Abstract
Infectious diseases, such as the most recent case of coronavirus disease 2019, have brought the prospect of point-of-care (POC) diagnostic tests into the spotlight. A rapid, accurate, low-cost, and easy-to-use test in the field could stop epidemics before they develop into full-blown pandemics. Unfortunately, despite all the advances, it still does not exist. Here, we critically review the limited number of prototypes demonstrated to date that is based on a polymerase chain reaction (PCR) and has come close to fulfill this vision. We summarize the requirements for the POC-PCR tests and then go on to discuss the PCR product-detection methods, the integration of their functional components, the potential applications, and other practical issues related to the implementation of lab-on-a-chip technologies. We conclude our review with a discussion of the latest findings on nucleic acid-based diagnosis.
Keywords: COVID-19 diagnoses, Polymerase chain reaction, Microfluidics, Miniaturization, Point of care, Future of PCR
1. Introduction
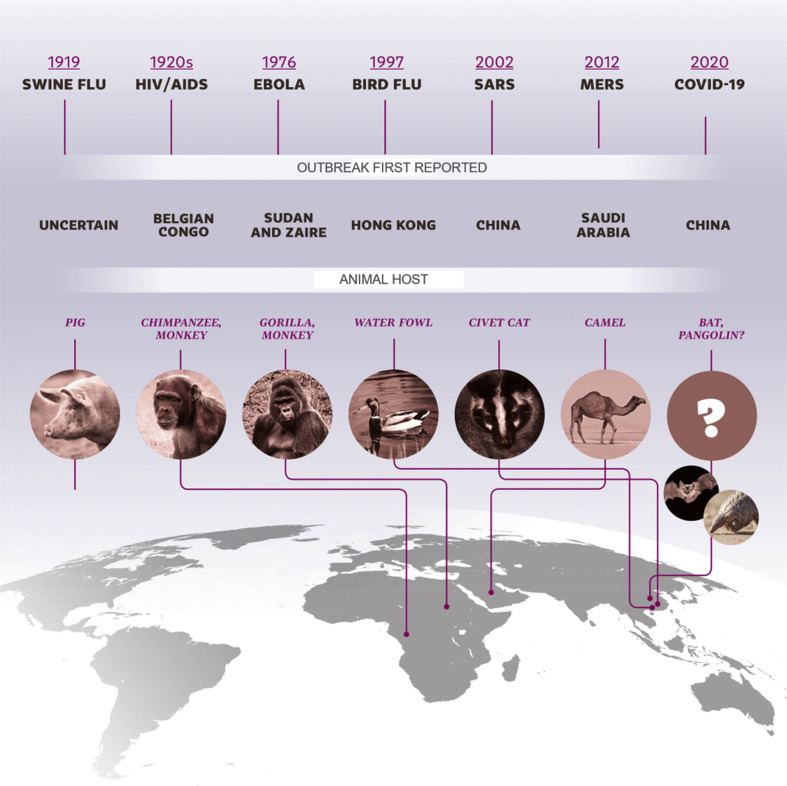
The 21st century has witnessed exponential growth in science and technology; however, sadly and ironically, it has succumbed to devastating coronavirus infections: severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and now coronavirus disease 2019 (COVID-19) [1,2]. As of July 7, 2020, a total of 11,500,302 COVID-19 cases had been reported across the world, together with 535,759 deaths [3]. The SARS and MERS coronaviruses were transmitted indirectly from bats to humans through civets and camels, respectively, as intermediate hosts [1], but the one responsible for COVID-19, has yet to be identified [4]. One of the main reasons that outbreaks such as these occur is the ability of viruses to evolve rapidly and change their animal hosts; this was seen with Spanish influenza, the single-largest pandemic of the last century, which was probably caused by a virus that was transmitted from ducks to domestic pigs and then to humans, in the densely populated region of southern China [5]. The virus was eventually transmitted to the USA and Europe after World War I. Recombination among viruses as they change their hosts is a natural process. There is, therefore, always a strong probability of new epidemics emerging from novel types of viruses, especially in densely inhabited regions (Fig. 1 ). The emergence of a coronavirus epidemic in the southern provinces of China has been predicted for some time, given the presence of multiple bat species that are infected with many different coronaviruses, in these highly populated areas [6].
Fig. 1.
A timeline of the major viral infections over the past 100 years, showing the geographical location of the first reported case and the interspecies transmission.
Coronaviruses and influenza viruses tend to be diagnosed through the detection of their specific genomes, in particular their ribonucleic acid (RNA) sequences. Rapid and precise screening of the public is essential to identify and quarantine infected people, thereby limiting or potentially eliminating the spread of the virus. The viral load of coronaviruses is high, making it easily detectable in symptomatic patients. Unfortunately, there is also a high percentage of asymptomatic individuals, making an accurate disease diagnosis of the population a challenging job unless the entire population is tested [7]. A low level of limit-of-detection (LOD) is crucial to shift the diagnostic window of opportunity toward the start of the infection process, to detect newly infected individuals. The test can be based on immunoassays, using antibodies to detect a specific antigen produced by the body's immune system or polymerase chain reactions (PCR) to detect a viral genome sequence. The PCR method is capable of multiplying the DNA with specific sequences in a short time, greatly enhancing the capability of infectious disease diagnostics. Since its invention in 1986, variants of PCR, such as the standard PCR (end-point PCR), the quantitative PCR (qPCR), and the digital PCR have been developed and subsequently applied in the field of molecular diagnostics. For coronaviruses, as with other RNA viruses, a reverse transcription step precedes the PCR (RT-PCR) and transcribes the viral RNA into complementary deoxyribonucleic acid (cDNA). The PCR test becomes the method of choice due to its sensitivity, as well as its specificity, and it is, in principle, capable of detecting a single copy of the virus, resulting in a shortening of the diagnostic window in comparison with immunoassays.
Technically, once a disease emerges, researchers identify the infectious agent by sequencing its genome, after which the newly obtained sequence is compared against sequences in the databases to evaluate its similarity to other known viruses. Determination of a sequence that is truly unique to the newly characterized virus represents a crucial step in the development of an effective diagnostic procedure. Furthermore, such a sequence should be selected from a region of the viral genome that is not variable between its different isolates and is not expected to evolve and mutate quickly. When a suitable sequence is determined, the design and synthesis of the relevant PCR primers can be performed routinely. In the case of RNA viruses, the reverse transcription that converts viral RNA to cDNA represents a crucial step, allowing the subsequent application of all methodologies based on PCR (Fig. 2 ).
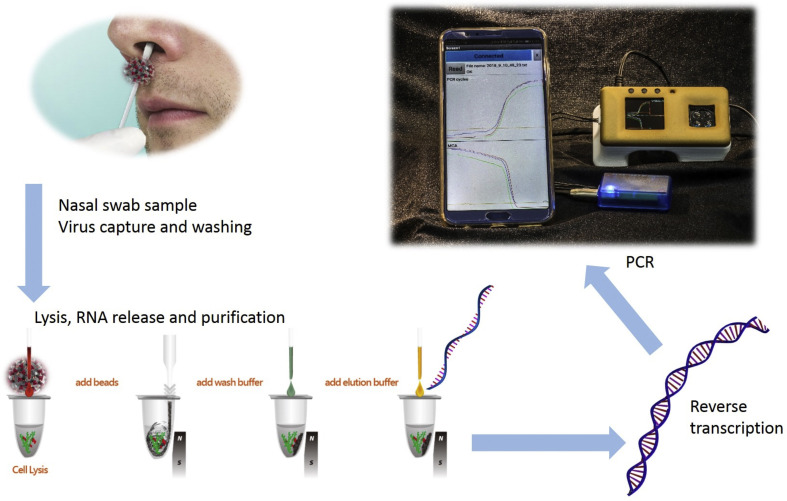
Fig. 2.
A conceptual demonstration of the detection of viruses in the field, based on real-time RT-PCR at POC. The process starts with a nasal swab to obtain a sample that might contain the virus. The sample is purified using paramagnetic beads and viruses are captured, followed by virus lysis to release the RNA, and purification Then, the RNA is reverse transcribed to cDNA; the number of cDNA molecules is multiplied by the PCR, and the results are displayed and possibly transferred to a centralized laboratory via a mobile device [19].
The raw sample and the reagents used in the sample preparation often contain a significant level of PCR-inhibiting factors; thus, proper sample preparation—whether manual or automated—is an essential step for successful detection [8]. The success of virus detection in real-world biological samples is highly dependent on the volume of the analyzed sample. During the initial phases of infection, the viral loads in patients may be near or below the test LOD [9]. The analysis of larger volumes of biological fluids with a low viral load is likely to reduce the risk of false-negative results, but microfluidic point-of-care (POC) devices are typically unable to handle mL-scale volumes in a short time due to the requirement of slow flow rates. Multiple approaches have been developed to address this problem. Flow-through capture membranes [10] have been used to capture nucleic acids in a short period of time, allowing the direct amplification of the captured material without any elution step, detecting as few as ≈10 copies of DNA with flow rates as high as ≈1 mL min−1. Magnetic beads [11,12] were used to isolate the RNA of avian influenza H5N1 and SARS from a ≈40 μL sample of blood; this is an insufficient volume on which to perform reliable coronavirus diagnostics. Carbon nanotubes [13] or aptamers [14] have been used to capture RNA from influenza A and H5N1 from avian flu viruses, respectively, and to promote their enrichment in samples. Here, we look back and identify those technologies that appeared since the emergence of SARS in late 2002. It is our aim to put landmark developments under the spotlight, critically evaluate them, and raise questions to identify the gaps that are currently hindering a commercial POC-PCR test. There have also been other recent outbreaks of infectious diseases that have threatened global human health, such as Ebola virus disease in the Democratic Republic of the Congo in 2018, measles in Burundi, and yellow fever. Additionally, the lower respiratory infections, tuberculosis, and human immunodeficiency virus (HIV) disease were listed among the top ten global causes of death in 2016 [15]. The PCR test on samples prepared from blood and urine specimens is capable of detecting the virus in the early stages of the disease, resulting in early diagnosis and subsequent isolation of infected patients to block transmission. For a comprehensive snapshot of the field, we refer the reader to in-depth reviews on the topic that have appeared in recent years [[16], [17], [18]].
2. PCR and its technology
2.1. The principles and brief history of PCR
Single-step RT-PCR is a technique derived from the original PCR [20] to perform a reverse transcription followed by the PCR temperature cycling. It requires a mixture of reverse transcriptase, DNA polymerase, short oligonucleotides complementary to the target cDNA, called primers, free nucleotides, and bivalent salts, such as MgCl2. The RT-PCR starts with reverse transcription, typically conducted at ≈60°C, followed by a hot start performed at ≈93°C, destroying the reverse transcriptase and activating the polymerase. Typically, there are 35–45 cycles consisting of a double stranded DNA (dsDNA) denaturation at 93°C, primer annealing at a temperature ranging between 55°C and 65°C, and extension at 72°C to finish the dsDNA. The three temperature protocols can be simplified by a two-step temperature protocol, whereby the annealing and elongation are conducted at the same temperature [21]. The classical endpoint detection system, which required an external PCR product detection by hybridization or electrophoresis, was modified by adding a fluorescent agent [22,23], either intercalating dye or a fluorescent probe that converts the endpoint PCR into a real-time PCR, also known as a qPCR.
PCR-based systems are, in principle, capable of detecting a single copy of RNA or DNA, with the assumption that the single DNA/RNA copy is presented in the sample and not lost during the sample preparation. The PCR/RT-PCR is a far superior technique in comparison with antibody-based assays due to the much smaller diagnostic window, i.e., it stops providing false-negative results earlier than the antibody-based assays. An alternative to PCR is a ligase chain reaction with short templates, whereby the products are only one base pair longer than the combined length of both the primers [24].
2.2. Other techniques based on the amplification of nucleic acids
A group of new techniques, called isothermal amplification of nucleic acids, has been introduced with loop-mediated isothermal amplification (LAMP) [21] and nucleic acid sequence-based amplification (NASBA) [25] as its main representative, with an important aim of bypassing PCR-related patents. Papers that have described these techniques start with the claim that LAMP is far superior to PCR because complicated temperature cycling is not required for isothermal amplification. However, temperature cycling is a very simple engineering method that helps to improve the PCR specificity. Proportional-integral-derivative controllers that include temperature sensors and solid-state relays are cheap and readily available. In addition to that, LAMP still needs a closed-feedback loop system heater to maintain the system's temperature in a range of 50°C–60°C. Both real-time PCR and real-time LAMP require a fluorescence detection system, and due to the rather weak fluorescent output signal the fluorescent signal related electronic processing circuits is far more demanding than those for temperature control. Although there is no requirement for temperature cycling, LAMP is a complicated process. Given that nothing comes for free, a simple LAMP requires six primers, but a PCR needs only two; thus, it does not seem to be correct to assume the simplicity of LAMP. Nevertheless, the LAMP system has its merits, which include the fact that thermal cycling is not required, resulting in better thermal isolation and, therefore, less power consumption compared with PCR. Also, LAMP can be designed to simplify the readout by colorimetry or nephelometry, instead of conventional fluorescence detection. [26] There are other isothermal nucleic acid amplification techniques using different principles and temperatures, such as strand displacement amplification [27], rolling circle amplification [28], nicking enzyme amplification reaction [29], recombinase polymerase amplification [30], and catalytic hairpin amplification [31].
2.3. Microfluidics and PCR miniaturization
Following in the footsteps of microelectromechanical systems (MEMS), the field of microfluidics—or lab-on-a-chip—emerged in the early 1990s, with the grand vision of producing a micro-total-analysis system (μTAS) [32]. Two analytical techniques pioneered the field: capillary electrophoresis (CE) [33] and PCR [34]. The first demonstrations of both techniques independently on the microchip format led to the PCR-CE integration [35] later on by implementation of a real-time PCR to a portable unit for a POC-PCR [36]. Since then, important innovations have taken place in terms of materials [37], as well as the method of implementation [38], extending the boundaries of this field. However, nearly three decades on, these efforts have yet to bear fruit in daily life outside the laboratory. One cannot help but wonder: why has the development and deployment of practical POC-PCR testing taken so much time? This is a pressing question, especially at a dire time like the present, when the world is battling another once-in-a-century pandemic, COVID-19 [39]. This is the raison d’être of μTAS. This background should not be taken as a criticism of the slow development because the task of making portable systems for viral diagnostics with minimal false-positive and -negative results is complex, and great progress has indeed been made in recent years.
PCR runs on a platform which cycles the temperature of the reaction between two or three set points for over 30 to 40 cycles. The reaction time is often limited by heating and cooling rate within each cycle determined by power dissipation (P) and system thermal time constant (τ). It is reduced with a reduced thermal mass (inclusive of the sample and the chamber) and with decreased isolation of this thermal mass from surrounding. Both features can be fulfilled by miniaturization, through MEMS technology and microfluidics, which has lowered the reaction time from >30 min to several min in the past decade [40] or even less [41] as:
| (1) |
where is the time taken by the system to change the temperature from one PCR step to another (), and C is its thermal capacitance. The total reaction time depends on a series of parameters, such as the chip size, the PCR master mix volume determining the value of C, the thermal conductivity of the substrate (G), and the temperature cycling rate. Here, the miniaturization of the PCR results in a smaller value of C, which proportionally lowers the Δt. Recently, an extremely fast PCR with a 0.4 s·cycle−1 has been demonstrated and resulted in an incredible total reaction time of less than 15 s [41]. This fast PCR could be combined with a fast melting curve analysis to achieve a total detection time of less than 1 min [42].
The PCR can also utilize passive cooling, with a cooling rate determined by its τ value:
| (2) |
Increasing the value of G leads, ultimately, to a faster system, unfortunately, the cooling-rate increase comes with a cost, making the system power demanding as:
| (3) |
and power demanding systems are not suitable for battery-operated POC devices. Nevertheless, the system heating/cooling rate can still be increased without compromising the power increase by minimizing the sample volume. Isothermal amplification systems benefit from the absence of a cooling requirement, making the τ value irrelevant. Then, there is an option to thermally isolate the heated part with a G value as low as possible to consume very little energy—a valuable feature of portable, battery-operated systems.
3. Temperature control (contact and non-contact heating)
POC-PCR requires fast thermal cycling and precise temperature control. There are demanding requirements for the design of temperature modules for miniaturized PCR devices.
Heating methods determine the temperature ramping rate and are currently divided into contact and non-contact heating. The contact heating method utilizes embedded/external heat sources, such as deposited thin films or external Peltier elements [43]; this method increases thermal mass, inevitably hindering fast thermal transitions during reactions. The non-contact heating method was developed using hot air [44] or infrared radiation as a heat source and directly heats the sample in small volumes [45]. These contact and non-contact heating methods employed for microchips depend on the material of the substrate, the reaction volume, and the structure of the chip.
Thermal isolation must also be considered in a miniaturized PCR system because thermal cross-talk deteriorates the chip's performance, especially in multifunctional reaction chambers on a single chip [46]. Furthermore, poor thermal isolation results in heat loss from the temperature zones to the surroundings, while the suspended PCR chamber enables good thermal isolation. A hollow reaction chamber has been made out of micromachined silicon (Si) that is integrated with both the heater and the sensor and is connected by a narrow beam to the substrate to achieve thermal isolation [47]. In addition, isolated layers with high thermal resistance have been applied to provide excellent thermal isolation [48]; however, they result in more complex fabrication and additional stress in different layers. Materials with low thermal conductivity, such as glass or plastics, could provide excellent thermal isolation for PCR chips, thereby avoiding the temperature cycling of the entire device [49].
4. Materials
The implementation of microfluidics has led to the development of miniaturized PCR systems that offer portability and save time, thereby facilitating applications in the realm of POC diagnostics. Researchers have developed a family of miniaturized devices based on different types of PCR. Materials such as Si [50], glass [51], polydimethylsiloxane (PDMS) [52], polycarbonate [53], and polymethylmethacrylate (PMMA) [54], used for fabricating microfluidic chips, are also available for the substrate of PCR chips. Each material has a number of merits and demerits, based on its individual properties.
Glass is used to make PCR chips due to its transparency and low auto-fluorescence. A multifunctional platform suitable for the on-chip detection of biomolecules consists of different thin films integrated on a single glass substrate that can also be optically and thermally coupled with another glass [51]. However, the low thermal conductivity of glass leads to an uneven temperature distribution and slow heating/cooling rates.
PDMS is currently one of the most widely used polymers for microfluidic devices due to its biocompatibility, low cost, and simple processing [55]. A disposable PDMS–glass bonding chamber has been utilized to perform PCR with an intercalating reusable electrode part for thermal cycling [52]. However, the evaporation of the PCR solution, located in the PDMS chamber, at a denaturation step may lead to cross-talk because PDMS is porous. PMMA is another popular candidate [56]. Inexpensive and versatile PMMA PCR chips can be fabricated by laser ablation technology, followed by bonding methods such as low-temperature bonding that uses optically clear adhesive film and liquid optically clear adhesive [54]. Additionally, the nonspecific adsorption between PMMA and DNA is minimal.
5. Surface treatment
The surface properties of microfluidic channels/chambers must be considered because the interaction between biomolecules in the PCR solution and the channels/chambers affects PCR efficiency. A high surface-to-volume ratio of microchannels/chambers leads to the non-specific adsorption of the enzymes, limiting or even prohibiting the reaction [57]. Two methods have been employed to solve this problem. One involves the use of silanization to modify the surface permanently. Silanization is accomplished by filling the channel with a silanizing agent and heating the filled chip for a period of time, then removing the agent and washing the chip. Alternatively, solutions such as bovine serum albumin (BSA) [58], polyvinylpyrrolidone [59], glycerol [60], polyethylene glycol 8000 [59], and Tween 20 [61] are commonly added into the PCR solution to reduce the non-specific adsorption, thereby improving the PCR efficiency.
Chips made of Si and glass can be cleaned using solutions such as piranha (H2SO4/H2O2), since the solution performs the mineralization of organic materials, and no RNA/DNA can be left behind. Nevertheless, general practitioners prefer disposable systems because they are safer in terms of cross-contamination. In the future, PCR chips could well be able to be cleaned and reused, once the cleaning process has shown its consistency.
6. Types of PCR
6.1. Time domain
The first conventional thermal cycler, called Mr. Cycle, was a time-domain system, as are most current PCR devices. Samples are placed at fixed locations on a heater whose temperature is modulated according to the required PCR protocol. The heaters can be based on dissipating Joule heat with active heating and passive cooling, or by utilizing thermoelectric coolers with active heating, as well as cooling.
The PCR master mix is dispensed into a stationary location, such as droplets or microwells, and microheaters are then used to conduct the thermal cycling beneath the droplets or wells. There are several techniques that can be employed to split samples to form droplets. A few μL of PCR master mix is pipetted onto a hydrophobic/oleophobic glass coverslip and covered by mineral oil, forming a virtual reaction chamber (VRC) [47].
Alternatively, the droplets, including cells separated by mineral oil, form an emulsion and are loaded into the microfluidic channel and are then transferred to different positions within the chip, after which they are subjected to a series of procedures such as cell lysis, DNA extraction, and purification [62]. Then, the thermal cycling is conducted by heating this emulsion or VRC, and cameras capture the subsequent fluorescence.
A variety of chip structures are fabricated by micromachining with different wells or microfluidic channels in materials such as Si, glass, and polymers, typically PDMS or PMMA. Samples are loaded into either wells or channels for subsequent DNA amplification by PCR [63].
6.2. Space domain
The PCR master mix is pumped into a channel or chamber in the chip, typically with two/three zones maintained at constant temperatures [34]. The reaction is performed when the solution is passing through these temperature areas by denaturation, annealing, and extension. This is an old technique that was used in the early days of PCR systems, with three baths with different temperatures and a laboratory technician or robot to move a basket with samples from bath to bath. Currently, this technique is still utilized in miniaturized systems, such as flow-through and rotational types. The flow-through method is a typical space-domain PCR method. The solution is pumped into a microfluidic channel made of polymers with different temperature regions, and a number of microheaters are applied to provide different temperature zones for denaturation, annealing, and elongation when performing the PCR. These zones maintain constant temperatures, and only the temperature of the samples is changed while it is moved through the zones with different temperatures [49].
6.3. Integration with sample preparation
The nucleic acids to be detected are first isolated from the bacteria or viruses using technologies such as lysis, pre-concentration, and purification. These procedures can be fulfilled on-chip or off-chip using bench-top instruments.
A number of fully integrated systems have so far been developed, starting with the non-portable GeneXpert from Cepheid, which was first utilized for anthrax detection by the United States Postal Service [64], and then with different primers to perform HIV and tuberculosis diagnostics in South Africa [65]. Despite being non-portable, it is a true sample-to-answer system that uses injection-molded plastic preloaded cartridges with lyophilized compounds, including a PCR master mix, as well as infectious agent-specific primers. Their prohibitive capital cost has unfortunately restricted the spread of these systems, given that those employed in South Africa were heavily subsidized.
We demonstrated the first monolithic Si-based chip integrating a micromixer for diluting blood sample, a microfilter for isolating and chemical lysing of leukocytes, a binder for capturing and purifying nucleic acids, and microvalves for fluidic control [66,67]. The captured DNA was amplified in a PCR chip followed by the product detection using Si nanowires [68].
A large sample volume to be processed needed to be stored in a plastic cartridge next to the Si chip, and the Si chip experienced processing problems due to its small channel cross section. The first step is to filter out red blood cells, while effectively capturing white blood cells for subsequent lysis. The DNA-capturing system was based on a SiO2 surface inside the chip, was orders of magnitude smaller than the one based on microfiber membranes or silica beads. Finally, the use of Si nanowires as DNA sensors offers somewhat questionable repeatability. The replacement of a conventional PCR with a qPCR would make life easier because there would be no sample manipulation after PCR, and qPCR is well established technique. Nevertheless, the system had limited portability.
6.4. Integration with detection methods
Standard PCR (end-point) typically utilizes electrophoresis of either a gel or capillary type to detect PCR products after thermal cycling. A PCR master mix is pumped into a microchannel fabricated in a microfluidic chip, and PCR is performed using microheaters. After the reaction, the solution is taken out, and gel electrophoresis (GE) is conducted to check the presence and specificity of the amplicons [69]. Alternatively, the solution is transferred to a CE column on a chip with electrodes on both sides, and the products are detected by CE [70].
Detection by GE/CE made the chip or system design complicated, hindering the system miniaturization for POC diagnostics. Inspired by commercial real-time PCR instruments, optical detection modules, such as optical fibers [71] and lock-in amplifiers [72], were then integrated into miniaturized qPCR systems while intercalating dyes such as SYBR Green or probe were added into the PCR master mix, and the reaction was monitored in real time. Furthermore, additional types of qPCR systems combined with other technologies have been developed to improve performance. A pico-liter droplet array generated by double-inkjet printing was demonstrated to be capable of performing parallel qPCR with an enhanced throughput [73]. A complementary metal oxide semiconductor was integrated with the PCR chip used for multiplexing detection, which enhanced detection sensitivity [74]. Electrowetting on dielectric technology is well adapted for single-cell isolation, mRNA purification, and subsequent multiplex qPCR at the single-cell level [75].
7. The vision and the reality
The miniaturized PCR was first envisioned in the early 1990s, and since then numerous PCR microchips have emerged. Most of these PCR microchips have required off-chip sample preparation and even off-chip detection. These undesired features hinder complete freedom from laboratory settings and give rise to the tongue-in-cheek expression for these devices: chip-in-a-lab [76]. The sample preparation, which involves the extraction and purification of nucleic acids from bodily fluids, plays a determining role in the accuracy of the test, which heavily depends on the effective removal of PCR inhibitors such as hemoglobin. However, the diversity and complexity of bodily fluid samples, together with the sample volume requirement, make it difficult to combine sample preparation with PCR on a microchip or in a cartridge. Few studies have undertaken the challenge of demonstrating a fully integrated PCR system, where sample preparation, amplification, and detection occur in one platform. In this section, we describe these integrated solutions and the issues preventing their POC use.
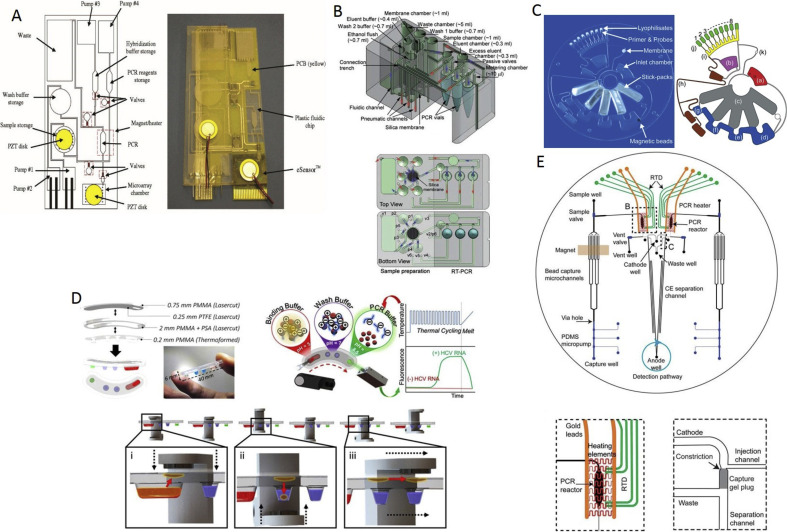
The first of these was arguably a unit reported ≈15 years ago based on the eSensor microarray chip (Fig. 3 A) [77]. The eSensor was a printed circuit board featuring a thin-film metal electrode array (4 × 4 array), with DNA capture probes to hybridize the PCR products for multiplexed electrochemical detection. The overall unit was a fully integrated system made up of pumps, valves, and fluidic micromixers, all made free from any fabricated moving parts and based on thermopneumatic expansion and electrolysis for pumping, cavitation microstreaming for mixing, and phase transition for fluidic valving. Eliminating such parts simplified not only the design but also the fabrication, thus lowering the overall cost. The cost was further reduced by patterning the electrodes through the standard printed circuit board (PCB) process and forming the polycarbonate channels and chambers through computerized numerical control-machining. Cavitation mixing was induced by a pair of piezoelectric disks mounted on the sample chamber and the microarray detection chamber, while electrolytic pumping was activated by platinum wires inserted into the pumping chambers. A magnet was mounted on the PCR chamber to trap and enrich the immunomagnetic capture beads that carried target pathogens or cells from the sample chamber for subsequent thermal lysis and asymmetric PCR. The unit was shown to work for pathogenic bacteria detection from whole rabbit blood by spiking it with Escherichia coli, after which a single nucleotide polymorphism analysis was performed. Despite the device's simple fabrication process, it entailed post-fabrication steps, such as introducing phase-changing material wax into the valves and conditioning the channel's surface. Despite the claim that it is completely self-contained in terms of reagents and waste storage, the reagents were loaded at the time of sample introduction, which added to the in-use handling. Other technical issues included the low efficiency of the PCR due to the immunomagnetic beads (≈50%) and their low cell-capture efficiency of ≈40%, as well as the low sensitivity of electrochemical detection. Furthermore, the overall assay time was relatively long as sample preparation, amplification, and detection took ≈1 h each.
Fig. 3.
The cores of the fully integrated PCR systems. (A) (left) Schematic of the plastic fluidic cartridge using three electrochemical and one thermopneumatic pump; (right) Photograph of the fabricated unit, consisting of a plastic fluidic cartridge, the PCB, and the eSensor microarray chip [82]. (B) (up) Drawing of the all-in-one cartridge. (down) The top and bottom views of the all-in-one cartridge [83]. (C) (left) Photograph of the LabDisk. (right) Schematic of the LabDisk [84]. (D) (top left) Schematic of the cartridge; (top right) Principle of the platform; (down) Schematic of the particle transfer [85]. (E) (up) Schematic of the microdevice; (bottom left) View of the PCR chamber; (bottom right) View of the gel-based capture part [86].
A biochip featuring an integrated PCR with solid-phase extraction (SPE) and CE detection has been proposed and tested [78]. Bacillus anthraces (anthrax) and Bordetella pertussis have been detected within ≈30 min. While this is impressive, the biochip was not a self-contained unit with reagents and waste storage. Reagents had to be externally supplied during each test and the sample had to be lysed off-chip. Both these steps have to be integrated when providing a biochip or a cartridge for a portable diagnostic system. The possible inhibition of PCR by reagent traces of SPE was avoided through the integration of membrane valves that decoupled the two process sites. The valves, however, involved multiple structural layers, adding further complexity to the fabrication. Moreover, the valve activation required compressed air from an external line, which increased the hardware requirement, in addition to the infrared mediated thermal cycling, high-voltage product separation, and laser-induced fluorescence detection. The SPE site had to be packed with silica beads, which added to the post-fabrication handling. Despite all these technical issues, however, the study demonstrates the feasibility of a highly rapid sample-in answer-out capability.
A portable and affordable real-time PCR system has been developed. The core of the system is a micro-machined Si chip integrated with a thin-film metal heater and a resistive temperature detector type of sensor [47]. The basic philosophy the researchers had in mind was to develop a cheap system with disposable parts in contact with the sample to avoid sample-to-sample cross-contamination, and, therefore, the MEMS chip was separated from the sample by a disposable microscope coverslip. The sample was placed together with a volume of a few μL, covered with mineral oil to prevent evaporation, forming a VRC. The first device was rather slow with sample and mineral oil volumes of 5 μL and 10 μL, respectively. The temperature was controlled externally via a personal computer, and the fluorescence was captured by an external microscope. Separate to the PCR system, the same group also developed a miniaturized fluorescent system with the vision of integrating it later on with the PCR system [79]. Then, the MEMS system was redesigned, and the sample volume was lowered to increase the speed to perform 40 PCR cycles in less than 6 min [40], after which everything was integrated into a single system, demonstrating real-time RT-PCR detection in a sample containing the RNA of the H5N1 avian flu virus in a single VRC—which is clearly insufficient for actual testing [80]. The system was evolved into a world-smallest real-time PCR capable of detecting four samples at a time and demonstrated the ability to perform real-time PCR of cDNA from the H7N9 avian influenza virus [81], RT-PCR from the Ebola virus RNA [72], and finally, it was integrated with a Bluetooth communication system that detected cDNA from the dengue fever virus [19]. The system was even combined with the sample preparation step to detect RNA from the H5N1 avian flu [11] or SARS [12], although they were far from perfect and required further improvement. Furthermore, the VRC looked like an open system, but the system should be converted into a closed system to increase user comfort and avoid the possibility of accidental contamination.
A sample-to-answer system that runs an automated sample preparation and real-time RT-PCR in an all-in-one cartridge has been developed and tested for influenza (H1N1), achieving an LOD of 100 copies·mL−1 [83]. The cartridge was a three-dimensional acrylic block with chambers that extended between the top and bottom surfaces and featured millimeter-sized interconnecting channels (Fig. 3B). One chamber contained a silica membrane for nucleic acid adsorption, while another was designated for waste storage. All the remaining chambers were preloaded with liquid reagents, and the cartridge was completely sealed to reduce the risk of viral exposure or sample-to-sample cross-contamination. A nasopharyngeal swab sample, diluted in viral transport media, was injected, together with a lysis buffer, into the silica membrane chamber from a syringe equipped with a needle by punching the seal. The sample and reagent liquids were pneumatically manipulated within the cartridge under pressure, and a vacuum was applied from syringe pumps through the seal-piercing connectors. Further fluidic control was provided by built-in surface tension valves. The amplification took place in three separate vials, each containing a frozen PCR master mix, which were inserted into the cartridge prior to the test. All the hardware was built in a desktop system that completed a fully automated test within 2.5 h. The test duration could be further shortened through miniaturization, which could also minimize the large dead volume introduced by the commercial silica membrane, as well as avoid diluting the viral RNA due to a large eluent volume for elution consistency. Moreover, the absence of mixing strategies implemented within the PCR vials led to a slightly lower PCR efficiency. Otherwise, the cartridge appears to be simple and amenable to injection molding for mass dissemination.
The fully automated sample-to-answer detection of influenza type H3N2 was based on a centrifugal microfluidic disk [84] named the LabDisk (Fig. 3C). The disk was fully enclosed with preloaded reagents comprising liquid stick packs for nucleic acid extraction, air-dried specific PCR primers with fluorescence probes, and a lyophilized RT-PCR master mix. After loading the sample, the disk started to rotate creating a centrifugal force, triggering a series of events. First, the content of each stick pack was released into a designated chamber at a well-defined number of revolutions per min, breaking the fragile seals of the stick packs. Then, the silica-coated magnetic beads captured and transported the nucleic acids from the lysis chamber into subsequent chambers for washing and release, with the aid of an external stationary magnet. The subsequent eluent was aliquoted into eight reaction cavities, dissolving the lyophilized RT-PCR master mix before thermal cycling through convective heating. The entire test took less than ≈3.5 h, with an LOD of ≈2.4 × 104 of copies⋅mL-1 . All the hardware was packed into a lightweight shoebox-sized unit suitable for POC use, with the only manual step being the sample loading. The centrifugal pumping, together with the surface tension valving, simplified the overall design, allowing the disk to be fabricated by microthermoforming a polymer foil via hot embossing. The thermoformed foil was then sealed by a pressure-sensitive adhesive tape. Although the LOD shown is fourfold lower than the clinically relevant level, the disk needs to be further developed to process viscous clinical specimens, such as sputum, through sample liquefaction and homogenization. A further reduction in analysis time and a demonstration of the ability to conduct multiplexing for sub-typing viral strains would be likely to bring the disk closer to practical POC use.
The detection of the hepatitis C virus (HCV) in serum was demonstrated using a system based on a thermoformed plastic cartridge [85] where the reagents were confined into discrete wells isolated from each other by a layer of oil. The first well contained an acidic binding buffer that induced a positive surface charge on the pH-responsive magnetic particles mixed into a serum sample before being introduced. Accordingly, the nucleic acids were captured by the charged particles and transported from well to well as the cartridge went through a series of translations and rotations between a pair of opposing stationary magnets (Fig. 3D). The surface charge was neutralized by an alkaline PCR solution, and the nucleic acid release was triggered. Following the removal of the particles from the well, rapid thermal cycling took place, administered by the surrounding heat block through the thermoformed thin layer of plastic. A fluorescent signal was captured via a confocal detector placed above the well. More than a dozen clinical serum samples were tested, each analyzed within an hour and generating LODs as low as 45 IU per 10 μL. While these results are encouraging, the unit gave rise to some of the same concerns as the others. First, the RT-PCR enzymes required refrigeration; this issue might be difficult to address by keeping the enzymes lyophilized because the PCR solution itself served as the RNA eluate necessary for rehydration. Second, the unit analyzed blood serum, which must be extracted beforehand, while the unit must be evaluated for other bodily samples relevant to respiratory infections such as sputum or a nasopharyngeal swab. Third, the dynamic range should match the entire range of viral loads encountered clinically. Fourth, the unit faced the issue of equally allocating particles among the wells, each of which has a distinct set of primers specific to the target serotype. Fifth, the greater spread of the data was an issue relevant to the retrieval of viral RNA but could be addressed by increasing the eluate volume by an order of magnitude, at the expense of test sensitivity.
The processes of DNA purification, PCR, post-PCR clean up, and inline injection, as well as CE separation, were all combined into a single microchip for forensic human identification (Fig. 3E) [86]. The microchip included a miniaturized PDMS pump and valves, a fluidized bed of immunomagnetic beads in bifurcating channels, a 250 nL PCR chamber, and a capillary electrophoresis channel with a double-T junction. The DNA purification was realized through a sequence-specific capture of DNA by a fluidized bed of streptavidin-coated magnetic beads. This, however, required off-chip handling, fragmenting the genomic DNA, and then modifying the fragments with biotin-labeled capture probes. Moreover, the DNA capture efficiency was rather low (5.4%). An external magnet was applied to arrest the beads in the bifurcating channels and then manually displaced to transport the beads into the PCR chamber. The PCR master mix, including biotin-labeled primers, was introduced, and the PCR products were then concentrated into a narrow band within a streptavidin-modified gel plug inside the double-T junction. The thermal release of the band led the CE separation and detection. The microchip was accompanied by a small-footprint, battery-powered instrument capable of a full nineplex short tandem repeat (STR) typing from 2.5 ng input DNA, as well as forensic STR analysis from oral swabs within 3 h. Nevertheless, the high level of integration creates complex fabrication and post-fabrication handling, including the alignment and bonding of multiple layers of glass and a PDMS membrane. All reagents had to be externally supplied and the sample lysed outside the microchip.
Table 1 lists the prototypes discussed above and compares their important features. The LabDisk stands out as the sole cartridge that is fully enclosed with reagents and a waste storage. It can be stored at room temperature owing to the lyophilized PCR master mix. Reagent evaporation and cross-mixing are addressed by the individually sealed stick packs that keep the reagents isolated. This isolation, along with the plastic housing, renders the cartridge robust against handling and transportation. The use of centrifugal microfluidics and silica-coated magnetic beads facilitates on-chip pumping, valving, and fluidic mixing that are realized in a simple design that can be produced through thermoforming. Moreover, the cartridge runs on an instrument very similar to the CD or DVD players of the past. However, several challenges remain. The cartridge must be further developed to handle clinical specimens. The overall test duration should be reduced to below 1 h. Finally, all these prototypes, albeit underdeveloped and very limited in number, are encouraging. However, it is imperative that many more studies are geared towards a POC-PCR test, instead of stand-alone PCR microchips, so that the next outbreak can be contained promptly. The time is running out for the vision before it fades away into a dream and a legacy for future generations.
Table 1.
A comparison between the different systems currently available or under development. “cfu” stands for “colony forming unit,” “c” for “copy,” “HCV” for “hepatitis c virus,” and “STR” for “short tandem repeat,” “NA” for “not applicable,” “NS” for “not stated,” “LOD” for “limit of detection”, “MA” for “microarray”, “CE” for “capillary electrophoresis,” and “IU” for “infection unit”.
| On-chip manipulation |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref | Reagents | Waste Storage | Fully Enclosed | Refrigeration | On-chip lysis | SPE column/membrane | Pumping | Valving | Magnetic beads | Fluidic Mixer | Multiplexed PCR | Sample | Target | On-chip Detection | Volume (μL) | RT-PCR/PCR (min) | Sample-to-answer (min) | LOD | # of units tested | Hardwareb | Fabricationc |
| [77] | Y | Y | Y | N | Y | N | Y | Y | Y | Y | Y | Blood | DNA (E. coli) | MA | 1000 | 90 | 210 | 103 cells | NS | ∗∗ | $$ |
| [78] | N | N | N | Y | N | Y | N | Y | N | N | N | Blood/Nasal | DNAa | CE | 0.75/1 | 11 | 30 | 1500-2000 cfu | NS | ∗ | $$$ |
| [83] | Y | Y | Y | Y | N | Y | Y | Y | N | N | Y | Nasal swab | RNA (H1N1) | qPCR | 200 | 130 | 150 | 100 c·μL−1 | NS | ∗ | $ |
| [84] | Y | Y | Y | N | Y | N | Y | Y | Y | N | Y | Viral stock | RNA (H3N2) | qPCR | 200 | 68.7 | 210 | 2.4·104 c·mL−1 | 18 | ∗∗∗∗ | $$ |
| [85] | Y | Y | Y | N | NA | N | NA | NA | Y | NA | N | Serum | RNA (HCV) | qPCR | 10 | 20 | 60 | 45 IU/10 μL | NS | ∗ | $$ |
| [86] | N | N | N | Y | N | N | Y | Y | Y | N | Y | Oral swab | DNA (STR) | CE | 10 | 40 | 180 | 2.5 ng | NS | ∗∗∗∗ | $$$$ |
| [11] | N | N | N | Y | Y | Y | N | N | Y | N | N | Throat swab | RNA (H5N1) | qPCR | 24.5 | 21.7 | 28 | 5 c | NS | ∗∗ | $ |
Blood and nasal samples were tested for B. anthracis and B. pertussis, respectively.
∗∗/∗∗∗∗ means the system requires more peripheral instruments.
$$/$$$/$$$$ means more expensive.
8. Quo vadis portable viral detection?
One of the most important practical challenges to be resolved for any POC molecular diagnostic device is sample preparation. Ideally, it should be a sample-to-answer system with no human interaction; this is especially important for POC diagnostics, such as for SARS in the past and currently for COVID-19. During the SARS outbreak in Singapore, which was one of the worst-hit countries, the Tan Tock Seng Hospital was almost the only clinic capable of carrying out routine SARS diagnostics. Prospective patients' only option for testing was to get there, leading to a high chance of cross-infection. It was seen to be greatly beneficial to have a system for POC to ensure that prospective patients did not have to go anywhere, but rather stay at home and call health care providers to come to their location to conduct infectious disease diagnostics, and not simply collect a sample. This would demand a small system that is easily portable, with samples sealed to avoid sample-to-sample cross-contamination, thus suppressing the number of false-negative results.
Sample preparation is definitely a problem, and many attempts have been made to simplify these crucial steps. There are too many PCR inhibitors in human body fluid samples, such as blood and saliva. Over the years, there has been continuous research into the development of PCR reagents, and especially into methods aimed at simplifying the sample preparation steps and fully integrating the sample preparation with PCR. Single-step direct PCR has also been developed, greatly simplifying the sample preparation process; only hemoglobin has to be removed before performing PCR to diagnose malaria infection [87]. From the standpoint of practicality, the extraction and isolation of the DNA/RNA of pathogens from saliva are most desirable for POC applications. Several studies have explored this area, including standard labor-intensive techniques [88].
PCR systems that offer multiplexing capability are preferred because they can reduce the test time for multiple diseases. This can be particularly beneficial during the flu season because different virus strains often present during one season. The differential diagnostics of community-acquired pneumonias can also benefit from this methodological approach, allowing not only the detection of multiple viruses and bacteria in one PCR reaction but also their relative quantification in the examined sample. Such a quantification may lead to the correct determination of the disease-causing agent by selecting the most suitable therapy [93], thus tackling the spread of a pandemic disease. The application of the broad-range PCR in multiplex arrangements can be used to detect a suspect group of viruses, giving rise to more detailed analyses in the initial phases of outbreaks [94,95].
The PCR for POC and other applications should fulfill the essential requirements of portability, operability by untrained personnel, fast and adequate accuracy for screening reliability, and giving the minimum information for the publication of quantitative real-time PCR experimental guidelines [96]. Miniaturization enjoys a big advantage in POC applications due to its portability and excellent accuracy and specificity; however, the complications arising from system operation outside the specialized laboratory environment as well as by untrained people pose the biggest challenge for the development of such devices.
Developments in the past have led to the commercialization of sample-to-answer systems that are more or less portable. One of the early birds in this regard was GeneXpert [97], a cartridge-based system that performed a fully automated nucleic acid amplification technique. It was designed for the detection of healthcare-associated infections, critical infectious diseases, sexual health, viruses, and cancers. Its development was accelerated by concerns over anthrax in the USA [64] and via the call for a proposal by The Bill and Melinda Gates Foundation to tackle HIV and tuberculosis in South Africa [65]. The system simplifies the testing procedure as it integrates sample preparation with qPCR/RT-PCR. The system is designed to be employed in areas lacking sophisticated facilities to conduct conventional testing. It is not easily portable but it does offer the option of multicolor multiplexing.
There was another push to release PCR systems for POC applications and this has contributed to disease diagnosis during pandemics such as the 2014 Ebola outbreak. Starting in early 2014, an unprecedented Ebola outbreak occurred in West Africa. On August 8, 2014, the World Health Organization (WHO) declared it a public health emergency of international concern [98] and initiated an emergency use assessment and listing (EUAL) mechanism to encourage research and development (R&D) of early access to unregistered products of in-vitro diagnostics [99]. Cepheid received a $3.3 million grant, co-financed by the Paul G. Allen Family Foundation and the Bill & Melinda Gates Foundation, to develop the GeneXpert Ebola Assay based on the GeneXpert system. It took only ≈5 months from development for it to get listed in EUAL by WHO. The GeneXpert Ebola Assay is fully automated and only requires the placing of the patient sample into the cartridge and inserting the cartridge into a compact desktop-level instrument. It takes ≈2 h for an entire assay, from sample to answer, and features high detection accuracy [100]. FilmArray BioThreat-E is another product that was given emergency use authorization from both the food and drug administration (FDA) and WHO. The assay is also a cartridge-based automatic RT-PCR system with a turnaround time of only 1 h [101].
The COVID-19 global pandemic has greatly boosted the determination and motivation of researchers worldwide to speed up the development of POCT systems, as well as the number of companies prepared to fund such work. Some have adapted existing devices, while others have made new ones. The systems that were available in March 2020 mostly contained immunoassays [102]; one was PCR-based, involving clusters of regularly interspaced short palindromic repeats, known as clustered regularly interspersed short palindromic repeats (CRISPR), with a CRISPR-associated protein, known as Cas, performing the detection of the isothermal amplification products. Other companies that have developed systems to diagnose seasonal flu have made use of different primers to convert these systems into POCT for COVID-19.
The Eiken Chemical Co. in Japan has developed a system for multiple testing that combines sample preparation with LAMP, and the optical detection of the results with RNA purification is based on magnetic force (Fig. 4 A) [89]. It employed a microfluidic cartridge with 25 wells, allowing for multiplexed detection.
Fig. 4.
The systems currently available for COVID-19 POC testing (POCT). (A) A system performing 25 reactions at a time using magnetic beads for sample preparation, followed by a LAMP [89]. (B) Cobas® Liat® System by Roche originally designed as a universal molecular biology platform for diagnoses based on a real-time PCR [90]. (C) ID NOW, made by Abbott Laboratories, originally sought to diagnose seasonal flu; it has currently been converted into a COVID-19 system, based on an isothermal DNA amplification originally developed for the seasonal flu test [91]. (D) Bosch's new microfluidic system is capable of virus diagnostics; the sample preparation is integrated with a multiplexed end-point PCR [92].
Roche created a POCT system called Cobas® Liat®, a fast and fully automated sample-to-answer system based on a PCR capable of testing samples in 20 min or less (Fig. 4B). It utilized a sealed-tube design that eliminated the operators' contact with the reagents. Its closed system with multiple controls also reduces the potential for human error [90].
ID NOW, from Abbott Laboratories, is a fully integrated sample-to-answer system that was originally developed to diagnose a seasonal flu (Fig. 4C) [91], and is currently available with modified primers to diagnose the COVID-19 virus [103]. It uses the isothermal amplification of nucleic acid, and it is capable of processing two samples at a time, with a claimed 5 min wait required for positive and 15 min for negative results.
Recently, Bosch began R&D of a POCT system; the result of their efforts is the Vivalytic System (Fig. 4D), a molecular diagnostic device based on a microwell array that is capable of detecting the coronavirus in less than 2.5 h, using a fully sealed cartridge. The Vivalytic system performs sample preparation, after which it performs a multiplex PCR, followed by microarray-detection, to allow the identification of SARS-CoV-2 [92].
Currently, the large-scale practical deployment of the above-mentioned devices to combat the pandemic spread of COVID-19 will allow their advantages and disadvantages to be quickly determined, after which further development can be encouraged. More systems will emerge in the near future in the context of the COVID-19 pandemic, after which companies are likely to switch their interest to systems that are more viable from an economic perspective. As of July 7, 2020, there were 104 approved test kits and systems approved by the U.S. FDA for emergency use authorization, with the vast majority being based on a real-time RT-PCR technique with or without sample preparation [104].
Given the development of communication technology, more and more devices are envisioned to be connected to form a network called the Internet of things (IoT), a burgeoning industry that is projected to change human society tremendously by bringing about convenience, ubiquitous information, and intelligence to improve our lives in a fundamental way; the health care industry can greatly benefit from this technological advancement. There have also been developments in personal healthcare monitoring devices linked with wearable healthcare monitoring, such as electrocardiogram and blood O2 measurement devices. The data collected can be used for remote and/or real-time disease monitoring and predictive model building. Furthermore, it is easy to predict that a diagnostic device such as PCR, with high connectivity and data transmission capabilities, can become highly impactful in pandemic management. We have demonstrated such a PCR in a pseudo dengue fever test [19]; however, in order to make it a true IoT device we have to improve the compatibility of its communication module and its back-end data analysis capability.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a W099109 from SAFEA and 2018YFE010900 project from Ministry of Science and Technology, both P.R. China. The work was also supported by the Grant Agency of the Czech Republic (17-20716S) and the Ministry of Education, Youth and Sports of the Czech Republic under the project OP VVV CEITEC Nano+ (CZ.02.1.01/0.0/0.0/16_013/0001728). LY and SN acknowledge the funding from the Research Grant Council of Hong Kong, received under Grant Number 16209316 and 16201017. MK was supported by the grants no. LTACH19005 and Progres Q25/LF1 of the Ministry of Education, Youth and Sport of the Czech Republic, and by the grant no. RVO-VFN 64165 of the Ministry of Health of the Czech Republic.
References
- 1.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.W.H.O. 7 July 2020. Coronavirus Disease (COVID-19) Situation Report-169.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200707-covid-19-sitrep-169.pdf?sfvrsn=c6c69c88_2 [Google Scholar]
- 4.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak, Emerg. Microb. Infect. 2020;9:313. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Mol. Biol. Rev. 1992;56:152. doi: 10.1128/mr.56.1.152-179.1992. https://doi.org/10146-0749/92/010152-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Tawfiq J.A. Asymptomatic coronavirus infection: MERS-CoV and SARS-CoV-2 (COVID-19) Travel med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies, Emerg. Microb. Infect. 2020;9:275. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh M.-d., Park W.B., Choe P.G., Choi S.-J., Kim J.-I., Chae J., Park S.S., Kim E.-C., Oh H.S., Kim E.J. Viral load kinetics of MERS coronavirus infection. N. Engl. J. Med. 2016;375:1303. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 10.Schlappi T.S., McCalla S.E., Schoepp N.G., Ismagilov R.F. Flow-through capture and in situ amplification can enable rapid detection of a few single molecules of nucleic acids from several milliliters of solution. Anal. Chem. 2016;88:7647. doi: 10.1021/acs.analchem.6b01485. [DOI] [PubMed] [Google Scholar]
- 11.Pipper J., Inoue M., Ng L.F.P., Neuzil P., Zhang Y., Novak L. Catching bird flu in a droplet. Nat. Med. 2007;13:1259. doi: 10.1038/nm1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pipper J., Zhang Y., Neuzil P., Hsieh T.-M. Clockwork PCR including sample preparation. Angew. Chem. Int. Ed. 2008;47:3900. doi: 10.1002/anie.200705016. [DOI] [PubMed] [Google Scholar]
- 13.Yeh Y.-T., Gulino K., Zhang Y., Sabestien A., Chou T.-W., Zhou B., Lin Z., Albert I., Lu H., Swaminathan V. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. U. S. A. 2020;117:895. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Wang R. Aptasensors for detection of avian influenza virus H5N1. Biosensors Biodetection. 2017;379 doi: 10.1007/978-1-4939-6911-1_25. [DOI] [PubMed] [Google Scholar]
- 15.W.H.O. The top 10 causes of death. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death
- 16.Caygill R.L., Blair G.E., Millner P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta. 2010;681:8. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Huang H.-S., Tsai C.-L., Chang J., Hsu T.-C., Lin S., Lee C.-C. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin. Microbiol. Infect. 2018;24:1055. doi: 10.1016/j.cmi.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H., Fohlerová Z., Pekárek J., Basova E., Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020;153:112041. doi: 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H., Podesva P., Liu X., Zhang H., Teply T., Xu Y., Chang H., Qian A., Lei Y., Li Y., Niculescu A., Iliescu C., Neuzil P. IoT PCR for pandemic disease detection and its spread monitoring. Sensor. Actuator. B Chem. 2020;303:127098. doi: 10.1016/j.snb.2019.127098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Specific enzymatic amplification of DNA invitro - the polymerase chain-reaction. Cold Spring Harbor Symp. Quant. Biol. 1986;51:263. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Li H., Zhu H., Pekárek J., Podešva P., Chang H., Neužil P. Revealing the secrets of PCR. Sensor. Actuator. B Chem. 2019;298:126924. doi: 10.1016/j.snb.2019.126924. [DOI] [Google Scholar]
- 22.Higuchi R., Dollinger G., Walsh P.S., Griffith R. Simultaneous amplification and detection of specific DNA sequences. Bio Technol. 1992;10:413. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi R., Fockler C., Dollinger G., Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnol. 1993;11:1026. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto M., Barany F., Soper S.A. Polymerase chain reaction/ligase detection reaction/hybridization assays using flow-through microfluidic devices for the detection of low-abundant DNA point mutations. Biosens. Bioelectron. 2006;21:1915. doi: 10.1016/j.bios.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Deiman B., van Aarle P., Sillekens P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA) Mol. Biotechnol. 2002;20:163. doi: 10.1385/MB:20:2:163. [DOI] [PubMed] [Google Scholar]
- 26.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 27.Walker G.T., Fraiser M.S., Schram J.L., Little M.C., Nadeau J.G., Malinowski D.P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M.M., Li F., Zhang Z., Zhang K., Kang D.-K., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324. doi: 10.1039/C3CS60439J. [DOI] [PubMed] [Google Scholar]
- 29.Tan E., Erwin B., Dames S., Ferguson T., Buechel M., Irvine B., Voelkerding K., Niemz A. Specific versus nonspecific isothermal DNA amplification through thermophilic polymerase and nicking enzyme activities. Biochemistry. 2008;47:9987. doi: 10.1021/bi800746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010;10:887. doi: 10.1039/B921140C. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y., Li B., Milligan J.N., Bhadra S., Ellington A.D. Real-time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly. J. Am. Chem. Soc. 2013;135:7430. doi: 10.1021/ja4023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz A., Graber N., Widmer H.M. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sensor. Actuator. B Chem. 1990;1:244. doi: 10.1016/0925-4005(90)80209-I. [DOI] [Google Scholar]
- 33.Harrison D.J., Fluri K., Seiler K., Fan Z., Effenhauser C.S., Manz A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science. 1993;261:895. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- 34.Kopp M.U., De Mello A.J., Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 35.Woolley A.T., Hadley D., Landre P., deMello A.J., Mathies R.A., Northrup M.A. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 1996;68:4081. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 36.Belgrader P., Young S., Yuan B., Primeau M., Christel L.A., Pourahmadi F., Northrup M.A. A battery-powered notebook thermal cycler for rapid multiplex real-time PCR analysis. Anal. Chem. 2001;73:286. doi: 10.1021/ac000905v. [DOI] [PubMed] [Google Scholar]
- 37.Potrich C., Lunelli L., Forti S., Vozzi D., Pasquardini L., Vanzetti L., Panciatichi C., Anderle M., Pederzolli C. Effect of materials for micro-electro-mechanical systems on PCR yield. Eur. Biophys. J. 2010;39:979. doi: 10.1007/s00249-009-0466-5. [DOI] [PubMed] [Google Scholar]
- 38.Haeberle S., Mark D., von Stetten F., Zengerle R. Springer; 2012. Microfluidic Platforms for Lab-On-A-Chip Applications; p. 853. [DOI] [Google Scholar]
- 39.Gates B. Responding to Covid-19—a once-in-a-century pandemic? N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 40.Neuzil P., Zhang C., Pipper J., Oh S., Zhuo L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006;34:e77. doi: 10.1093/nar/gkl416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrar J.S., Wittwer C.T. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin. Chem. 2015;61:145. doi: 10.1373/clinchem.2014.228304. [DOI] [PubMed] [Google Scholar]
- 42.Myrick J.T., Pryor R.J., Palais R.A., Ison S.J., Sanford L., Dwight Z.L., Huuskonen J.J., Sundberg S.O., Wittwer C.T. Integrated extreme real-time PCR and high-speed melting analysis in 52 to 87 seconds. Clin. Chem. 2020;65:263. doi: 10.1373/clinchem.2018.296608. [DOI] [PubMed] [Google Scholar]
- 43.Christensen T.B., Bang D.D., Wolff A. Multiplex polymerase chain reaction (PCR) on a SU-8 chip. Microelectron. Eng. 2008;85:1278. doi: 10.1016/j.mee.2008.01.066. [DOI] [Google Scholar]
- 44.Wittwer C.T., Fillmore G.C., Hillyard D.R. Automated polymerase chain reaction in capillary tubes with hot air. Nucleic Acids Res. 1989;17:4353. doi: 10.1093/nar/17.11.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W., Zhang M., Liu X., Sharma A., Ding X. A Point-of-Need infrared mediated PCR platform with compatible lateral flow strip for HPV detection. Biosens. Bioelectron. 2017;96:213. doi: 10.1016/j.bios.2017.04.047. [DOI] [PubMed] [Google Scholar]
- 46.Zou Z.-Q., Chen X., Jin Q.-H., Yang M.-S., Zhao J.-L. A novel miniaturized PCR multi-reactor array fabricated using flip-chip bonding techniques. J. Micromech. Microeng. 2005;15:1476. doi: 10.1088/0960-1317/15/8/014. [DOI] [Google Scholar]
- 47.Neuzil P., Pipper J., Hsieh T.M. Disposable real-time microPCR device: lab-on-a-chip at a low cost. Mol. Biosyst. 2006;2:292. doi: 10.1039/b605957k. [DOI] [PubMed] [Google Scholar]
- 48.Yoon D.S., Lee Y.-S., Lee Y., Cho H.J., Sung S.W., Oh K.W., Cha J., Lim G. Precise temperature control and rapid thermal cycling in a micromachined DNA polymerase chain reaction chip. J. Micromech. Microeng. 2002;12:813. doi: 10.1088/0960-1317/12/6/312. [DOI] [Google Scholar]
- 49.Moschou D., Vourdas N., Kokkoris G., Papadakis G., Parthenios J., Chatzandroulis S., Tserepi A. All-plastic, low-power, disposable, continuous-flow PCR chip with integrated microheaters for rapid DNA amplification. Sensor. Actuator. B Chem. 2014;199:470. doi: 10.1016/j.snb.2014.04.007. [DOI] [Google Scholar]
- 50.Li H., Zhang H., Xu Y., Tureckova A., Zahradník P., Chang H., Neuzil P. Versatile digital polymerase chain reaction chip design, fabrication, and image processing. Sensor. Actuator. B Chem. 2019;283:677. doi: 10.1016/j.snb.2018.12.072. [DOI] [Google Scholar]
- 51.Petrucci G., Caputo D., Lovecchio N., Costantini F., Legnini I., Bozzoni I., Nascetti A., De Cesare G. Multifunctional system-on-glass for lab-on-chip applications. Biosens. Bioelectron. 2017;93:315. doi: 10.1016/j.bios.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 52.Cui F., Chen W., Wu X., Guo Z., Liu W., Zhang W., Chen W. Design and experiment of a PDMS-based PCR chip with reusable heater of optimized electrode. Microsyst. Technol. 2017;23:3069. doi: 10.1007/s00542-016-3064-3. [DOI] [Google Scholar]
- 53.Yang J., Liu Y., Rauch C.B., Stevens R.L., Liu R.H., Lenigk R., Grodzinski P. High sensitivity PCR assay in plastic micro reactors. Lab. Chip. 2002;2:179. doi: 10.1039/B208405H. [DOI] [PubMed] [Google Scholar]
- 54.Liu K., Xiang J., Ai Z., Zhang S., Fang Y., Chen T., Zhou Q., Li S., Wang S., Zhang N. PMMA microfluidic chip fabrication using laser ablation and low temperature bonding with OCA film and LOCA. Microsyst. Technol. 2017;23:1937. doi: 10.1007/s00542-016-2924-1. [DOI] [Google Scholar]
- 55.Mata A., Fleischman A.J., Roy S. Characterization of polydimethylsiloxane (PDMS) properties for biomedical micro/nanosystems. Biomed. Microdevices. 2005;7:281. doi: 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- 56.van Midwoud P.M., Janse A., Merema M.T., Groothuis G.M.M., Verpoorte E. Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal. Chem. 2012;84:3938. doi: 10.1021/ac300771z. [DOI] [PubMed] [Google Scholar]
- 57.Kodzius R., Xiao K., Wu J., Yi X., Gong X., Foulds I.G., Wen W. Inhibitory effect of common microfluidic materials on PCR outcome. Sensor. Actuator. B Chem. 2012;161:349. doi: 10.1016/j.snb.2011.10.044. [DOI] [Google Scholar]
- 58.Qin K., Lv X., Xing Q., Li R., Deng Y. A BSA coated NOA81 PCR chip for gene amplification. Anal. Methods. 2016;8:2584. doi: 10.1039/C5AY03233D. [DOI] [Google Scholar]
- 59.Crabtree H.J., Lauzon J., Morrissey Y.C., Taylor B.J., Liang T., Johnstone R.W., Stickel A.J., Manage D.P., Atrazhev A., Backhouse C.J. Inhibition of on-chip PCR using PDMS–glass hybrid microfluidic chips. Microfluid. Nanofluid. 2012;13:383. doi: 10.1007/s10404-012-0968-9. [DOI] [Google Scholar]
- 60.Trung N.B., Saito M., Takabayashi H., Viet P.H., Tamiya E., Takamura Y. Multi-chamber PCR chip with simple liquid introduction utilizing the gas permeability of polydimethylsiloxane. Sensor. Actuator. B Chem. 2010;149:284. doi: 10.1016/j.snb.2010.06.013. [DOI] [Google Scholar]
- 61.Tachibana H., Saito M., Shibuya S., Tsuji K., Miyagawa N., Yamanaka K., Tamiya E. On-chip quantitative detection of pathogen genes by autonomous microfluidic PCR platform. Biosens. Bioelectron. 2015;74:725. doi: 10.1016/j.bios.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Kim S.C., Clark I.C., Shahi P., Abate A.R. Single-cell RT-PCR in microfluidic droplets with integrated chemical lysis. Anal. Chem. 2018;90:1273. doi: 10.1021/acs.analchem.7b04050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao H., Chung J., Lee K., Balaj L., Min C., Carter B.S., Hochberg F.H., Breakefield X.O., Lee H., Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ulrich M., Christensen D., Coyne S., Craw P., Henchal E., Sakai S., Swenson D., Tholath J., Tsai J., Weir A. Evaluation of the Cepheid GeneXpert® system for detecting Bacillus anthracis. J. Appl. Microbiol. 2006;100:1011. doi: 10.1111/j.1365-2672.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 65.Gilbert J.A., Long E.F., Brooks R.P., Friedland G.H., Moll A.P., Townsend J.P., Galvani A.P., Shenoi S.V. Integrating community-based interventions to reverse the convergent TB/HIV epidemics in rural South Africa. PloS One. 2015;10 doi: 10.1371/journal.pone.0126267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hui W.C., Yobas L., Samper V.D., Heng C.-K., Liw S., Ji H., Chen Y., Cong L., Li J., Lim T.M. Microfluidic systems for extracting nucleic acids for DNA and RNA analysis. Sensors Actuat. A-Phys. 2007;133:335. doi: 10.1016/j.sna.2006.06.031. [DOI] [Google Scholar]
- 67.Ji H.M., Samper V., Chen Y., Heng C.K., Lim T.M., Yobas L. Silicon-based microfilters for whole blood cell separation. Biomed. Microdevices. 2008;10:251. doi: 10.1007/s10544-007-9131-x. [DOI] [PubMed] [Google Scholar]
- 68.Yobas L., Ji H., Hui W.-C., Chen Y., Lim T.-M., Heng C.-K., Kwong D.-L. Nucleic acid extraction, amplification, and detection on Si-based microfluidic platforms. IEEE J. Solid State Circ. 2007;42:1803. doi: 10.1109/JSSC.2007.900232. [DOI] [Google Scholar]
- 69.Ha M.L., Lee N.Y. Miniaturized polymerase chain reaction device for rapid identification of genetically modified organisms. Food Contr. 2015;57:238. doi: 10.1016/j.foodcont.2015.04.014. [DOI] [Google Scholar]
- 70.Beyor N., Yi L., Seo T.S., Mathies R.A. Integrated capture, concentration, polymerase chain reaction, and capillary electrophoretic analysis of pathogens on a chip. Anal. Chem. 2009;81:3523. doi: 10.1021/ac900060r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J.-H., Chien L.-J., Hsieh T.-M., Luo C.-H., Chou W.-P., Chen P.-H., Chen P.-J., Lee D.-S., Lee G.-B. A miniaturized quantitative polymerase chain reaction system for DNA amplification and detection. Sensor. Actuator. B Chem. 2009;141:329. doi: 10.1016/j.snb.2009.06.034. [DOI] [Google Scholar]
- 72.Ahrberg C.D., Manz A., Neuzil P. Palm-sized device for point-of-care Ebola detection. Anal. Chem. 2016;88:4803. doi: 10.1021/acs.analchem.6b00278. [DOI] [PubMed] [Google Scholar]
- 73.Sun Y., Zhou X., Yu Y. A novel picoliter droplet array for parallel real-time polymerase chain reaction based on double-inkjet printing. Lab Chip. 2014;14:3603. doi: 10.1039/C4LC00598H. [DOI] [PubMed] [Google Scholar]
- 74.Hassibi A., Singh R., Manickam A., Sinha R., Kuimelis B., Bolouki S., Naraghi-Arani P., Johnson K., McDermott M., Wood N., Savalia P., Gamini N. A fully integrated CMOS fluorescence biochip for multiplex polymerase chain-reaction (PCR) processes. IEEE Int. Solid State Circ. Conf. (ISSCC) 2017;68 doi: 10.1109/ISSCC.2017.7870264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rival A., Jary D., Delattre C., Fouillet Y., Castellan G., Bellemin-Comte A., Gidrol X. An EWOD-based microfluidic chip for single-cell isolation, mRNA purification and subsequent multiplex qPCR. Lab. Chip. 2014;14:3739. doi: 10.1039/c4lc00592a. [DOI] [PubMed] [Google Scholar]
- 76.Neuzil P., Campos C.D.M., Wong C.C., Soon J.B.W., Reboud J., Manz A. From chip-in-a-lab to lab-on-a-chip: towards a single handheld electronic system for multiple application-specific lab-on-a-chip (ASLOC) Lab Chip. 2014;14:2168. doi: 10.1039/c4lc00310a. [DOI] [PubMed] [Google Scholar]
- 77.Liu R.H., Yang J., Lenigk R., Bonanno J., Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal. Chem. 2004;76:1824. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 78.Easley C.J., Karlinsey J.M., Bienvenue J.M., Legendre L.A., Roper M.G., Feldman S.H., Hughes M.A., Hewlett E.L., Merkel T.J., Ferrance J.P. A fully integrated microfluidic genetic analysis system with sample-in–answer-out capability. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19272. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Novak L., Neuzil P., Pipper J., Zhang Y., Lee S. An integrated fluorescence detection system for lab-on-a-chip applications. Lab Chip. 2007;7:27. doi: 10.1039/b611745g. [DOI] [PubMed] [Google Scholar]
- 80.Neuzil P., Novak L., Pipper J., Lee S., Ng L.F.P., Zhang C. Rapid detection of viral RNA by a pocket-size real-time PCR system. Lab Chip. 2010;10:2632. doi: 10.1039/c004921b. [DOI] [PubMed] [Google Scholar]
- 81.Ahrberg C.D., Ilic B.R., Manz A., Neuzil P. Handheld real-time PCR device. Lab Chip. 2016;16:586. doi: 10.1039/c5lc01415h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reed M.R., Coty W.A. Springer; 2009. eSensor® A Microarray Technology Based on Electrochemical Detection of Nucleic Acids and its Application to Cystic Fibrosis Carrier Screening; p. 247. [DOI] [Google Scholar]
- 83.Xu G., Hsieh T.-M., Lee D.Y., Ali E.M., Xie H., Looi X.L., Koay E.S.-C., Li M.-H., Ying J.Y. A self-contained all-in-one cartridge for sample preparation and real-time PCR in rapid influenza diagnosis. Lab Chip. 2010;10:3103. doi: 10.1039/C005265E. [DOI] [PubMed] [Google Scholar]
- 84.Stumpf F., Schwemmer F., Hutzenlaub T., Baumann D., Strohmeier O., Dingemanns G., Simons G., Sager C., Plobner L., Von Stetten F. LabDisk with complete reagent prestorage for sample-to-answer nucleic acid based detection of respiratory pathogens verified with influenza A H3N2 virus. Lab Chip. 2016;16:199. doi: 10.1039/C5LC00871A. [DOI] [PubMed] [Google Scholar]
- 85.Shin D.J., Trick A.Y., Hsieh Y.-H., Thomas D.L., Wang T.-H. Sample-to-answer droplet magnetofluidic platform for point-of-care hepatitis C viral load quantitation. Sci. Rep. 2018;8:1. doi: 10.1038/s41598-018-28124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu P., Li X., Greenspoon S.A., Scherer J.R., Mathies R.A. Integrated DNA purification, PCR, sample cleanup, and capillary electrophoresis microchip for forensic human identification. Lab Chip. 2011;11:1041. doi: 10.1039/C0LC00533A. [DOI] [PubMed] [Google Scholar]
- 87.Echeverry D.F., Deason N.A., Davidson J., Makuru V., Xiao H., Niedbalski J., Kern M., Russell T.L., Burkot T.R., Collins F.H. Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene. Malar. J. 2016;15:128. doi: 10.1186/s12936-016-1185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.To K.K.-W., Tsang O.T.-Y., Yip C.C.-Y., Chan K.-H., Wu T.-C., Chan J.M.-C., Leung W.-S., Chik T.S.-H., Choi C.Y.-C., Kandamby D.H. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yonekawa T., Watanabe H., Hosaka N., Semba S., Shoji A., Sato M., Hamasaki M., Yuki S., Sano S., Segawa Y. Fully automated molecular diagnostic system “simprova” for simultaneous testing of multiple items. Sci. Rep. 2020;10:1. doi: 10.1038/s41598-020-62109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Melchers W.J., Kuijpers J., Sickler J.J., Rahamat-Langendoen J. Lab-in-a-tube: real-time molecular point-of-care diagnostics for influenza A and B using the cobas® Liat® system. J. Med. Virol. 2017;89:1382. doi: 10.1002/jmv.24796. [DOI] [PubMed] [Google Scholar]
- 91.Kanwar N., Michael J., Doran K., Montgomery E., Selvarangan R. Comparison of the ID Now influenza A & B 2, Cobas influenza A/B, and Xpert Xpress Flu point-of-care nucleic acid amplification tests for influenza A/B virus detection in children. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01611-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang H., Chen Z., Cao X., Li Z., Stavrakis S., Choo J., deMello A.J., Howes P.D., He N. A sample-in-digital-answer-out system for rapid detection and quantitation of infectious pathogens in bodily fluids. Anal. Bioanal. Chem. 2018;410:7019. doi: 10.1007/s00216-018-1335-9. [DOI] [PubMed] [Google Scholar]
- 93.Gadsby N.J., Russell C.D., McHugh M.P., Mark H., Conway Morris A., Laurenson I.F., Hill A.T., Templeton K.E. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin. Infect. Dis. 2016;62:817. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 95.Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 97.Nash M., Ramapuram J., Kaiya R., Huddart S., Pai M., Baliga S. Use of the GeneXpert tuberculosis system for HIV viral load testing in India. Lancet Glob. Health. 2017;5:e754. doi: 10.1016/S2214-109X(17)30247-4. [DOI] [PubMed] [Google Scholar]
- 98.Malvy D., McElroy A.K., de Clerck H., Günther S., van Griensven J. Ebola virus disease. Lancet. 2019;393:936. doi: 10.1016/s0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 99.Organization W.H. Norms and standards: assessing new medical products in health emergencies: the EUAL procedures. WHO Drug Inf. 2015;29:305. [Google Scholar]
- 100.Semper A.E., Broadhurst M.J., Richards J., Foster G.M., Simpson A.J., Logue C.H., Kelly J.D., Miller A., Brooks T.J., Murray M., Pollock N.R. Performance of the GeneXpert Ebola assay for diagnosis of Ebola virus disease in Sierra Leone: a field evaluation study. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gay-Andrieu F., Magassouba N., Picot V., Phillips C.L., Peyrefitte C.N., Dacosta B., Dore A., Kourouma F., Ligeon-Ligeonnet V., Gauby C., Longuet C., Scullion M., Faye O., Machuron J.L., Miller M. Clinical evaluation of the BioFire FilmArray((R)) BioThreat-E test for the diagnosis of Ebola virus disease in Guinea. J. Clin. Virol. 2017;92:20. doi: 10.1016/j.jcv.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 102.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 103.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg. Microb. Infect. 2020;9:747. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.U.S.F.D. Administration . Approved kits and tests for SARS; 2020. Emergency Use Authorizations. [Google Scholar]