Abstract
Introduction: Diverse microbiotas which have some contributions to gene expression reside in human skin. To identify the protective role of the skin microbiome against UV exposure, proteinprotein interaction (PPI) network analysis is used to assessment gene expression alteration.
Methods: A microarray dataset, GEO accession number GSE117359, was considered in this respect. Differential expressed genes (DEGs) in the germ-free (GF) and specific pathogen-free (SPF) groups are analyzed by GEO2R. The top significant DEGs were assigned for network analysis via Cytoscape 3.7.2 and its applications.
Results: A total of 28 genes were identified as significant DEGs and the centrality analysis of the network indicated that only one of the seven hub-bottlenecks was from queried genes. The gene ontology analysis of Il6, Cxcl2, Cxcl1, TNF, Il10, Cxcl10, and Mmp9 showed that the crucial genes were highly enriched in the immune system.
Conclusion: The skin microbiome plays a significant role in the protection of skin against UV irradiation and the role of TNF and IL6 is prominent in this regard.
Keywords: Microbiome, UV radiation, Gene expression, Protein-protein interaction network
Introduction
While skin is a barrier for pathogens, it is also an environment for no harmful micro-organisms.1 Bacteria, fungi, archaea, mites, and viruses are the natives of human skin that have some role in the gene expression of skin.2 Moreover, each individual has specific microbiome flora that is diverse based on that person’s genetic blueprint and way of living.3 On the other hand, the imbalance and changes of these microbes’ communities on skin could promote some sorts of skin disorders.4 In addition, it has been mentioned that these dermal microbiotas could have some influence on the activation and reaction of the immune system.5
UV radiation biological interference in humans alters since the wavelength ranges with different subdivisions as A, B, and C. UV light could be beneficial and harmful for humans.6, 7 There are pieces of evidence about the role of UV radiation in skin damage. It is reported that the primary cause of benign and malignant skin tumors such as malignant melanoma may be solar radiation including UV exposure.8 Scientists recommend many sun-protective substances, especially nutritional elements such as carotenoids and flavonoids.9, 10 Microbiome alteration in association with skin damage from UV exposure is a subject investigated by researchers.11 In another document, the role of the skin microbiome in the modulation of UV irradiation is investigated based on gene expression alteration.2
Protein-protein interaction (PPI) network analysisis anapproach to clarifying the molecular mechanism of corresponding organism condition.12 In this approach, a set of differentially expressed genes or proteins are screened for finding the critical individuals based on centrality parameters or the other aspects of the constructed network.13, 14 Gene ontology assessment is also a suitable method for finding the characteristics of the studied genes, such as the biological processes and biochemical pathways in which the genes are involved.15 In the present study, the Differential expressed genes (DEGs) after UV irradiation in the mouse skin in the absence and presence of the microbiome are analyzed via network analysis to find the possible positive role of the microbiome in the skin protection against UV irradiation.
Materials and Methods
In the study entitled “Modulatory role of skin microbiome against UV” available in 2019, 12 samples are available in the GEO database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117359. These groups are germ-free (GF) and specific pathogen-free (SPF) Mus musculus which are treated and untreated with UV irradiation. This study is with the GEO accession of GSE117359 and available in the platform of GPL16570. This study is published as “Skin microbiome modulates the effect of ultraviolet radiation on cellular response and immune function” .2 The samples were collected from dorsal skin with the related protocol mentioned in the main study.2 Differentially expressed genes of this study were obtained by comparison of the UV treated GF samples with the UV treated SPF individuals via GEO2R, an online analyzer (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE117359). This analysis provides 250 top spots with differential expression patterns which are ranked based on statistical significance. Among them, those with fold change (FC) cut-off ≥2 were assigned to differentially expressed genes (DEGs). To convert microarray IDs to gene names and descriptions, we used “Ensembl Genome Browser” (http://www.ensembl.org/biomart/martview). Some settings regarding the properties of the designated dataset were adjusted as follows: Dataset: Ensembl Gene 99, Mouse genes (GRC m38.p6); Filter: Input microarray probes/probesets ID list, [MoGene-2_0-st] Affymetrix Mouse Gene 2.0 ST Array; and Attributes: gene description, gene name.
Cytoscape 3.7.2 was used to make a PPI network from these DEGs.16 The string database was the source for network construction and a kappa cut-off score of ≥0.5 was applied in this regard.17 The network analyzer examined the topological parameters including degree and betweenness centrality. The genes with the highest degree and betweenness, which are called hubs and bottlenecks, were determined.18 Hub-bottlenecks which are common between both hubs and bottlenecks were identified.
ClueGO v2.5.6+CluePedia v1.5.6 was applied to analyze the biological processes and biochemical pathways that were associated with the hub bottlenecks. The action roles of the hub-bottleneck nodes were investigated to find the prominent regulatory genes.19, 20
Results
A graphically statistical depiction of the study groups in terms of expression data analysis is provided in Figure 1. It is aimed to analyze whether the groups are comparable for differential expression recognition or not. Data analysis via box-plot in Figure 1 indicates that the samples of the designated groups are median-centered (middle of the dataset) since their horizontal bar is in the center of the boxes. Therefore, the samples are comparable statistically.
Figure1.

Boxplot Analysis of 2 Groups of UV Radiated GF (in Blue) Versus UV Radiated SPF (in Pink).
A paired-samples t test analysis was run with the GEO2R application for two groups of study; 250 top-ranked spots based on statistical significance were retrieved. Among them, those that did not meet the designated cut-off criteria (FC ≥2) were omitted for further explorations. 41 DEGs were identified as the significant DEGs and were a candidate for further processing in terms of gene name availability. Finally, 28 characterized DEGs were included in the network study after the sequence of data processing. Seven of these genes were down-regulated while the rest (21 individuals) were up-regulated.
Among the 28 queried DEGs, 26 individuals were recognized by the STRING database (see Figure 2). There were poor interactions between the queried DEGs so most of the assigned genes were isolated and were not included in the network.
Figure 2.
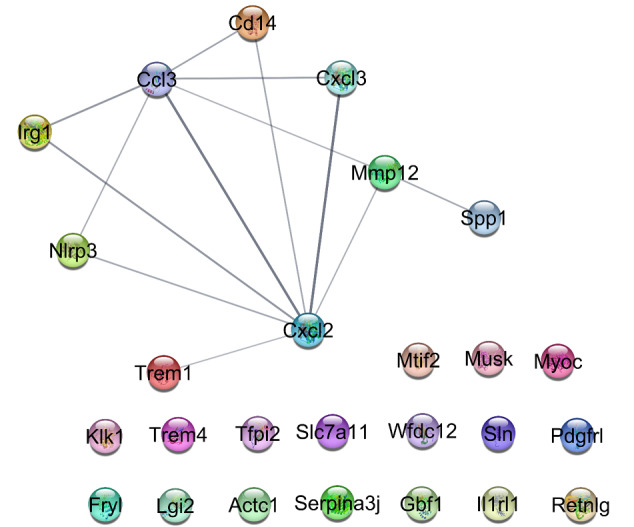
A PPI Network Including the 26 (17 Isolated Ones and 9 Interacted Individuals) Recognized Queried DEGs. A statistical kappa cut-off score ≥ 0.5 is applied.
To maximize the connections between the nodes, 50 first neighbors were added to the 26 nodes and a network of 76 nodes and 1134 edges was constructed. In this case, 12 DEGs were isolated and 14 individuals were included in the main connected component. To analyze the central genes of the network, the top 20% of the nodes based on degree value were selected as hubs and in a similar action, the bottlenecks based on betweenness centrality were chosen. Among them, common ones were then introduced as hub-bottlenecks as it is shown in Table 1. Seven hub-bottlenecks were recognized in this study. The list shows that the highest degree value is 58 for IL6 and the lowest belong to MMP9 which is 47. The highest betweenness value is 0.03 for both IL6 and Mmp9 and the lowest BC is for CXCL10. IL6 as the potent hub-bottleneck is characterized by the highest amounts of degree and betweenness centrality.
Table 1. The List of Hub-Bottlenecks of the Constructed PPI Network .
| Display Name | K | BC |
| IL6 | 58 | 0.03 |
| CXCL2 | 57 | 0.02 |
| CXCL1 | 56 | 0.02 |
| TNF | 53 | 0.02 |
| IL10 | 51 | 0.02 |
| CXCL10 | 51 | 0.01 |
| MMP9 | 47 | 0.03 |
Note . K and BC refer to degree and betweenness centrality respectively. The asterisked node is a queried DEG and the others are the added first neighbor genes.
ClueGO plug-in, in terms of biological process and biochemical pathway process annotations, revealed biological terms which were associated with the 7 hub-bottlenecks (see Figure 3). The statistical setting is as follows: gene per term: 2; gene percentage: 2; grouping cut-off score ≥ 0.5; and P value ≤ 0.05. Six clusters including “positive regulation of smooth muscle cell proliferation”, “amoebiasis”, “legionellosis”, “regulation of mononuclear cell migration”, “IL17 signaling pathway”, and “IL10 signaling” were identified. The determined biological terms can be considered as a tool to screen the related genes. The action roles of the 7 central nodes are illustrated in Figure 4. Except for CXCLs, activation is the prominent action between the other genes. Associated cellular components for the 7 central genes are shown in Figure 5. As it is depicted in Figure 5, the critical genes are related to the extracellular, plasma membrane, and intracellular components. TNF and IL6 are linked to all components while IL10 and CXCL2 are related to the extracellular component.
Figure 3.

Six clusters including 82 biological terms associated with the hub-bottlenecks which are obtained from KEGG, REACTOME pathways, and the GO-biological process are presented.
Figure 4.

Action roles of the hub-bottleneck nodes are presented. Green and yellow refer to activation and expression relationships. Binding, catalysis, and reaction are shown by blue, purple, and black respectively. The bar tips refer to down-regulation while the round ones consist of up-regulation action.
Figure 5.

The related cellular components for the 7 critical genes are presented. The genes are shown on the left side and the related cellular components are displayed on the right side.
Discussion
There are several documents about the role of the skin microbiome in gene expression and the related processes such as cell differentiation, the alteration of immune system function, and the wound healing process.21 The data source of this study is related to the effect of the skin microbiome on skin response to the exposure to UV irradiation from gene expression alteration analysis. Thus, the present study was an attempt to find the key affected genes which were deregulated in the absence of the skin microbiome. Network analysis revealed that among the assigned DEGs, CXCL2 was a central gene the function of which can be investigated in more detail. On the other hand, PPI network examination provided useful information; 6 first neighbors were introduced as critical genes in this regard. Among the crucial first neighbors, CXCL1 and CXCL10 like CXCL2 belong to the family of CXCLs. As it is reported, 17 ligands that bind to the CXC family of chemokines are identified; these ligands are known as CXCL1 to CXCL17.22 The close relationship between these CXCLs is shown in Figures 3 and 4. The introduced CXCLs are linked mostly to the legionellosis, the collection of infections that are caused by Legionella penumophila .23 As it is depicted in Figure 4, TNF and IL6 activate the collection of CXCLs. The role of TNF in activation action is more significant compared to IL6; it activates all other genes. The expression regulatory connection between IL10 and CXCL10 is another interesting finding that is illustrated in Figure 4. Maingat et al published a document about the regulation of CXCL10 by IL10. Based on their investigation, the amounts of CXCL10 in the brain of examined animals were suppressed by IL10 24. MMP9 is the other central node that is regulated mostly by the other genes. It has been indicated that the up-regulation of MMP9 is associated with promoting tumorigenesis.25 Kim et al explored the role of the MMP9/TIMP1 axis in the control of differentiation and function status of myelin-forming Schwann cells in the nerve regeneration process.26 Considering the regulation role of the central genes, it seems that TNF and IL6 are the two essential genes among the identified central nodes. As it is shown in Figure 5, both IL6 and TNF are linked to extracellular, intracellular, and plasma membrane components.
The largest cluster of biological terms is “positive regulation of smooth muscle cell (SMC) proliferation”. There are several common biological terms between this cluster and the other two clusters, namely legionellosis and amoebiasis. This cluster is related directly to the four central genes (all central genes except CXCLs). It is reported that the enhanced growth of SMC plays an essential role in atherosclerotic wound development as well as post-angioplasty restenosis. This process is known as a characteristic feature in arteries of hypertensive patients and animals.27 “IL10 signaling” and “IL17 signaling pathway” are the other two clusters that appeared in the gene ontology examination. The role of these two interleukins in the regulation of the immune system in the case of diseases is well-known in detail.28, 29 The last introduced cluster is “regulation of mononuclear cell migration” which is directly connected to TNF and CXCL10 genes. Scott and Mann published a piece of data about the regulation of mononuclear phagocytes function by the microbiota at mucosal sites.30
Conclusion
In conclusion, the skin microbiome plays a protective role against UV irradiation. This effect is associated mostly with the immune system. The significant role of TNF and IL6 was highlighted in the present study. The prominent cluster of biological terms entitled “positive regulation of smooth muscle cell proliferation” was identified as a protected individual by the skin microbiome against UV irradiation.
Ethical Considerations
Not applicable.
Conflict of Interests
The authors declare no conflict of interest.
Acknowledgment
Shahid Beheshti University of Medical Sciences supports this research.
Please cite this article as follows: Heidari MH, Razzaghi M, Akbarzadeh Baghban A, Rostami-Nejad M, Rezaei-Tavirani M, Zamanian Azodi M, et al. Assessment of the microbiome role in skin protection against UV irradiation via network analysis. J Lasers Med Sci. 2020;11(3):238-242. doi:10.34172/jlms.2020.40.
References
- 1.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patra V, Wagner K, Arulampalam V, Wolf P. Skin microbiome modulates the effect of ultraviolet radiation on cellular response and immune function. iScience. 2019;15:211–22. doi: 10.1016/j.isci.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L. et al. Universal dermal microbiome in human skin. mBio. 2020;11(1):e02945–19. doi: 10.1128/mBio.02945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunjani N, Hlela C, O’Mahony L. Microbiome and skin biology. Curr Opin Allergy Clin Immunol. 2019;19(4):328–33. doi: 10.1097/ACI.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 5.Ubags ND, Trompette A, Nibbering B, Pernot J, Pattaroni C, Rapin A. et al. The skin microbiome drives immune maturation and exacerbation of both skin and airway inflammation. ERJ Open Res. 2019;5 doi: 10.1183/23120541.lungscienceconference-2019.PP228. [DOI] [Google Scholar]
- 6. Mohania D, Chandel S, Kumar P, Verma V, Digvijay K, Tripathi D, et al. Ultraviolet radiations: skin defense-damage mechanism. In: Ahmad SI, editor. Ultraviolet Light in Human Health, Diseases and Environment. Cham, Switzerland: Springer; 2017. p. 71-87. 10.1007/978-3-319-56017-5_7 [DOI] [PubMed]
- 7.Diffey BL. Sources and measurement of ultraviolet radiation. Methods. 2002;28(1):4–13. doi: 10.1016/s1046-2023(02)00204-9. [DOI] [PubMed] [Google Scholar]
- 8.Ichihashi M, Ueda M, Budiyanto A, Bito T, Oka M, Fukunaga M. et al. UV-induced skin damage. Toxicology. 2003;189(1-2):21–39. doi: 10.1016/s0300-483x(03)00150-1. [DOI] [PubMed] [Google Scholar]
- 9.Sies H, Stahl W. Nutritional protection against skin damage from sunlight. Annu Rev Nutr. 2004;24:173–200. doi: 10.1146/annurev.nutr.24.012003.132320. [DOI] [PubMed] [Google Scholar]
- 10.Stahl W, Sies H. Carotenoids and flavonoids contribute to nutritional protection against skin damage from sunlight. Mol Biotechnol. 2007;37(1):26–30. doi: 10.1007/s12033-007-0051-z. [DOI] [PubMed] [Google Scholar]
- 11.Patra V, Byrne SN, Wolf P. The skin microbiome: is it affected by UV-induced immune suppression? Front Microbiolog. 2016;7:1235. doi: 10.3389/fmicb.2016.01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zali MR, Zamanian Azodi M, Razzaghi Z, Heydari MH. Gallbladder cancer integrated bioinformatics analysis of protein profile data. Gastroenterol Hepatol Bed Bench. 2019;12(Suppl 1):S66–S73. [PMC free article] [PubMed] [Google Scholar]
- 13.Rezaei-Tavirani M, Rezaei Tavirani M, Zamanian Azodi M, Moravvej Farshi H, Razzaghi M. Evaluation of skin response after erbium: yttrium–aluminum–garnet laser irradiation: a network analysis approach. J Lasers Med Sci. 2019;10(3):194–99. doi: 10.15171/jlms.2019.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rostami-Nejad M, Rezaei-Tavirani M, Zadeh-Esmaeel MM, RezaeiTavirani S, Akbari Z, Esmaeili S. et al. Assessment of cytokine-mediated signaling pathway dysregulation in arm skin after CO2 laser therapy. J Lasers Med Sci. 2019;10(4):257–63. doi: 10.15171/jlms.2019.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalkhal E, Razzaghi M, Rostami-Nejad M, Rezaei-Tavirani M, Heidari Beigvand H, Rezaei Tavirani M. Evaluation of laser effects on the human body after laser therapy. J Lasers Med Sci. 2020;11(1):91–7. doi: 10.15171/jlms.2020.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2018;18(2):623–32. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rezaei Tavirani M, Zamanian Azodi M, Rostami-Nejad M, Morravej H, Razzaghi Z, Okhovatian F. et al. Introducing Serine as cardiovascular disease biomarker candidate via pathway analysis. Galen. 2020;9:e1696. doi: 10.31661/gmj.v9i0.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–3. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J. et al. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6(1):20. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boshagh MA, Foroutan P, Moloudi MR, Fakhari S, Malakouti P, Nikkhoo B. et al. ELR positive CXCL chemokines are highly expressed in an animal model of ulcerative colitis. J Inflamm Res. 2019;12:167–74. doi: 10.2147/JIR.S203714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartram J, Chartier Y, Lee JV, Pond K, Surman-Lee S. Legionella and the Prevention of Legionellosis. Geneva: World Health Organization; 2007.
- 24.Maingat F, Viappiani S, Zhu Y, Vivithanaporn P, Ellestad KK, Holden J. et al. Regulation of lentivirus neurovirulence by lipopolysaccharide conditioning: suppression of CXCL10 in the brain by IL-10. J Immunol. 2010;184(3):1566–74. doi: 10.4049/jimmunol.0902575. [DOI] [PubMed] [Google Scholar]
- 25.Demers M, Magnaldo T, St-Pierre Y. A novel function for galectin-7: promoting tumorigenesis by up-regulating MMP-9 gene expression. Cancer Res. 2005;65(12):5205–10. doi: 10.1158/0008-5472.CAN-05-0134. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, Remacle AG, Chernov AV, Liu H, Shubayev I, Lai C. et al. The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS One. 2012;7(3):e33664. doi: 10.1371/journal.pone.0033664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens GK, Vernon SM, Madsen CS. Molecular regulation of smooth muscle cell differentiation. J Hypertens Suppl. 1996;14(5):S55–64. [PubMed] [Google Scholar]
- 28.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19(1):683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 29.Onishi RM, Gaffen SL. Interleukin‐17 and its target genes: mechanisms of interleukin‐17 function in disease. Immunology. 2010;129(3):311–21. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott NA, Mann ER. Regulation of mononuclear phagocyte function by the microbiota at mucosal sites. Immunology. 2020;159(1):26–38. doi: 10.1111/imm.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]


