-transfection of miR-421 mimics, accompanied by the increased expression of Bcl-2 and the decreased levels of Bax, cleaved caspase-3 and PDCD4. We also found that miR-421 restoration blocked the inhibitory role of circ-ITCH overexpression on the proliferation of SCC6 and HN4 cells, as evidenced by the results of MTT assay (Figure 5C).
Figure 5.
MiR-421 blocks the tumor suppressive role of circ-ITCH in OSCC. (A) Flow cytometry analysis for the apoptosis of OSCC cells after transfection. (B) Western blot analysis for the expression levels of PDCD4 protein and other apoptosis-related proteins in OSCC cells after transfection. (C) MTT assay for the proliferation of OSCC cells after transfection. *P<0.05 vs. circ-ITCH+miR-NC-transfected cells.
Discussion
OSCC is one of the most common tumors worldwide. In recent years, an increasing number of studies have indicated that abnormally expressed circRNAs are closely associated with the induction and progression of OSCC.10 For example, Xia et al reported that a circRNA derived from MMP9 facilitated OSCC metastasis,11 while Chen et al showed that upregulation of circATRNL1 enhanced the radiosensitivity of OSCC cells.12
Circ-ITCH, a novel tumor suppressor in many cancers,13 aroused our great interest. Our study first identified the decreased expression of circ-ITCH in OSCC, whereas low expression of circ-ITCH is closely correlated with malignant progression and poor survival in OSCC patients. Cell proliferation and apoptosis are two key hallmarks of tumors. We then performed a series of gain-of-function assays, and the results showed that circ-ITCH overexpression suppressed cell proliferation, but promoted apoptosis of OSCC cells in vitro. These data indicated that circ-ITCH also plays a tumor-suppressive role in OSCC.
It has been widely reported that circRNA can serve as competing endogenous RNAs (ceRNAs) by competitively binding with related miRNA sequences to regulate target mRNAs. CircRNA-miRNA-mRNA axis are involved in a series of diseases, including cancers.14 This study provided several lines of evidence implicating that circ-ITCH was primarily located in the cytoplasm of OSCC cells, and could directly bind to miR-421, which has been previously identified as an oncogenic miRNA in several types of cancers. In this study, miR-421 was also confirmed to play an oncogenic role in OSCC cells and exert inhibitory effect on PDCD4, a well-known tumor suppressor protein in OSCC.15,16 Rescue experiments further validated that restoration of miR-421 blocked the tumor suppressive role of circ-ITCH in OSCC.
In conclusion, this study showed that circ-ITCH is downregulated in OSCC and serves as a tumor suppressor in OSCC partly by acting as a ceRNA to sponge miR-421 and subsequently increased PDCD4 expression. We therefore suggested that circ-ITCH might be a potential therapeutic target for OSCC.
Acknowledgments
This work was supported by Key Research and Development Project of Hainan (ZDYF2019216), Key Scientific Research Projects of Higher Education in Hainan Province (Hnky2019ZD-22), and Hainan Natural Science Foundation (817320 and 20168281).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Glazer CA, Chang SS, Ha PK, Califano JA. Applying the molecular biology and epigenetics of head and neck cancer in everyday clinical practice. Oral Oncol. 2009;45(4–5):440–446. doi: 10.1016/j.oraloncology.2008.05.013 [DOI] [PubMed] [Google Scholar]
- 3.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 5.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Xiang H, Sun B, Feng F, Chen P. Circular RNA-ITCH inhibits the proliferation of ovarian carcinoma by downregulating lncRNA HULC. Reprod Sci. 2020;27(1):375–379. doi: 10.1007/s43032-019-00049-w [DOI] [PubMed] [Google Scholar]
- 8.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10(6):e0131225. doi: 10.1371/journal.pone.0131225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 10.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 11.Xia B, Hong T, He X, Hu X, Gao Y. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant. 2019;28(12):1614–1623. doi: 10.1177/0963689719875409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen G, Li Y, He Y, et al. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Nucleic Acids. 2020;19:961–973. doi: 10.1016/j.omtn.2019.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Ge YZ, Xu L, Jia R. Circular RNA ITCH: a novel tumor suppressor in multiple cancers. Life Sci. 2019:117176. [DOI] [PubMed] [Google Scholar]
- 14.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.19154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZH, Gao W, Ho W-K, et al. The clinical association of programmed cell death protein 4 (PDCD4) with solid tumors and its prognostic significance: a meta-analysis. Chin J Cancer. 2016;35(1):95. doi: 10.1186/s40880-016-0158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis PP, Tomenson M, Cervigne NK, et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer. 2010;9:238. doi: 10.1186/1476-4598-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]