Abstract
Quantitative assessment of spatial relations between tumor and tumor-infiltrating lymphocytes (TIL) is increasingly important in both basic science and clinical aspects of breast cancer research. We have developed and evaluated convolutional neural network analysis pipelines to generate combined maps of cancer regions and TILs in routine diagnostic breast cancer whole slide tissue images. The combined maps provide insight about the structural patterns and spatial distribution of lymphocytic infiltrates and facilitate improved quantification of TILs. Both tumor and TIL analyses were evaluated by using three convolutional neural network networks (34-layer ResNet, 16-layer VGG, and Inception v4); the results compared favorably with those obtained by using the best published methods. We have produced open-source tools and a public data set consisting of tumor/TIL maps for 1090 invasive breast cancer images from The Cancer Genome Atlas. The maps can be downloaded for further downstream analyses.
Among women worldwide, invasive breast cancer is the most common cancer and the second most common cause of cancer-related deaths.1 This finding is despite decreasing mortality rates in recent years due to early diagnosis and current therapeutic options that significantly prolong survival. Invasive breast cancers are a heterogeneous category of disease phenotypes2,3 that are histologically classified into subtypes based on: growth patterns; the expression of estrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER2); and the Ki-67 proliferation index.
The role of tumor-infiltrating lymphocytes (TILs) in invasive breast cancer has become increasingly important as a biomarker that can predict clinical outcomes, as well as treatment response in the neoadjuvant and adjuvant settings.4, 5, 6, 7, 8, 9, 10, 11 TILs are a readily available biomarker, and their evaluation is likely to expand with the emergence of immunotherapy. Elevated concentrations of TILs in HER2-positive12 and triple-negative (ER–/PR–/HER2–)13 breast cancers are associated with prolonged overall and disease-free survival, whereas elevated concentrations of TILs in luminal HER2-negative breast cancer have been associated with poor overall survival.4 TILs can also serve as a predictive biomarker because a significant part of the cytotoxic effects of systemic chemotherapy and radiation therapy is actually mediated by activating the immune system to kill cancer cells instead of directly targeting the tumor cells.14 Targeted therapies against HER2 and vascular endothelial growth factor are mediated by both antibody-dependent and complement-mediated cytotoxicity in cancer cells through lymphocytes and other immune cells in the tumor microenvironment.15 Recent studies suggest the potential for synergistic effects between targeted and immune therapies in multiple disease sites.16,17
Current practice routinely includes manual assessments of hematoxylin and eosin (H&E)–stained tissue sections by surgical pathologists to identify and classify invasive breast cancer. Such diagnostic evaluation provides insight about clinical management, treatment selection, survival, and recurrence. Because H&E–stained tissue sections are readily available, there is a sustainable opportunity to provide potentially actionable data about TILs without the need for additional tissue samples [eg, immunohistochemical (IHC) testing]. H&E–stained tissue also permits the interpretation of the lymphocyte infiltrate within and proximal to the tumor in the context of histology to provide insight about the spatial relations between tumor regions and TILs. The published guidelines for the histologic assessment of TILs in invasive breast cancer9,18,19 require pathologists to select the region of tumor and to delineate stromal areas to assess the percentage of TILs in these regions as a continuous variable from 0% to 100% within the boundaries of the entire tumor; this is used to classify the lymphocyte infiltrate as low, intermediate, and high, respectively. However, this evaluation is intrinsically qualitative and often subject to interobserver variability; previous research has articulated these concerns20 in an attempt to clearly state the need for automated methods to evaluate the percentage of TILs in H&E–stained breast cancer tissue sections.
Computationally calculating the percentage of TILs intrinsically provides spatial information about how TILs are distributed in whole slide images (WSIs), where it is likely that the distinction between intratumoral and stromal TIL infiltrates is important. Although some relatively small studies have examined intratumoral and stromal TILs,21 the predictive power of the spatial distribution of TILs within tumor and tumor-associated stroma must be better elucidated. Automated evaluation of TILs in H&E–stained WSIs fundamentally requires tumor segmentation linked with the detection of lymphocyte infiltrates. Automation of H&E–stained tumor-TIL analyses will make it possible to conduct large-scale correlative studies that quantitatively describe TIL distributions in well-characterized clinical populations. Computer analysis of high-resolution images of whole slide tissue specimens can enable a data-driven and quantitative characterization of TIL patterns.
With the recent success of deep learning22 and the availability of public data sets,23, 24, 25, 26 several research groups have proposed deep learning–based algorithms to detect or segment cancer/tumor regions in breast cancer WSIs.27, 28, 29, 30 Previous methods developed classification models from customized convolutional neural networks (CNNs)27,28 or from limited training data.29,30
In this work, standard state-of-the-art deep learning models are used along with a large-scale data set to detect invasive breast cancer regions in WSIs. This approach automates breast cancer detection at intermediate- to high-resolution to generate detailed probability-based heatmaps of the tumor bed. It achieves an F1 score of 0.82, a positive predictive value (PPV) of 79%, and a negative predictive value of 98% in terms of pixel-by-pixel evaluation in an unseen and independent test data set consisting of 195 WSIs from The Cancer Genome Atlas (TCGA) repository. These performance numbers are better than those achieved by the models in the previous studies.27, 28
Moreover, our study combines tumor detection with lymphocyte detection to identify tumor-TIL patterns in a large number of publicly accessible WSIs. TIL prediction models were trained using training data sets from a previously published deep learning approach31 to generate high-resolution TIL maps. The cancer detection results were then combined with the TIL results. The combined results represent regions of tumor with intratumoral and peritumoral TILs in publicly available 1090 WSIs from the TCGA repository. We expect that the availability of high-resolution spatial tumor-TIL maps will allow quantitative estimation and characterization of the relation between tumor cells and TILs. The ability to quantify and visualize the spatial relations between tumor and TILs can be a very practical and useful way to further elucidate intriguing observations in previous studies. It will also further our collective understanding of the biological behavior of invasive breast cancers within the context of cancer–immune interactions in the tumor microenvironment.4, 6
Materials and Methods
Datasets and Data Availability
High-resolution WSIs from the Surveillance, Epidemiology, and End Results (SEER, https://seer.cancer.gov, last accessed February 13, 2020) cancer registry system and from TCGA (https://portal.gdc.cancer.gov, last accessed February 13, 2020) were used to train and evaluate the deep learning models and generate cancer region maps. The WSIs from TCGA are de-identified and publicly available for research use. The WSIs from SEER came from a pilot program examining the feasibility of and best practices for a Virtual Tissue Repository (ie, the VTR Pilot). Because all data in the VTR Pilot, including the WSIs, had been de-identified before receipt, the NIH Office of Human Subjects Research Protection determined that the study was excluded from NIH Institutional Review Board review. Each of the SEER registries supplying the de-identified WSIs has obtained institutional review board approval from their respective institutions. The Stony Brook Institutional Review Board has classified the data set as being a non–human subjects research data set.
The training, validation, and test data sets for the breast cancer detection models consisted of image patches extracted from 102, 7, and 89 SEER WSIs, respectively. All of the images were scanned at 40× magnification and manually segmented by an expert pathologist (R.G.) into cancer and non-cancer regions using a Web-based application.32 In addition, the deep learning models were evaluated with 195 TCGA WSIs (referred to here as Ttcga), which had been manually annotated in work done by Cruz-Roa et al.28 The details of the training, validation, and test data sets for tumor region segmentation are presented (Table 1). The trained models were applied to 1090 diagnostic WSIs from TCGA invasive breast cancer cases.
Table 1.
Data Statistics of the Training, Validation, and Testing Data Sets for the Breast Cancer Detection Models
| Source | Purpose | ID | WSIs, n | Patches, n | Cancer-positive, n | Cancer-negative, n |
|---|---|---|---|---|---|---|
| SEER | Training | Dtr | 102 | 333,604 | 99,889 | 233,715 |
| Validation | Dval | 7 | 10,224 | 4953 | 5271 | |
| Testing | Tseer | 89 | - | - | - | |
| TCGA | Testing | Ttcga | 195 | - | - | - |
-, not applicable; ID, referred name used in the texts; SEER, Surveillance, Epidemiology, and End Results; T, test; TCGA, The Cancer Genome Atlas; WSIs, whole slide images.
The same set of 1090 WSIs was also analyzed by using the TIL classification models trained with data generated by Saltz et al.31 These data consisted of 86,154 and 653 image patches for training and validation, respectively. A test data set of 327 patches extracted from TCGA invasive breast cancer WSIs was created to evaluate the trained TIL models. The details of the training, validation, and test data sets for TIL classification are presented (Table 2).
Table 2.
Data Statistics of the Lymphocyte Data Set
| Source | Purpose | Patches, n | TIL-positive, n | TIL-negative, n |
|---|---|---|---|---|
| TCGA | Training | 86,154 | 21,773 | 64,381 |
| Validation | 653 | 295 | 357 | |
| Testing | 327 | 174 | 153 |
Data set provided in Saltz et al.31
TIL, tumor-infiltrating lymphocyte; TCGA, The Cancer Genome Atlas.
The SEER images used in the training data set were gathered in research conducted with the SEER consortium. At the time of the writing of the current article, the images were not publicly accessible. The SEER team is working with The Cancer Imaging Archive to make them public for use in future research. The invasive breast cancer images are publicly available and provided by TCGA (http://cancergenome.nih.gov) and the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov, last accessed February 13, 2020). The cancer and TIL heatmaps for TCGA can be found at https://app.box.com/s/1qux9ub21zcvpwao1cf81ar4milxl25x (last accessed February 28, 2020).
The cancer detection pipelines implemented in this work are available at https://github.com/SBU-BMI/quip_cancer_segmentation (last accessed February 13, 2020).
Patch Extraction for Breast Cancer Detection Models
Image patches were extracted at the highest image resolution within and outside cancer regions, which were manually segmented and annotated by a pathologist (R.G.) using an open source library called OpenSlide33 to train breast cancer detection models. Each patch was labeled cancer positive (ie, it intersected or was in a cancer/tumor region) or cancer negative (ie, it was outside cancer/tumor regions). An example of the pathologist's annotations (R.G.) is shown (Figure 1).
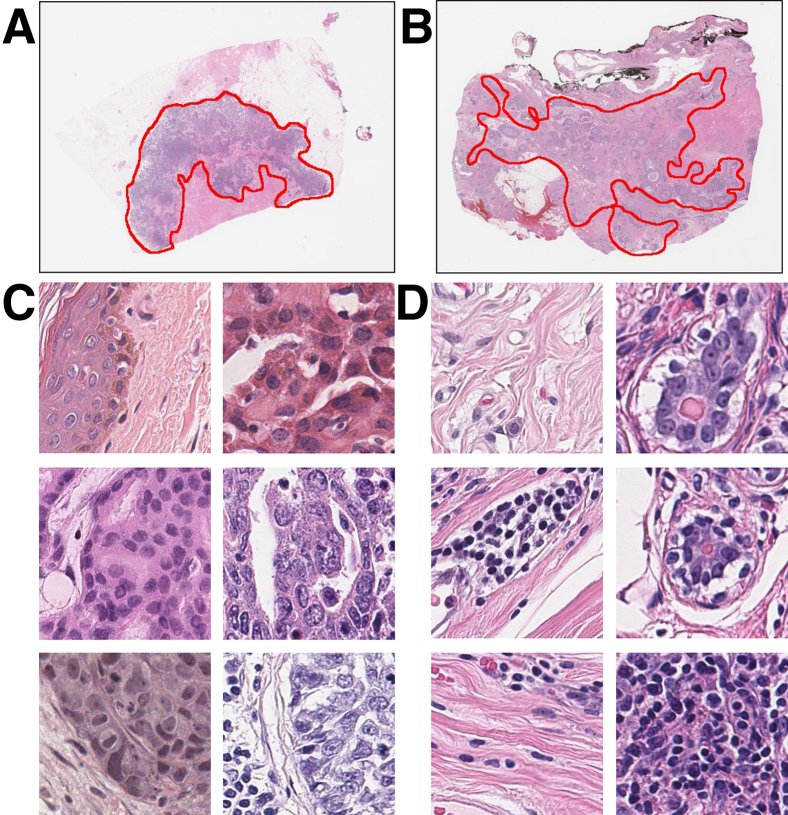
Figure 1.
Annotation example from pathologist (R.G.) (A and B) and image patches extracted from whole slide images (C and D). Red lines in A and B indicate cancer regions. Regions outside the annotated regions are noncancer regions. Patches inside C are positive samples that contain invasive cancer cells. Patches inside D are negative samples that do not contain invasive cancer cells. Original magnification, ×40 (C and D).
Patches of 350 × 350 pixels at 40× magnification (equivalent to 88 μm × 88 μm) were used to create the training data sets. The patch size was determined as follows: multiple patch sets were extracted from the images in the training set, and the patch size was then varied across the sets (patches in the same set were of the same size). Multiple cancer detection models were trained using the patch sets. As was expected, patch sets with larger patches generated more accurate models but resulted in coarser segmentations of cancer regions. To achieve a good balance of model accuracy-versus-segmentation resolution, a pathologist (R.G.) reviewed the results from each model and chose the minimum patch size that provided acceptable cancer detection (segmentation) accuracy.
Previous research has shown that it is beneficial to have more negative samples than positive samples in a training data set for image classification in digital pathology.28,34, 35, 36 A good ratio of negative to positive patches will increase the generalization of a CNN model and decrease the false-positive rate. We experimented with a range of ratios of cancer-negative patches to cancer-positive patches with the same validation data set. The final training, validation, and test data sets are presented (Table 1).
Convolutional Neural Networks
We investigated and adapted multiple state-of-the-art deep learning architectures, namely the 16-layer VGG (VGG16),37 the 34-layer ResNet (ResNet34),38 and the Inception-v4 network.39 These CNNs are widely used in an extensive range of application domains. VGG16 and ResNet34 are designed to process 224 × 224-pixel patches. Inception-v4 accepts 299 × 299-pixel image patches. Our tumor data set consists of 350 × 350-pixel patches at 40× magnification. The lymphocyte training data sets contain 100 × 100-pixel patches at 20× magnification; this patch size is the same patch size used in a previous study.31 Input patches in these data sets were resized to the desired input size for each network by using standard image resizing functions in PyTorch version 0.4.40 In addition, for ResNet34 and Inception-v4, the dimension of the output layer was changed from 1000 classes to two classes, because each patch in our case is labeled positive or negative.
For VGG16, the size of the intermediate features of the classification layer was reduced from 4096 to 1024 and only the first four layers in the classification layer were kept. This modification reduced the number of trainable parameters of this network from 138 million to 41 million. Our modifications to the classification layers of the CNN architectures are presented (Table 3). The CNN networks were implemented by using PyTorch version 0.4.40
Table 3.
Modifications to the Classification Layers of the CNNs
| VGG16 |
ResNet34 |
Inception-v4 |
|||
|---|---|---|---|---|---|
| Original | Modified | Original | Modified | Original | Modified |
| Linear (25,088, 4096) ReLU → Dropout Linear (4096, 4096) ReLU → Dropout Linear (4096, 1000) |
Linear (25,088, 4096) ReLU → Dropout Linear (1024, 2) |
Linear (512, 1000) | Linear (512, 2) | Linear (1536, 1000) | Linear (1536, 2) |
CNN, convolutional neural network; ResNet34, 34-layer ResNet; VGG16, 16-layer VGG.
Earlier work41,42 showed that refining a CNN pretrained on the ImageNet data set43 is a good approach to boosting image classification performance in digital pathology. The pretrained CNN models were refined with our training data. The pretrained CNN models were trained with natural images after the RGB channels of the images had been normalized. This study applied the same approach and normalized the RGB channels of the image patches in our training data set. Without normalization, the ranges of RGB values in our data set would be different from those in the data sets used for the pretrained models, significantly reducing the effectiveness of model refinement.
The same training procedure was used for all of the networks. At the beginning of the training, the weights of the networks were initially fixed except for the classification layer. The networks were trained in this state for N epochs (N is three for the cancer models and N is five for the lymphocyte models) with a batch size of B (B is 256 for the cancer models and 128 for the lymphocyte models), an initial learning rate of 0.01, a momentum of 0.9, and a weight decay of 0.0001. After N epochs, the training process turned on updates to the initially fixed weights. The network was then trained for total of 20 epochs, updating all of the weights. The training process used a stochastic gradient descent method44 to minimize a cross entropy loss function.
The color profiles of WSIs may vary from image to image because of variations in staining and image acquisition.45, 46, 47 The R, G, and B channels of each patch were normalized to a mean of 0.0 and an SD of 1.0. In addition, data augmentation was used to further reduce the effects of color/intensity variability and data acquisition artifacts. The data augmentation operations included random rotation between 0 and 22.5 degrees, random vertical and horizontal flipping, and perturbations in patch brightness, contrast, and saturation. In the prediction (test) phase, no data augmentation was applied except for the normalization of the color channels. Each patch was assigned a value between 0.0 and 1.0 by the trained model, indicating the probability of the patch being positive.
Experiments
In the experimental evaluation, accuracy, F1 score, and area under the receiver-operating characteristic curve (AUC) were used as performance metrics. Accuracy is the ratio of correctly classified patches to the total number of patches in the ground truth test data set. Because a data set is not always balanced between classes, the F1 score that considers both precision and recall was used to compute a score. Mathematically, the F1 score is equal to 2 × (precision × recall)/(precision + recall). Lastly, AUC was used to evaluate the prediction performance of the models at different threshold settings. AUC shows the relation between the true-positive rate and the false-positive rate of a model. It is a widely used metric to assess model performance for binary classification tasks.
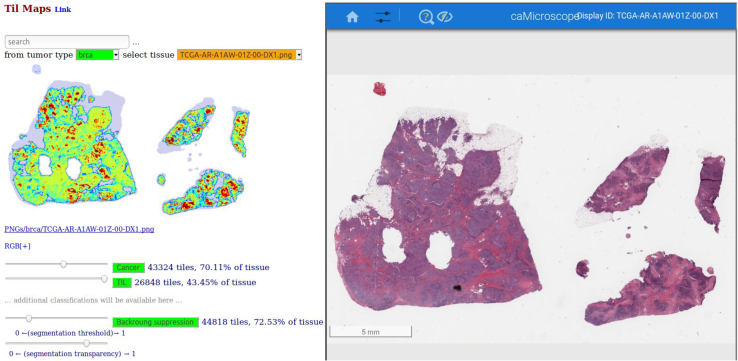
The cancer region segmentation and TIL classification performances of the different CNNs are presented (Tables 4 and 5) (Results). The best models were applied to the 1090 WSIs from TCGA invasive breast cancer cases to generate prediction probability maps for cancer regions and TILs. A prediction probability map is constructed by uniformly partitioning a WSI into image patches in each image dimension. The image patches are analyzed by a trained model and assigned a label probability between 0.0 and 1.0. For cancer region segmentation, the label of a patch was either cancer positive (ie, the patch predicted to be within or intersect a cancer region) or cancer negative (ie, the patch is predicted to be outside the cancer regions in the WSI). For TIL classification, the label of a patch was either TIL positive (ie, the patch was predicted to contain lymphocytes) or TIL negative. A Web-based application was implemented to visualize and interact with the prediction probability maps as heatmaps (Figure 2) (Materials and Methods).
Table 4.
Performance Comparison of the Cancer Detection Task between the ConvNet and Our Models
| Method | F1 score | PPV | NPV | TPR | TNR | FPR | FNR |
|---|---|---|---|---|---|---|---|
| ConvNet28 | 0.76 ± 0.20 | 0.72 ± 0.22 | 0.97 ± 0.05 | 0.87 ± 0.16 | 0.92 ± 0.08 | 0.08 ± 0.08 | 0.13 ± 0.16 |
| ConvNet-ours | 0.75 ± 0.18 | 0.69 ± 0.22 | 0.96 ± 0.09 | 0.87 ± 0.18 | 0.91 ± 0.09 | 0.09 ± 0.07 | 0.12 ± 0.16 |
| ConvNet-ours∗ | 0.77 ± 0.21 | 0.73 ± 0.23 | 0.97 ± 0.09 | 0.87 ± 0.23 | 0.92 ± 0.09 | 0.08 ± 0.09 | 0.13 ± 0.22 |
| C-VGG16 | 0.80 ± 0.20 | 0.78 ± 0.20 | 0.97 ± 0.05 | 0.88 ± 0.21 | 0.94 ± 0.06 | 0.06 ± 0.06 | 0.12 ± 0.21 |
| C-ResNet34 | 0.82 ± 0.18 | 0.79 ± 0.20 | 0.98 ± 0.04 | 0.89 ± 0.18 | 0.95 ± 0.05 | 0.05 ± 0.05 | 0.11 ± 0.18 |
| C-Incepv4 | 0.81 ± 0.19 | 0.79 ± 0.20 | 0.97 ± 0.05 | 0.88 ± 0.19 | 0.94 ± 0.06 | 0.06 ± 0.06 | 0.12 ± 0.19 |
Data are expressed as means ± SEM. ConvNet-ours indicates our implementation of the ConvNet28 that was trained on the Surveillance, Epidemiology, and End Results (SEER) data set. The ConvNet-ours results are reported without applying the postprocessing method (Materials and Methods). All of the models were trained on the SEER data set (Dtr) and evaluated on 195 whole slide images of The Cancer Genome Atlas (Ttcga). Positive predictive value (PPV), negative predictive value (NPV), true-positive rate (TPR), true negative-rate (TNR), false-positive rate (FPR), and false-negative rate (FNR) were used to measure the performance. Bold numbers indicate the best results.
∗ConvNet-ours: Our implemented version of the ConvNet28 that was trained on the SEER data set. The ConvNet-ours results are reported after the postprocessing step is executed. The last three rows show the performances of our convolutional neural networks.
C, cancer detection models; ResNet34, 34-layer ResNet; VGG16, 16-layer VGG.
Table 5.
Performance Comparison of the Lymphocytes Detection Task between Saltz et al and Our Models
| Method | F1 score | Accuracy | AUC |
|---|---|---|---|
| Saltz et al31 | 0.770 | 74.9% | 0.808 |
| L-VGG16 | 0.891 | 88.4% | 0.943 |
| L-ResNet34 | 0.893 | 89.0% | 0.950 |
| L-Incepv4 | 0.879 | 87.5% | 0.938 |
Bold numbers indicate the best results. L indicates the lymphocyte detection models. AUC, area under the receiver-operating characteristic curve; ResNet34, 34-layer ResNet; VGG16, 16-layer VGG.
Figure 2.
User interface of Web-based application to study the spatial relation between cancer regions and lymphocyte regions. Left panel shows the tumor-infiltrating lymphocyte (TIL) heatmap; invasive breast cancer detection is denoted in yellow with superimposed lymphocyte detection denoted in red. Right panel is the caMicroscope32 that displays the regions of the whole slide image. Users can click on the TIL map to zoom in the corresponding regions on the caMicroscope. Readers can access our tool at https://mathbiol.github.io/tcgatil (last accessed March 19, 2020). Scale bar = 5 mm.
Postprocessing Step for Cancer Heatmaps
Most patch-based classification algorithms48,49 predict the label of a patch independent of other patches in an image. They do not take into account the characteristics and labels of neighbor patches. Invasive cancer regions in breast cancer tend to be close to each other. Thus, the probability of a patch to be cancer positive is correlated with its surrounding patches. To incorporate this information into our analysis pipeline, a simple, yet effective, aggregation approach was used as a postprocessing step. This approach takes per-patch classification probability values, converts them into a probability map, called H, and produces an aggregated probability map, called A. The classification probability value of a patch in A is computed by an aggregation operation over neighbor patches within a specific distance of the patch in H. The relation between A and H can be formulated as follows:
| (1) |
Here, H(m,n) is the probability values of a patch at location (m,n) in H; A(i, j) is the probability value of the aggregated patch at location (i,j) in A; and f is the aggregation function over a set of patches in a window of
| (2) |
where w is the window size. In our aggregation approach, all patches within the window will have the same prediction score after the aggregation operation. ⌊x⌋ is the floor operation which takes x as an input and returns the largest integer that is less than or equal to x. Different aggregation functions such as Average, Median, and Max were explored. The experiments were conducted by using Tseer. The best aggregation method from these experiments was used to generate aggregated probability maps for Ttcga. Empirically, the Max function and a window of 4 × 4 resulted in the best performance with Tseer. These settings were applied to postprocess predictions in Ttcga.
Combined Tumor-TIL Maps
Each pair of cancer and lymphocyte heatmaps were merged into a single heatmap as an RGB image. The R channel stores the lymphocyte probabilities quantized from 0 to 255; the G channel stores the cancer probabilities quantized from 0 to 255; and the B channel stores 0 or 255 to indicate if a patch is glass background or tissue, respectively.
Software Support for Analysis Workflow
QuIP32 and caMicroscope32 software were used to support the data management and visualization requirements in this study. A typical whole slide tissue image can be several gigabytes in size. Even a modest cohort of 100 subjects can result in one terabyte of image data. It is a nontrivial task to efficiently store, manage, and index a data set of this size and to provide interactive capabilities for visualization of images and analysis results for evaluation, validation, and additional downstream analyses. Examination of the analysis results (ie, the probability maps) requires their interactive interrogation through visual analytic tools that link the probability maps with the underlying images. Our software converts probability maps into heatmaps for visualization purposes. We have developed a Web-based application, called FeatureMap, and a database, called PathDB, in QuIP. PathDB manages and indexes metadata about whole slide tissue images and metadata regarding heatmaps. It links the heatmaps with the images for query and retrieval. FeatureMap implements a browser-based multivariate visualization library that is sufficiently lightweight to run on a mobile device. It interacts with PathDB to query and retrieve heatmaps and then display them as low-resolution images so that a user can rapidly go through multiple images and probability maps. The low-resolution image representations of the probability maps are linked to full-resolution images and high-resolution heatmaps. The user can switch to the high-resolution view for more detailed and interactive examination of a probability map and the source image.
Results
Evaluation of Cancer Detection Models
Three cancer detection and segmentation models, C-VGG16, C-ResNet34, and C-Incepv4, were trained by using VGG16, ResNet34, and Inception-v4, respectively. The performances of the models were compared with each other as well as with another network, called ConvNet, which was developed by Cruz-Roa et al.27,28 ConvNet was trained on a different training data set, called HUP (from the Hospital of the University of Pennsylvania) and UHCMC/CWRU (from University Hospitals Case Medical Center/Case Western Reserve University), in previous work.27 To use our training data sets, ConvNet was implemented by using PyTorch40 by precisely following the network description in the original paper. We call our implementation ConvNet-ours.
An average F1 score was computed across all of the test images by varying the threshold value from 0.0 to 1.0 in steps of 0.01. At each threshold value, prediction probability maps were computed for the 195 test images by the model under evaluation. The patch labels were assigned by applying the threshold value to the corresponding probability maps. The label maps and the ground-truth masks28 were then used to compute average F1 score, PPV, a negative predictive value, true-positive rate, true-negative rate, false-positive rate, and false-negative rate. The performance comparison between our models, the original ConvNet model,27,28 and our implementation of the ConvNet model (ConvNet-ours) is presented (Table 4). We report the performance of ConvNet-ours both with and without applying our postprocessing step (described in the Methods section) because the original ConvNet model did not include a postprocessing step. In addition, the ConvNet-ours model outperformed the original ConvNet model in all metrics. Furthermore, the postprocessing step improved the average F1 score from 0.75 to 0.77 and PPV from 0.69 to 0.73. Given that the postprocessing step is relatively simple and inexpensive, we recommend the inclusion of this step in the implementations of the proposed approach in research and clinical settings.
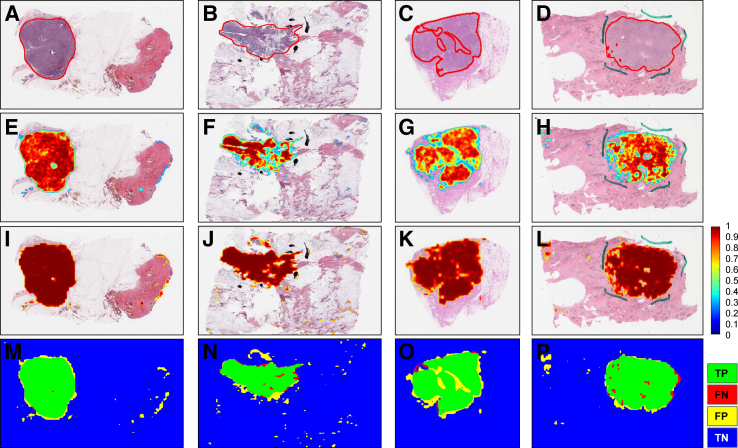
Probability maps from the C-ResNet34 model for a set of representative WSIs in Ttcga are shown (Figure 3). Visual inspection of the maps and the respective WSIs showed that the model was able to detect and segment cancer regions well.
Figure 3.
Prediction map of representative slides from The Cancer Genome Atlas whole slide images. A–D: Whole slide images with ground truth generated by an expert pathologist (R.G.). E–H: The corresponding prediction heatmap generated by our cancer detection algorithm (C-ResNet34) before applying any aggregation methods. I–L: The corresponding prediction map after applying the Max aggregation function with window size of 4, then applying a threshold of 0.6 to exclude prediction scores that are <0.6. The shades of red in the map images indicate the probability of a patch being cancer positive as predicted by the model. M–P: Results of our algorithm in terms of true positives (TP; green), false negatives (FN; red), false positives (FP; yellow), and true negatives (TN; blue) regions. Red lines in A–D indicate cancer regions. Blue lines in D, H, and L and the black lines in B, F, and J are ink marks that are artifacts during the staining process. C, cancer detection model; ResNet34, 34-layer ResNet.
Evaluation of Lymphocyte Classification Models
Three lymphocyte detection models, L-VGG16, L-ResNet34, and L-Incepv4, were trained by using VGG16, ResNet34, and Inception-v4, respectively. A training data set, containing 2912 image patches from invasive breast cancer WSIs only, was created from the original TIL training data set in work performed by Saltz et al.31 The 86,154 patches in the original training data set had been selected from multiple cancer types. Our experiments showed that the smaller training data set resulted in more accurate classification models than the full original data set.
The trained models were tested with a set of image patches extracted from TCGA invasive breast cancer WSIs. The performance comparison between our models versus the model developed in the previous work is presented (Table 5).31 The new models consistently outdid the previous model in all of the performance metrics.
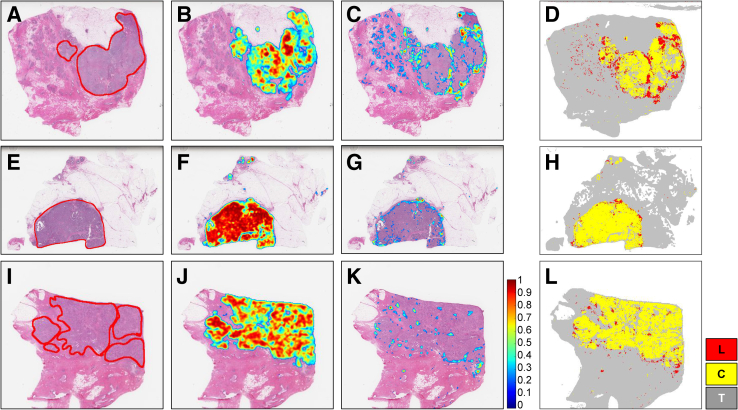
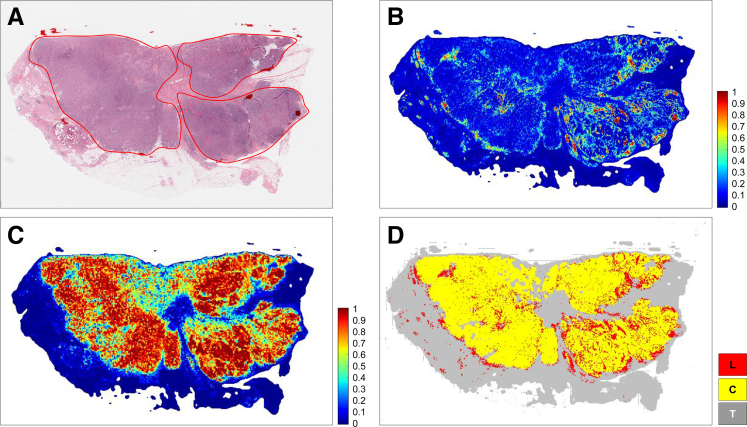
The experimental evaluation showed that the cancer region segmentation and lymphocyte classification models achieved very good performance with respect to the F1 score, accuracy, and AUC metrics and performed better than the previous models. The best of these models were applied to 1090 TCGA invasive breast cancer WSIs and generated tumor, TIL, and combined tumor-TIL maps. These maps will be made publicly available (Datasets and Data Availability). Examples of the tumor-TIL combined maps overlaid on WSIs as heatmaps are shown (Figures 4 and 5).
Figure 4.
A–C: Cancer and lymphocyte probability maps along with a map of cancer and lymphocyte labels generated through analysis of representative slides from The Cancer Genome Atlas (TCGA) whole slide images (A: TCGA-A2-A0CL-01Z-00-DX1; B: TCGA-A2-A04X-01Z-00-DX1; C: TCGA-A2-A0CW-01Z-00-DX1). Figures in a given row are results generated from the whole slide image depicted in the first column. A, E, and I: Whole slide images with ground truth generated by an expert pathologist (R.G.). Red lines in A, E, and I indicate the cancer regions annotated by pathologist (R.G.). B, F, and J: The corresponding cancer probability maps generated by our cancer detection models (C-ResNet34). C, G, and K: The corresponding lymphocyte probability maps generated by the lymphocyte classification models (L-ResNet34). D, H, and L: A combined heatmap of cancer and lymphocytes. Invasive breast cancer detection is denoted in yellow with superimposed lymphocyte detection denoted in red. These figures visualize the spatial relations between lymphocytes and tumor regions. The lymphocyte patches in these examples show the TILs and tumor-associated lymphocytes (TALs) that surround the cancer regions. These visual representations of TILs, TALs, and cancer regions provide valuable information for further analyses. C, cancer detection model; L, lymphocyte detection model; ResNet34, 34-layer ResNet; T, tissue region.
Figure 5.
Enlarged example of a cancer and lymphocyte probability map and cancer along with map of cancer and lymphocyte labels for The Cancer Genome Atlas (TCGA) whole slide image (case ID: TCGA-E9-A248-01Z-00-DX1) generated by our algorithms (C-ResNet34 and L-ResNet34). A: Whole slide image of an invasive breast cancer hematoxylin and eosin–stained tissue section. Red line indicates the viable tumor region. B and C: Lymphocyte probability map and cancer probability map predicted by our algorithm, respectively. The probabilities range from 0 to 1. D: Invasive breast cancer detection denoted in yellow with superimposed lymphocyte detection denoted in red. Gray areas outside of the yellow tumor region denote nontumor connective and adipose tissues. C, cancer detection model; L, lymphocyte detection model; ResNet34, 34-layer ResNet; T, tissue region.
Assessment of Interrater and Machine versus Human Scoring of TIL Patches
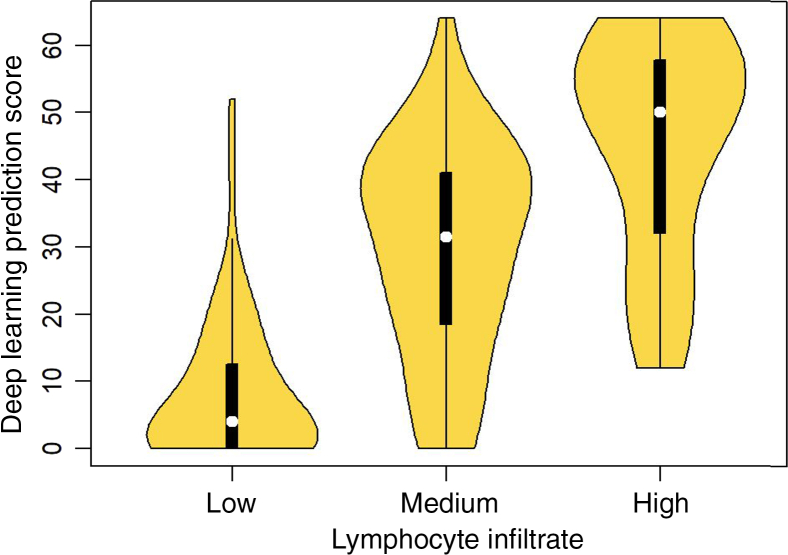
A direct comparison of TIL predictions according to the trained models with labeling of patches was performed by experienced pathologists (R.G., R.B., T.Z.) by scoring 8 × 8 super-patches for TIL content. Three pathologists assessed 500 super-patches as having low, medium, or high TIL content. Machine-derived scores were assigned to a super-patch by counting TIL-positive patches in the super-patch; thus, the scores range from 0 to 64. To assess concordance between the human pathologists, the polychoric correlation coefficient, designed for comparing ordinal variables, was used.50 The polyserial coefficient was used for comparison of continuous valued TIL counts estimated by the deep learning models versus the ordinal scores of the experienced pathologists (as having low, medium, or high TIL content). The performance comparison between human raters with the models developed is shown (Table 6). A somewhat consistent improvement in the quality of concordance between human experts and machine predictions was observed, even perhaps slightly better than human–human concordance. Also, the deep learning models permit a lower variability relative to human raters, as evidenced by the width of the corresponding CIs. The concordance between the summarized scores (using median) across pathologists vis-a-vis machine-derived predictions also generally improves relative to concordance measures of individual experts against the machine predictions. The median machine-derived score is distinct between the three ordinal bins (Figure 6).
Table 6.
Interrater Concordance (Between Human Raters: A, R1, and R2) and Human versus Machine Models
| Rater | Human | VGG16 | ResNet34 | Incepv4 |
|---|---|---|---|---|
| A | R1: 0.62 (0.48, 0.76) | 0.85 (0.81, 0.88) | 0.82 (0.78, 0.86) | 0.85 (0.82, 0.88) |
| R1 | R2: 0.74 (0.64, 0.85) | 0.73 (0.68, 0.79) | 0.73 (0.67, 0.79) | 0.72 (0.66, 0.78) |
| R2 | A: 0.73 (0.62, 0.84) | 0.73 (0.68, 0.79) | 0.74 (0.69, 0.80) | 0.76 (0.70, 0.81) |
| Median | NA | 0.77 (0.70, 0.83) | 0.74 (0.67, 0.81) | 0.76 (0.70, 0.83) |
Point estimate of correlation coefficients and CIs are provided.
NA, not applicable; ResNet34, 34-layer ResNet; VGG16, 16-layer VGG.
Figure 6.
Comparison of tumor-infiltrating lymphocyte scores of super-patches between pathologists (R.G. R.B., and T.Z.) and computational stain. x axis: median scores from three pathologists (R.G. and T.Z.) assessing 500 super-patches as having low, medium, or high lymphocyte infiltrate. y axis: scores from deep learning predictions on a scale from 0 to 64.
TIL Infiltration as a Predictor of Survival
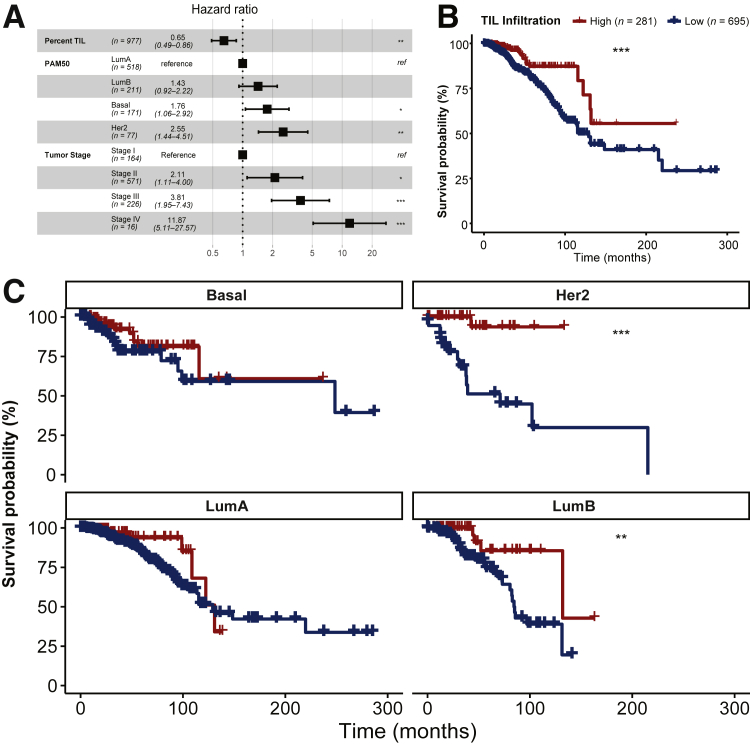
To validate the potential clinical relevance of our system, the relation between overall survival and TIL infiltration into the tumor was investigated. Continuous patch likelihoods from tumor (C-ResNet34 model) or lymphocyte (L-ResNet34 model) predictions were binarized such that a prediction >50% was counted as predicted to contain tumor or lymphocytes, respectively. The proportion of pixels in our image that were predicted as containing tumor as well as lymphocytes was counted: (number of pixels predicted as lymphocyte AND tumor)/(number of pixels predicted as tumor). For associations with survival, clinical information including stage was obtained from Genomic Data Commons using the TCGAbiolinks package in R statistical software version 3.6.0 (R Foundation for Statistical Computing, Vienna, Austria). PAM50 labels were obtained.51 TIL fractions were analyzed as both continuous and dichotomous variables. The distribution of TIL infiltration fraction across samples was right-skewed, and the mean of this distribution (6.4%) served as a natural inflection point separating the majority (n = 695) of cases with low infiltration from the minority group (n = 281) with high infiltration. First, it was checked if TIL infiltration fraction was predictive of survival as a continuous variable when correcting for major factors known to predict survival: stage (aggregated into stage I, II, III, and IV) as well as gene expression subtype (PAM50 Basal, Luminal A, Luminal B, and HER2). Finally, it was established that binarized TIL infiltration fraction was still predictive of survival even after subdividing cases according to PAM50 subtype or stage. Detail analysis is shown (Figure 7).
Figure 7.
A: Forest plot of hazard ratios (estimates and 95% CIs) from a multivariate Cox proportional hazards model of overall survival. Concordance Index: 0.73 # Events: 131; Global P value (log-rank): 3.4809 × 10−11. Tumor-infiltrating lymphocyte (TIL) infiltration is a continuous variable calculated as the fraction of area predicted to contain tumor that is also predicted to contain lymphocytes (range, 0% to 63%), and then rescaled in terms of SDs (9.4%). The dotted line indicates a hazard ratio of 1. B: Kaplan-Meier plot of survival probability after dividing patients into high and low TIL infiltration groups around the mean TIL infiltration fraction for all cases (6.4%). C: Kaplan-Meier plot subdivided by PAM50 tumor subtype shows that TIL infiltration is prognostic within at least two groups defined by gene expression, Her2 and Luminal B (LumB). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 versus low. LumA, Luminal A; ref, reference.
Discussion
Studies have shown that TILs can be used as a biomarker to predict clinical outcomes, including treatment response, in patients with invasive breast cancer.9, 10, 11 With the emergence of immunotherapy for breast cancer treatment, evaluation of the concentration of TILs as a readily available biomarker is increasingly important. The cancer detection algorithm indicates that the cancer region occupies approximately 50% to 60% of the total tissue area in the WSI (Figure 3). The lymphocyte detection algorithm shows high probability areas with TILs. The tumor-TIL method provides insight regarding scattered TILs that occupy approximately 20% to 30% of the cancer region, consistent with a low percentage of TIL categorization, with additional spatial information that indicates a sparse multifocal distribution. Combined breast cancer tumor–TIL maps like the one shown in this example have been generated for 1090 TCGA breast cancer WSIs, and they will be made publicly available in our custom Web-based application.
The evaluation of TILs in invasive breast cancer is likely to expand due to the accumulating evidence showing how TILs can be used to predict treatment response in the settings of neoadjuvant and adjuvant chemotherapy. However, the routine evaluation of TILs has not achieved widespread adoption even though the methodology established by the International Immuno-Oncology Biomarker Working Group18 is relatively straightforward, uncomplicated, and based on the examination of TILs on standard H&E–stained tissue sections TILs and a focal area with peritumoral TALs as a surrogate computational biomarker that is similar to how IHC is routinely used by pathologists to highlight cells and structures are identified (Figure 4). However, IHC is not routinely performed to identify and classify subsets of TILs in breast cancer due to the time constraints of pathologists, desire to preserve diagnostic tissue, and additional costs, whereas this kind of insight can be made readily available in a low-cost and scalable manner to achieve the goals of the International Immuno-Oncology Biomarker Working Group. With emerging methods such as our breast cancer tumor–TIL detection tool, pathologists will be able to add the evaluation of TILs to the standard IHC panel to determine ER, PR, and HER2 expression status.
In previous work, several research groups conducted image analyses focused on detection of metastatic breast cancer52, 53, 54 and mitosis55, 56, 57 using highly curated but relatively small data sets from algorithm evaluation challenges.23, 24, 25, 26 Cruz-Roa et al27,28 used deep learning approaches for detecting invasive breast cancer in WSIs. The deep learning models were trained by using WSIs from the Hospital of the University of Pennsylvania and from University Hospitals Case Medical Center/Case Western Reserve University and evaluated with 195 WSIs from TCGA. Kwok29 and Dong et al30 proposed methods to classify breast cancer regions in WSIs by using data sets provided by the 2018 International Conference on Image Analysis and Recognition Grand Challenge on Breast Cancer Histology Images.26 The conference data set contains two subsets of training data: Part A consists of 400 images of 2048 × 1536 pixels at 0.42 μm × 0.42 μm resolution; and Part B comprises 10 WSIs with manual annotations from pathologists. Kwok implemented a two-stage training approach in which a basic CNN network is trained in the first stage to mine hard examples on data from part B. These examples were then used to train a deep learning model in the second stage. Dong et al used deep reinforcement learning to decide whether regions of interest should be processed for segmentation at high or low image resolutions. Most recently, Amgad et al58 proposed a fully convolutional framework for semantic segmentation of histology images via structured crowdsourcing. This was the first work using crowdsourcing in pathology tasks that involved a total of 25 participants at different expertise levels, from medical students to expert pathologists, to generate training data for a deep learning algorithm. The authors solely focused on segmenting triple-negative breast cancer, an aggressive genomic subtype that comprises 15% of breast cancer cases, into five distinct classes: tumor, stroma, inflammatory infiltration, necrosis, and other. Using a training data set of 151 representative regions of interest (mean region of interest size, 1.18 mm2) selected from 151 H&E–stained TCGA WSIs with detailed curated annotations, a fully convolutional VGG16-FCN-8 network was able to achieve an AUC of 0.941 for tumor region.
The current methods for assessing TILs in individual patients are still subjective, laborious, and may be difficult to quantify. More rigorous, objective, and efficient methods are needed. This is especially true for precision medicine applications because the tumor microenvironment in breast cancer is heterogeneous and composed of malignant cells, premalignant lesions, adjacent normal tissue, stroma, immune cell infiltrates, vessels, nerves, and fat. Therefore, to help further our understanding of breast cancer biology for research and clinical applications, we developed a tumor-TIL spatial mapping tool to automatically detect breast cancer in H&E–stained WSIs to quantitatively estimate and characterize the relation between tumor cells and TILs.
In the current state, the breast cancer tumor–TIL maps can be used to identify spatial patterns of distributions of TILs within intratumoral and peritumoral regions of invasive cancer, as well as lymphocyte infiltrates in adjacent tissues beyond the borders of the tumor. This tool can also be adapted for practical uses that include improving the reproducibility and precision in reporting tumor size and features of the tumor boundary for radiologic–pathologic correlation. As a potential clinical application to quantify TILs and identify spatial patterns of distribution of TILs, this tool can help guide management and select treatment in conjunction with existing molecular subtyping platforms; the goal is to predict survival and recurrence, as the TILs have been shown to be reliable prognostic and predictive biomarkers in invasive breast cancer. Another potential application of this tool is to screen candidates who may benefit from immunotherapy in primary, refractory, and recurrent disease because such treatments are not expected to be useful if a significant amount and distribution of TILs are not present.
Most existing software algorithms for TILs assessments are proprietary, expensive, and cannot be customized by the user. Therefore, we are making our invasive breast cancer TCGA tumor–TIL data set publicly available with an interface to visually interact with the data. The interface permits quantification of TILs in tumor areas and the ability to rapidly spot check and evaluate true-positive and false-positive predictions by the deep learning models. The invasive breast cancer TCGA–TIL maps are displayed side-by-side with an interactive H&E slide viewer to permit a high level of exploration within the entire data set. We also intend to further combine this tumor-TIL method to characterize tumor immune heterogeneity and spatially characterize local patterns of the lymphocytic infiltrate in different parts of the tumor (eg, center of the tumor, invasive margins, metastases). The tumor-TIL heatmaps can also be combined with other types of digital pathology–based image analyses that extract object-level information such as size, shape, color, and texture (collectively known as pathomics) to generate an unprecedented quantitative examination of invasive breast cancer. Such analytic data can complement traditional histopathologic evaluation, which can be correlated with clinical information, radiologic imaging, molecular studies, survival, and treatment response. We believe that the availability of tumor-TIL maps along with software that allows interactive viewing of the computational analysis will improve reproducibility and precision in reporting tumor size, tumor boundary features, TIL assessment, and extraction of relevant nuclear and cellular features. These improvements will in turn enhance clinical and pathology decision support in guiding management, treatment selection, and predicting survival and recurrence, in conjunction with existing molecular subtyping platforms.
The need to quantify spatial interrelations between tumor regions and infiltrating lymphocytes is becoming increasingly important in invasive breast cancer. Tumor-TIL maps generated from H&E–stained images can be used to perform a wide range of correlative studies in the context of clinical trials, epidemiologic investigations, and surveillance studies. Our methods leverage open-source CNNs; the programs we have developed are also being made public and freely available. In summary, this study has produced a reliable and robust methodology, data sets of TIL and cancer region predictions, and programs that can be used to conduct tumor-TIL tissue image analyses of invasive breast cancers. Although our analysis approach has been implemented and evaluated with breast cancer cases, the proposed approach is not specific to breast cancer. Indeed, this approach has been used for the detection and segmentation of cancer regions in prostate and pancreatic cancer cases.
Our approach enables detection and segmentation of cancer regions in whole slide tissue images. Once a cancer region is identified, a more focused, higher resolution analysis within those regions can be executed. In addition, we expect that output from our approach could be used to improve the performance of other cell-level or subregion-level analysis methods. For example, an analysis method that detects and classifies cancer cells could use the output of our approach to check if a cell, which it labels as a cancer cell, actually is within the cancer region. We also expect that with sufficient training data that include regions of necrosis, deep learning models can be taught to differentiate between cancer regions, normal regions, and regions of necrosis. We should note that this claim requires further study and evaluation. Another approach could be to train a necrosis-specific model that can detect and segment regions of necrosis with high accuracy, as used in our previous work.32 We trained a necrosis-specific model to segment out necrosis regions and improve the accuracy of the overall TIL analysis pipeline.
In future studies, we will further refine our methodology and tools to differentiate between invasive and in situ premalignant lesions and explore methods that can facilitate faster predictions for practical real-time clinical applications.
Footnotes
Supported by National Cancer Institute (NCI) grants 1U24CA180924-01A1, 3U24CA215109-02, and 1UG3CA225021-01; and U.S. National Library of Medicine grants R01LM011119-01 and R01LM009239.
This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation grant ACI-1548562. Specifically, it used the Bridges system, which is supported by National Science Foundation award ACI-1445606, at the Pittsburgh Supercomputing Center (PSC). NCI Surveillance Research Program oversaw the Virtual Tissue Repository (VTR) Pilot Program, from which participating Surveillance, Epidemiology, and End Results (SEER) cancer registries (Greater California, Connecticut, Hawaii, Iowa, Kentucky, and Louisiana) supplied the whole slide images used in algorithm development and testing. The SEER VTR Pilot Program is supported by the Division of Cancer Control and Population Sciences at the NCI of the NIH.
A guest editor acted as the Editor-in-Chief for this manuscript. No one at Stony Brook University was involved in the peer review process or final disposition for this article.
H.L. and R.G. contributed equally to this work.
Disclosures: None declared.
Author Contributions
Conceptualization, J.S., T.K., R.G., H.L., A.L.V.D., D.S., D.F., T.Z., R.B.; methodology, J.S., T.K., H.L., A.R., L.H., S.A., D.S., R.A.M., L.T.-H.; data curation, R.G.; running experiments, H.L., S.A.; writing–original draft, H.L., R.G., T.K., J.S.A., A.S.; writing–review and editing, H.L., R.G., T.K., J.S., A.R., A.L.V.D; formal analysis, J.S., R.G., A.R., A.L.V.D, R.A.M., L.T.-H.; training CNNs, R.G. and H.L.; supervision, J.S., T.K., D.S.; visualization, H.L., J.S.A., E.B.; and software, T.K., E.B., J.S.A., A.S.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Anderson W.F., Matsuno R. Breast cancer heterogeneity: a mixture of at least two main types? J Natl Cancer Inst. 2006;98:948–951. doi: 10.1093/jnci/djj295. [DOI] [PubMed] [Google Scholar]
- 3.Heselmeyer-Haddad K., Garcia L.Y.B., Bradley A., Ortiz-Melendez C., Lee W.J., Christensen R., Prindiville S.A., Calzone K.A., Soballe P.W., Hu Y., Chowdhury S.A., Schwartz R., Schaffer A., Ried T. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of myc during progression. Am J Pathol. 2012;181:1807–1822. doi: 10.1016/j.ajpath.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denkert C., Von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E., Budczies J., Huober J., Klauschen F., Furlanetto J., Schmitt W.D., Blohmer J.U., Karn T., Pfitzner B., Kummel S., Engels K., Schneeweiss A., Hartmann A., Noske A., Fasching P.A., Jackisch C., Mackelenbergh M.V., Sinn P., Schem C., Hanusch C., Untch M., Loibl S. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 5.Loi S., Sirtaine N., Piette F., Salgado R., Viale G., Van Eenoo F., Rouas G., Francis P., Crown J., Hitre E., Azambuja E.D., Quinaux E., Leo A.D., Michiels S., Piccart M.J., Sotiriou C. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 6.Mao Y., Qu Q., Zhang Y., Liu J., Chen X., Shen K. The value of tumor infiltrating lymphocytes (TILs) for predicting response to neoadjuvant chemotherapy in breast cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e115103. doi: 10.1371/journal.pone.0115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruneri G., Vingiani A., Bagnardi V., Rotmensz N., De Rose A., Palazzo A., Colleoni A., Goldhirsch A., Viale G. Clinical validity of tumor-infiltrating lymphocytes analysis in patients with triple-negative breast cancer. Ann Oncol. 2015;27:249–256. doi: 10.1093/annonc/mdv571. [DOI] [PubMed] [Google Scholar]
- 8.Denkert C., Loibl S., Noske A., Roller M., Muller B., Komor M., Budczies J., Darb-Esfahani S., Kronenwett R., Hanusch C., Torne C.V., Weichert W., Engels K., Solbach C., Schrader I., Dietel M., Minckwitz G.V. Tumor associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 9.Denkert C., Wienert S., Poterie A., Loibl S., Budczies J., Badve S. Standardized evaluation of tumor-infiltrating lymphocytes in breast cancer: results of the Ring studies of the International Immuno-Oncology Biomarker Working Group. Mod Pathol. 2016;29:1155–1164. doi: 10.1038/modpathol.2016.109. [DOI] [PubMed] [Google Scholar]
- 10.Loi S., Michiels S., Salgado R., Sirtaine N., Jose V., Fumagalli D., Ellokumpu-Lehtinen P.L., Bono P., Kataja V., Desmedt C., Piccart M.J., Loibl S., Denkert C., Smyth M.J., Joensuu H., Sotiriou C. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the finHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 11.Ingold Heppner B., Untch M., Denkert C., Pfitzner B.M., Lederer B., Schmitt W., Eidtmann H., Fasching P.A., Tesch H., Solbach C., Rezai M., Zahm D.M., Holms F., Glados M., Krabisch P., Heck E., Ober A., Lorenz P., Diebold K., Habeck J.O., Loibl S. Tumor-infiltrating lymphocytes: a predictive and prognostic biomarker in neoadjuvant-treated HER2-positive breast cancer. Clin Cancer Res. 2016;22:5747–5754. doi: 10.1158/1078-0432.CCR-15-2338. [DOI] [PubMed] [Google Scholar]
- 12.Salgado R., Denkert C., Campbell C., Savas P., Nuciforo P., Aura C., De Azambuja E., Eidtmann H., Ellis C.E., Baselga J., Piccart-Gebhart M.J., Michiels S., Bradbury I., Sotiriou C., Loi S. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the neoALTTO trial. JAMA Oncol. 2015;1:448–455. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams S., Gray R.J., Demaria S., Goldstein L., Perez E.A., Shulman L.N., Martino S., Wang M., Jones V.E., Saphner T.J., Wolff A.C., Wood W.C., Davidson N.E., Sledge G.W., Sparano J.A., Badve S.S. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West N.R., Kost S.E., Martin S.D., Milne K., Deleeuw R.J., Nelson B.H., Watson P.H. Tumour-infiltrating FOXP3+ lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br J Cancer. 2013;108:155. doi: 10.1038/bjc.2012.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strome S.E., Sausville E.A., Mann D. A mechanistic perspective of monoclonal antibodies in cancer therapy beyond target related effects. Oncologist. 2007;12:1084–1095. doi: 10.1634/theoncologist.12-9-1084. [DOI] [PubMed] [Google Scholar]
- 16.Sharma P., Allison J.P. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotwals P., Cameron S., Cipolletta D., Cremasco V., Crystal A., Hewes B., Mueller B., Quaratino S., Sabatospeyton C., Petruzzelli L., Engelman J.A., Dranoff G. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17:286. doi: 10.1038/nrc.2017.17. [DOI] [PubMed] [Google Scholar]
- 18.Dieci M.V., Radosevic-Robin N., Fineberg S., Van Den Eynden G., Ternes N., Penault-Llorca F., Pruneri G., D’Alfonso T.M., Demaria S., Castaneda C., Sanchez J., Badve S., Michiels S., Bossuyt V., Rojo F., Singh B., Nielsen T., Viale G., Kim S.-R., Hewitt S., Wienert S., Loibl S., Rimm D., Symmans F., Denkert C., Adams S., Loi S., Salgado R. Update on tumor-infiltrating lymphocytes (TILS) in breast cancer, including recommendations to assess TILS in residual disease after neoadjuvant therapy and in carcinoma in situ: a report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol. 2018;52:16–25. doi: 10.1016/j.semcancer.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van Den Eynden G., Baehner F.L., Penault-Llorca F., Perez E.A., Thompson E.A., Symmans W.F., Richardson A.L., Brock J., Criscitiello C., Bailey H., Ignatiadis M., Floris G., Sparano J., Kos Z., Nielsen T., Rimm D.L., Nad Allison K.H., Reisfilho J.S., Loibl S., Sotiriou C., Viale G., Badve S., Adams S., Willard-Gallo K., Loi S. The evaluation of tumor-infiltrating lymphocytes (TILS) in breast cancer: recommendations by an International TILS Working Group 2014. Ann Oncol. 2014;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klauschen F., Muller K.R., Binder A., Bockmayr M., Hagele M., Seegerer P., Wienert S., Pruneri G., De Maria S., Badve S., Michiels S., Nielsen T., Adams S., Savas P., Symmans F., Willis S., Gruosso T., Park M., Haibe-Kains B., Gallas B., Thompson A., Cree I., Sotiriou C., Solinas C., Preusser M., Hewitt S., Rimm D., Viale G., Loi S., Loibl S., Salgado R., Denkert C. Scoring of tumor-infiltrating lymphocytes: from visual estimation to machine learning. Semin Cancer Biol. 2018;52:151–157. doi: 10.1016/j.semcancer.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Catacchio I., Silvestris N., Scarpi E., Schirosi L., Scattone A., Mangia A. Intratumoral, rather than stromal, CD8+ t cells could be a potential negative prognostic marker in invasive breast cancer patients. Transl Oncol. 2019;12:585–595. doi: 10.1016/j.tranon.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodfellow I., Bengio Y., Courville A. MIT Press; Cambridge, MA: 2016. Deep Learning. [Google Scholar]
- 23.Bejnordi B.E., Veta M., Van Diest P.J., van Ginneken B., Karssemeijer N., Litjens G. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandi P., Geessink O., Manson Q., Van Dijk M., Balkenhol M., Hermsen M. From detection of individual metastases to classification of lymph node status at the patient level: the CAMELYON17 challenge. IEEE Trans Med Imaging. 2019;38:550–560. doi: 10.1109/TMI.2018.2867350. [DOI] [PubMed] [Google Scholar]
- 25.Litjens G., Bandi P., Ehteshami Bejnordi B., Geessink O., Balkenhol M., Bult P., Halilovic A., Hermsen M., Van De Loo R., Vogels R., Manson Q.F., Stathonikos N., Baidoshvili A., Van Diest P., Wauters C., Van Dijk M., Van Der Laak J. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience. 2018;7:giy065. doi: 10.1093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aresta G., Araujó T., Kwok S., Chennamsetty S.S., Safwan M., Alex V. BACH: grand challenge on breast cancer histology images. Med Image Anal. 2019;56:122–139. doi: 10.1016/j.media.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Cruz-Roa A., Gilmore H., Basavanhally A., Feldman M., Ganesan S., Shih N.N., Tomaszewski J., Gonzalez´ F.A., Madabhushi A. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Roa A., Gilmore H., Basavanhally A., Feldman M., Ganesan S., Shih N., Tomaszewski J., Madabhushi A., González F. High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: application to invasive breast cancer detection. PLoS One. 2018;13:e0196828. doi: 10.1371/journal.pone.0196828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok S. International Conference Image Analysis and Recognition. Springer; Cham, Switzerland: 2018. Multiclass classification of breast cancer in whole-slide images; pp. 931–940. [Google Scholar]
- 30.Dong N., Kampffmeyer M., Liang X., Wang Z., Dai W., Xing E. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; Cham, Switzerland: 2018. Reinforced auto-zoom net: towards accurate and fast breast cancer segmentation in whole-slide images; pp. 317–325. [Google Scholar]
- 31.Saltz J., Gupta R., Hou L., Kurc T., Singh P., Nguyen V., Samaras D., Shroyer K.R., Zhao T., Batiste R., Van Arnam J., Cancer Genome Atlas Research Network. Shmulevich I., Rao A.U.K., Lazar A.J., Sharma A., Thorsson V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saltz J., Sharma A., Iyer G., Bremer E., Wang F., Jasniewski A., Diprima T., Almeida J.S., Gao Y., Zhao T., Saltz M., Kurc T. A containerized software system for generation, management, and exploration of features from whole slide tissue images. Cancer Res. 2017;77:e79–e82. doi: 10.1158/0008-5472.CAN-17-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goode A., Gilbert B., Harkes J., Jukic D., Satyanarayanan M. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagos Y.B., Merida A.G., Teuwen J. Image Analysis for Moving Organ, Breast, and Thoracic Images. Springer; Cham, Switzerland: 2018. Improving breast cancer detection using symmetry information with deep learning; pp. 90–97. [Google Scholar]
- 35.Liu Y., Kohlberger T., Norouzi M., Dahl G.E., Smith J.L., Mohtashamian A., Olson N., Peng L.H., Hipp J.D., Stumpe M.C. Artificial intelligence–based breast cancer nodal metastasis detection: Insights into the black box for pathologists. Arch Pathol Lab Med. 2019;143:859–868. doi: 10.5858/arpa.2018-0147-OA. [DOI] [PubMed] [Google Scholar]
- 36.Lee B., Paeng K. 2018. A robust and effective approach towards accurate metastasis detection and pn-stage classification in breast cancer. International Conference on Medical Image Computing and Computer-Assisted Intervention; pp. 841–850. MICCAI. [Google Scholar]
- 37.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv. 1409.1556. [Google Scholar]
- 38.He K., Zhang X., Ren S., Sun J. 2016. Deep residual learning for image recognition; pp. 770–778. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- 39.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. 2017. Inception-v4, Inception-ResNet and the impact of residual connections on learning. Thirty-First AAAI Conference on Artificial Intelligence. [Google Scholar]
- 40.Paszke A., Gross S., Chintala S., Chanan G. 2017. Automatic differentiation in PyTorch. NeurIPS Autodiff Workshop. [Google Scholar]
- 41.Xu Y., Jia Z., Ai Y., Zhang F., Lai M., Eric I., Chang C. 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; 2015. Deep convolutional activation features for large scale brain tumor histopathology image classification and segmentation; pp. 947–951. [Google Scholar]
- 42.Hou L., Singh K., Samaras D., Kurc T.M., Gao Y., Seidman R.J., Saltz J.H. 2016. Automatic histopathology image analysis with CNNs. Proceedings of the New York Scientific Data Summit. [Google Scholar]
- 43.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. [Google Scholar]
- 44.Bengio Y., Courville A., Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 45.Macenko M., Niethammer M., Marron J.S., Borland D., Woosley J.T., Guan X., Schmitt C., Thomas N.E. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009. ISBI’09. IEEE; 2009. A method for normalizing histology slides for quantitative analysis; pp. 1107–1110. [Google Scholar]
- 46.Reinhard E., Adhikhmin M., Gooch B., Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21:34–41. [Google Scholar]
- 47.Vahadane A., Peng T., Albarqouni S., Baust M., Steiger K., Schlitter A.M., Sethi A., Esposito I., Navab N. 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) IEEE; 2015. Structure-preserved color normalization for histological images; pp. 1012–1015. [DOI] [PubMed] [Google Scholar]
- 48.Hou L., Samaras D., Kurc T.M., Gao Y., Davis J.E., Saltz J.H. 2016. Patch-based convolutional neural network for whole slide tissue image classification; pp. 2424–2433. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X., Yao J., Zhu F., Huang J. 2017. Wsisa: making survival prediction from whole slide histopathological images; pp. 7234–7242. IEEE Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- 50.Drasgow F. Polychoric and polyserial correlations. Encycl Stat Sci. 2004;9 [Google Scholar]
- 51.Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., Mclellan M., Yau C., Kandoth C., Bowlby R., Shen H., Hayat S., Fieldhouse R., Lester S., Tse G., Factor R., Collins L., Allison K., Chen Y., Jensen K., Johnson N., Oesterreich S., Mills G., Cherniack A., Robertson G., Benz C., Sander C., Laird P., Hoadley K., King T., Network T.R., Perou C. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D., Khosla A., Gargeya R., Irshad H., Beck A.H. 2016. Deep learning for identifying metastatic breast cancer. arXiv. 1606.05718. [Google Scholar]
- 53.Nazeri K., Aminpour A., Ebrahimi M. International Conference Image Analysis and Recognition. Springer; 2018. Two-stage convolutional neural network for breast cancer histology image classification; pp. 717–726. [Google Scholar]
- 54.Golatkar A., Anand D., Sethi A. International Conference Image Analysis and Recognition. Springer; 2018. Classification of breast cancer histology using deep learning; pp. 837–844. [Google Scholar]
- 55.Albarqouni S., Baur C., Achilles F., Belagiannis V., Demirci S., Navab N. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35:1313–1321. doi: 10.1109/TMI.2016.2528120. [DOI] [PubMed] [Google Scholar]
- 56.Veta M., Van Diest P.J., Willems S.M., Wang H., Madabhushi A., Cruz-Roa A., Gonzalez F., Larsen A.B., Vestergaard J.S., Dahl A.B., Ciresan D.C., Schimidhuber J., Giusti A., Gambardella L.M., Boray T.F., Walter T., Wang C.-W., Kondo S., Matuszewski B.J., Precioso F., Snell V., Kittler J., Campos T.E.D., Khan A.M., Rajpoot N.M., Arkoumani E., Lacle M.M., Viergever M.A., Pluim J.P. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Rao S. 2018. Mitos-rcnn: a novel approach to mitotic figure detection in breast cancer histopathology images using region based convolutional neural networks. arXiv. :1807.01788. [Google Scholar]
- 58.Amgad M., Elfandy H., Khallaf H.H., Atteya L.A., Elsebaie M.A., Elnasr L.S.A., Sakr R.A., Salem H.S., Ismail A.F., Saad A.M., Ahmed J., Elsebaie M.A.T., Rahman M., Ruhban I.A., Elgazar N.M., Alagha Y., Osman M.H., Alhusseiny A.M., Khalaf M.M., Younes A.A.F., Abdulkarim A., Younes D.M., Gadallah A.M., Elkashash A.M., Fala S.Y., Zaki B.M., Beezly J., Chittajallu D.R., Manthey D., Gutman D.A., Cooper L.A.D. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics. 2019;35:3461–3467. doi: 10.1093/bioinformatics/btz083. [DOI] [PMC free article] [PubMed] [Google Scholar]