Graphical abstract
Keywords: Molecular biology, Molecular dynamics, Non-canonical amino acid, Phosphomimetic, Protein phosphorylation, Protein structure
Abstract
Post-translational modifications of proteins expand their functional diversity, regulating the response of cells to a variety of stimuli. Among these modifications, phosphorylation is the most ubiquitous and plays a prominent role in cell signaling. The addition of a phosphate often affects the function of a protein by altering its structure and dynamics. However, these alterations are often difficult to study and the functional and structural implications remain unresolved. New approaches are emerging to overcome common obstacles related to the production and manipulation of these samples. Here, we summarize the available methods for phosphoprotein purification and phosphomimetic engineering, highlighting the advantages and disadvantages of each. We propose a general workflow for protein phosphorylation analysis combining computational and biochemical approaches, building on recent advances that enable user-friendly and easy-to-access Molecular Dynamics simulations. We hope this innovative workflow will inform the best experimental approach to explore such post-translational modifications. We have applied this workflow to two different human protein models: the hemeprotein cytochrome c and the RNA binding protein HuR. Our results illustrate the usefulness of Molecular Dynamics as a decision-making tool to design the most appropriate phosphomimetic variant.
1. Introduction
The complexity of the human proteomes goes far beyond the ca. 20,000 ORFs (open reading frames) contained in the genome. A myriad of processes give rise to the structural and functional variety of proteins without increasing the genetic background. Alternative promoters and splicing, as well as mRNA editing yield a five-fold increase in the transcriptome. Post-translational modifications (PTMs) represent another tool by which cells can expand the functional diversity of proteins. These modifications may occur at any step in the “life cycle” of a protein and alter either its function or stability. We can sort PTMs into five groups: 1) addition of complex molecules, 2) addition of proteins and peptides, 3) amino acid modifications, 4) addition of chemical groups and 5) cleavage of part of the protein sequence [1]. This work will focus on phosphorylation, which is the main PTM involved in cell regulatory processes [2], [3], [4], [5], [6].
Metabolic regulation by protein phosphorylation is notable for its rapid reversibility, an essential feature in cell signaling [7]. This PTM consists of the addition of a phosphoryl group either to a hydroxyl-containing residue through an ester bond, or to a nitrogen-containing one to from an amidate, this process is generally mediated by kinases. The energy of this phosphate ester linkage is similar to that of the ATP β–γ anhydride bond. In fact, under certain experimental conditions, it is possible for kinases to act in the opposite direction, thereby generating ATP. Thus, high ATP:ADP ratios forestall dephosphorylation by protein kinases [6], [8], [9]. The residues most prone to phosphorylation are Ser (ca. 60%), followed by Thr (ca. 25%) and Tyr (ca. 15%) [6], [10]. Phosphorylation of other residues—such as histidine and arginine—is less stable and infrequent in eukaryotes [11], [12], and is thus excluded from this work.
Phosphorylation sites can be found within intrinsically disordered regions (IDRs) or structured domains. Analyses suggest that serine and threonine phosphorylation takes place predominantly in IDRs, while tyrosine phosphorylation occurs both in IDRs and structured regions [13]. Phosphorylation induces both disorder to order and order to disorder transitions. An example of order to disorder transition is nucleophosmin (NPM1), sequential phosphorylation of solvent exposed Ser and Thr regulates the thermodynamic stability of the NPM1 oligomerization domain. This phosphorylation event promotes the exposure of other sites buried within the structure for subsequent phosphorylation and drives the switch from structured pentamer to disordered monomer [14]. K-homology splicing regulator protein (KSRP) contains four K-homology domains (KH1-4), in which KH1 phosphorylation acts as a conformational switch between folded (non-phosphorylated protein) and essentially unfolded (phosphorylated protein) states [15]. Unfolding of KH1 domain creates a 14-3-3ζ binding site which prevents the recruitment of the exosome [16]. On the other hand, phosphorylation of IDR eukaryotic translation initiation factor 4E (eIF4E)-binding protein 2 (4E-BP2) protein constitutes an example of disorder to order transition by inducing folding into a four-stranded β-domain, which blocks the accessibility of eIF4E [17].
1.1. Overview of phosphoprotein detection and purification methods
Phosphoproteomic investigations usually require purification and often enrichment of endogenous proteins from cells or tissues. The substitution of the hydroxyl group of Ser, Thr and/or Tyr by a phosphate group affects the physicochemical and immunogenic properties of any protein, and is consequently used for its isolation and analysis. Purification methods for phosphorylated proteins usually take advantage of 1) changes in the isoelectric point (pI), 2) metal cation chelating capacity of phosphate groups, and 3) unique immunogenicity of isoforms [18].
The main purification techniques used for isolation of phosphorylated proteins are chromatofocusing (CF), ion exchange chromatography (IEX), immobilized metal ion affinity chromatography (IMAC), metal oxide affinity chromatography (MOAC) and immunoaffinity chromatography (IAC). CF and IEX are based on the downshift in protein pI caused by phosphorylation [19]. The magnitude of this shift depends strongly on amino acid composition, secondary structure and number of concurrently-phosphorylated residues [18], [19]. There are several pI prediction algorithms which calculate the influence of different PTMs, including phosphorylation, but such theoretical approaches are not exempt from errors and their accuracy for phosphorylated proteins depends on neighboring phosphoresidues and protein folding [20], [21].
Many phosphoprotein purification strategies utilize the tendency of phosphate groups to chelate metal ions and oxides. IMAC consists of phosphoproteins binding to metal-functionalized resins, usually containing Fe3+, and its elution by either a pH gradient or competitive displacement by chelating ligands, typically imidazole [22], [23]. To improve the selectivity and efficacy of IMAC, Kinoshita et al. [24] designed a phosphate chelating group, the so-called Phos-tag, which is an alkoxide-bridged binuclear metal (Zn2+ or Mn2+) complex. Phos-tag and its conjugated derivatives bind to phosphorylated residues in an amino acid-independent manner [25]. This technology has been extensively used for phosphoprotein detection by SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) and western blot analyses [25], [26], [27], and has not yet been surpassed by other types of IMAC developed over the last few years [22], [23], [28]. As an alternative to this technique, MOAC employs metal oxides, most commonly TiO2, to selectively isolate phosphoproteins [29]. As with IMAC, phosphoproteins are complexed with metal oxides under acidic conditions; however, complete recovery of phosphorylated proteins in MOAC requires an alkaline pH gradient from 8.5 to 11.5 for elution of multiple phosphorylated isoforms [30].
When purifying phosphoproteins, IAC is more selective than IMAC or MOAC, though it still suffers from low specificity. IAC exploits the unique immunogenic properties of phosphorylated proteins, utilizing antibodies that detect specific phosphoresidues in a sequence-independent manner. Antibodies raised against kinase consensus sequences can also be used [31]. Currently, only anti-phosphorylated Tyr (pTyr) antibodies are widely used in phosphoproteomics, despite the lower abundance of phosphorylated Tyr residues, compared with Ser and Thr [32]. The relatively low specificity of anti-phosphorylated Ser (pSer) and anti-phosphorylated Thr (pThr) antibodies favors the use of more cost-efficient approaches such as IMAC/MOAC [31]. In fact, the best asset of IAC, i.e. the high affinity of antigen–antibody interactions, is offset by the high cost of antibody production [18]. Recent papers report combined purification strategies using IEX, TiO2-MOAC and/or IAC, either sequentially or as complementary techniques, to optimize protein enrichment into phosphotyrosine [33], [34], [35].
Unlike anti-pTyr antibodies, the Src homology 2 (SH2) domain has recently emerged as an affinity reagent in the field of phosphoproteomics. SH2 modular domains are present in many signaling proteins such as growth factor receptor-bound and signal transducers and activators of transcription (STAT) protein family members. Single SH2 domain expression and covalent coupling to chromatographic resins allows the purification of phosphotyrosine proteins [36], [37]. For selectivity improvement, Kaneko et al. [38] designed high-affinity SH2 domains by directed evolution methods. They discovered that the substitution of three key residues in the SH2 binding site of the tyrosine kinase Fyn notably increases its affinity for Tyr-phosphorylated proteins.
Obtaining phosphoproteins from natural sources has limitations in terms of sample quantity and possible downstream applications. Many researchers dealing with endogenous phosphoproteins conduct quantitative and temporal phosphoproteomic profiling of the components of cellular signaling networks under certain stress conditions, including disease [39], [40], [41]. The methodology used in these studies includes chromatographic techniques for phosphoprotein isolation and enrichment so far described, as well as typical mass spectrometry analyses [37], [42]. Nonetheless, natural sources of endogenous phosphoproteins are not optimal since large quantities are needed for structural and/or biophysical assays.
The simplest and most common strategy to produce high amounts of any protein is the use of overexpression vectors for bacterial (usually E. coli) or yeast systems. They allow the expression of the construct of interest, normally a gene encoding a fusion protein with a tag sequence—e.g. polyhistidine, glutathione S-transferase (GSH), or maltose-binding protein—to facilitate purification by affinity chromatography (Ni2+-IMAC, GSH-sepharose or amilose resin). Assuming that the expression and purification of the recombinant protein provides good yields, both in terms of quality and quantity of the sample, it is still necessary to phosphorylate in vitro the target residues using the appropriate kinase enzyme. However, there are very few examples in the literature of preparative methods for in vitro phosphorylation of proteins used in binding assays, i.e. to study the effect of protein phosphorylation on an interaction with a third protein [43], [44]. In most papers, the in vitro kinase reactions are intended to characterize the phosphorylation event itself – to determine parameters such as kinetics, kinase substrate specificity and target-site identification [45], [46], [47], [48].
Baudin and Müller [45], [46], [47], [48] took a different approach and in bacteria co-expressed both a kinase domain and a STAT protein for in vivo phosphorylation. Using this approach, they purified Tyr-phosphorylated STAT proteins, which were used to assess the binding of STAT proteins to specific DNA sequences by electrophoretic mobility shift assay (EMSA). This successful strategy could potentially be applied to other kinase-phosphoprotein pairs. Although, if any given kinase phosphorylates more than one residue, it will be necessary to resort to point (e.g. Tyr to Phe) and/or deletion mutants of the target protein to study the effect of each phosphoresidue separately.
The lack of information on specific kinases for the vast majority of the identified phosphorylation targets demands the adoption of other methodological approaches. Thus, the use of phosphomimetic mutants, either classic variants (e.g. Ser/Thr/Tyr to Asp or Glu) or those incorporating non-canonical amino acids (ncAAs), is a good alternative for studying the structure and interactions of phosphorylated proteins.
1.2. Classical phosphomimetic variants using standard amino acids
As discussed above, the use of endogenous phosphorylated protein is frequently not feasible because of its low yield when purified from cell extracts or the absence of information on specific kinases. In this case, the easiest approach is the use of phosphomimetic proteins. Since phosphorylation is a post-translational event, cells lack native tRNAs to carry phospho-residues and to transfer them to nascent polypeptides. Classically, Ser- and Thr-phosphorylation have been mimicked by mutations to aspartate and glutamate. These substitutions provide the negative charge of each phosphoresidue and are of a similar volume. Therefore, they constitute the simplest methodology to introduce canonical amino acids [49], [50]. Glu is a more suitable phosphomimetic replacement as it matches the phosphate oxygens of pSer and pThr, while Asp is less suitable as its chain is shorter. However, a major drawback of these mutations is they fail to feature the hydration layer and formal charges of phosphate [7].
Despite their utility, residue substitutions are not always possible. In some cases, it is necessary to introduce a pair of acidic residues to generate the double charge and mimic the behavior of the natural phosphoprotein [51], [52], [53]. However, this strategy may lead to local stress on protein structure due to side chain repulsion.
Glutamate is often used to mimic pTyr [54], [55]. However, the glutamate residue is unable to mimic either the charge or the volume of pTyr. Indeed, there are examples in the literature wherein Tyr phosphorylation could not be emulated by this mutation [56], [57]. Alternatively, several ncAAs can be used to mimic the abovementioned properties of the phosphate group (Table 1).
Table 1.
Non-canonical amino acids mimicking phosphoresidues.
| Phosphorylated residues | Unnatural amino acid | Citation |
|---|---|---|
| Phosphotyrosine | 4′-Carboxymethyloxy-3′-phosphonophenylalanine | [85] |
| N-[4-(2-{2-[3-(2-Bromo-acetylamino)-propionylamino]-3-hydroxy-propionylamino}-ethyl)-phenyl]-oxalamic acid | [86] | |
| 3-(4-{2-[2-(2-Bromo-acetylamino)-ethyldisulfanyl]-ethylcarbamoyl}-cyclohexylcarbamoyl)-pyrazine-2-carboxylic acid | [86] | |
| Photocaged phosphotyrosine | [73] | |
| Sulfotyrosine | [87] | |
| p-Carboxymethyl-l-phenylalanine | [88] | |
| p-(Phosphonoamino)-phenylalanine | [89] | |
| O-phosphotyrosine | [90] | |
| Benzyl 2-benzyloxycarbonylamino-3-[4-bis(dimethylamino)phosphoryloxyphenyl] propanoate | [91] | |
| 4-Phosphomethyl-L-phenylalanine | [77] | |
| Phosphothreonine | Photocaged phosphothreonine | [73] |
| O-phospho-l-threonine | [92] | |
| Phosphoserine | Photocaged phosphoserine | [73] |
| Phosphonomethylene alanine | [161] | |
1.3. Incorporation of unnatural amino acid to obtain phosphomimetic proteins
The use of ncAAs in recombinant protein expression enables modification of the chemical properties of one or more residues [58]. Moreover, the use of technologies to selectively and co-translationally incorporate phosphomimetic amino acids into proteins can increase sample preparation yields to enable biophysical analysis. There are two current options for introducing ncAAs into polypeptides: genetic code expansion and reprogramming. Both strategies rely on codon reassignment. The expansion approach targets stop or quadruplet codons, whereas reprogramming involves reassigning sense codons to ncAAs [59]. The most popular approach utilizes the evolved tRNA and aminoacyl-tRNA synthetase (aa-RS) pair system, which consists of a suppressor tRNA, normally targeting the low-usage amber (TAG) triplet, and an orthogonal aa-RS recognizing the tRNA bearing a specific ncAA. Thus, recombinant expression of aa-RS/tRNA pairs and the protein construct of interest, together with the addition of the ncAA to the culture medium, constitute the basis for phosphomimetic protein biosynthesis [60], [61], [62]. The aa-RS/tRNA pair system has successfully been used with a wide variety of cells, both prokaryotic and eukaryotic, including yeast, plants and animals [58]. However, the effectiveness of ncAA incorporation into proteins is variable. It depends on multiple factors, such as expression levels and performance of a given orthogonal aa-RS/tRNA pair, sequence context of repurposed codons within a gene, number and/or diversity of ncAAs present in the same protein, and cellular bioavailability of ncAAs [63]. Nehring et al. [64] examined the incorporation efficiency of several Tyr analogs by cognate aa-RS/tRNA pairs, at a specific position of a target protein in yeast. Using western blot analysis followed by mass spectrometry characterization, the authors reported significant differences in the performance of the aa-RSs tested [64]. Pott et al. [65] studied the influence of nucleotide context in amber codon suppression efficiency. They generated a library of sequences around the TAG triplet and performed expression assays, identifying hit sequence contexts with relative protein yields of 70–110% (compared to the levels of control proteins without ncAAs), three to five fold higher than the average yield obtained for random sequences [65]. Schwark et al. [63] assessed the role of the release factor-1 (RF-1) in the efficiency of amber codon reassignment. The results of an expression screening with ncAA-containing green fluorescent protein (GFP) mutants, both in wild type and RF-1 deleted E. coli cells, suggested that the contribution of RF-1 in translation efficiency is highly variable in single amber codon constructs, but has a consistent and significant negative effect on multisite mutants [63].
These few examples illustrate the convenience, even necessity, of optimizing the orthogonal translation systems. This can be achieved with high-yield protein expression vectors such as pEVOL, or selectivity-enhanced aa-RS, tRNA and/or bacterial elongation factor Tu mutants for stronger interactions [58], [66], [67], [68], [69]. It is also possible to use a knock-out of the tRNA competitor RF-1 for improved binding to amber/quadruplet codons, or engineered ribosomes for higher decoding efficiency [70], [71], [72]. Moreover, the ncAA bioavailability can also be optimized through protection against phosphatases or gene deletion of these enzymes for elevated intracellular levels of phosphoresidues [73], [74].
A common issue encountered when introducing phosphorylated residues, is the lack of permeability of cell membranes to highly-charged ncAAs [62], [75]. Two main strategies have been developed to increase cellular uptake of phosphoresidues and ncAAs: 1) site-directed mutagenesis of periplasmic transporters for ncAA-specific internalization [76], and 2) addition of ncAAs to the culture medium as dipeptides with any natural amino acid, which are imported by the ATP-dependent DppA transporter and eventually hydrolyzed, releasing the ncAAs inside the cell [77]. Alternatively, the use of cell-free protein synthesis (CFPS) for ncAA incorporation has gained popularity in recent years. This approach seems to be particularly suitable for solving the intrinsic cellular bioavailability constraints of phosphoresidues, among other issues [78]. Protein expression with this method occurs in an open reaction environment to which crude cell extracts containing an overexpressed tRNA/aa-RS pair are added along with the ncAA, which is perpetually accessible to the translation machinery. There are several reports of increased production yield due to the implementation of CFPS protocols, as corroborated by western blot and mass spectrometry analyses [78], [79]. Oza et al. [80] developed a CFPS-based method to enhance the synthesis efficiency of site-specific Ser-phosphorylated constructs. Notably, the application of this system allowed them to increase the relative yield (i.e. percentage of wild type protein expression) of doubly phosphorylated human MEK1 kinase by ca. 80% compared to previously reported data for the same construct using an in vivo approach [81]. Wakamori et al. [82] took advantage of the CFPS platform to boost the efficiency of the simultaneous incorporation of acetyl-lysine into four specific positions of histone H4, achieving virtually 100% expression of the tetra-acetylated species. Yanagisawa et al. [83] also resorted to CFPS to improve the ratio of Lys-methylated Histone H3 synthesized to ncAA consumed by 10- to 20-fold compared to in vivo expression, which is particularly relevant when working with expensive and/or difficult to synthesize ncAAs [78].
Despite the evident strengths of CFPS, a great variability in reaction performance has also been documented, the causes of which can be multiple (such as ncAA sequence location in the protein construct) and unmanageable [84]. Some authors have developed strategies to obtain pSer, pThr, and pTyr by adding a protective “hood” against the action of phosphatases [73]. This protective “hood” is a photocleavable o-nitrophenylethyl group, which uncaps the phosphorylated residue upon irradiation with 350 nm light.
There is a great variety of ncAAs developed to mimic both the charge and the volume of phosphoresidues (Table 1). pTyr residue is the amino acid for which the greatest variety of ncAAs are available, in an attempt to mimic the volume and properties of its aromatic ring [73], [77], [85], [86], [87], [88], [89], [90], [91]. However, most authors choose the ncAA p-carboxy-methyl-L-phenylalanine (pCMF) because of the relatively low cost of its synthesis—it is even commercially available—and the efficient development of the technique employed to introduce it in a desired position (see below). Recently, further strategies based on the pEVOL technique have been designed to allow the synthesis and incorporation of pTyr and pThr residues in vivo, thus preventing the action of phosphatases [90], [92] (Table 1).
1.4. Case study
Many researchers tend to picture how proteins or biomolecules function by using static atomic-level structures while neglecting the dynamics of the molecule. Nonetheless, Molecular Dynamics (MD) simulations have been used increasingly over the past few years to study macromolecular systems due to the constant progress in computing power. Recently, Hollingsworth and Dror (2018) published an extensive review on MD simulations highlighting their potential application in molecular biology and drug discovery [93]. For example, MD simulations can be used to figure out how a biological system will respond to certain perturbations, such as amino acid mutation [94] or post-translational modification [95].
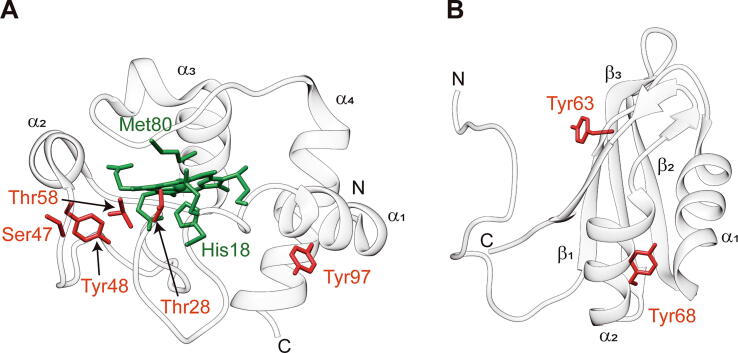
We herein provide a methodological article to offer a guide for readers who want to study phosphorylated proteins by combining computational and biochemical approaches. To illustrate this, two human proteins—for which phosphorylation in several residues has been studied—have been chosen as models: cytochrome c (Cc) and human antigen R (HuR) (Fig. 1). These two proteins have been selected as their number of phosphorylation sites is known and their relatively small size reduces the computational time required for simulations.
Fig. 1.
Phosphorylation sites of human cytochrome c and HuR. Richardson representations of human Cc (A; PDB ID: 2N9I; [142], [113]) and HuR1-101 (B; PDB ID: 5SZW; [118], [114]). Heme group and axial ligands (His18 and Met80) of Cc are colored in green. Phosphorylation sites described in the literature for mammalian Cc and HuR are highlighted in red. Thr at position 58 is the most conserved amino acid from bacteria to mammals, but it is not present in human Cc. N-terminal and C-terminal of the polypeptide chains are marked with N and C, respectively. Secondary structure elements (α-helix and β -sheet) are depicted. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Cc is an essential metalloprotein for mitochondrial metabolism and homeostasis, playing a dual-purpose role in cell life and death. Under homeostasis, Cc acts as an electron carrier within the respiratory chain in the mitochondria. However, stress stimuli induce the release of Cc to the cytosol and nucleus where it interacts with several targets [96], [97], [98], [99], [100], [101], [102]. In the cytosol, Cc triggers programmed cell death (PCD) by activating the caspase cascade [103], [104], [105]. In the nucleus, Cc is involved in DNA damage response through its interaction with histone chaperones [99], [100]. All of these Cc functions are modulated by post-translational modifications [106], [107], [108]. In mammals, phosphorylation of residues 28, 47, 48, 58 and 97 has been reported [4], [109], [110], [111], [112], [113] (Fig. 1A). Tyr48 is the folding nucleus of Cc, and its phosphorylation alters the dynamics of the hemeprotein, thus regulating oxidative phosphorylation and PCD-related functions [114].
HuR is an RNA-binding protein that modulates the processing of mRNAs involved in cell survival, proliferation and apoptosis. HuR is composed of three RNA recognition motifs (RRMs) connected by flexible regions. The modular structure of HuR means the function of the RRM domains can be studied independently and/or in tandem [115], [116], [117], [118]. For this reason, computational and biochemical analyses of single RRMs can provide information of molecular changes caused by PTMs. Its function is highly regulated by the phosphorylation catalyzed by several kinases [119]. For example, Janus kinase 3 (JAK3) can phosphorylate Tyr63, Tyr68 and Tyr200 [120]. Tyr63 and Tyr68 are located at the first motif (RRM1), which comprises residues 1–101 (HuR1-101) (Fig. 1B). On the other hand, Tyr200 is located at the linker region between RRM2 and RRM3. Functional data suggests that Tyr200 phosphorylation impairs HuR binding to target transcripts by either shifting its localization or altering the conformation of RRMs [120]. However, the effect of phosphorylation at RRM1 remains unveiled. Understanding conformational or dynamic changes induced by this PTM may provide a rationale for changes in functionality.
2. Methodology
2.1. In silico approach to study protein phosphorylation
2.1.1. Prediction of phosphorylation sites
Tyr-phosphorylation is the most frequent PTM found in cell signaling, due to the highly conserved pTyr binding domains and the low basal phosphorylation levels of kinase protein targets, essential for a signaling mechanism [3], [121]. However, there may be several residues in a protein that can be phosphorylated. Hence, as a first approach, it is useful to check the information on phosphorylation sites provided by database servers, such as PhosphoSite ([122]), Phospho.ELM ([123]) or NetworKIN ([124]) (Table 2). Several machine learning methods have been used to generate prediction tools for phosphorylation sites. These prediction tools are based on artificial neural networks (NetPhos [125], NetPhosBac [126], NetPhosYeast[127]), convolutional neural networks (NetPhospan[128]), position specific scoring matrices (GPS [129], ScanSite [130]), hidden Markov models (KinasePhos 1.0 [131], Predikin [132]), support vector machines (PHOSIDA [133], KinasePhos 2.0 [134]), logistic regression (DISPHOS [13]) or structural analysis (pKaPS [135]) (Table 2).
Table 2.
Databases and prediction tools to study protein phosphorylation.
| Databases | ||
|---|---|---|
| Phosphosite | https://www.phosphosite.org | [122] |
| Phospho.ELM | http://phospho.elm.eu.org | [123] |
| NetworKIN | http://www.networkin.info | [124] |
| Prediction tools | ||
| DISPHOS | http://www.dabi.temple.edu/disphos/ | [13] |
| GPS 5.0 | http://gps.biocuckoo.cn | [129] |
| Scansite 4.0 | http://scansite.mit.edu | [130] |
| pKaPS | http://mendel.imp.ac.at/sat/pkaPS | [135] |
| KinasePhos 2.0 | http://KinasePhos2.mbc.nctu.edu.tw | [134] |
| NetPhos | https://services.healthtech.dtu.dk/service.php?NetPhos-3.1 | [125] |
| NetPhosBac | https://services.healthtech.dtu.dk/service.php?NetPhosBac-1.0 | [126] |
| NetPhosYeast | https://services.healthtech.dtu.dk/service.php?NetPhosYeast-1.0 | [127] |
| NetPhospan | https://services.healthtech.dtu.dk/service.php?NetPhospan-1.0 | [128] |
| PHOSIDA | http://phosida.de | [133] |
| Predikin | http://predikin.biosci.uq.edu.au | [132] |
2.1.2. Analysis of phosphorylation-induced perturbations on protein structure and dynamics
Comparison between MD simulations on wild-type (WT) and phosphorylated protein species enables us to anticipate possible perturbations in the target structure and dynamics induced by phosphorylation at certain positions. MD simulations allows the evaluation of the motions of residues and atoms within a protein structure. This in silico information may be useful in assessing the behavior of phosphomimetic variants (Fig. 2). The chosen phosphomimetic variant should present similar dynamic properties and conformational changes as the phosphorylated protein.
Fig. 2.
Proposed experimental approach for protein phosphorylation analysis. The decision-making at each step is the result of assessing in silico approaches with wet-lab techniques.
Due to the extensive development of MD simulations, there is a broad range of software packages that can be used to perform the simulation. Some examples include GROMACS, AMBER, CHARMM, NAMD and OpenMM [136], [137], [138], [139], [140]. Latest versions of MD software packages usually work “out of the box” with detailed manuals and tools to enable first-time use of the software. All MD simulations performed in this work were carried out on a small workstation equipped with a i7-6700 Intel CPU processor and NVIDIA 1050-Ti GPU. With this setup, we achieved a simulation speed of 50 ns/day for a 30,000 atoms system, which could be hugely increased with a high-end workstation and high-performance computing clusters. In our case, we performed MD simulations using the OpenMM [140] software in an OpenCL platform with AMBER 14SB force field [141] and particle mesh Ewald (PME) electrostatics with an Ewald summation cutoff of 10 Å.
The initial structures were based on the nuclear magnetic resonance (NMR) structures of reduced human Cc (PDB ID: 2N9I; [142]), reduced human Y48pCMF Cc mutant (PDB ID: 2N3Y; [114]) and HuR1-101 (PDB ID: 5SZW; [118]). All mutants and phosphorylated species structural models were built using the WT protein as a template. The force field parameters for phosphorylated tyrosine [143], unnatural amino acid pCMF [144] and heme group [145] were used to generate the topology and coordinate files required for the simulation, with the TLEAP module of AMBER [139].
For each simulation, the system was neutralized with sodium and chlorine counter-ions according to the total charge of the protein, and solvated with optimal 3-charge, 4-point rigid water model (OPC) water molecules [146]. The whole system was subjected to 2500 steps of energy minimization at 298 K. Langevin thermostat was used to control the temperature with a friction coefficient of 1 ps−1 and a step size of 0.002 ps. Each system was subjected to 500 ns MD simulation. To study the stability of the phosphomimetic variants, similar MD simulations were carried out at different temperatures (400 K and 500 K) and the system was subjected to 10 ns simulation.
There are several software analysis frameworks available such as MDTRAJ [147], CPPTRAJ [148] or GROMACS [138]. We opted to use the CPPTRAJ module of AMBER for trajectory analysis [148]. For each simulation, root mean square deviation of both atomic positions (RSMD, rms command on CPPTRAJ), atomic fluctuations (RMSF, atomicfluct command on CPPTRAJ), structure content (secstruct command on CPPTRAJ), hydrogen bonds (hbond command on CPPTRAJ), surface accessible to solvent (surf command on CPPTRAJ) and native contacts (nativecontact command on CPPTRAJ) were analyzed to evaluate the perturbation induced by the addition of a phosphate group or a phosphomimetic variant at a specific position of the protein. Statistics of simulated proteins throughout the simulations are listed in Table 3.
Table 3.
Statistics of simulated proteins along MD trajectories.
| Average RMSD (Å)a | Average RG (Å)a | Average SAS (Å2)b | Average RMSF (Å)c | |
|---|---|---|---|---|
| Cc WT | 0.85 ± 0.10 | 12.96 ± 0.04 | 39.20 ± 10.66 | 0.43 ± 0.07 |
| Cc Y48E | 0.73 ± 0.07 | 12.87 ± 0.04 | 61.33 ± 13.42 | 0.45 ± 0.07 |
| Cc Y48pCMF | 1.93 ± 0.08 | 13.24 ± 0.05 | 128.10 ± 18.21 | 0.65 ± 0.14 |
| Cc pY48 | 2.13 ± 0.11 | 13.20 ± 0.05 | 123.61 ± 22.67 | 0.66 ± 0.12 |
| HuR1-101 WT | 1.59 ± 0.18 | 12.14 ± 0.07 | −17.25 ± 3.34 | 0.47 ± 0.05 |
| HuR1-101 Y68E | 1.73 ± 0.19 | 12.21 ± 0.07 | 34.96 ± 12.93 | 0.83 ± 0.13 |
| HuR1-101 Y68pCMF | 2.23 ± 0.25 | 12.33 ± 0.07 | −4.71 ± 8.46 | 1.26 ± 0.43 |
| HuR1-101 pY68 | 2.35 ± 0.29 | 12.40 ± 0.08 | 102.76 ± 28.16 | 1.57 ± 0.53 |
| HuR1-101 WT | 1.59 ± 0.18 | 12.14 ± 0.07 | 18.84 ± 10.54 | 0.87 ± 0.52 |
| HuR1-101 Y63E | 1.56 ± 0.10 | 12.20 ± 0.07 | 66.36 ± 10.54 | 1.10 ± 0.70 |
| HuR1-101 Y63pCMF | 1.66 ± 0.12 | 12.18 ± 0.06 | 140.11 ± 10.29 | 0.67 ± 0.32 |
| HuR1-101 pY63 | 1.92 ± 0.10 | 12.23 ± 0.06 | 102.27 ± 21.22 | 0.65 ± 0.30 |
Average RMSD/RG values were calculated for the plateau (last 100 ns). For HuR1-101 trajectories, values were calculated for the structured domain (residues 22–97). For all simulations, RMSD drift was smaller than 0.001 Å/ns.
Average SAS values were calculated for the plateau (last 100 ns). Values were calculated for the heme group (Cc trajectories), position 68 of HuR1-101 (Tyr68 mutants’ trajectories) and position 63 of HuR1-101 (Tyr63 mutants’ trajectories).
Average RMSF values of Cc were calculated for the loop 80–91, for the stretch 42–49 of HuR1-101 Tyr68 mutants, and for the stretch 52–69 of HuR1-101 Tyr63 mutants.
2.2. Overexpression of phosphomimetic variants
2.2.1. Site-directed mutagenesis
Incorporation of the unnatural amino acid pCMF was accomplished by co-expressing the unnatural tRNA/aminoacyl‐tRNA synthetase pair specific for pCMF (encoded by pEVOL-pCMF plasmid) along with the protein of interest, the DNA sequence of which was mutated to a TAG amber codon at the desired incorporation site. The DNA encoding HuR1-101 was cloned into the pGEX expression vector with an N-terminal 6xHis-tag and was used as a template for the mutagenic polymerase chain reaction (PCR). AMBER mutation was performed on a pBTR1 plasmid comprising the CYCS gene encoding human Cc, along with the CYC3 gene of the yeast Cc heme lyase. CYC3 is required for proper cytoplasmic maturation of human Cc, as it covalently attaches the heme group to the Cc apoprotein. Primers used for mutagenic PCR are detailed in Table S1.
2.2.2. Protein expression
The expression of WT Cc and its phosphomimetic variants (Y48E and Y48pCMF) were performed as previously described [144], [149] in E. coli BL21 (DE3) strain. Briefly, ncAA-containing protein was expressed as follows: pre-culture was incubated at 37 °C at 150 rpm in minimal medium M9 supplemented with ampicillin (100 μg /mL) and chloramphenicol (20 μg/mL). Pre-induction of the pre-culture with 0.02% arabinose is necessary to ensure the presence of the unnatural tRNA/aaRS pair. Protein expression was induced at an optical density (OD600) of 0.6 with 0.02% arabinose and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG); pCMF and δ-aminolevulinic acid were added after induction at a final concentration of 1 mM each. Cells were incubated for 20 h under agitation (150 rpm) at 30 °C before harvesting by centrifugation.
WT HuR1-101 and its phosphomimetic variants were expressed in the E. coli BL21 (DE3) strain. For WT HuR1-101 and non-containing pCMF mutants, the cells were cultured at 37 °C under agitation (150 rpm) in LB medium supplemented with 100 μg/mL ampicillin. When OD600 reached 0.6, protein expression was induced with 1 mM IPTG for 20 h under agitation (150 rpm) at 30 °C. For the HuR1-101 mutants Y63pCMF and Y68pCMF, cells were cultured at 37 °C at 150 rpm in minimal medium M9, supplemented with 100 μg /mL ampicillin and 20 μg/mL chloramphenicol. Protein expression was induced at OD600 of 0.6 with 0.02% arabinose and 1 mM IPTG, as described previously [144]. pCMF was added after induction at a final concentration of 1 mM. Cells were harvested by centrifugation, resuspended in the lysis buffer 20 mM Tris-HCl (pH 8.0), containing 800 mM NaCl, 10 mM imidazole, 1 mM phenylmethanesulfonyl fluoride, 0.2 mg/mL lysozyme and 0.02 mg/mL DNase, and ruptured by sonication. Afterwards, cellular debris was separated by centrifugation at 14,000 × g for 30 min at 4 °C.
Protein expression was monitored by SDS-PAGE and Western-Blot. Proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% fat-free dry milk in phosphate buffered saline (PBS)-0.1% Tween-20. Immunoblotting was performed with specific mouse primary antibody against HuR (catalog number 32160702, Sigma) or 6xHis-tag (catalog number 11922416001, Roche) and horseradish peroxidase (HRP)-conjugated secondary antibody against mouse IgG (catalog number A4416, Sigma). The immunoreactive bands were detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
3. Results and discussion
3.1. General workflow
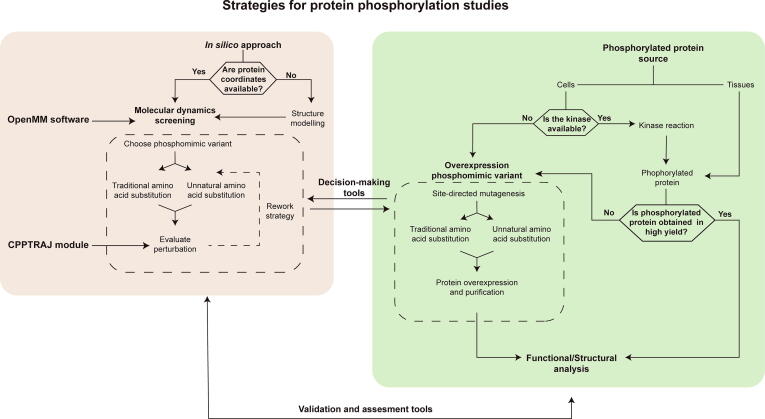
A general workflow for protein phosphorylation studies is proposed in Fig. 2. The first decision to be made before carrying out any functional and/or structural analysis is choosing the source of phosphorylated protein. If the phosphorylated protein is obtained with a high yield, either by direct purification from tissues/cells extracts or further purification after a kinase in vitro reaction, the desired physicochemical analysis can be performed.
However, as pointed out in Section 1.1, obtaining large amounts of phosphorylated protein is not always feasible. For this reason, it is necessary to overexpress a phosphomimetic variant. At this stage, an in silico approach can help to indicate which variant is the most suitable for your system.
To perform the MD simulation screening, the 3D atomic coordinates for the desired protein (preferably from the crystallographic or NMR structure) must be available at the PDB. Homology modelling software such as SWISS-MODEL (https://swissmodel.expasy.org; [150]), MODELLER (https://salilab.org/modeller; [151]) or ROBETTA (http://robetta.bakerlab.org; [152]) can be used to generate the protein coordinates if they are not available. If X-ray diffraction data from homologous proteins is available crystallographic water molecules can be included in the model. Once structural models have been generated, a comparison of phosphorylation-induced perturbations should be performed by analyzing the MD simulations of the WT protein versus the phosphorylated species (see Section 2.1.2). Subsequent analysis of multiple MD simulations of candidate phosphomimetic variants a helps to identify which molecule best mirrors the conformational changes induced by phosphorylation. Computational-based interaction network analysis may also be helpful to evaluate phosphomimetic constructs [153].
As soon as the selection of the phosphomimetic variant has been completed based on the MD data (decision-making tool), overexpression and purification of the chosen protein may be performed. This stage requires optimization of the expression conditions as mutations may affect protein folding and stability. Mimicking phosphorylation of residues exposed to the solvent – which are the most common phosphorylation sites – is often a straight forward process. However, if the phosphoresidue is located in the protein core (i.e. Tyr68 of HuR), the addition of negative charges may induce conformational changes or impair correct protein folding. MD simulations at varying conditions (temperature, ionic force or pH) can inform and detect possible difficulties in protein expression and folding.
Finally, MD simulations, in a feedback-like loop, can validate and assess the gathered functional/structural information of the phosphorylated/phosphomimetic protein.
3.2. Cytochrome c phosphorylation
A clear example where it was possible to verify the poor choice of glutamate to mimic pTyr is in Cc. The specific Cc kinase is unknown and a broad study on the functional implications of phosphorylation is unachievable with purified phosphorylated Cc because of low yields obtained from cell extracts [106], [113], [144], [154], [155]. The proposed methodology (see Section 3.1) was used to analyze different phosphomimetic variants (mutation by glutamate or substitution with pCMF) for the study of Cc Tyr48 phosphorylation (pY48) [144].
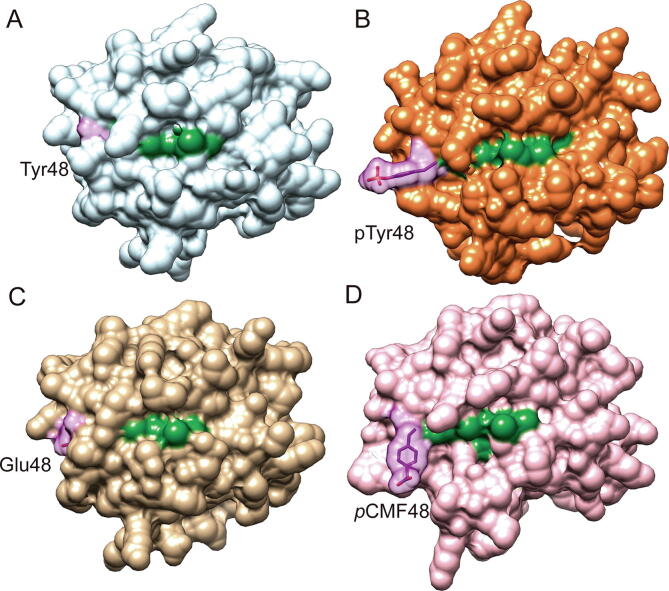
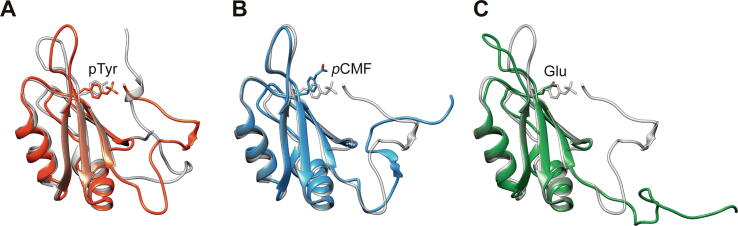
Initial MD calculations of WT Cc and modelled mutants Y48E, Y48pCMF and pY48 Cc revealed that the Y48pCMF mutation induces a localized distortion of the structure similar to that computed for the phosphorylated species. Radius of gyration (RG) values of phosphomimetic Y48pCMF and phosphorylated Cc displayed a similar behavior, while Y48E profile resembles that of WT Cc (Fig. S2). Indeed, surface accessible to solvent (SAS) analysis revealed that the solvation of the heme group in the ncAA mutant (128.1 ± 18.2 Å2) mirrors more closely the solvation experienced by the phosphorylated variant (123.6 ± 22.7 Å2) as compared with the Y48E mutant (61.3 ± 13.4 Å2) (Fig. 3 and Fig. S2). The loop containing residues 20–30 showed the highest root mean square fluctuations (RMSF) for all mutants, and were similar to values previously reported for Thr28 and Thr58 mutants [113], [155], [156] (Fig. S2). RMSF values of residues surroundings position 48 from Y48pCMF Cc were slightly higher than those from pY48 Cc, and RMSF values from Y48E Cc were slightly lower in comparison with those from WT Cc. Notably, the RMSF values at the loop between residues 80–91, where one of the axial ligands (Met80) of the heme group is located, were similar for both phosphomimetic Y48pCMF and phosphorylated pY48 Cc species, whereas WT and Y48E mutant displayed analogous values. In summary, the Y48pCMF variant showed a behavior closer to the one displayed by phosphorylated cytochrome c with respect to the overall properties calculated by MD simulations.
Fig. 3.
Cytochrome c conformational changes induced by Tyr48 phosphorylation. Surface representation of human Cc (A, WT in white; B, phosphorylated at Tyr48 in orange; C, Tyr-to-Glu mutant in gold; D, Tyr-to-pCMF mutant in pale rose) after MD simulation. Heme group is colored in green and position 48 in purple. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
This data was used as a starting point to characterize the Y48pCMF Cc mutant [114]. This mutant impairs Cc diffusion between respiratory complexes, boosts hemeprotein peroxidase and reactive oxygen species (ROS) scavenging activities, and hampers caspase-dependent apoptosis [114]. Further experimental NMR relaxation measurements and hydrogen–deuterium exchange experiments on the Y48pCMF variant revealed that the structure of the ncAA mutant exhibits enhanced dynamics around the pCMF-containing loop (also observed in MD simulations), which could explain the changes observed in its functionality (Table 4). Upon cardiolipin binding, Cc undergoes conformational changes which allows H2O2 to access the heme crevice [157]. In the absence of cardiolipin, Y48pCMF Cc displayed higher peroxidase activity compared to the Y48E phosphomimetic variant. This disparity in activity may be explained by the difference in the heme group SAS, which is two-times higher for the ncAA variant. Another explanation could be the increased dynamics of the Met80-containing loop for the Y48pCMF variant. The Y48H Cc variant (naturally occurring mutation related to thrombocytopenia) also showed a higher peroxidase activity in the absence of cardiolipin with heightened dynamics in the Met80-containing loop [158].
Table 4.
Comparison of biophysical and functional properties of two phosphomimetic cytochrome c mutants with respect to the native phosphorylated protein.
|
in vivo pY48 Cc [110], [96] |
Y48E Cc [154], [126] |
Y48pCMF Cc [144], [116] |
|
|---|---|---|---|
| Detection by anti-pTyr | Yes | No [154], [126] |
No [144], [116] |
| E’0, pH 7.0 (mV) | – | 192.0 ± 5.0 [154], [126] 187.3 ± 0.8 [162], [130] |
208.5 ± 0.3 [144], [116] |
| Alkaline transition (pKa) | 7.0 [162], [130] |
6.3 [144], [116] |
|
| Oxygen consumption, Vmax (% of diminished capacity) | 50% | 30% [154], [126] |
60% [114], [100] |
| CcO activity, Km values (μM) | 3.0 | 3.7 [154], [126] |
– |
| Caspase activity | – | Unable to activate caspase-3 [154], [126] Decrease by 60% (ca.) compared to WT [162], [130] |
Decrease by 47.7% compared to WT [114], [100] |
| Bound to cardiolipin | – | 30% lower affinity [154], [126] |
27% more affinity [114], [100] |
| Peroxidase activity | – | Cc peroxidase activity only inducible at high cardiolipin concentration [154], [126] Larger than WT in the absence of CL [162], [130] |
In the absence of CL, the mutant shows 3 times more activity than WT [114], [100] In the presence of CL, the peroxidase activity increases twice as much as in the absence of CL [114], [100] |
– : Not available.
3.3. HuR phosphorylation
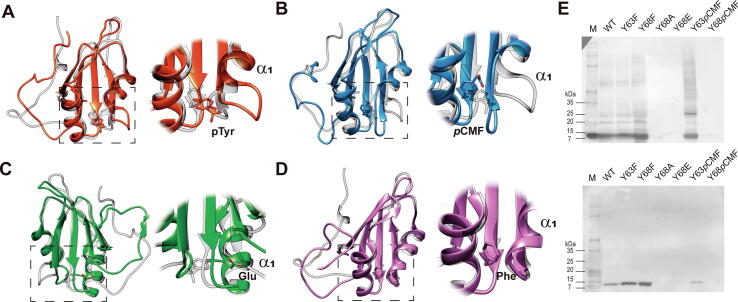
We also applied the workflow detailed above to study HuR1-101 phosphorylation. Perturbation analysis of the MD simulation of phosphorylated Tyr68 (pY68) HuR1-101 revealed that phosphorylation at this position induces conformational changes at the α1 helix (residues 33–47), thereby increasing Tyr68 solvent accessibility (Fig. 4A, Fig. S3-4). SAS of position 68 in phosphorylated HuR1-101 (102,7 ± 28.2 Å2) was higher than WT HuR1-101 (−17.3 ± 3.3 Å2). Analogous MD simulations were performed to evaluate the behavior of phosphomimetic variants (Fig. S3-4). Substitution of Tyr68 by pCMF slightly destabilizes the α1 helix, as does Tyr68 phosphorylation (Fig. 4B, Fig. S3). RMSF values for this stretch are comparable between pY68 and Y68pCMF HuR1-101 (Fig. S4). However, the solvent accessibility does not increase (−4.7 ± 8.5 Å2), displaying similar values as the WT protein (Fig. S4). On the other hand, Tyr68-by-Glu68 substitution does increase the solvent accessibility of the Glu residue (35.0 ± 13.0 Å2) without affecting the secondary structure of HuR1-101 nor inducing significant conformational changes (Fig. S4). Interestingly, the Glu side-chain flips with respect to its initial conformation, popping out of the hydrophobic pocket (Fig. 4C). MD simulations of the Y68F mutant, in which Tyr68 is replaced with Phe, was carried out as a control to check that this substitution does not alter HuR1-101 structure (Fig. 4D, Fig. S3).
Fig. 4.
HuR conformational changes induced by Tyr68 phosphorylation. A, D) Superimposition of ribbon representation of Tyr68 phosphorylated (red) or mutated by Phe (purple) and non-phosphorylated (light gray) HuR1-101 structures after 500 ns of MD simulation. B, C) Overlay of the structures mutated in Tyr68 by unnatural amino acid pCMF (blue) or by canonical Glu (green) amino acids with respect to the phosphorylated Tyr68 (light gray) HuR1-101 after 500 ns of MD simulation. E) Western blots using anti-HuR (upper panel) and anti-6xHis-tag (lower panel) antibodies from E. coli cell extracts previously transformed with plasmids encoding different HuR1-101 mutants at positions 63 and 68. Molecular mass markers are also depicted (lane M). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Based on the findings of this data, we decided to study Tyr68 phosphorylation using ncAA substitution, as it better mirrors the generated perturbation. Nonetheless, HuR1-101 Y68pCMF, like Y68A and Y68E mutants, could not be expressed in vitro. In fact, the only mutant at position 68 that was successfully expressed was Y68F HuR1-101 recombinant variant (Fig. 4E). Evolutionary conservation analysis performed with ConSurf web server [159] revealed that the most conserved residue at position 68 is Tyr (88%) followed by Phe (12%). Intriguingly, ncAA HuR1-101 mutant at position 63 (Y63pCMF) and the variant Y63F were all well expressed (Fig. 4E). A plausible explanation is that Tyr68 is located at the hydrophobic core of HuR1-101, and as an aromatic amino acid key in protein folding may further stabilize the protein. To test this hypothesis, we performed several MD simulations of WT, Y68F and Y68A HuR1-101 variants at two different temperatures (400 and 500 K) to analyze their respective stability (Fig. S5). Trajectory analyses indicated that the most stable mutant at position 68 is Y68F—in fact, the only one can be successfully expressed in vitro—and that Y68F correlation between native contacts and radius of gyration throughout the whole simulation is similar to that of WT HuR1-101. Altogether, these findings suggest a crucial role for the aromatic ring (provided by either Tyr or Phe) at position 68 for correct HuR1-101 folding.
A recent NMR analysis of HuR1-101 binding to an RNA oligonucleotide derived from adenine and uracil-rich element (ARE) I of the cyclooxygenase 2 mRNA revealed that the RNA binding site comprises Tyr63 among other residues in the β 1 and β 3 strands [114]. Moreover, Tyr63 has been found phosphorylated in Hodgkin’s lymphoma cell lines [160]. Further analysis performed by Yoon et al. [120] detected Y63 phosphorylation after in vitro kinase assays with recombinant purified JAK3 kinase and HuR. As the Y63pCMF mutant is recombinantly expressed, we wanted to explore how effectively it mirrors phosphorylation at this position. For Tyr63, similar MD simulations on phosphorylated (pY63) and phosphomimetic variants (Y63E and Y63pCMF) of HuR1-101 were carried out (Figs. S6–7). Tyr63 is located at the surface of the protein, where it contacts the unstructured N-terminal tail. Perturbation analysis of the Y63E HuR1-101 mutant revealed that substitution by Glu undermines the contacts between the N-terminal tail and the protein core. This mutant displayed higher values of RG, which is consistent with partial unfolding of the N-terminal (Fig. S6). This perturbation is not induced by phosphorylation nor substitution by ncAA pCMF (Fig. 5). Moreover, RMSF values of the area surrounding position 63 are higher than those for WT, pY63 and Y63pCMF HuR1-101. Analysis of the Y63E mutant could thus led to an overestimation of the effects of phosphorylation. For this reason, the functional implications of Tyr63 phosphorylation will be assessed using the Y63pCMF HuR1-101 mutant in future studies.
Fig. 5.
HuR Tyr63 phosphorylation is mimicked better by pCMF than Glu substitution. A) Superimposition of ribbon representation of Tyr63 phosphorylated (red) and non-phosphorylated (light gray) HuR1-101 structures after 500 ns of MD simulation. B-C) Overlay of the structures mutated in Tyr63 by the unnatural amino acid pCMF (blue) or by canonical amino acid Glu (green) with respect to the phosphorylated Tyr68 (light gray) HuR1-101 after 500 ns of MD simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4. Concluding remarks
The structural and functional analysis of phosphorylated proteins can be achieved by several approaches. The purpose of the herein presented workflow is to help researchers choose the most suitable alternative when an extensive study of the functional effects of phosphorylation is unfeasible with phosphorylated species purified from cells and/or tissues. MD simulations have become more accessible due to the development of user-friendly software packages and the increase in computing power at a modest cost. However, computational approaches are often ignored by molecular biologists. For this reason, we believe that a combination of computational and biochemical approaches will help to better understand how phosphorylation events affect protein structure and function.
CRediT authorship contribution statement
Gonzalo Pérez-Mejías: Conceptualization, Methodology, Software, Visualization, Investigation, Writing - original draft, Writing - review & editing. Alejandro Velázquez-Cruz: Writing - original draft, Writing - review & editing. Alejandra Guerra-Castellano: Investigation, Writing - original draft, Writing - review & editing. Blanca Baños-Jaime: Resources. Antonio Díaz-Quintana: Software, Writing - review & editing. Katiuska González-Arzola: Writing - review & editing. Miguel Ángel De Rosa: Supervision, Writing - review & editing, Project administration, Funding acquisition. Irene Díaz-Moreno: Conceptualization, Supervision, Writing - review & editing, Project administration, Funding acquisition.
Acknowledgments
Acknowledgements
This work was supported by the Ministry of Science and Innovation (PGC2018-096049-B-I00), European Regional Development Fund (FEDER), Andalusian Government (BIO-198, US-1254317 and US-1257019) and TA Instruments. G.P.-M. and A.V.-C. were awarded a PhD fellowship from the Spanish Ministry of Education, Culture and Sport (FPU17/04604 and FPU16/01513). Molecular graphics and analyses were performed with the UCSF Chimera software, developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Conflict of interest
All authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.06.043.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Duan G., Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanderson T.H., Mahapatra G., Pecina P. Cytochrome c is tyrosine 97 phosphorylated by neuroprotective insulin treatment. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0078627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X., Wang X., Wang Z. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–212. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardito F., Giuliani M., Perrone D., Troiano G., Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review) Int J Mol Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc B Biol Sci. 2012;367:2513–2516. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukami Y., Lipmann F. Reversal of Rous sarcoma-specific immunoglobulin phosphorylation on tyrosine (ADP as phosphate acceptor) catalyzed by the src gene kinase. Proc Natl Acad Sci U S A. 1983;80:1872–1876. doi: 10.1073/pnas.80.7.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kole H.K., Abdel-Ghany M., Racker E. Specific dephosphorylation of phosphoproteins by protein-serine and -tyrosine kinases. Proc Natl Acad Sci U S A. 1988;85:5849–5853. doi: 10.1073/pnas.85.16.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornbeck P.V., Chabra I., Kornhauser J.M., Skrzypek E., Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 11.Puttick J., Baker E.N., Delbaere L.T.J. Histidine phosphorylation in biological systems. Biochim Biophys Acta, Proteins Proteomics. 2008;1784:100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Cieśla J., Frączyk T., Rode W. Phosphorylation of basic amino acid residues in proteins: important but easily missed. Acta Biochim Pol. 2011;58:137–148. [PubMed] [Google Scholar]
- 13.Iakoucheva L.M., Radivojac P., Brown C.J. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrea D.M., Grace C.R., Buljan M. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc Natl Acad Sci U S A. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Moreno I., Hollingworth D., Kelly G. Orientation of the central domains of KSRP and its implications for the interaction with the RNA targets. Nucleic Acids Res. 2010;38:5193–5205. doi: 10.1093/nar/gkq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Díaz-Moreno I., Hollingworth D., Frenkiel T.A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat Struct Mol Biol. 2009;16:238–246. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bah A., Vernon R.M., Siddiqui Z. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt S.R., Schweikart F., Andersson M.E. Current methods for phosphoprotein isolation and enrichment. J Chromatogr, B: Anal Technol Biomed Life Sci. 2007;849:154–162. doi: 10.1016/j.jchromb.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhu K., Zhao J., Lubman D.M., Miller F.R., Barder T.J. Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal Chem. 2005;77:2745–2755. doi: 10.1021/ac048494w. [DOI] [PubMed] [Google Scholar]
- 20.Halligan B.D. ProMoST: a tool for calculating the pI and molecular mass of phosphorylated and modified proteins on two-dimensional gels. Methods Mol Biol. 2009;527(283–98):ix. doi: 10.1007/978-1-60327-834-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjerrum E.J., Jensen J.H., Tolborg J.L. pICalculax: improved prediction of isoelectric point for modified peptides. J Chem Inf Model. 2017;57:1723–1727. doi: 10.1021/acs.jcim.7b00030. [DOI] [PubMed] [Google Scholar]
- 22.Block H., Maertens B., Spriestersbach A. Immobilized-metal affinity chromatography (IMAC): a review. Methods Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- 23.Ruprecht B., Koch H., Domasinska P. Optimized enrichment ofpPhosphoproteomes by Fe-IMAC column chromatography. Methods Mol Biol. 2017;1550:47–60. doi: 10.1007/978-1-4939-6747-6_5. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Horinouchi T., Terada K., Higashi T., Miwa S. Using Phos-Tag in Western Blotting analysis to evaluate protein phosphorylation. Methods Mol Biol. 2016;1397:267–277. doi: 10.1007/978-1-4939-3353-2_18. [DOI] [PubMed] [Google Scholar]
- 26.Kumar G. A simple method for detecting phosphorylation of proteins by using Zn(2+)-Phos-Tag SDS-PAGE at neutral pH. Methods Mol Biol. 2018;1853:223–229. doi: 10.1007/978-1-4939-8745-0_25. [DOI] [PubMed] [Google Scholar]
- 27.Nagy Z., Comer S., Smolenski A. Analysis of protein phosphorylation using Phos-Tag gels. Curr Protoc Protein Sci. 2018;93 doi: 10.1002/cpps.64. [DOI] [PubMed] [Google Scholar]
- 28.Yang C., Zhong X., Li L. Recent advances in enrichment and separation strategies for mass spectrometry-based phosphoproteomics. Electrophoresis. 2014;35:3418–3429. doi: 10.1002/elps.201400017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner A. Enrichment strategies in phosphoproteomics. Methods Mol Biol. 2016;1355:105–121. doi: 10.1007/978-1-4939-3049-4_7. [DOI] [PubMed] [Google Scholar]
- 30.Simon E.S., Young M., Chan A., Bao Z.-Q., Andrews P.C. Improved enrichment strategies for phosphorylated peptides on titanium dioxide using methyl esterification and pH gradient elution. Anal Biochem. 2008;377:234–242. doi: 10.1016/j.ab.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins M.O., Yu L., Choudhary J.S. Analysis of protein phosphorylation on a proteome-scale. Proteomics. 2007;7:2751–2768. doi: 10.1002/pmic.200700145. [DOI] [PubMed] [Google Scholar]
- 32.Paradela A., Albar J.P. Advances in the analysis of protein phosphorylation. J Proteome Res. 2008;7:1809–1818. doi: 10.1021/pr7006544. [DOI] [PubMed] [Google Scholar]
- 33.Francavilla C., Hekmat O., Blagoev B., Olsen J.V. SILAC-based temporal phosphoproteomics. Methods Mol Biol. 2014;1188:125–148. doi: 10.1007/978-1-4939-1142-4_10. [DOI] [PubMed] [Google Scholar]
- 34.Wang M.-C., Lee Y.-H., Liao P.-C. Optimization of titanium dioxide and immunoaffinity-based enrichment procedures for tyrosine phosphopeptide using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Bioanal Chem. 2015;407:1343–1356. doi: 10.1007/s00216-014-8352-0. [DOI] [PubMed] [Google Scholar]
- 35.Possemato A.P., Paulo J.A., Mulhern D. Multiplexed phosphoproteomic profiling using titanium dioxide and immunoaffinity enrichments reveals complementary phosphorylation events. J Proteome Res. 2017;16:1506–1514. doi: 10.1021/acs.jproteome.6b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buhs S., Gerull H., Nollau P. Identification of tyrosine phosphorylated proteins by SH2 domain affinity purification and mass spectrometry. Methods Mol Biol. 2017;1555:407–418. doi: 10.1007/978-1-4939-6762-9_23. [DOI] [PubMed] [Google Scholar]
- 37.Ke M., Chu B., Lin L., Tian R. SH2 Domains as affinity reagents for phosphotyrosine protein enrichment and proteomic analysis. Methods Mol Biol. 2017;1555:395–406. doi: 10.1007/978-1-4939-6762-9_22. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko T., Huang H., Cao X. Superbinder SH2 domains act as antagonists of cell signaling. Sci Signal. 2012;5:ra68. doi: 10.1126/scisignal.2003021. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey S.J., Azimifar S.B., Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 40.Casado P., Hijazi M., Britton D., Cutillas P.R. Impact of phosphoproteomics in the translation of kinase-targeted therapies. Proteomics. 2017;17 doi: 10.1002/pmic.201600235. [DOI] [PubMed] [Google Scholar]
- 41.Yang W., Freeman M.R., Kyprianou N. Personalization of prostate cancer therapy through phosphoproteomics. Nat Rev Urol. 2018;15:483–497. doi: 10.1038/s41585-018-0014-0. [DOI] [PubMed] [Google Scholar]
- 42.Minic Z., Dahms T.E.S., Babu M. Chromatographic separation strategies for precision mass spectrometry to study protein-protein interactions and protein phosphorylation. J Chromatogr, B: Anal Technol Biomed Life Sci. 2018;1102–1103:96–108. doi: 10.1016/j.jchromb.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 43.Sommerfeld M.R., Metzger S., Stosik M., Tennagels N., Eckel J. In vitro phosphorylation of insulin receptor substrate 1 by protein kinase C-zeta: functional analysis and identification of novel phosphorylation sites. Biochemistry. 2004;43:5888–5901. doi: 10.1021/bi049640v. [DOI] [PubMed] [Google Scholar]
- 44.Baudin F., Müller C.W. Bacterial expression, purification, and crystallization of tyrosine phosphorylated STAT proteins. Methods Mol Biol. 2013;967:301–317. doi: 10.1007/978-1-62703-242-1_21. [DOI] [PubMed] [Google Scholar]
- 45.Shin S.-B., Woo S.-U., Lee Y.-J., Yim H. Comparative analysis of a FRET-based PLK1 kinase assay to identify PLK1 inhibitors for chemotherapy. Anticancer Res. 2017;37:1177–1183. doi: 10.21873/anticanres.11431. [DOI] [PubMed] [Google Scholar]
- 46.Duchemin A.-M., Neff N.H., Hadjiconstantinou M. Aromatic L-amino acid decarboxylase phosphorylation and activation by PKGIalpha in vitro. J Neurochem. 2010;114:542–552. doi: 10.1111/j.1471-4159.2010.06784.x. [DOI] [PubMed] [Google Scholar]
- 47.Babon J.J., Murphy J.M. In vitro JAK kinase activity and inhibition assays. Methods Mol Biol. 2013;967:39–55. doi: 10.1007/978-1-62703-242-1_3. [DOI] [PubMed] [Google Scholar]
- 48.Loeffler D.A., Smith L.M., Klaver A.C., Martić S. Effects of antibodies to phosphorylated and non-phosphorylated tau on in vitro tau phosphorylation at Serine-199: Preliminary report. Exp Gerontol. 2015;67:15–18. doi: 10.1016/j.exger.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Thorsness P.E., Koshland D.E.J. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- 50.Pearlman S.M., Serber Z., Ferrell J.E.J. A mechanism for the evolution of phosphorylation sites. Cell. 2011;147:934–946. doi: 10.1016/j.cell.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W., Erikson R.L. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc Natl Acad Sci U S A. 1994;91:8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng L., Dominski Z., Yang X.-C. Phosphorylation of Stem-Loop Binding Protein (SLBP) on Two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strickfaden S.C., Winters M.J., Ben-Ari G. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dissmeyer N., Schnittger A. Use of phospho-site substitutions to analyze the biological relevance of phosphorylation events in regulatory networks. Methods Mol Biol. 2011;779:93–138. doi: 10.1007/978-1-61779-264-9_6. [DOI] [PubMed] [Google Scholar]
- 55.Stateva S.R., Salas V., Benaim G. Characterization of phospho-(tyrosine)-mimetic calmodulin mutants. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0120798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zisch A.H., Pazzagli C., Freeman A.L. Replacing two conserved tyrosines of the EphB2 receptor with glutamic acid prevents binding of SH2 domains without abrogating kinase activity and biological responses. Oncogene. 2000;19:177–187. doi: 10.1038/sj.onc.1203304. [DOI] [PubMed] [Google Scholar]
- 57.Jonson P.H., Petersen S.B. A critical view on conservative mutations. Protein Eng. 2001;14:397–402. doi: 10.1093/protein/14.6.397. [DOI] [PubMed] [Google Scholar]
- 58.Fang K.Y., Lieblich S.A., Tirrell D.A. Incorporation of non-canonical amino acids into proteins by global reassignment of sense codons. Methods Mol Biol. 2018;1798:173–186. doi: 10.1007/978-1-4939-7893-9_13. [DOI] [PubMed] [Google Scholar]
- 59.Terasaka N., Iwane Y., Geiermann A.-S., Goto Y., Suga H. Recent developments of engineered translational machineries for the incorporation of non-canonical amino acids into polypeptides. Int J Mol Sci. 2015;16:6513–6531. doi: 10.3390/ijms16036513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young T.S., Ahmad I., Yin J.A., Schultz P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 61.Chatterjee A., Sun S.B., Furman J.L., Xiao H., Schultz P.G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis inEscherichia coli. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leisle L., Valiyaveetil F., Mehl R.A., Ahern C.A. Incorporation of Non-Canonical Amino Acids. Adv Exp Med Biol. 2015;869:119–151. doi: 10.1007/978-1-4939-2845-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwark D.G., Schmitt M.A., Fisk J.D. Dissecting the contribution of release factor interactions to amber stop codon reassignment efficiencies of the methanocaldococcus jannaschii orthogonal pair. Genes (Basel) 2018;9:546. doi: 10.3390/genes9110546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nehring S., Budisa N., Wiltschi B. Performance analysis of orthogonal pairs designed for an expanded eukaryotic genetic code. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0031992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pott M., Schmidt M.J., Summerer D. Evolved sequence contexts for highly efficient amber suppression with noncanonical amino acids. ACS Chem Biol. 2014;9:2815–2822. doi: 10.1021/cb5006273. [DOI] [PubMed] [Google Scholar]
- 66.Guo J., Melançon C.E., 3rd, Lee H.S., Groff D., Schultz P.G. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew Chem Int Ed Engl. 2009;48:9148–9151. doi: 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohtsuki T., Yamamoto H., Doi Y., Sisido M. Use of EF-Tu mutants for determining and improving aminoacylation efficiency and for purifying aminoacyl tRNAs with non-natural amino acids. J Biochem. 2010;148:239–246. doi: 10.1093/jb/mvq053. [DOI] [PubMed] [Google Scholar]
- 68.Wang N., Ju T., Niu W., Guo J. Fine-tuning interaction between aminoacyl-tRNA synthetase and tRNA for efficient synthesis of proteins containing unnatural amino acids. ACS Synth Biol. 2015;4:207–212. doi: 10.1021/sb500195w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwok H.S., Vargas-Rodriguez O., Melnikov S.V., Söll D. Engineered aminoacyl-tRNA synthetases with improved selectivity toward noncanonical amino acids. ACS Chem Biol. 2019;14:603–612. doi: 10.1021/acschembio.9b00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang K., Neumann H., Peak-Chew S.Y., Chin J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 71.Johnson D.B.F., Xu J., Shen Z. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu P., Gafken P.R., Mehl R.A., Cooley R.B. A highly versatile expression system for the production of multiply phosphorylated proteins. ACS Chem Biol. 2019;14:1564–1572. doi: 10.1021/acschembio.9b00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rothman D.M., Petersson E.J., Vázquez M.E. Caged phosphoproteins. J Am Chem Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- 74.Steinfeld J.B., Aerni H.R., Rogulina S., Liu Y., Rinehart J. Expanded cellular amino acid pools containing phosphoserine, phosphothreonine, and phosphotyrosine. ACS Chem Biol. 2014;9:1104–1112. doi: 10.1021/cb5000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smolskaya S., Andreev Y.A. Site-specific incorporation of unnatural amino acids into Escherichia coli recombinant protein: methodology development and recent achievement. Biomolecules. 2019:9. doi: 10.3390/biom9070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ko W., Kumar R., Kim S., Lee H.S. Construction of bacterial cells with an active transport system for unnatural amino acids. ACS Synth Biol. 2019;8:1195–1203. doi: 10.1021/acssynbio.9b00076. [DOI] [PubMed] [Google Scholar]
- 77.Luo X., Fu G., Wang R.E. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat Chem Biol. 2017;13:845–849. doi: 10.1038/nchembio.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Venkat S., Chen H., Gan Q., Fan C. The application of cell-free protein synthesis in genetic code expansion for post-translational modifications. Front Pharmacol. 2019;10:248. doi: 10.3389/fphar.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin X., Park O.-J., Hong S.H. Incorporation of non-standard amino acids into proteins: challenges, recent achievements, and emerging applications. Appl Microbiol Biotechnol. 2019;103:2947–2958. doi: 10.1007/s00253-019-09690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oza J.P., Aerni H.R., Pirman N.L. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat Commun. 2015;6:8168. doi: 10.1038/ncomms9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park H.S., Hohn M.J., Umehara T. Expanding the genetic code of Escherichia coli with phosphoserine. Science (80-) 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wakamori M., Fujii Y., Suka N. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep. 2015;5:17204. doi: 10.1038/srep17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yanagisawa T., Takahashi M., Mukai T. Multiple site-specific installations of Nε-monomethyl-L- lysine into histone proteins by cell-based and cell-free protein synthesis. ChemBioChem. 2014;15:1830–1838. doi: 10.1002/cbic.201402291. [DOI] [PubMed] [Google Scholar]
- 84.Chemla Y., Ozer E., Shaferman M. Simplified methodology for a modular and genetically expanded protein synthesis in cell-free systems. Synth Syst Biotechnol. 2019;4:189–196. doi: 10.1016/j.synbio.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawahata N., Yang M.G., Luke G.P. A novel phosphotyrosine mimetic 4’-carboxymethyloxy-3’-phosphonophenylalanine (Cpp): exploitation in the design of nonpeptide inhibitors of pp60(Src) SH2 domain. Bioorg Med Chem Lett. 2001;11:2319–2323. doi: 10.1016/s0960-894x(01)00446-2. [DOI] [PubMed] [Google Scholar]
- 86.Erlanson D.A., McDowell R.S., He M.M. Discovery of a new phosphotyrosine mimetic for PTP1B using breakaway tethering. J Am Chem Soc. 2003;125:5602–5603. doi: 10.1021/ja034440c. [DOI] [PubMed] [Google Scholar]
- 87.Liu C.C., Schultz P.G. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 88.Xie J., Supekova L., Schultz P.G. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol. 2007;2:474–478. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 89.Serwa R., Wilkening I., Del Signore G. Chemoselective Staudinger-phosphite reaction of azides for the phosphorylation of proteins. Angew Chem Int Ed Engl. 2009;48:8234–8239. doi: 10.1002/anie.200902118. [DOI] [PubMed] [Google Scholar]
- 90.Fan C., Ip K., Söll D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 2016;590:3040–3047. doi: 10.1002/1873-3468.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoppmann C., Wong A., Yang B. Site-specific incorporation of phosphotyrosine using an expanded genetic code. Nat Chem Biol. 2017;13:842–844. doi: 10.1038/nchembio.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang M.S., Brunner S.F., Huguenin-Dezot N. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat Methods. 2017;14:729–736. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hollingsworth S.A., Dror R.O. Molecular Dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cordero-Morales J.F., Jogini V., Lewis A. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14:1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 95.Fields J.B., Németh-Cahalan K.L., Freites J.A. Calmodulin gates Aquaporin 0 permeability through a positively charged cytoplasmic loop. J Biol Chem. 2017;292:185–195. doi: 10.1074/jbc.M116.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martínez-Fábregas J., Díaz-Moreno I., González-Arzola K. New Arabidopsis thaliana cytochrome c partners: a look into the elusive role of cytochrome c in programmed cell death in plants. Mol Cell Proteomics. 2013;12:3666–3676. doi: 10.1074/mcp.M113.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martínez-Fábregas J., Díaz-Moreno I., González-Arzola K. Structural and functional analysis of novel human cytochrome c targets in apoptosis. Mol Cell Proteomics. 2014;13 doi: 10.1074/mcp.M113.034322. 1439 LP – 1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martínez-Fábregas J., Díaz-Moreno I., González-Arzola K., Díaz-Quintana A., De la Rosa M.A. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.González-Arzola K., Díaz-Moreno I., Cano-González A. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc Natl Acad Sci U S A. 2015;112:9908–9913. doi: 10.1073/pnas.1508040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.González-Arzola K., Díaz-Quintana A., Rivero-Rodríguez F. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 2017;45:2150–2165. doi: 10.1093/nar/gkw1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.González-Arzola K., Velázquez-Cruz A., Guerra-Castellano A. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett. 2019;593:3101–3119. doi: 10.1002/1873-3468.13655. [DOI] [PubMed] [Google Scholar]
- 102.Díaz-Moreno I., Velázquez-Cruz A., Curran-French S., Díaz-Quintana A., De la Rosa M.A. Nuclear cytochrome c - a mitochondrial visitor regulating damaged chromatin dynamics. FEBS Lett. 2018;592:172–178. doi: 10.1002/1873-3468.12959. [DOI] [PubMed] [Google Scholar]
- 103.Desagher S., Martinou J.C. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 104.Jiang X., Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 105.Elena-Real C.A., Díaz-Quintana A., González-Arzola K. Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition. Cell Death Dis. 2018;9:365. doi: 10.1038/s41419-018-0408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalpage H.A., Bazylianska V., Recanati M.A. Tissue-specific regulation of cytochrome c by post-translational modifications: respiration, the mitochondrial membrane potential, ROS, and apoptosis. FASEB J. 2019;33:1540–1553. doi: 10.1096/fj.201801417R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Díaz-Moreno I., García-Heredia J.M., Díaz-Quintana A., Teixeira M., De La Rosa M.A. Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim Biophys Acta - Bioenerg. 2011;1807:1616–1623. doi: 10.1016/j.bbabio.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 108.Ly H.K., Utesch T., Díaz-Moreno I. Perturbation of the redox site structure of cytochrome c variants upon tyrosine nitration. J Phys Chem B. 2012;116:5694–5702. doi: 10.1021/jp302301m. [DOI] [PubMed] [Google Scholar]
- 109.Lee I., Salomon A.R., Yu K. New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry. 2006;45:9121–9128. doi: 10.1021/bi060585v. [DOI] [PubMed] [Google Scholar]
- 110.Yu H., Lee I., Salomon A.R., Yu K., Hüttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta - Bioenerg. 2008;1777:1066–1071. doi: 10.1016/j.bbabio.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abriata L.A., Cassina A., Tórtora V. Nitration of solavent-exposed tyrosine 74 on cytochrome c triggers heme iron-methionine 80 bond disruption nuclear magnetic resonance and optical spectroscopy studies. J Biol Chem. 2009;284:17–26. doi: 10.1074/jbc.M807203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao X., León I.R., Bak S. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics. 2011;10(M110) doi: 10.1074/mcp.M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wan J., Kalpage H.A., Vaishnav A. Regulation of respiration and apoptosis by cytochrome c Threonine 58 phosphorylation. Sci Rep. 2019;9:15815. doi: 10.1038/s41598-019-52101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moreno-Beltrán B., Guerra-Castellano A., Díaz-Quintana A. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc Natl Acad Sci U S A. 2017;114:E3041–E3050. doi: 10.1073/pnas.1618008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pieper D., Schirmer S., Prechtel A.T. Functional characterization of the HuR:CD83 mRNA interaction. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pabis M., Popowicz G.M., Stehle R. HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res. 2019;47:1011–1029. doi: 10.1093/nar/gky1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Díaz-Quintana A., García-Mauriño S.M., Díaz-Moreno I. Dimerization model of the C-terminal RNA recognition motif of HuR. FEBS Lett. 2015;589:1059–1066. doi: 10.1016/j.febslet.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 118.Lixa C., Mujo A., de Magalhães M.T.Q. Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution. J Biomol NMR. 2018;72:179–192. doi: 10.1007/s10858-018-0217-y. [DOI] [PubMed] [Google Scholar]
- 119.Grammatikakis I., Abdelmohsen K., Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoon J.-H., Abdelmohsen K., Srikantan S. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res. 2014;42:1196–1208. doi: 10.1093/nar/gkt903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu B.A., Nash P.D. Evolution of SH2 domains and phosphotyrosine signalling networks. Philos Trans R Soc B Biol Sci. 2012;367:2556–2573. doi: 10.1098/rstb.2012.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hornbeck P.V., Zhang B., Murray B. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dinkel H., Chica C., Via A. Phospho.ELM: A database of phosphorylation sites-update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Linding R., Jensen L.J., Pasculescu A. NetworKIN: A resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 126.Miller M.L., Soufi B., Jers C. NetPhosBac - A predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics. 2009;9:116–125. doi: 10.1002/pmic.200800285. [DOI] [PubMed] [Google Scholar]
- 127.Ingrell C.R., Miller M.L., Jensen O.N., Blom N. NetPhosYeast: Prediction of protein phosphorylation sites in yeast. Bioinformatics. 2007;23:895–897. doi: 10.1093/bioinformatics/btm020. [DOI] [PubMed] [Google Scholar]
- 128.Fenoy E., Izarzugaza J.M.G., Jurtz V., Brunak S., Nielsen M. A generic deep convolutional neural network framework for prediction of receptor-ligand interactions-NetPhosPan: application to kinase phosphorylation prediction. Bioinformatics. 2019;35:1098–1107. doi: 10.1093/bioinformatics/bty715. [DOI] [PubMed] [Google Scholar]
- 129.Xue Y., Ren J., Gao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang H.D.A., Lee T.Y.I., Tzeng S.W. Incorporating hidden Markov models for identifying protein kinase-specific phosphorylation sites. J Comput Chem. 2005;26:1032–1041. doi: 10.1002/jcc.20235. [DOI] [PubMed] [Google Scholar]
- 132.Saunders N.F.W., Brinkworth R.I., Huber T., Kemp B.E., Kobe B. Predikin and PredikinDB: A computational framework for the prediction of protein kinase peptide specificity and an associated database of phosphorylation sites. BMC Bioinf. 2008;9:245. doi: 10.1186/1471-2105-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gnad F., Ren S., Cox J. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wong Y.H., Lee T.Y., Liang H.K. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–W594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Neuberger G., Schneider G., Eisenhaber F. pkaPS: Prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol Direct. 2007;2:1. doi: 10.1186/1745-6150-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Phillips J.C., Braun R., Wang W. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brooks B.R., Brooks C.L., 3rd, Mackerell A.D.J. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abraham M.J., Murtola T., Schulz R. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 139.Case DA, Belfon K, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham III TE, et al. DMY and PAK. AMBER 2020 n.d.
- 140.Eastman P., Swails J., Chodera J.D. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maier J.A., Martinez C., Kasavajhala K. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Imai M., Saio T., Kumeta H. Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c. Biochem Biophys Res Commun. 2016;469:978–984. doi: 10.1016/j.bbrc.2015.12.079. [DOI] [PubMed] [Google Scholar]
- 143.Homeyer N., Horn A.H.C., Lanig H., Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12:281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 144.Guerra-Castellano A., Díaz-Quintana A., Moreno-Beltrán B. Mimicking tyrosine phosphorylation in human cytochrome c by the evolved tRNA synthetase technique. Chem - A Eur J. 2015;21:15004–15012. doi: 10.1002/chem.201502019. [DOI] [PubMed] [Google Scholar]
- 145.Autenrieth F., Tajkhorshid E., Baudry J., Luthey-Schulten Z. Classical force field parameters for the heme prosthetic group of cytochrome c. J Comput Chem. 2004;25:1613–1622. doi: 10.1002/jcc.20079. [DOI] [PubMed] [Google Scholar]
- 146.Izadi S., Anandakrishnan R., Onufriev A.V. Building water models: A different approach. J Phys Chem Lett. 2014;5:3863–3871. doi: 10.1021/jz501780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McGibbon R.T., Beauchamp K.A., Harrigan M.P. MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roe D.R., Cheatham T.E., 3rd. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 149.Guerra-Castellano A., Díaz-Quintana A., Pérez-Mejías G. Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc Natl Acad Sci U S A. 2018;115:7955–7960. doi: 10.1073/pnas.1806833115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Waterhouse A., Bertoni M., Bienert S. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 152.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.O’Rourke K.F., Gorman S.D., Boehr D.D. Biophysical and computational methods to analyze amino acid interaction networks in proteins. Comput Struct Biotechnol J. 2016;14:245–251. doi: 10.1016/j.csbj.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pecina P., Borisenko G.G., Belikova N.A. Phosphomimetic substitution of cytochrome c tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry. 2010;49:6705–6714. doi: 10.1021/bi100486s. [DOI] [PubMed] [Google Scholar]
- 155.Mahapatra G., Varughese A., Ji Q. Phosphorylation of cytochrome c threonine 28 regulates electron transport chain activity in kidney: Implications for amp kinase. J Biol Chem. 2017;292:64–79. doi: 10.1074/jbc.M116.744664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guerra-Castellano A., Díaz-Moreno I., Velázquez-Campoy A. Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47. Biochim Biophys Acta - Bioenerg. 2016;1857:387–395. doi: 10.1016/j.bbabio.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 157.Díaz-Quintana A., Pérez-Mejías G., Guerra-Castellano A., De la Rosa M.A., Díaz-Moreno I. Wheel and deal in the mitochondrial inner membranes: the tale of cytochrome c and cardiolipin. Oxid Med Cell Longev. 2020;2020:6813405. doi: 10.1155/2020/6813405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deacon O.M., Karsisiotis A.I., Moreno-Chicano T. Heightened dynamics of the oxidized Y48H variant of human cytochrome c increases its peroxidatic activity. Biochemistry. 2017;56:6111–6124. doi: 10.1021/acs.biochem.7b00890. [DOI] [PubMed] [Google Scholar]
- 159.Ashkenazy H., Abadi S., Martz E. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gu T.-L., Cherry J., Tucker M. Identification of activated Tnk1 kinase in Hodgkin’s lymphoma. Leukemia. 2010;24:861–865. doi: 10.1038/leu.2009.293. [DOI] [PubMed] [Google Scholar]
- 161.Rogerson D.T., Sachdeva A., Wang K. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.García-Heredia J.M., Díaz-Moreno I., Díaz-Quintana A. Specific nitration of tyrosines 46 and 48 makes cytochrome c assemble a non-functional apoptosome. FEBS Lett. 2012;586:154–158. doi: 10.1016/j.febslet.2011.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.