Abstract
Abstract
Purpose
We aimed to investigate the diagnostic performance of chest CT compared with first RT-PCR results in adult patients suspected of COVID-19 infection in an ED setting. We also constructed a predictive machine learning model based on chest CT and additional data to improve the diagnostic accuracy of chest CT.
Methods
This study’s cohort consisted of 319 patients who underwent chest CT and RT-PCR testing at the ED. Patient characteristics, demographics, symptoms, vital signs, laboratory tests, and chest CT results (CO-RADS) were collected. With first RT-PCR as reference standard, the diagnostic performance of chest CT using the CO-RADS score was assessed. Additionally, a predictive machine learning model was constructed using logistic regression.
Results
Chest CT, with first RT-PCR as a reference, had a sensitivity, specificity, PPV, and NPV of 90.2%, 88.2%, 84.5%, and 92.7%, respectively. The prediction model with CO-RADS, ferritin, leucocyte count, CK, days of complaints, and diarrhea as predictors had a sensitivity, specificity, PPV, and NPV of 89.3%, 93.4%, 90.8%, and 92.3%, respectively.
Conclusion
Chest CT, using the CO-RADS scoring system, is a sensitive and specific method that can aid in the diagnosis of COVID-19, especially if RT-PCR tests are scarce during an outbreak. Combining a predictive machine learning model could further improve the accuracy of diagnostic chest CT for COVID-19. Further candidate predictors should be analyzed to improve our model. However, RT-PCR should remain the primary standard of testing as up to 9% of RT-PCR positive patients are not diagnosed by chest CT or our machine learning model.
Electronic supplementary material
The online version of this article (10.1007/s10140-020-01821-1) contains supplementary material, which is available to authorized users.
Keywords: Emergency Department, COVID-19, Real-time reverse transcription polymerase chain reaction (RT-PCR), Chest computed tomography, CO-RADS classification, Prediction model, Machine learning
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus which causes coronavirus disease 2019 (COVID-19), was first reported in Wuhan, China, in December 2019 [1]. Since then, the virus spread around the world and on February 27, the first patient with COVID-19 was identified in the Netherlands. As of the 1 May, 39,791 Dutch patients tested positive for COVID-19, 10,854 patients were admitted to hospitals, and 4893 patients died due to SARS-CoV-2 [2]. Although the Dutch government enforced measurements to enter a mitigation phase, to prevent a peak of demand which could exceed the capacity of hospital treatment, increasing numbers of patients visited the emergency department (ED).
Patients suspected of COVID-19 are recognized and triaged mainly based on presenting clinical characteristics and symptoms [1, 3]. The definitive diagnosis is performed by real-time reverse transcription polymerase chain reaction (RT-PCR) on samples obtained from oropharyngeal and nasopharyngeal swabs [4]. Recent studies reported that the gold standard, RT-PCR has a sensitivity of 59–71% when compared with repeated testing with the same RT-PCR method [5, 6]. This means that if the clinical suspicion is high, repeated testing is necessary. RT-PCR is thereby a time-consuming procedure and resources are or can become scarce during an outbreak.
Large-scale RT-PCR testing can be challenging, and during an outbreak peak, it can be unfeasible to wait hours for RT-PCR results to identify, isolate, and treat patients at an ED. Recent studies suggest that chest computed tomography (CT) could be leading in triage and aid in the early diagnosis of patients suspected of COVID-19 infection [7]. Recent studies from China reported high sensitivities (97–98%) for chest CT in the early detection of COVID-19 in patients who later had a positive RT-PCR test result [5, 6]. The National Health Commission of the People’s Republic of China even stated that diagnosis of COVID-19 could solely be based on chest CT findings [8]. Typical abnormalities seen on chest CT in COVID-19 patients [9–11] are also seen in patients with an initial false negative RT-PCR result [12].
Earlier attempts were also made to create a machine learning prediction model for COVID-19, based on chest X-ray and additional data in order to improve diagnostic accuracy of conventional chest X-ray alone [13]. For chest CT, this has not been done yet.
This study was initiated at the start of an outbreak peak in the Netherlands when RT-PCR testing capacity was considered to be a potential bottleneck in the flow of patients at an ED. The aim of this study was to investigate the diagnostic performance of chest CT compared with the first RT-PCR test in adult patients suspected of COVID-19 infection in the setting of the ED. In addition, we also constructed and internally validated a predictive machine learning model based on chest CT and additional data sources such as laboratory data for early prediction of COVID-19 infection. The ultimate goal was to assess whether chest CT could be used to substitute RT-PCR testing in triage during COVID-19 outbreaks where scarcity of RT-PCR tests would hinder efficient and rapid diagnosis, isolation, and treatment of patients.
Methods
Population and study design
This single-center prospective cohort study was performed at both locations of the Franciscus Gasthuis & Vlietland hospital in Rotterdam and Schiedam, the Netherlands, which has a level 2 trauma center with 48,000 visits annually at the ED’s. Waiver of informed consent was obtained by the medical ethical commission (MEC-U, W20.076).
Consecutive patients who visited the ED between March 27–April 20, 2020, and who met the following inclusion criteria for this study were included (a) age ≥ 18 years; (b) suspected infection with COVID-19 in combination with at least one of the following: (1) new respiratory symptoms persisting for ≤ 2 weeks and present during the last 24 h, (2) saturation ≤ 94% and/or respiration rate ≥ 20/min and/or abdominal complaints, and (3) a high clinical suspicion even in the absence of symptoms; and (c) RT-PCR and chest CT performed within 24 h after each other.
Exclusion criteria for this study were (a) previously confirmed COVID-19 infection; (b) instability defined as a peripheral oxygen saturation < 92% despite 5 l of oxygen and/or a systolic blood pressure < 90 mmHg; (c) principle presentation due to high energetic trauma, thrombolysis, or acute coronary syndrome; (d) pregnancy; and (e) non-interpretable first RT-PCR result.
Study procedures
COVID-19 suspected patients were triaged by a nurse in a triage tent especially set-up for the crisis as part of routine care. The Manchester Triage System [14] was used to triage patients; additional symptoms related to possible COVID-19 were assessed and vital signs were registered. Thereafter, arterial blood gas, blood samples, and nasopharyngeal swabs were obtained from all patients. Nasopharyngeal swabs were sampled from the oral cavity and subsequently from the nasal cavity using the same swab. After swabs were obtained, patients immediately underwent a chest CT, after which the physician took the medical history and performed a physical examination.
Chest CT and blood results followed within 60 min. Based on the clinical performance of the patient and the test results, a decision was made to admit the patient or not. RT-PCR results would mostly follow after 5–12 h. Due to scarce resources, repeated RT-PCR testing, preferably from sputum samples, was only performed in admitted patients in case of a persisting high suspicion for COVID-19 despite earlier negative RT-PCR results.
Patient data
Data of additional variables was extracted from electronic medical records of all patients included in the study. These variables encompassed demographic information, information about ED triage, COVID-19 presenting symptoms and vital signs, laboratory, microbiology, and CT results, and treatment-related variables (Online Resource 1).
Chest CT, RT-PCR, and laboratory assays
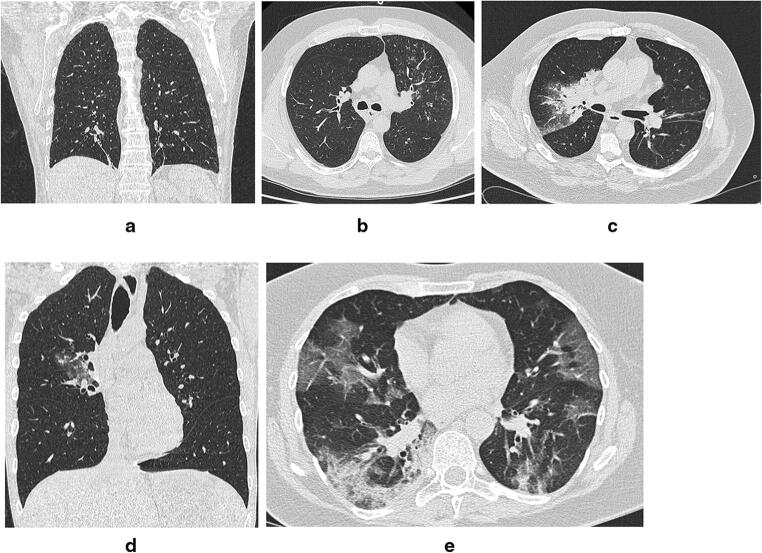
All chest CT images were obtained with patients in supine position in one of five CT scanners (Canon Aquilion One Genesis 320 slices, Canon Aquilion Prime 80 slices, Philips Brilliance 64 64 slices, Philips Big Bore 16 slices, Siemens Symbia T16 16 slices). Twenty board-certified radiologists were trained to read the CT images and classify them according to the CO-RADS classification, which was recently created by the Dutch Association of Radiologists (NVVR) [15]. Examples of CT images of all 5 CO-RADS categories are shown in Fig. 1. A CO-RADS score of 1–3 was classified as non-COVID-19,whereas a CO-RADS score of 4 or 5 was classified as COVID-19 positive (Online Resource 2). A standardized reporting format was developed. Two independent radiologists were consulted in case of any doubt about the classification.
Fig. 1.
a CO-RADS 1: A few fibrotic bands in the lower lobes. No evidence of infection. RT-PCR−. b CO-RADS 2: Bronchial wall thickening, small centrilobular nodules, and tree in bud abnormalities in the left upper lobe. Consistent with bronchiolitis. RT-PCR−. c CO-RADS 3: Consolidation with surrounding ground glass opacity in right upper lobe. RT-PCR−. d CO-RADS 4: Bilateral areas of patchy ground glass opacity with associated small peribronchovascular consolidations. Predominantly central distribution. RT-PCR+. e CO-RADS 5: Bilateral peripheral ground glass abnormalities with areas of associated consolidation. RT-PCR+
RT-PCR was performed according to the national reference method that was established after international collaboration [16], or by the CE-IVD kit GeneFinderTM COVID-19 Plus RealAmp Kit using the sample-to-result platform ELITe InGenius®.
Hematology tests were performed on Beckman Coulter DxH-800 and Sysmex XN-10 analyzers. Clinical chemistry tests were performed on Abbott Architect c-8000 and i-2000 platforms. Blood gas analyses were performed using Werfen GEM-4000 and Siemens RAPIDlab 1265 analyzers. Siemens CS-2100i and CS-5100 analyzers were used to determine fibrinogen concentrations.
Groups
With RT-PCR as reference, four groups were identified based on combining chest CT and RT-PCR outcomes, namely, true positives (TP) with a positive RT-PCR and a positive chest CT, false positives (FP), true negatives (TN), and false negatives (FN).
Statistical analysis
Statistical analysis was performed using SPSS version 26 and the scikit-learn machine learning library for Python version 3.7.
Characteristics of patients were summarized using the mean (± SD) or median (IQR) for continuous variables and counts and percentages for categorical variables.
Normality of variables was tested using Shapiro–Wilk tests. Normally distributed variables were compared using unpaired t-tests, while non-normally distributed variables were compared using Mann–Whitney U-test. Nominal variables were tested using Pearson’s chi-square tests.
Diagnostic performance of chest CT was assessed as diagnostic accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), and positive likelihood ratio (LR+) and negative likelihood ratio (LR-), taking the first RT-PCR result as reference. These measures are summarized as proportions with (exact) 95% confidence intervals (CI). A ROC was constructed using logistic regression.
A prediction model for COVID-19 diagnosis based on radiological, laboratory, and clinical data was constructed using logistic regression while using scikit in order to improve the diagnostic accuracy of CT. Again, the first RT-PCR was taken as a reference. Regression analysis was performed on complete cases (no missing variables). Due to missing variables in some patients, fewer patients could be used to train the model than could be used to evaluate chest CT performance. Variables were checked for multicollinearity by calculation of variance inflation factors (VIF). A VIF higher than 4 was determined to be unsatisfactory due to high collinearity [17]. The model was validated using 10-fold cross validation, and a ROC curve was obtained from logistic regression probabilities.
Results
Demographical and clinical patient characteristics
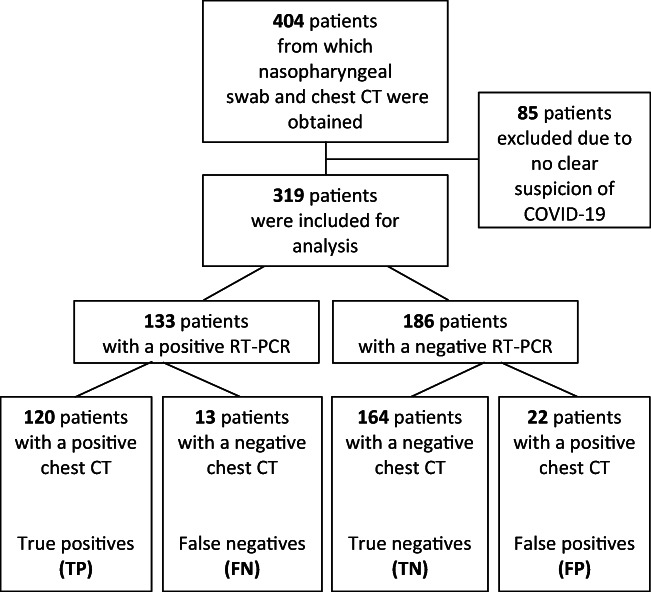
Approximately 2100 patients were presented to the ED during our inclusion period. Both a nasopharyngeal swab and a chest CT were obtained from 404 of these patients. Of these, 85 patients retrospectively did not meet the case definition and were subsequently excluded leaving 319 patients eligible for analyses (Fig. 2). Of these patients, 186 had a negative RT-PCR result and 133 had a positive RT-PCR result. There were no significant differences in the presence of comorbidities (Table 1). Fever, coughing, dyspnea, myalgia, malaise, and diarrhea were the symptoms experienced most often in RT-PCR positive patients, whereas RT-PCR negative patients more often experienced a sore throat, were more often current smokers, and had fewer moments of contact with confirmed COVID-19 patients. No differences were found for risk factors like obesity, recent travel to high-risk areas, or employment in the healthcare sector. At admission, average temperature and respiratory rates were higher and median saturation levels were lower in patients with a positive RT-PCR result. Finally, patients with a positive RT-PCR result were more likely to be admitted, before RT-PCR results were known, compared with patients with a negative RT-PCR result.
Fig. 2.
Flowchart of study with included and excluded patients
Table 1.
Characteristics of COVID-19 suspected patients at the ER
| Characteristic | RT-PCR+ | RT-PCR- | TP | FN | TN | FP |
|---|---|---|---|---|---|---|
| Number of patients | 133 | 186 | 120 | 13 | 164 | 22 |
| Age | ||||||
| Median (IQR)—yr | 59 (50–68) | 62 (44–75) | 60 (50–68) | 54 (50–73) | 63 (45–75) | 69 (54–77) |
| Distribution—% | ||||||
| 18–40 yr | 7.9 | 23.2 | 9.2 | 15.4 | 22.0 | 9.1 |
| 41–60 yr | 34.8 | 29.3 | 42.5 | 46.2 | 26.2 | 18.2 |
| 61–80 yr | 32.9 | 45.7 | 42.5 | 23.1 | 37.2 | 59.1 |
| ≥ 81 yr | 5.5 | 15.2 | 5.8 | 15.4 | 13.4 | 13.6 |
| Female sex % | 44.4 | 55.4 | 43.4 | 53.8 | 54.9 | 59.1 |
| Comorbidities no./total no. (%) | ||||||
| Chronic heart disease | 18.0 | 17.3 | 20.0 | 0.0 | 16.5 | 22.7 |
| Chronic lung disease*** | 11.3 | 22.2 | 10.8 | 15.4 | 22.0 | 22.7 |
| Chronic kidney disease | 6.1 | 4.9 | 5.9 | 7.7 | 4.9 | 4.5 |
| Asthma | 15.8 | 18.4 | 16.7 | 7.7 | 17.1 | 27.3 |
| Malignancy** | 4.5 | 11.4 | 5.0 | 0.0 | 12.2 | 4.5 |
| Diabetes | 23.3 | 22.7 | 24.2 | 15.4 | 22.6 | 22.7 |
| Hypertension | 33.8 | 23.2 | 33.3 | 38.5 | 22.6 | 27.3 |
| Other comorbidities | 31.6 | 35.1 | 30.8 | 38.5 | 36.0 | 27.3 |
| Days after onset of symptoms | ||||||
| Median (IQR) | 7 (5–10) | 7 (3–14) | 7 (5–10) | 7 (4–10) | 7 (3–14) | 11 (3–14) |
| Symptoms % | ||||||
| Fever*** | 60.2 | 37.6 | 59.2 | 69.2 | 37.2 | 40.9 |
| Coughing* | 76.5 | 62.9 | 74.8 | 92.3 | 63.4 | 59.1 |
| Sore throat* | 12.8 | 21.5 | 13.3 | 7.7 | 23.2 | 9.1 |
| Runny nose | 19.5 | 20.4 | 20.0 | 15.4 | 22.0 | 9.1 |
| Chest pain | 24.1 | 31.4 | 20.8 | 53.8 | 32.9 | 18.2 |
| Myalgia** | 36.2 | 21.6 | 36.7 | 30.8 | 19.5 | 36.4 |
| Malaise** | 52.6 | 38.9 | 51.7 | 61.5 | 38.4 | 40.9 |
| Dyspnea | 70.7 | 63.4 | 69.2 | 84.6 | 61.6 | 77.3 |
| Stomach ache | 15.0 | 16.2 | 16.7 | 0.0 | 15.2 | 22.7 |
| Nausea | 18.0 | 15.1 | 17.5 | 23.1 | 14.0 | 22.7 |
| Diarrhea* | 30.8 | 21.1 | 31.7 | 23.1 | 20.7 | 22.7 |
| Loss of taste*** | 19.5 | 14.1 | 18.3 | 30.8 | 12.8 | 22.7 |
| Loss of smell*** | 14.5 | 10.8 | 13.6 | 23.1 | 10.4 | 13.6 |
| Risk factors no./total no. (%) | ||||||
| Current smoker*** | 20/123 (16.3) | 52/162 (32.1) | 17/112 (15.2) | 3/11 (27.3) | 46/144 (31.9) | 6/19 (31.6) |
| Obesity (BMI ≥ 25) | 59/112 (52.7) | 74/142 (52.1) | 51/100 (51.0) | 8/12 (66.7) | 65/125 (52.0) | 9/17 (52.9) |
| Recent travel to high-risk area | 2/101 (2.0) | 6/117 (5.1) | 0/90 (0.0) | 2/11 (18.2) | 6/108 (5.6) | 0/10 (0.0) |
| Exposure to source of transmission*** | 24/102 (23.5) | 14/108 (13.0) | 21/92 (22.8) | 3/10 (30.0) | 11/96 (11.5) | 3/13 (23.1) |
| Healthcare worker | 11/91 (12.1) | 11/120 (9.2) | 9/80 (11.3) | 2/11 (18.2) | 10/110 (9.1) | 1/11 (9.1) |
| Radiological findings*** | ||||||
| CORADS % | ||||||
| CORADS 1 | 4.5 | 50.0 | - | 46.1 | 56.7 | - |
| CORADS 2 | 2.3 | 15.6 | - | 23.1 | 17.7 | - |
| CORADS 3 | 3.0 | 22.6 | - | 30.8 | 25.6 | - |
| CORADS 4 | 15.0 | 5.9 | 16.7 | - | 50.0 | |
| CORADS 5 | 75.2 | 5.9 | 83.3 | - | 50.0 | |
| Vitals | ||||||
| Average temperature °C ± SD*** | 37.5 ± 0.9 | 36.8 ± 0.8 | 37.6 ± 1.0 | 36.9 ± 0.8 | 36.8 ± 0.8 | 36.8 ± 0.8 |
| Median respiratory rate/min (IQR) * | 24 (21–30) | 23 (20–28) | 24 (21–30) | 25 (22–29) | 23 (20–28) | 24 (20–27) |
| Median saturation % (IQR)** | 96 (94–98) | 97 (95–99) | 96 (94–98) | 98 (97–99) | 97 (95–99) | 95 (94–97) |
| Median heart rate/min (IQR) | 97 (87–110) | 93 (82–109) | 96 (85–109) | 104 (94–115) | 93 (82–109) | 89 (87–100) |
| Oxygen required %*** | 63.2 | 37.8 | 65.8 | 69.2 | 37.8 | 36.3 |
| Median oxygen requirement (IQR) | 2 (0–2) L | 0 (0–2) L | 2 (0–2) L | 2 (0–5) L | 0 (0–2) L | 0 (0–2) L |
| Laboratory findings median (IQR) | ||||||
| CRP (mg/L)*** | 65 (30–123) | 15 (5–80) | 65 (33–124) | 39 (3–104) | 14 (4–68) | 27 (13–162) |
| Procalcitonin (ng/mL)*** | 0.09 (0.04–0.17) | 0.04 (0.02–0.09) | 0.09 (0.05–0.17) | 0.04 (0.02–0.74) | 0.04 (0.02–0.09) | 0.06 (0.04–0.23) |
| Ferritin (ng/mL)*** | 659 (254–1220) | 125 (62–286) | 712 (279–1309) | 293 (86–433) | 125 (60–253) | 185 (82–518) |
| LDH (U/L)*** | 319 (255–386) | 211 (183–252) | 328 (276–395) | 217 (186–247) | 208 (181–252) | 244 (207–294) |
| CK (IU/L)*** | 110 (59–237) | 67 (44–106) | 118 (59–240) | 89 (57–125) | 67 (43–104) | 77 (46–185) |
| Platelet count (× 109/L)*** | 192 (144–247) | 262 (215–322) | 177 (141–236) | 247 (202–315) | 265 (219–321) | 226 (202–327) |
| Leucocyte count (× 109/L)*** | 6.3 (4.8–8.5) | 9.7 (7.6–12.8) | 6.3 (4.7–8.5) | 6.7 (5.4–9.1) | 9.7 (7.6–12.8) | 9.1 (7.8–13.7) |
| Neutrophil count (× 109/L)*** | 4.5 (3.3–6.7) | 6.2 (4.8–9.4) | 4.5 (3.3–6.7) | 5.3 (3.4–6.5) | 6.2 (4.7–9.3) | 6.8 (5.5–11.2) |
| Lymfocyte count (×109/L)*** | 1.0 (0.7–1.3) | 1.7 (1.1–2.4) | 1.0 (0.7–1.3) | 1.2 (1.0–1.8) | 1.8 (1.2–2.4) | 1.2 (0.9–1.8) |
| Admitted to hospital %*** | 80.5 | 46.2 | 80.8 | 76.9 | 59.1 | 63.6 |
The result of the first nose/throat swab PCR test was used to categorize patients. Confidence intervals are given for the significance of difference between RT-PCR+ and RT-PCR- patients (first two columns).
******, , and represent p ≤ 0.05, ≤0.0,1 and ≤ 0.001, respectively
Performance of CO-RADS score in the diagnosis of COVID-19 compared with first RT-PCR
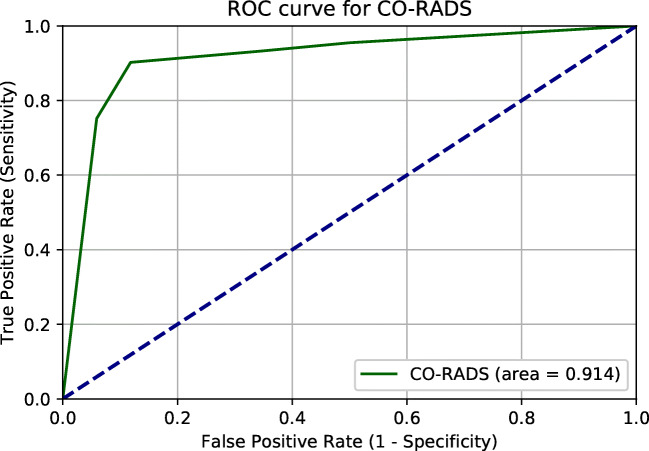
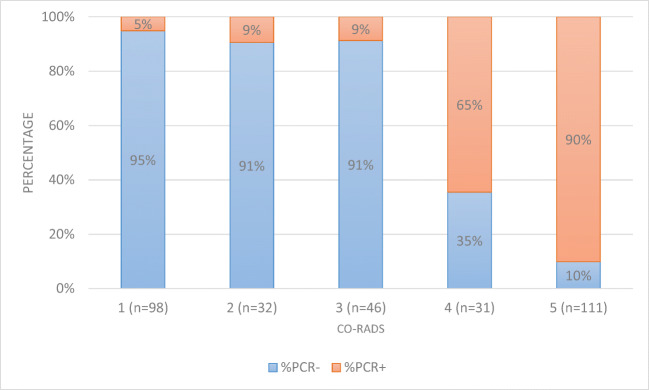
Table 2 shows the performance characteristics of chest CT using the CO-RADS score, compared with the first RT-PCR. Figure 3 shows the ROC curve with an AUC of 0.914 (0.879–0.949), and Fig. 4 illustrates the percentage of RT-PCR positive and negative patients per CO-RADS subgroup: 95% of patients with a CO-RADS score of 1 had a first RT-PCR which was negative, whereas 90% of patients with a CO-RADS score of 5 had positive RT-PCR results. Of all patients, 4.1% tested false negative on CT and 6.9% tested false positive. In this cohort with a COVID-19 prevalence of 41.7%, CT scan using the CO-RADS scoring yielded a PPV of 84.5% and an NPV of 92.7%.
Table 2.
Performance of CORADS score in classification of COVID-19 patients
| TP | FP | TN | FN | Sensitivity (%) | Specificity (%) | Prevalence (%) | PPV (%) | NPV (%) | LR+ | LR- | Accuracy (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CORADS | 120 | 22 | 164 | 13 | 90.2 (83.9–94.7) | 88.2 (82.6–92.4) | 41.7 (36.2–47.3) | 84.5 (78.6–89.0) | 92.7 (88.3–95.5) | 7.63 (5.13–11.34) | 0.11 (0.07–0.19) | 89.0 (85.1–92.2) |
Patients with CORADS 1–3 were classified as negative for COVID-19, CORADS 4–5 were classified as positive. Numbers in brackets are 95% confidence intervals. The first RT-PCR for SARS-CoV-2 was used to determine true positives and negatives
Fig. 3.
ROC curve of CO-RADS score for COVID-19 diagnosis, taking the first RT-PCR result as a reference. AUC 0.914 (0.879–0.949)
Fig. 4.
PCR results for each separate CO-RADS category
Differences between groups as classified using chest CT (CO-RADS) scoring
FN patients experienced fever, coughing, dyspnea, chest pain, and malaise more often compared with TN (Table 1). The FN group showed the highest number of obese patients. Most current smokers were TP. Patients in the FP group had a median longer duration of complaints compared with patients from the other three groups (11 vs 7 days). Interestingly, false negative patients presented more often with CO-RADS 1 than CO-RADS 2 or CO-RADS 3.
Vital signs and laboratory results on admission also differed between groups. Both true positive and false negative patients needed oxygen more often, and also in higher levels. Elevated levels of ferritin, procalcitonin, lactate dehydrogenase (LDH), C-reactive protein (CRP), and creatine kinase (CK), as well as leucopenia, lymphopenia, and neutropenia were most often seen in TP patients and least seen in TN patients. Of true positives, 80.8% were admitted, followed by FN (76.9%), FP (63.6%), and TN (59.1%).
A predictive machine learning model based on chest CT and additional data sources
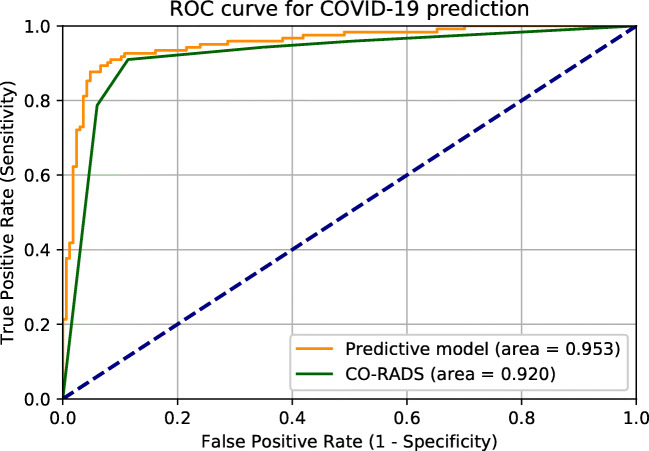
Our prediction model is based on the CO-RADS score from chest CT complemented with the following additional variables: ferritin, leucocyte count, CK, the presence of diarrhea, and the number of days since onset of disease. Figure 5 shows the ROC curves of CO-RADS alone and the prediction model. For performance characteristics see Online Resource 3, and for VIF scores of the variables see Online Resource 4. The prediction model has a significantly higher specificity than the CORADS score alone in categorizing COVID-19 patients (0.934 vs 0.886), with comparable sensitivity (0.910 vs 0.893). K-fold cross validation (k = 10) was performed on this prediction model to check for overfitting. This resulted in an accuracy of 0.91 ± 0.10 and an adjusted R2 of 0.652.
Fig. 5.
ROC curves of CO-RADS alone (green line) and the prediction model (yellow line). Accuracy 10-fold cross validation is 0.91 ± 0.10. AUC for CO-RADS alone is 0.920 and 0.953 for the prediction model
Discussion
In this prospective observational study during the peak of the COVID-19 outbreak in the Netherlands, 319 patients with suspected COVID-19 presenting to the ED were assessed by chest CT and nasopharyngeal swabs. With a sensitivity of 90.2% and specificity of 88.2%, the chest CT performed relatively well as a diagnostic modality compared with the first RT-PCR performed in COVID-19 suspected ED patients. Additionally, we constructed a machine learning prediction model by using variables which were readily available during ED visit. Ferritin, leucocyte count, creatine kinase, presence of diarrhea, and the number of days after start of complaints were predictors of COVID-19 infection in addition to the CO-RADS classification. With an AUC-ROC of 0.953, our model showed excellent performance implying that this model could possibly further aid clinicians in early recognition and diagnosis of COVID-19.
We found a sensitivity of chest CT using CO-RADS scores which was only slightly lower (90.2%) as compared with the first European study from Caruso et al. who reported a sensitivity, specificity, and diagnostic accuracy of 97%, 56%, and 72%, respectively [18] for chest CT for COVID-19 and used repeat RT-PCR as a reference. Other studies performed in China, who classified chest CT as positive based on consensus among radiologists, reported higher sensitivities for chest CT of 97% [6] and 98% [5]. Our slightly lower sensitivity could be an underestimation as 4/22 false positive patients later tested positive for COVID-19 (repeated RT-PCR or SARS-CoV-2 serology) during admission. It is plausible that these 4 patients were initially misdiagnosed due to the fact that RT-PCR tests are prone to sampling errors related to the quality and method of sampling [16].Repeated testing is recommended to decrease the number of FN results as previous studies showed that it is possible for patients with initial negative RT-PCR results to turn positive over time [12, 19, 20]. Due to limited RT-PCR testing capacity during the peak of the outbreak, repeated testing was only available and deemed meaningful for admitted patients with a high clinical suspicion of COVID-19 infection meaning that the remaining 18 FP patients were never retested.
Our FN group consisted of 13 patients. In the setting of this study, we decided not to perform repeat chest CTs unless necessary for other medical reasons. This renders us unable to account if, and how many patients, later developed chest CT features matching a CO-RADS 4 or 5 classification. Bernheim et al. [21] studied possible causes of misinterpretations of chest CTs in COVID-19 positive (RT-PCR) patients and found that 56% of patients who received a scan during early onset of symptoms (0–2 days) and 9% of patients within the intermediate group (3–5 days symptoms) had a normal chest CT at that moment without any abnormalities. Another study found that chest CT abnormalities are moreover found after symptoms persisting for about 10 days [22]. The fact that 4/13 FN patients had complaints shorter than 6 days could therefore account for a part of the FN results.
The added value of our prediction model, compared with chest CT alone, is mainly the reduction of false positives, thus increasing specificity, PPV, and LR+, probably due to the fact that certain features of COVID-19 (e.g., high ferritin levels, leukopenia, and high CK) are not present in other pathologies that result in high CO-RADS scores. In this study, the accuracy of the predictive model was higher compared with chest CT alone (93.1% vs 90.4%). In addition, 30% of RT-PCR negative patients and 21% of RT-PCR positive patients are correctly classified without generating FP or FN results using cutoff thresholds with 100% sensitivity and 100% specificity, respectively.
A strength of our study is that we constructed a machine learning predictive model by using readily available ED variables and CT characteristics. Something previously done with chest X-rays instead of chest CT [13]. Conventional chest X-ray could be limited since pulmonary lesions can be ambiguous or absent while they are already there on chest CT the same day [23]. Our prediction model showed that in addition to the CO-RADS score, ferritin, leucocyte count, creatine kinase, diarrhea as a symptom, and the number of days since onset of disease enhanced the prediction capacity for COVID-19 infection. An association between the laboratory parameters we included in our model and COVID-19 has also been reported in other studies [1, 3] and our findings confirm these previous observations. Our model’s parameters were found to have acceptable VIF’s (see Table, Supplemental Digital Content 5); the model has an adjusted R2 of 0.652 and has a k-fold cross validation accuracy of 0.91 ± 0.10.
Finally, our study has several limitations. Firstly, this was a single-center study. Secondly, only the first RT-PCR result was used as a reference as repeated RT-PCR was only performed on admitted patients with persistent high suspicion of COVID-19. Thirdly, we did not perform repeated chest CT’s in FN patients unless necessary for other medical reasons.
Conclusion
Chest CT, using the CO-RADS scoring system, is a sensitive and specific method that can aid in the diagnosis of COVID-19 at the ED during the peak of an outbreak, especially if PCR tests are scarce. Combining a predictive machine learning model could further improve the accuracy of diagnostic chest CT for COVID-19. Large prospective studies should further analyze additional candidate predictors that could be used to improve the performance of this machine learning prediction model. Still, 8–9% of patients in our cohort with RT-PCR positive results were classified as negative by chest CT and our prediction model. Therefore, RT-PCR is still indispensable in the diagnosis of COVID-19 and should remain the primary standard of testing.
Electronic supplementary material
(DOCX 17 kb)
Authors’ contributions
Joep J.R. Hermans and Joost Groen contributed equally as first authors.
Conceptualization: J.J.R. Hermans, J. Groen, E. Zwets, and V. Brown;
Methodology: J.J.R. Hermans, J. Groen, E. Zwets, B.M. Boxma–de Klerk, J.M. Werkhoven, D.S.Y. Ong, W.E.J.J. Hanselaar, L. Waals–Prinzen, and V. Brown;
Formal analysis and investigation: J.J.R. Hermans, J. Groen, and V. Brown;
Writing—original draft preparation: J.J.R. Hermans, J. Groen, and V. Brown;
Writing—review and editing: J.J.R. Hermans, J. Groen, E. Zwets, B.M. Boxma–de Klerk, J.M. Werkhoven, D.S.Y. Ong, W.E.J.J. Hanselaar, L. Waals–Prinzen, and V. Brown;
Resources: J.J.R. Hermans, J. Groen, E. Zwets, B.M. Boxma–de Klerk, J.M. Werkhoven, D.S.Y. Ong, W.E.J.J. Hanselaar, L. Waals–Prinzen, and V. Brown;
Supervision: V. Brown.
Data availability
Available as excel document if necessary.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Waiver of informed consent was obtained by the medical ethical commission (MEC-U, W20.076).
Statement of informed consent
Awarded.
Consent for publication
Awarded.
Code availability
Available as separate text document if necessary.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Joep J. R. Hermans and Joost Groen contributed equally to this work.
References
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, DSC H, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for C Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.RIVM Actuele informatie over het coronavirus (COVID 19). https://www.rivm.nl/actuele-informatie-over-coronavirus
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, Chen JI, Gutierrez RA, Gwee SXW, Chua PEY, Yang Q, Ng XY, Yap RK, Tan HY, Teo YY, Tan CC, Cook AR, Yap JC, Hsu LY (2020) Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J Clin Med 9(3). 10.3390/jcm9030623 [DOI] [PMC free article] [PubMed]
- 5.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology:200432 doi:10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed]
- 6.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology:200642. doi:10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed]
- 7.Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Zeng B, Li Z, Li X, Li H. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yijiu X (2020) China’s Hubei reports jump in new cases of COVID-19 after diagnosis criteria revision. National Health Commission of the People’s Republic of China website. www.en.nhc.gov/cn/2020-02/13/c_15.htm. Accessed February 17, 2020
- 9.Zhou S, Wang Y, Zhu T, Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. 2020;214:1–8. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 10.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on COVID-19: an update-radiology scientific expert panel. Radiology:200527 doi:10.1148/radiol.2020200527 [DOI] [PMC free article] [PubMed]
- 11.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, Jacobi A, Li K, Li S, Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology:200343 doi:10.1148/radiol.2020200343 [DOI] [PMC free article] [PubMed]
- 13.Kurstjens SG, M. de Bel, T. Verheyen, R. Kusters, R. van der Horst, A. Thelen, M. Gottgens, E.L. Elisen, M. Kluiters-de Hingh, Y. Herpers, R. M Blaauw. (2020) Corona score. www.corona-score.com [DOI] [PubMed]
- 14.Manchester Triage Group. Emergency triage (1997). BMJ publishing group, Plymouth UK
- 15.Prokop M, van Everdingen W, van Rees Vellinga T, Quarles van Ufford J, Stoger L, Beenen L, Geurts B, Gietema H, Krdzalic J, Schaefer-Prokop C, van Ginneken B, Brink M, Society C-SRWGotDR (2020) CO-RADS - A categorical CT assessment scheme for patients with suspected COVID-19: definition and evaluation. Radiology:201473. doi:10.1148/radiol.2020201473 [DOI] [PMC free article] [PubMed]
- 16.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed]
- 17.Hair JF, Black WC, Babin BJ, Anderson RE, Tatham RL. Multivariate data analysis. 7. New York: Pearson; 2010. [Google Scholar]
- 18.Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, Rucci C, Guido G, Bracci B, de Dominicis C, Laghi A (2020) Chest CT features of COVID-19 in Rome, Italy. Radiology:201237 doi:10.1148/radiol.2020201237 [DOI] [PMC free article] [PubMed]
- 19.Xu J, Wu R, Huang H, Zheng W, Ren X, Wu N, Ji B, Lv Y, Liu Y, Mi R (2020) Computed tomographic imaging of 3 patients with coronavirus disease 2019 pneumonia with negative virus real-time reverse-transcription polymerase chain reaction test. Clin Infect Dis. 10.1093/cid/ciaa207 [DOI] [PMC free article] [PubMed]
- 20.Feng H, Liu Y, Lv M, Zhong J. A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Jpn J Radiol. 2020;38(5):409–410. doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M (2020) Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology:200463. doi:10.1148/radiol.2020200463 [DOI] [PMC free article] [PubMed]
- 22.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology:200370. doi:10.1148/radiol.2020200370 [DOI] [PMC free article] [PubMed]
- 23.Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim KH, Park CM, Kim YH. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J Radiol. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 17 kb)
Data Availability Statement
Available as excel document if necessary.