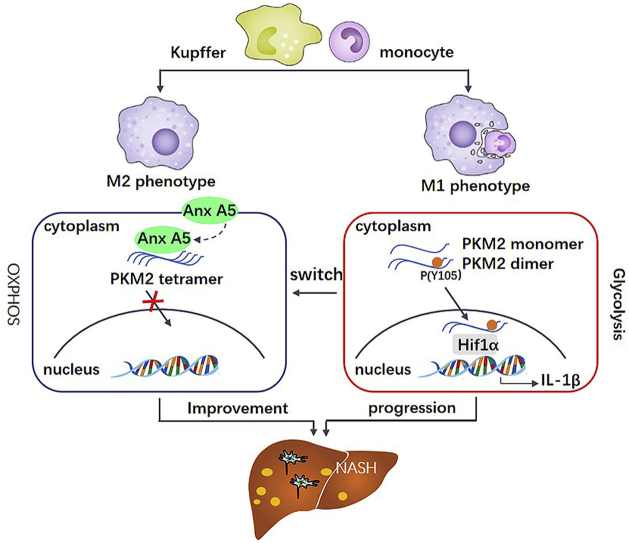
Abstract
Nonalcoholic steatohepatitis (NASH), the progressive form of nonalcoholic fatty liver disease (NAFLD), is becoming a common chronic liver disease with the characteristics of steatosis, inflammation and fibrosis. Macrophage plays an important role in the development of NASH. In this study, Annexin A5 (Anx A5) is identified with the special effect on hepatic macrophage phenotype shift from M1 to M2. And it is further demonstrated that Anx A5 significantly switches metabolic reprogramming from glycolysis to oxidative phosphorylation in activated macrophages. Mechanistically, the main target of Anx A5 in energy metabolism is confirmed to be pyruvate kinase M2 (PKM2). And we following reveal that Anx A5 directly interacts with PKM2 at ASP101, LEU104 and ARG106, inhibits phosphorylation of Y105, and promotes PKM2 tetramer formation. In addition, based on the results of PKM2 inhibitor (compound 3k) and the phosphorylated mutation (PKM2 (Y105E)), it is proved that Anx A5 exhibits the function in macrophage polarization dependently on PKM2 activity. In vivo studies also show that Anx A5 improves steatosis, inflammation and fibrosis in NASH mice due to specially regulating hepatic macrophages via interaction with PKM2. Therefore, we have revealed a novel function of Anx A5 in hepatic macrophage polarization and HFD-induced NASH, providing important insights into the metabolic reprogramming, which is important for NASH therapy.
Keywords: NASH, Macrophage polarization, Metabolic reprogramming, Annexin A5 (Anx A5), PKM2
Graphical abstract
1. Introduction
Nonalcoholic steatohepatitis (NASH), the progressive form of nonalcoholic fatty liver disease (NAFLD) with an estimated prevalence of 1.5%–6.5% in the general population, is characterized by inflammation and hepatocyte injury, and has an increased risk for liver fibrosis and progression to cirrhosis [1,2]. Inflammatory and fibrotic severity is the most essential histological feature of NASH [3]. Despite recent advances in understanding the mechanisms underlying the pathogenesis of NASH, no approved, disease-modifying treatments are currently available [4,5]. Abundant researches mainly focus on participation of hepatic macrophages in the process of initiation and development, and suggest that hepatic macrophages as the prospective therapeutic target to improve hepatic diseases [6].
Hepatic macrophages including resident Kupffer cells and infiltrated monocyte-derived macrophages (MDMs) [7], play a critical mediator in promotion of NAFLD and NASH. Kupffer cells as liver-specific cells are located in the liver sinusoids and closely contact with the liver sinusoidal endothelial cells, clear up gut-derived microbiomes or foreign pathogens from blood [8]. Liver excessive intake of fatty acids and lipids accumulation facilitate Kupffer cells activation and induce a pro-inflammatory state in liver, as well initiate massive influx of peripheral blood macrophages into liver [6,9]. The production of inflammatory cytokines activate hepatic stellate cells, leading to fibrosis and cirrhosis, as well further accelerate hepatocyte damage and increase inflammatory response [10]. Therefore, inhibiting excessive activation of hepatic macrophages to threat the vicious cycle in NASH probably is a superior method for treatment.
Diversity and plasticity are key features of macrophages. In response to various signals, macrophages undergo M1 macrophages (classically activated macrophages) and M2 macrophages (alternatively activated macrophages) [11]. M1 macrophages, stimulated by TLR ligands and IFN-γ, have a central role in initiate inflammatory responses, and release numerous pro-inflammatory mediators and reactive oxygen species (ROS) [7]. M2 macrophages, stimulated by IL-4/IL-13, are associated with responses to anti-inflammatory reactions and tissue remodeling [12]. The phenotypic plasticity of macrophages provides strategies for many kinds of inflammatory diseases [[13], [14], [15]]. Currently, precise control of hepatic macrophage polarization status remains difficult in NASH.
Among the signals involved in macrophage polarization, metabolic reprograming plays essential role [16,17]. The main metabolic characteristics of M1 macrophage is associated with increased glycolysis, weakened tricarboxylic acid (TCA) cycle and oxidative phosphorylation system (OXPHOS) [18]. On the contrary, the metabolism in M2 macrophage is greatly dependent on TCA cycle, fatty acids oxidation (FAO) and OXPHOS for energy generation [19]. Recent literatures report that pyruvate kinase M2 (PKM2) is a critical determinant of glycolytic reprogramming in macrophage [20]. Y105 of PKM2 regulates the glycolysis and oxidative phosphorylation via controlling the ration of inactive and active forms of PKM2 [21, 22]. Monomer and dimer forms of PKM2, with inactive enzymatic pyruvate kinase activity, promote macrophage glycolysis and inflammatory responses. While, tetramer form of PKM2, with active enzymic pyruvate kinase activity, induces TCA cycle, OXPHOS and anti-inflammatory responses [23]. In addition, PKM2 activation by both endogenous molecules [24] and synthetic compounds [20] can attenuate macrophage-induced inflammatory diseases. However, function of PKM2 in hepatic macrophages of NASH remains unclear.
At present, obeticolic acid (OCA) and cenicriviroc (CVC) are the promising compounds in the field of NASH treatment, both of which are in phase III clinical stage. However, OCA shows side effects on pruritus and high level of LDL cholesterol [25], and CVC fails to improve hepatic steatosis and insulin tolerance [26]. Annexin A5 (Anx A5) is a member of annexin family with the molecular weight of 36 kDa, binding to membrane in a Ca2+ dependent manner [27]. Anx A5 has been reported to play multiply role in tissue repair, antiphospholipid syndrome and inflammation [[28], [29], [30]]. AnxA5 participates the cellular inflammatory response, as it can reduce local vascular and systemic inflammation and enhance vascular function [30]. Nevertheless, effect of Anx A5 in NASH injury is unknown.
We provide evidence here that Anx A5 directly interacts with PKM2, resulting in metabolic reprogramming from glycolysis to OXPHOS, hepatic macrophage phenotype shift and NASH improvement. Notably, Anx A5 exhibits a strong probability for clinic use in NASH treatment.
2. Materials and methods
2.1. Regents
Annexin A5 (sc-4252) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Lipopolysaccharide (LPS)(L2630), DAPI (C1002), IV type collagenase (C1889), FITC (786-141) were purchased from Sigma-Aldrich (St. Louis, Mo, USA). Recombinant murine IFN-γ (cat#315-05), IL-4 (cat#214-14), and M-CSF (cat#315-02) were purchased from Peprotech (Rocky Hill, NJ, USA). DSS (21555) was purchased from ThermoFisher Scientific (Waltham, MA, USA). Compound 3k (S8616) was purchased from Selleck Chemicals (Shanghai, China). Protein A/G-agarose beads (sc-2003) were purchased from Santa Cruz Biotechnology. Recombinant PKM2 (7244-PK-020) were purchased from R&D Systems. Antibodies for α-Tublin (264675), β-Actin (303574), GAPDH (264160) were purchased from Abmart (Shanghai, China). Antibodies for p-P65 (3033), P65 (8242), STAT1 (9172), p-STAT1 (9167), p-STAT6, SOCS1 (3950), ERK (9102), p-ERK (9101), JNK (9252), p-JNK (4668), P38 (9212), p-P38 (9216), HK2 (2867T), HIF1α (36169T), PKM2 (4053T), LDH (3582), Fasn (3180S), Scd1 (2438S), HA (3724), Flag (2368), CPT1a (12252S), PPARγ (2443S), and Lamin B (12255S) were purchased from Cell Signaling Technology (Beverly, MA, USA). Anti-annexin A5 (sc-74438) and anti-PGC1b(H2916) were purchased from Santa Cruz Biotechnology, anti-p-AKT-S473(66444-I-Ig) was purchased from Proteintech (Shanghai, China), anti-PPARα(ab24509) was purchased from Abcam (Burlingame, CA, USA), anti-p-PKM2 (Y105)(abs137248) and anti-p-PKM2(Ser37)(abs137249) were purchased from Absin (shanghai, china). FITC-CD11c (117306), APC-CD206(141707), PE-ly6c(128007), FITC-ly6g(127605) were purchased from Biolend (San Diego, CA), PE-Cy7-F4/80 (25-4801-82), Alexa Fluor 488 Goat anti-Rabbit IG (A-11034), Alexa Fluor 594 Goat anti-Rabbit IG (A-11032) was purchased from ThermoFisher Scientific, (Waltham, MA, USA). ELISA kit for IL-1β (1210122), TNF-α (1217202), IL-6 (1210602) was purchased from Dakewe Biotech Co.Ltd. (Shenzhen, China). ELISA kit for Hyaluronic acid (MBS2886836) and Laminin (MBS2024735) was purchased from MyBioSource (San Diego, CA, USA), and ELISA kit for procollagen III (NBP2-75856) was purchased from Novus Biologicals (Centennial, CO, USA). All other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO).
2.2. Cell culture
Murine macrophage cell line Raw 264.7 cells was purchased from the American Type Culture Collection (Rockville, MD), HEK293T was purchased from the Institute of cell Biology, Chinese Academy of Sciences (Shanghai, China), and maintained in DMEM (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (GIBCO, Grand Island, NY), 100 U mL−1 penicillin, and 100 mg mL−1 streptomycin in 5% CO2 at 37 °C.
Bone marrow-derived macrophages (BMDMs) were obtained according to previously described [31]. In brief, C57BL/6 mice were killed and femurs were dissected, using a 21-ga needle flushed femurs with PBS. After centrifugation at 300g for 5 min at 4 °C, and cells were grown in RPMI 1640 (GIBCO, Grand Island, NY) medium containing 10% fetal bovine serum and 20 ng mL−1 M-CSF in 5% CO2 at 37 °C for 5 days. Differentiated macrophages were washed by PBS two times and cultured with fresh DMEM medium containing 10% fetal bovine serum.
For M1 macrophage polarization, Raw 264.7 cells and BMDMs were treated with 10 ng mL−1 LPS and 10 ng mL−1 IFN-γ for 6 h. For M2 macrophage polarization, Raw 264.7 cells and BMDMs were treated with 20 ng mL−1 IL-4 for 6 h.
2.3. Isolation of non-parenchymal cells and Kupffer cells from mouse liver
Non-parenchymal cells and Kupffer cells were isolated from NAFLD model C57BL/6 mice using liver perfusion collagenase system. The liver was perfused with EDTA and Hanks buffer for 5 min respectively and IV type collagenase for 10 min, chopped and filtered through 70 μm strainer. After centrifugation at 50g for 5 min, the supernatant was non-parenchymal cell fraction. Supernatant was carefully layered on the top of percoll gradients which composed of 25% and 50% percoll layers, and then centrifugation at 1800g 30 min. Kupffer cells fraction was isolated from between the two different percoll gradient layers [32]. Isolated Kupffer cells were polarized into M1 and M2 phenotype as described above.
2.4. Nuclear and cytosolic fractionation
Nuclear Extract Kit (Beyotime Biotechnology, P0027) was used to isolate cellular nuclear and cytoplasmic fractions according to manufacturer's recommendations. Each fraction was analyzed by SDS-PAGE western blotting.
2.5. Western blot analysis
Western blot was performed according to described previously [33]. Cells were collected and lysed using lysis buffer and quantitated by BCA assay, obtained protein lysates were degenerated at 100 °C for 5 min and separated by 10% SDS-PAGE and electrophoretically transferred onto poiyvinylidene fluoride (PVDF) membranes (Millipore Corp, Bedford, MA). The membranes were blocked with 3% BSA for 1 h at room temperature, then incubated with specific primary antibodies overnight at 4 °C, washed 5 times with washing buffer and then incubated with HRP-coupled secondary antibody. Protein bands were visualized using Western bolting detection system according to the manufacturer's instructions (Cell Signaling Technology, MA).
2.6. Quantitative PCR
Total RNA was extracted from cells or tissues by Trizol and 1 μg mRNA was reverse transcribed to cDNA and subjected to quantitative PCR, which was performed with the BioRad CFX96 ouchTM Real-Time PCR Detection System (Biorad, CA) using iQTM SYBR Green supermix (BioRad, CA), and threshold cycle numbers were obtained using BioRad CFX manager software. The program for amplification was 1 cycle of 95 °C for 2 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s. The primer sequences used in this study were described in Supplementary Materials Table 1. The relative expression of each gene was normalized to the expression of β-Actin, and then reported as fold change of basal level [14].
2.7. Anx A5 was labeled with FITC
The binding of FITC and Anx A5 was performed by using the label kit (786-141) purchased from G-Biosciences (St Louis, MO. USA), according to the manufacturer's instructions.
2.8. Immunofluorescent microscopy
Cells were seeded on coverslips, and pretreated with Anx A5 in the presence of LPS & IFN-γ. 6 h later cells were fixed in 4% paraformaldehyde (PFA) for 10 min at 37 °C, permeabilized with 0.5% Triton X-100 for 30 min and blocked with 3% BSA for 1 h at room temperature, and stained with P65, STAT1, STAT6, PKM2 antibody overnight at 4 °C, and then with specific fluorescent-coupled secondary antibody for 2 h at room temperature. The coverslips were stained with DAPI and imaged with a confocal laser scanning microscope (Olympus Lake Success, NY). The quantification of STAT1 and p65 location was carried out as described previously [14]. The quantification of CD11c, CD206, Ly6c and Ly6g positive cells in liver tissue were detected by immunofluorescence. Paraffin-embedded liver tissue were heat-fixed, deparaffinized, rehydrated, antigen retrieval before permeabilized, and subsequent process followed cells.
2.9. Flow cytometry
Cultured cells were harvested or separated from liver washed with cold PBS two times, stained with specific antibodies (CD11c, CD206, F4/80, Ly6c, Ly6g CD146, CD 163 and NK1.1) for 30 min at 4 °C in dark, washed three times with cold PBS, and analyzed by flow cytometry.
2.10. Measurement of mitochondrial membrane potential (ΔΨm)
To determine the changes in mitochondrial membrane potential, JC-1 dye (Beyotime Biotechnology, C2006) was used. Cells were seeded into 6-well plates with various concentrations of Anx A5 and 10 ng mL−1 LPS&IFN-γ or 20 ng mL−1 IL-4 for 6 h. Cells were collected and washed with PBS two times. Cells incubated with JC-1 working stock for 20 min at 37 °C, and then washed with JC-1 buffer solution for two times. The cells stained with JC-1 were detected using flow cytometry.
2.11. ELISA assay
IL-1β, IL-6 and TNF-α of culture supernatant or serum were measured by ELISA kits according to the manufacturer's instructions. Also, hyaluronic acid, laminin and procollagen III of serum were measured by ELISA kits.
2.12. Oxidative phosphorylation and glycolysis assay
Cellular glycolysis and OXPHOS in cells were monitored with Seahorse Bioscience XF-96 Extracellular Flux Analyzer (Seahorse Bioscience) by measuring the ECAR (extracellular acidification rates, indicative of glycolysis) and OCR (oxygen consumption rates, indicative of respiration). 10000 cells were seeded into 96-well plates designed for XF-96 in 80 μl growth media followed with overnight incubation, then treated with Anx A5 and LPS&IFN-γ or IL-4 for 6 h. To determine the ECAR, 10 mM glucose, 0.5 μM oligomycin, and 100 mM 2-deoxy-glucose (2-DG) were added into the solution. 0.5 μM oligomycin, 2.5 μM FCCP, and 5 μM rotenone/antimycin A were automatically and injected to measure the OCR. Detailed operation steps were followed according to instructions of Seahorse Bioscience.
2.13. Glucose uptake assay
Cells were seeded into a 96-well black wall bottom cell culture plate and treated with various concentrations of Anx A5 and 10 ng mL−1 LPS&IFN-γ or 20 ng mL−1 IL-4 for 6 h prior to measuring glucose uptake. Cells were grown in glucose-free DMEM medium (Gibco BRL, #11966–025) for additional 1 h. Glucose uptake was measured using glucose uptake assay kit (Abnova, KA4086) according to the manufacturer's instruction.
2.14. LDH activity
Cells were seeded into 6-well plates with various concentrations of Anx A5 and 10 ng mL−1 LPS&IFN-γ or 20 ng mL−1 IL-4 for 6 h, cells were collected and lysated. Lactate dehydrogenase enzymatic activity was assessed using a commercially available LDH Assay Kit (Beyotime Biotechnology, C0016) according to manufacturer's instructions.
2.15. Lactic acid production
In serum-free medium, cells were seeded into 96 well plates in the presence of Anx A5 and 10 ng mL−1 LPS&IFN-γ or 20 ng mL−1 IL-4, and incubated at 37 °C. After 6 h, the media was collected and measured the lactic acid production using commercial lactate assay kit purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The detailed protocol was performed according to the manufacturer's instructions.
2.16. Co-immunoprecipitation (Co-IP)
Harvested cell lysates were incubated with appropriate antibody at 4 °C overnight, and precipitated with protein A/G-agarose beads for another 4 h at 4 °C. The beads were washed by lysis buffer 5 times and centrifugated at 1000 g for 5 min at 4 °C and then immunoprecipitated proteins were detected by western blot.
2.17. Pyruvate kinase activity analysis of PKM2
Cells were seeded into 6-well plate and treated with various concentrations of Anx A5 and LPS&IFN-γ, after 6 h later, cells were collected and washed once with ice-cold PBS and lysed. The cells were incubated on ice for 30 min. After centrifugation at 12000 rpm for 10 min at 4 °C, PKM2 activity was measured in the supernatant using Pyruvate Kinase Assay Kit (Solarbio, BC0540) according to manufacturer's instructions.
2.18. Microscale thermophoresis (MST)
To determine solution equilibrium interaction constants between Anx A5 and PKM2, we used the microscale thermophoresis (MST) technology. Purified Anx A5 as ligand, and expressed EGFP-PKM2 or EGFP-PKM2 mutation (D101A, L104A, R106A) in Raw 264.7 cells as target, then we diluted AnxA5 to 16 different concentrations ranging from 0.037 ρM to 1.2 μM, and mixed both equivalently at room temperature for 20 min. The mixture samples were loaded into capillaries (Monolith NT.115 Capillary) and the thermophoresis signals were measured using Monolith NT.115 (NanoTemper) according to standard protocol of the manufacturer. The changes of the fluorescent thermophoresis and Kd values were analyzed using the Nano Temper analysis software.
2.19. Plasmids transient transfection in cells
Plasmids of Flag-PKM2, HA-Anx A5 and Flag-PKM2(Y105E) were manufactured by Genscript Biotechnology (Nanjing, China). Flag-PKM2 and HA-Anx A5 were transiently transfected to HEK293T cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufactures' recommendation, then incubated at 37 °C for 48 h. And Flag PKM2(Y105E) and vector were transiently transfected to Raw 264.7 cells. In the end, the cells were collected and prepared for the following experiments.
2.20. Cellular thermal shift assay (CETSA)
CETSA was performed as described previously reported [14]. Briefly, cells were incubated with Anx A5 for 6 h. Then cultured cell were harvested and equally divided. Heating with various temperature via PCR instrument and then repeating freeze-thawing with liquid nitrogen three times, and centrifuging at 20000 g for 20 min at 4 °C. Transferred supernatants to new microtubes and analyzed via western blot.
2.21. DSS crosslinking
Crosslinking was performed according to instructions of disuccinimidyl suberate (DSS) crosslinkers (Thermo Scientific) [20]. Cells were treated with 240 nM Anx A5 and 10 ng mL−1 LPS&IFN-γ or IL-4 for 6 h, cells were collected and washed two times with ice-cold PBS. Add the DSS solution to a final concentration of 500 μΜ, and incubate the reaction mixture for 30 min at room temperature. Add the Quench Solution to a final concentration of 10 mM Tris and incubate the quenching reaction for 15 min at room temperature. Lysates were analyzed by western blot.
2.22. Animals and ethical statement
Male C57BL/6 mice (6–8 weeks old, 18–22 g) were obtained from Model Animal Genetics Research Center of Nanjing University (Nanjing, China). They were maintained with free access to pellet food and water in plastic cages at 21 ± 2 °C and kept on a 12 h light/dark cycle in specific-pathogen-free (SPF) facilities.
Animal welfare and experimental procedures were carried out strictly in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, the United States) ‘The Detailed Rules and Regulations of Medical Animal Experiments Administration and Implementation’ (Order No. 1998–55, Ministry of Public Health, China) and approved by the Laboratory Animal Ethics Committee of School of Life Sciences, Nanjing University. The animal studies were reported in compliance with the ARRIVE guidelines [34,35]. All efforts were made to minimize animals' suffering and to reduce the number of animals used.
2.23. High fat diet-induced NASH model
Male C57BL/6 mice (6–8 weeks old, 18–22 g) were fed High-fat diet (HFD) contains 60% calories (D12492, Research Diets) for 16 weeks to induce NASH. And after 8 weeks of HFD, mice were randomly assigned to five experimental groups (9 mice per group): (a) normal diet, (b) HFD, (c) HFD + Anx A5 at 10 μg kg−1, (d) HFD + Anx A5 at 30 μg kg−1, (e) HFD + Anx A5 at 100 μg kg−1. Anx A5 was administrated once every 3 days by tail intravenous injection for 8 weeks. Mice were harvested by overdose sodium pentobarbital (300 μl·per 20 g body weight in 2% sodium pentobarbital intraperitoneally injection), serum, liver were collected after experiment completed. Data collection and analysis was performed blindly.
2.24. Serum assay
Serum was collected from mice orbital blood after experiment completed. Triglyceride (TG) and total cholesterol (TC) levels were measured using commercial reagent kits according to manufacturer's instructions. These reagent kits were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Hyaluronic acid, laminin, collagen III and inflammatory cytokines were measured by ELISA kits according to the instructions.
2.25. Tissue assay
Liver was collected, digested and homogenized. Protein expression was analyzed by western blot using antibodies. Hyp was analyzed by commercial kit purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Gene expression was analyzed by Real-time PCR.
2.26. Histology studies
Liver samples were fixed in 10% formalin solution, and prepared paraffin sections for H&E, immunohistochemistry, immunofluorescence and sirius red stain.
2.27. Statistical analysis
All experiments are randomized and blinded. All studies were performed at least three independent experiments with each experiment including triplicate sets in vitro, or nine animals per group in vivo. All data were presented as the mean ± SEM. GraphPad Prism 5.0 software (San Diego, CA, USA) was used for data statistical analysis. Student's t-test was used to test the difference between two groups. One-way ANOVA followed by Tukey's test was applied for multiple comparisons. P < 0.05 is considered to be statistically significant.
3. Results
3.1. Anx A5 promotes macrophage phenotype shift from pro-inflammatory M1 to anti-inflammatory M2
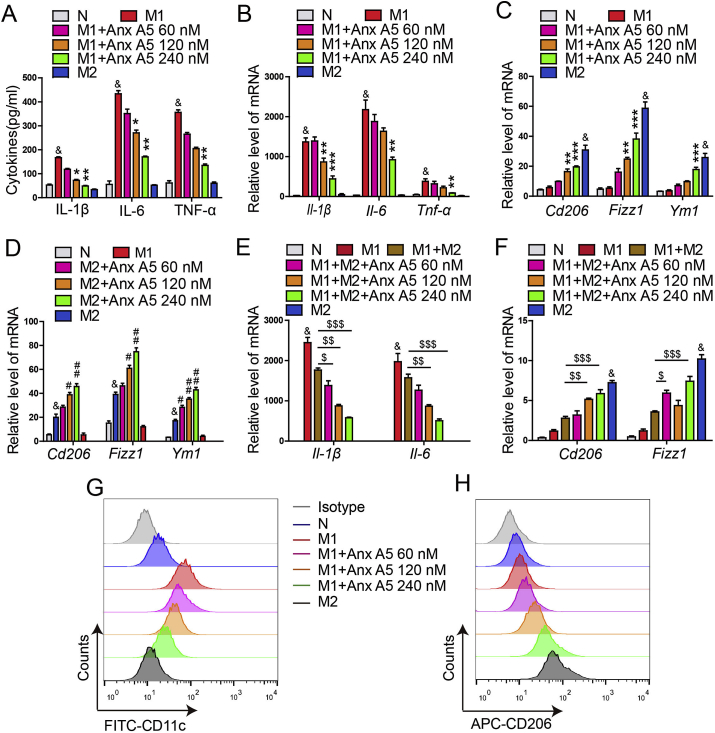
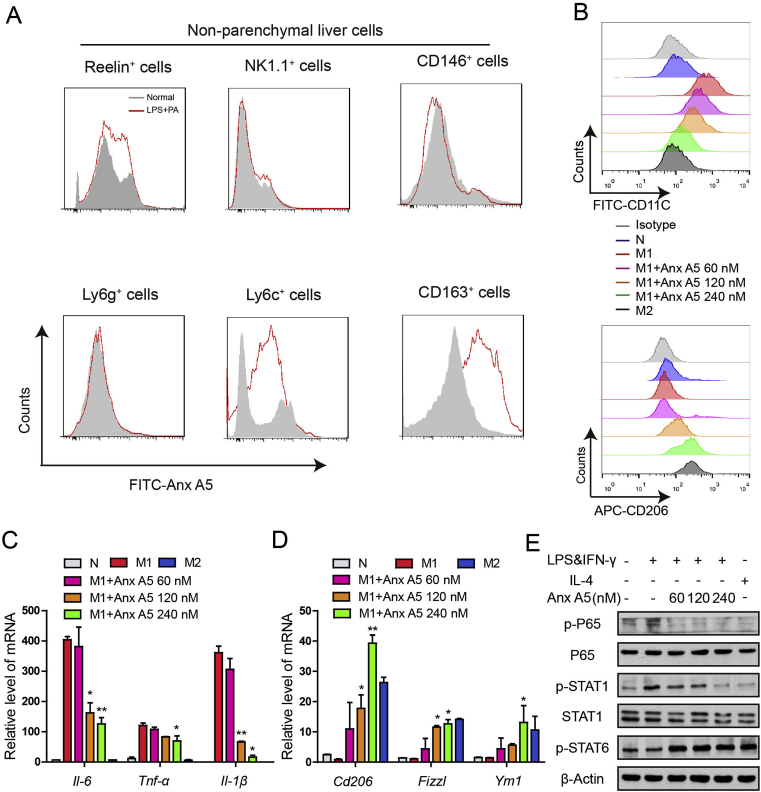
To evaluate the functional relevance of Anx A5 on macrophage, we established several polarization models, M1 phenotype was induced by 10 ng mL−1 LPS&IFN-γ, M2 phenotype was induced by 20 ng mL−1 IL-4, and LPS&IFN-γ together with IL-4 was used to induce the mixed phenotype. TUNEL assay showed there were no significant apoptosis in polarized M1 and M2 macrophages (Fig. S1). It was revealed that Anx A5 dose-dependently inhibited M1 macrophage related cytokines release (Il-6, Il-1β, Tnf-α) (Fig. 1A) and mRNA expression (Fig. 1B). Whilst, Anx A5 promoted M2 macrophage related mRNA expression (Cd206, Fizzl, Ym1) (Fig. 1C and D). Besides, Anx A5 also induced the mixed type macrophage preferable to M2 phenotype, not M1 phenotype (Fig. 1E and F). This was further supported by the result of flow cytometry shown in Fig. 1G and H, where CD11c (M1 surface marker) was progressively decreased and CD206 (M2 surface marker) was increased under the treatment of Anx A5. In addition, phagocytosis is also recognized as a characteristic marker of M1 macrophage, and Anx A5 exhibited the down-regulation in the phagocytosis of M1 macrophages (Fig. S2).
Fig. 1.
Anx A5 promoted macrophage phenotype shift from M1 to M2. (A) BMDMs were treated with various doses of Anx A5 (60, 120, 240 nM) in the presence of 10 ng/ml LPS and 10 ng/ml IFN-γ, or 20 ng/ml IL-4 for 6 h, cytokines release of Il-1β, Il-6, Tnf-α in supernatant were measured by ELISA. (B–F). mRNA levels of Il-1β, Il-6, Tnf-α, Cd206, Fizzl, Ym1 were determined by quantitative PCR. (B–C) Anx A5 decreased M1-related gene expression, and simultaneously increased M2-related gene expression in M1 macrophages. (D) Anx A5 increased M2-related gene expression in M2 macrophages. (E–F) Anx A5 promoted the mixed macrophage to M2 phenotype, not M1 phenotype. (G–H) CD11c+ and CD206+ cells were measured by flow cytometry. Date are represented as mean ± SEM of three independent experiments. &P < 0.05 vs normal group; *P < 0.05, **P < 0.01, ***P < 0.001 vs. M1 group. #P < 0.05, ##P < 0.01 vs. M2 group; $P < 0.05, $$P < 0.01, $$$P < 0.001 vs. M1 and M2 mixed group.
Meanwhile, we detected endogenous Anx A5 expression during M1 polarization procedure. Interestingly, endogenous Anx A5 was negatively correlated with the process of macrophage inflammation (Figs. S3A–B). Anx A5 expression was gradually decreased accompanied by the inflammation development (Fig. S3, from 0 to 8 h polarization), with the strongest inhibitory effect after 8-h polarization. Then, Anx A5 expression was progressively raised accompanied with inflammation resolution (Fig. S3, from 8 to 24 h polarization). Furthermore, endogenous Anx A5 was downregulated by siRNA in RAW 264.7 cells. Results exhibited that knockdown of endogenous Anx A5 induced moderate inflammation, which can be alleviated by supplement with exogenous Anx A5 (Fig. S4). These results confirmed Anx A5 participated M1 macrophage polarization, indicating the inhibitory effect.
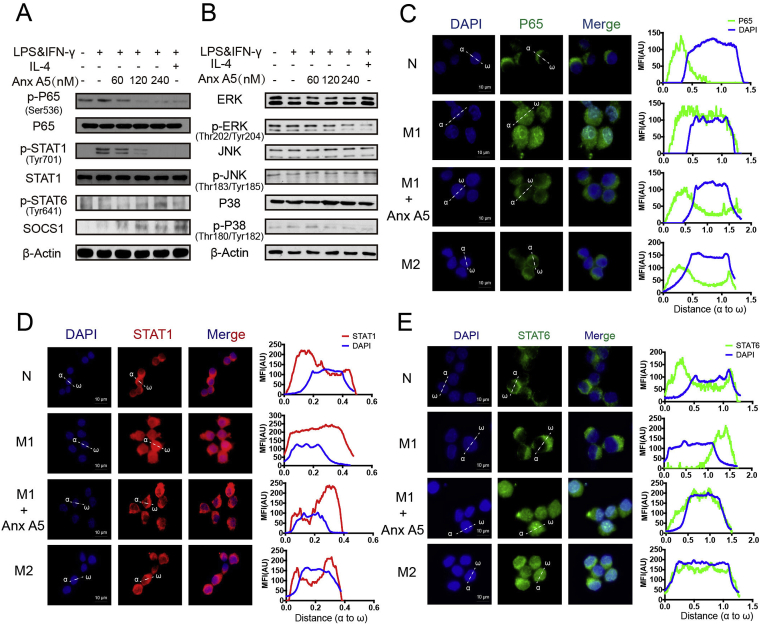
Since numerous studies have reported mechanisms of macrophage polarization, we also examined the classic signaling pathways. Anx A5 treatment significantly suppressed p-STAT1 and p-P65, enhanced p-STAT6/SOCS1 (Fig. 2A), while slightly affected MAPK pathway, including ERK, JNK and P38 (Fig. 2B). As transcription factors, STAT1, P65 and STAT6 need to translocate from cytoplasm to nucleus to mediate relevant genes expression. Immunofluorescence demonstrated that Anx A5 repressed LPS&IFN-γ induced translocation of STAT1 and P65 to nucleus (Fig. 2C and D), while encouraged STAT6 translocation in nucleus (Fig. 2E).
Fig. 2.
Anx A5 blocked NF-kB and STAT1 signals, and activated STAT6 signals. BMDMs were incubated with various doses of Anx A5 (60, 120, 240 nM) in the presence of 10 ng/ml LPS and 10 ng/ml IFN-γ, or 20 ng/ml IL-4 for 6 h. (A–B) Expression of p-P65, P65, p-STAT1, STAT1, p-STAT6, SOCS1, ERK, p-ERK, JNK, p-JNK, P38, p-P38 and β-actin was determined by Western blot. β-actin was a loading control. (C–E) Cells were treated with 240 nM Anx A5 in the presence of 10 ng/ml LPS and 10 ng/ml IFN-γ or 20 ng/ml IL-4 for 6 h, and the localization of P65 (C), STAT1 (D), STAT6 (E) were analyzed by immunofluorescence staining. The nuclei were stained with DAPI (blue). The line charts represent fluorescence intensity (MFI), which is presented the distance from α to ω in the images. Scale bar, 10 μm. Date of western blot are the representatives of three independent experiments. Pictures of immunofluorescence are the representatives of five different visual fields. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Above results depicted that Anx A5 dose-dependently facilitated macrophages turning from pro-inflammatory M1 phenotype to anti-inflammatory M2 phenotype.
3.2. Anx A5 attenuated glycolysis but enhanced OXPHOS in M1 macrophages
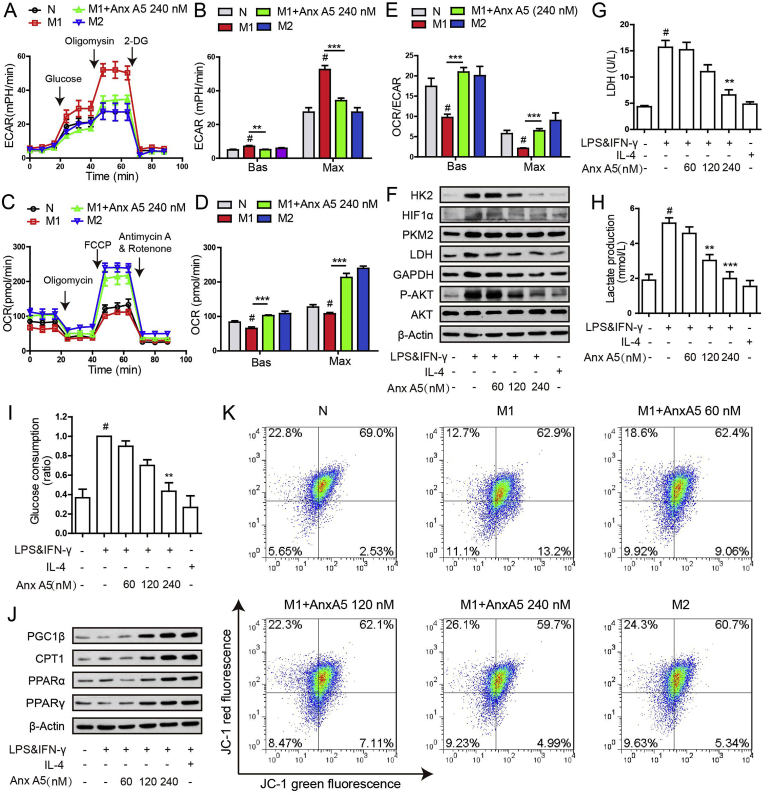
Metabolic reprogramming is the main factor of macrophage polarization. Then we measured macrophage ECAR (Extracellular acidification rate, represent glycolysis) and OCR (oxygen consumption rate, represent OXPHOS) level by using respirometry and metabolomics instrument seahorse XF96 extracellular flux analyzer. Results demonstrated that Anx A5 at 240 nM caused a significant decrease in ECAR (Fig. 3A and B), while led to a notable increase in OCR (Fig. 3C and D), of M1 macrophage. Basic and maximum of OCR/ECAR ratio revealed that Anx A5 induced a metabolic transition from glycolysis to mitochondrial oxidative phosphorylation in pro-inflammatory M1 macrophage (Fig. 3E). We further investigated glycolysis related signals by western blot (Fig. 3F), lactate production (Fig. 3H), lactate dehydrogenase (LDH) activity (Fig. 3G), and glucose consumption (Fig. 3I) by commercial kits. Results clearly revealed Anx A5 inhibited glycolysis capacity dose-dependently.
Fig. 3.
Anx A5 attenuated M1 macrophage glycolysis and enhanced OXPHOS and fatty oxidation. BMDMs were seeded in a Seahorse XF96 analyzer culture plates and treated with Anx A5 (240 nM) in the presence of 10 ng/ml LPS and 10 ng/ml IFN-γ or 20 ng/ml IL-4 for 6 h. Real-time rate changes in extracellular acidification rate (ECAR) (A) and oxygen consumption (OCR) (C) were analyzed under basal conditions and then with sequential additions of various agonist and inhibitors at the indicated times. Basic and maximum capacity of ECAR (B) and OCR (D) were shown. (E) OCR/ECAR was analyzed. (F) Cells were incubated with various dose of Anx A5 in the presence of 10 ng/ml LPS and 10 ng/ml IFN-γ or 20 ng/ml IL-4 for 6 h. Expression of HK2, HIF1α, PKM2, LDH, GAPDH, p-AKT, AKT, β-Actin were determined by western blot. (G) Lactate dehydrogenase (LDH) enzymatic activity was assessed. (H) Lactic acid production was measured. (I) Glucose uptake was analyzed. (J) Protein expression of some transcriptional factors about lipid metabolism (PGC1β, CTP1, PPARα, PPARγ) were determined by western blot. (K) BMDMs were incubated with JC-1 at working stock and mitochondrial membrane potential was measured by flow cytometry. Data of western blot and flow cytometry are the representatives of three independent experiments. Date of histograms are represented as mean ± SEM of three independent experiments. #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01, ***P < 0.001 vs. M1 group.
Fatty acid oxidation (FAO) is an important prerequisite for oxidative phosphorylation, and blots were indicated that FAO capacity was enhanced by Anx A5 treatment (PGC1β, CPT1, PPARα, PPARγ) (Fig. 3J). In addition, JC-1 staining (Fig. 3K) showed Anx A5 maintained high mitochondrial membrane potential, an event providing favorable conditions for oxidative phosphorylation. Taken together, Anx A5 evoked metabolic reprogramming in M1 macrophage, leading to phenotype shift and functional change.
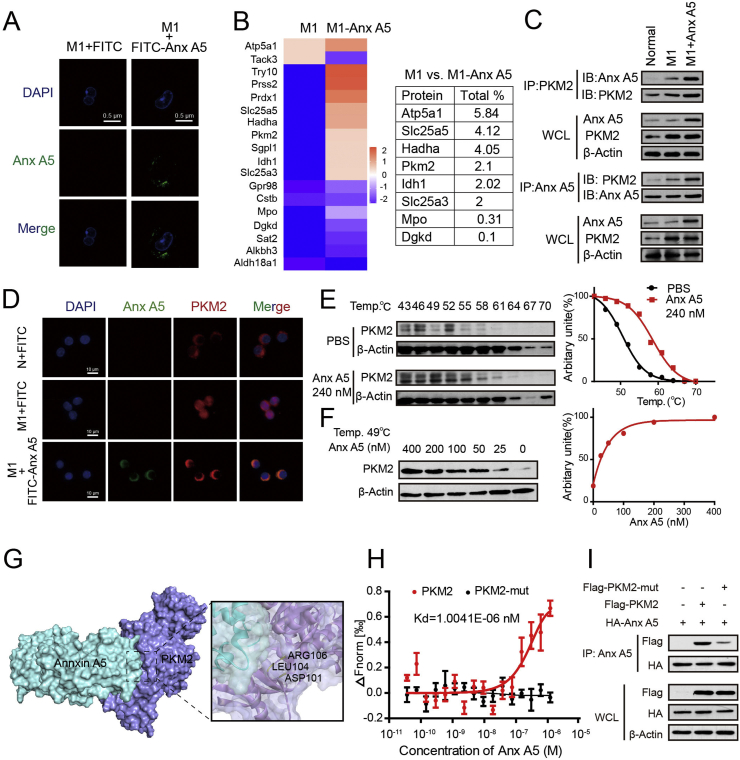
3.3. Anx A5 directly interacted with PKM2 at ASP101, LEU104 and ARG106
To investigate the mechanism of Anx A5 on regulating macrophage metabolic patterns and phenotype conversion, we used a series of experiments to seek the binding targets of Anx A5 in macrophage. First, Anx A5 was coupled with FITC, and FITC-Anx A5 was verified to have normal spatial structure and activity by TychoTM NT. 6 and flow cytometry assay (Fig. S5). Then, after 6-h incubation, ultrahigh resolution laser confocal showed most FITC-Anx A5 located in cytoplasm of M1 macrophage., predicting Anx A5 probably can incite the cellular signals (Fig. 4A). Next, co-immunoprecipitation and mass spectrometry were carried out to explore Anx A5-binding proteins. Among the candidates with TOP high scores, PKM2 is associated with glycolysis and OXPHOS (Fig. 4B). Previous literatures have reported that PKM2 plays a critical role in balancing glycolysis and OXPHOS by monomer/dimer and tetramer transformation, so we intensively studied the interaction of Anx A5 and PKM2. Co-immunoprecipitation clearly confirmed the binding activity of Anx A5 and PKM2 (Fig. 4C), and immunofluorescence revealed the binding reaction occurred in cytoplasm (Fig. 4D). Co-immunoprecipitation experiment was also conducted in the overexpression system with HA-Anx A5 and Flag-PKM2 plasmid transfection, and blots verified again the strong interaction of Anx A5 and PKM2 (Fig. S6). In addition, CETSA exhibited that Anx A5 significantly enhanced the thermal stability of PKM2 even at higher temperatures, compared to control group (PBS treatment) (Fig. 4E). And at a constant temperature (49 °C) and heating time, Anx A5 treatment preserved PKM2 stability in a dose-dependently manner (Fig. 4F). Next, molecular docking analysis demonstrated that Anx A5 (PDB ID: 2XO3) probably bound with PKM2 (PDB ID:1T5A) at ASP101, LEU104, ARG106 (Fig. 4G). To further confirm the binding strength and binding sites of Anx A5 with PKM2, microscale thermophoresis (MST) was carried out. The binding affinity for Anx A5 and PKM2 was shown with a Kd value of 1.0041E-06 nM. And more importantly, there was no binding reaction between Anx A5 and PKM2-mutant (D101A, L104A, R106A) (Fig. 4H). As well, Co-immunoprecipitation experiment proved that Anx A5 obviously bound with PKM2, not PKM2-mutant (D101A, L104A, R106A) (Fig. 4I).
Fig. 4.
Anx A5 directly interacted with PKM2 at ASP101, LEU104 and ARG106 (A) BMDMs were incubated with FITV-Anx A5 (240 nM) or FITC (as negative control) in the presence of 10 ng/ml LPS&IFN-γ for 6 h, and washed with PBS for 3 times. Cellular location FITC-Anx A5 were observed by ultrahigh resolution laser confocal. (B) Anx A5-interacted proteins were explored by co-immunoprecipitation and mass spectrometry, and some target proteins with higher score were about glycolysis and OXPHOS, as shown. (C) Cells were treated with Anx A5 (240 nM) in the presence of 10 ng/ml LPS & IFN-γ, the interation of Anx A5 and PKM2 was determined by co-immunoprecipitation. (D) The cellular location of Anx A5 and PKM2 complex was observed by ultrahigh resolution laser confocal. (E–F) Cells were incubated with PBS or Anx A5 for 6 h, and CETSA analyzed the thermal stabilization of PKM2 protein at different temperatures and concentrations. (G) Molecular docking showed binding domain of Anx A5 and PKM2. (H) The interaction of Anx A5 with PKM2, or Anx A5 with PKM2-mutant (D101A, L104A, R106A) was measured by microscale thermophoresis. The Kd value of Anx A5 and PKM2 interaction was determined with MO. Affinity Analysis Software. The graphs displayed data are represented as mean ± SEM of three independent experiments. (I) HA-Anx A5, Flag-PKM2 or Flag-PKM2 mutant (D101A, L104A, R106A) plasmid was transfected into HET293T cells, and protein-protein interaction was determined by co-immunoprecipitation. Immunofluorescence pictures are the representatives of five different visual fields.
Above results suggested Anx A5 directly interacted with PKM2, the key regulator of metabolic reprogramming.
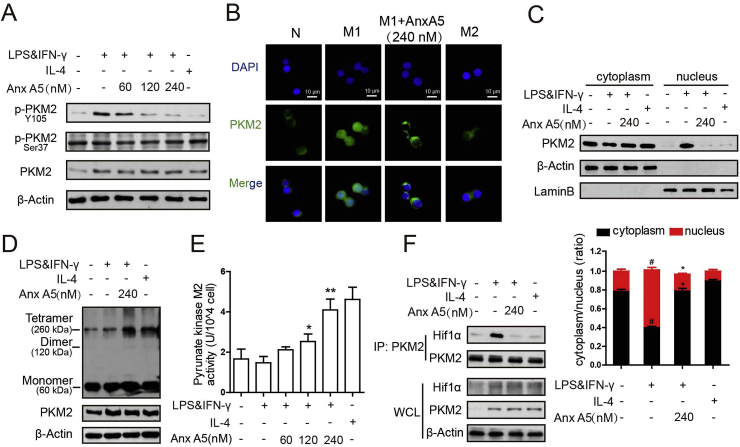
3.4. Anx A5 inhibited PKM2 Y105 phosphorylation and enhanced its enzymatic pyruvate kinase activity by promoting the formation of active PKM2 tetramers
PKM2 phosphorylation at different sites shows diverse function and different metabolic pathway. Y105 phosphorylation is indicative of monomer/dimer formation, as it prevents PKM2 tetramer configuration, further promoting the Warburg effect [36]. And Ser37 phosphorylation is responsible for recruitment of PIN1 to form PKM2/PIN1 complex [37]. As shown in Fig. 5A, LPS&IFN-γ induced high protein level of PKM2 in macrophages, which caused concurrent phosphorylation of Y105 phosphorylation (compared line 2 to line 1), and Anx A5 dose-dependently blocked Y105 phosphorylation (compared line 3–5 to line 2) of PKM2, while Ser37 phosphorylation had no obvious change. Furthermore, it was also depicted that Anx A5 prevented PKM2 transferring from cytoplasm to nucleus (Fig. 5B and C). PKM2 has monomer, dimer and tetramer forms, with different cellular location and diverse functions in glycolysis and OXPHOS, respectively. Subsequently, DSS cross-linking was carried out to support the nucleus exclusion of PKM2 by Anx A5 treatment. Cell lysates were cross-linked using DSS and endogenous PKM2 was detected. The blots revealed a high molecular weight of PKM2-containing complex at approximately 250 kDa by Anx A5 treatment, representing the tetramer formation of PKM2, and IL4, M2 phenotype inducer, exhibited the similar result (Fig. 5D). However, the monomer/dimer form of PKM2 did not obviously changed by Anx A5 (Fig. 5D). In addition, Data of Fig. 5E displayed an impressive increase of enzymatic pyruvate kinase activity of PKM2 with series concentration of Anx A5 treatment, again suggesting the promotion of tetramer form of PKM2. Then, co-immunoprecipitation experiment showed Anx A5 dampened the complex of PKM2 with Hif1α, which was occurred in nucleus and induced by the monomer/dimer form of PKM2 (Fig. 5F).
Fig. 5.
Anx A5 inhibited PKM2 tyrosine 105 (Y105) phosphorylation and enhanced its enzymatic pyruvate kinase activity (A) BMDMs were incubated with various concentrations of Anx A5 (60, 120 and 240 nM) in LPS&IFN-γ or IL-4 as described above, the expression of p-PKM (Y105), p-PKM2 (Ser37), PKM2 and β-Actin were determined by western blot. (B) Cellular localization of PKM2 was analyzed by immunofluorescence staining. The nuclei were stained with DAPI. Pictures are representative of five different visual fields. (C) Cells were incubated with Anx A5 (240 nM) with LPS&IFN-γ or IL-4, proteins of nuclear and cytoplasmic fractions were isolated respectively, and PKM2, Lamin B, β-Actin expression were determined by western blot and analyzed in histogram. (D) Cells were treated with Anx A5 (240 nM) with LPS&IFN-γ or IL-4, then collected and cross-linked with DSS. Tetramer, dimer and monomer form of PKM2 were analyzed by western blot. (E) Enzymatic pyrunate kinase activity was measured. (F) Association of PKM2 and Hif1α was detected by co-immunoprecipitation. Data of western blot are the representatives of three independent experiments. Date of histograms are represented as mean ± SEM of three independent experiments. #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01 vs. M1 group.
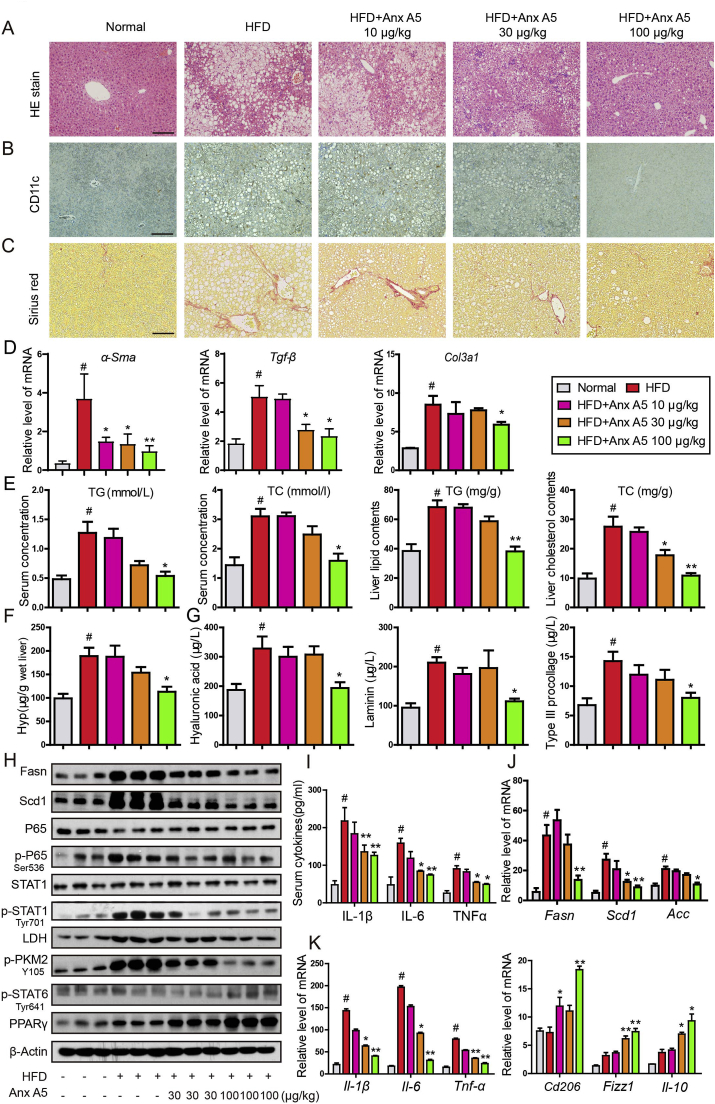
3.5. Anx A5 ameliorated HFD-induced NASH in mice via regulating hepatic macrophage polarization
Macrophage, including Kupffer cell and infiltrated monocyte, plays an essential role in the progression of NASH, and we evaluated the improvement of Anx A5 in HFD-induced NASH. Anx A5 effectively alleviated NASH symptoms, especially at high dosage of 100 μg/kg. HE staining reflected the descent of lipid accumulation, inflammatory cells infiltration and hepatocytes necrosis (Fig. 6A). Immunohistochemistry staining indicated the decrease of CD11c+ cells in liver (Fig. 6B). And sirius red staining showed the lessening of fibrosis (Fig. 6C). HFD-induced mice suffered high level of serum TC/TG and liver TC/TG, and treatment of Anx A5 at 100 μg/kg remarkably decreased TC/TG in serum and liver (Fig. 6E). Fibrosis is a risk factor for deterioration of NASH, hence we examined the fibrotic indexes in mice liver. QPCR results showed that Anx A5 obviously inhibited HFD-induced α-Sma, Tgf-β and Col3a1 mRNA (Fig. 6D). In addition, hepatic Hyp content was reduced (Fig. 6F), and serum hyaluronic acid, laminin and Type III procollagen were also decreased, with Anx A5 administration (Fig. 6G). Meanwhile, hepatic lipogenesis proteins and macrophage inflammatory pathways were measured by western blot, results showed that Anx A5 treatment attenuated HFD-induced lipid synthesis and inflammatory responses, with descent of Fasn, Scd1, p-P65, p-STAT1(Tyr701), LDH and p-PMK2(Y106) protein level and ascent of p-STAT6 and PPARγ protein level (Fig. 6H). Fans, Scd1 and Acc mRNA level was also inhibited by Anx A5 (Fig. 6J). In addition, Anx A5 was identified to suppress serum inflammatory cytokines, and concurrently increase anti-inflammatory cytokine (Fig. 6I and K).
Fig. 6.
Anx A5 ameliorated HFD-induced NASH in mice C57BL/6 mice were fed HFD for 16 weeks to induce NASH. After 8 weeks of HFD, mice were administrated with 10, 30, 100 μg/kg Anx A5 once every 3 days by tail intravenous injection for 8 weeks, and fasted for 12 h before harvest (n = 9 mice per group). (A) HE staining with liver paraffin sections, scale bar 20 μm. (B) Immunohistochemistry for CD11c expression. (C) Sirius red stain for fibrosis. (D) mRNA level of α-Sma, Tgf-β, Col3a1 in liver was measured by quantitative PCR. (E) TG, TC in serum and liver tissue was analyzed by commercial kit. (F) Liver Hyp concentration. (G) Liver fibrosis index analysis, including hyaluronic acid, laminin, Type III procollagen. (H) Protein level of Fasn, Scd1, p-STAT1/STAT1, p-P65/P65, LDH, p-PKM2 (Y105), p-STAT6, PPARγ and β-actin was determined by western blot. (I) Serum inflammatory cytokines of IL-1β, IL-6 and TNFα were determined by ELISA. (J–K) The mRNA level of Fasn, Scd1, Acc, Il-1β, Il-6, Tnf-α, Cd206, Fizzl, Il-10 was determined by quantitative PCR. Date are represented as mean ± SD, n = 9 mice for each experimental group. #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01 vs. HFD group. Pictures (A–C) were the representative of five field for per mice in every group. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
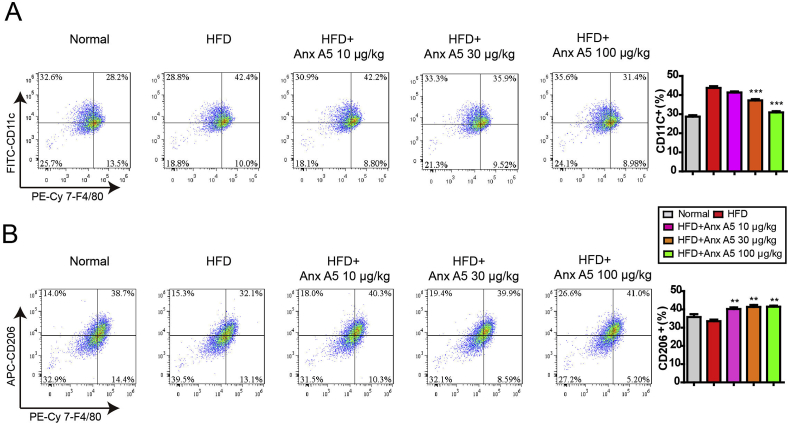
As mentioned above, Anx A5 regulated macrophage polarization in vitro, then we detected the hepatic macrophage functions in vivo. CD11c+F4/80+ is represented M1 phenotype and CD206+F4/80+ is indicated M2 phenotype in mouse liver. Anx A5 significantly inhibited CD11c+F4/80+ proportion and promoted CD206+F4/80+ proportion in HFD-induced NASH by flow cytometry analysis (Fig. 7A and B). Similarly, Immunofluorescent stain with antibodies of CD11c, Ly6c, Ly6g and CD206 confirmed again that Anx A5 decreased M1 macrophages, monocytes and neutrophils infiltration, and induced M2 macrophages proportion in NASH liver (Fig. S7).
Fig. 7.
Anx A5 inhibited hepatic macrophage M1 polarization and promoted M2 polarization. C57B/L6 mice were treated as above described. Hepatic macrophages were isolated from liver by using liver perfusion collagenase system and gradient centrifugation. F4/80+CD11c+ (A) and F4/80+CD206+ (B) macrophage proportion were analyzed by flow cytometry with respective antibodies. The positive cells are shown as a histogram of mean ± SEM, n = 4 mice for each experimental group. #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01 vs. HFD group.
In addition, we further explored the effect of Anx A5 in non-parenchymal cells of liver. Result demonstrated that Anx A5 selectively inhibited M1 macrophages (CD163+) and monocytes (Ly6c+), without influencing hepatic stellate cells (Retinol+), natural killer (NK) cells (NK1.1+) or liver sinusoidal endothelial cells (LSECs) (CD146+), as shown in Fig. 8A. Besides, Kupffer cells were isolated and polarized into M1 and M2 phenotype according to the description of Methods and Materials. Consistent with the results of RAW264.7 cells, Anx A5 also induced phenotype shift from M1 to M2 in Kupffer cells, as shown in CD11c+ and CD206+ proportion (Fig. 8B), classical inflammatory cytokines (Fig. 8C and D) and typical transcriptional factors (Fig. 8E, Fig. S8). These results suggested the in vivo transfer capacity of Anx A5 in hepatic macrophage phenotype shift from M1 to M2.
Fig. 8.
Anx A5 selectively affected hepatic macrophages. (A) Non-parenchymal liver cells of C57BL/6 mouse were isolated as described in Methods and Materials, and treated with 100 ng/ml LPS and 100 mM PA for 18 h (LPS plus PA was used to mimic NASH model in vitro), then incubated with FITC-Anx A5 (240 nM) for 6 h. FITC-Anx A5 in the non-parenchymal liver cells was analyzed by flow cytometry. (B) Kupffer cells from C57BL/6 mouse were isolated and incubated with various concentrations of Anx A5 (60, 120 and 240 nM) in the presence of LPS &IFN-γ. And the surface marker CD11c and CD206 were determined by flow cytometry. (C–D) The mRNA level of Il-6, Tnf-α, Il-1β, Cd206, Fizzl, Ym1 was determined by quantitative PCR. (E) p-P65, p65/P65, p-STAT1/STAT1, p-STAT6 and β-Actin was determined by western blot. Data are represented as mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 vs. M1 group. Data of western blot are the representatives of three independent experiments.
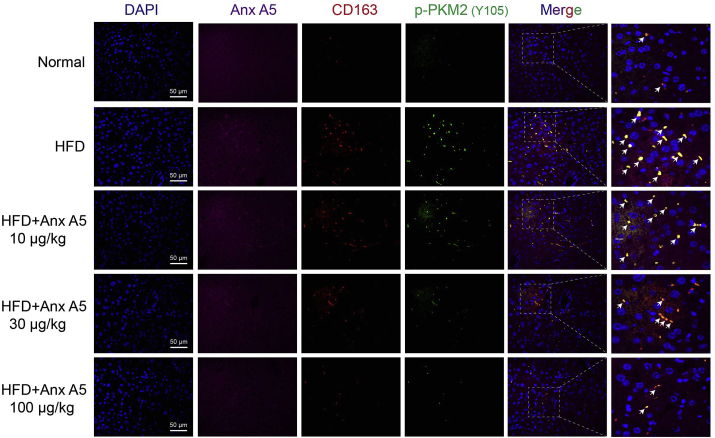
Furthermore, Anx A5 dose-dependently decreased PKM2(Y105) phosphorylation (shown in green) especially in hepatic inflammatory macrophages (CD163+, shown in red) in NASH mice (Fig. 9).
Fig. 9.
Anx A5 inhibited p-PKM2(Y105) expression in hepatic macrophages. C57BL/6 mice were treated as described in the legend of Fig. 6. Anx A5, CD163 and p-PKM2(Y105) expression in liver sections was detected by immunofluorescence. Scale bar 50 μm. Pictures are the representatives of five different fields for per mouse in every experimental group.
3.6. Effect of Anx A5 in NASH and hepatic macrophage phenotype shift was largely dependent on PKM2
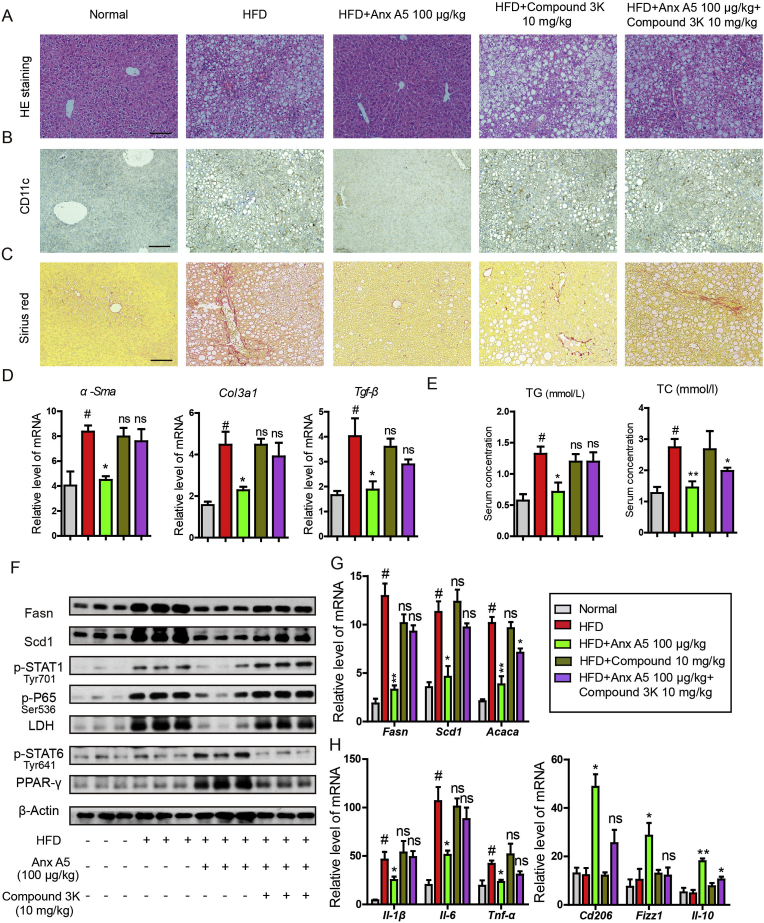
To further confirm the role of PKM2 in amelioration of Anx A5 in NASH, a PKM2 enzymic inhibitor, compound 3k, was used together with Anx A5 in NASH mice. Results indicated that co-treatment with compound 3k largely invalidated the therapeutic effect of Anx A5 in NASH. Compound 3k re-induced lipid accumulation (Fig. 10A), CD11c+ cells proportion (Fig. 10B) as well as liver fibrosis (Fig. 10C), events which were significantly attenuated by Anx A5 treatment. In addition, compound 3k greatly blocked the reduction of α-Sma, Col3a1 and Tgf-β mRNA level in liver which was verified by Anx A5 administration (Fig. 10 D), and similar results in serum TC and TG (Fig. 10E). Moreover, combination of compound 3k resisted the effect of Anx A5 on hepatic lipogenesis proteins, macrophage polarization and metabolic pathways, as shown in Fig. 10F and G. And compound 3k also reversed M1-and M2-related characteristic cytokines induced by Anx A5 in NASH mice (Fig. 10H). These results suggested effect of Anx A5 in NASH and hepatic macrophage phenotype shift can be effectively blocked by PKM2 enzymatic inhibitor, compound 3k. Besides, there were no significant changes in compound 3k alone group, comparing with HFD group.
Fig. 10.
PKM2 enzymatic inhibitor, compound 3k, abolished therapeutic effect of Anx A5 on NASH. C57BL/6 mice were fed with HFD for 16 weeks. After 8 weeks of HFD treatment, mice were administrated with Anx A5 (100 μg/kg) by tail intravenous injection and compound 3K by gavage (10 mg/kg) for 8 weeks, and both were once every 3 days (n = 9 mice per group). (A) HE staining with liver paraffin sections, scale bar 20 μm. (B) Immunohistochemistry for CD11c expression. (C) Sirius red stain for fibrosis. (D) mRNA level of α-Sma, Tgf-β, Col3a1 in liver was measured by quantitative PCR. (E) Serum TC and TG assay by commercial kits. (F) Hepatic Fasn, Scd1, p-STAT1, p-P65, LDH, p-STAT6, PPARγ, and β-actin expression was determined by western blot. (G–H) mRNA level of Fasn, Scd1, Acaca, Il-1β, Il-6, Tnf-α, Cd206, Fizzl, Il-10 was determined by quantitative PCR. Date are represented as mean ± SD. n = 9 mice for every experimental group.; #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01 vs. HFD group; ns (non-significant), P > 0.05 vs HFD group. Pictures (A–C) were the representative of five different fields for per mouse in every group. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
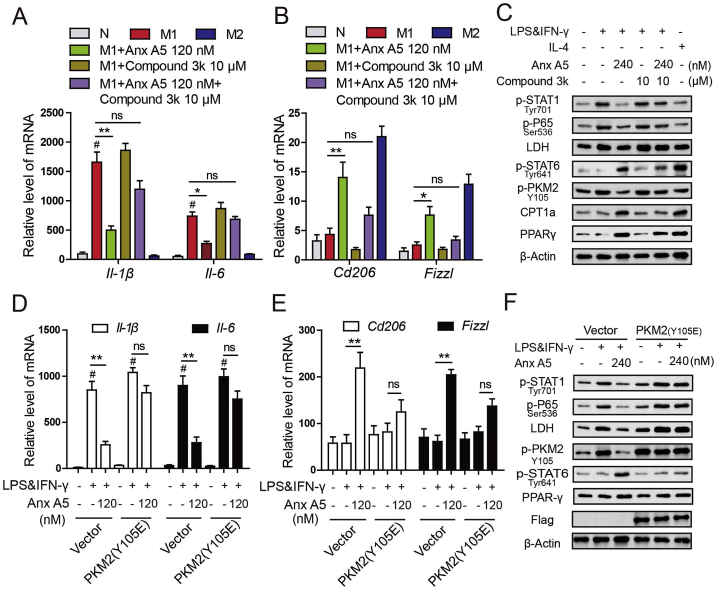
Then, consistent with the in vivo data, compound 3k greatly blocked effect of Anx A5 on macrophage phenotype shift in RAW264.7 cells. Anx A5 alone treatment significantly inhibited M1-related factors and promoted M2-related factors, however, Anx A5 plus compound 3k treatment led to non-significance change compared with LPS&IFNγ group (Fig. 11A–C). In addition, Flag-PKM2 (Y105E), the mutation plasmid to mimic phosphorylation in Y105 of PKM2, also displayed the similar results as compound 3k, weakened the function of Anx A5 in macrophage polarization (Fig. 11D–F). Above data confirmed that the regulation of Anx A5 in macrophage phenotype shift from M1 to M2 was obviously dependent on PKM2.
Fig. 11.
Regulation of Anx A5 in macrophage phenotype shift was dependent on PKM2(A–C) Raw 264.7 cells were incubated with Anx A5 (120 nM) and compound 3k (10 μM) under LPS&IFN-γ or IL-4 treatment. (A–B) mRNA level of Il-1β, Il-6, Tnf-α, Cd206 and Fizzl were determined by quantitative PCR. (C) Expression of p-STAT1, p-P65, LDH, p-STAT6, p-PKM2 (Y105), CPT1, PPARγ, and β-actin were determined by western blot. (D–F) Raw 264.7 cells were transfected with PKM2 (Y105E) plasmids and vector for 48 h, and then incubated with Anx A5 (240 nM) for 6 h in the presence of LPS&IFN-γ or not. (D–E) mRNA level of Il-1β, Il-6, Tnf-α, Cd206 and Fizzl were determined by quantitative PCR. (F) Expression of p-STAT1, p-P65, LDH, p-STAT6, p-PKM2 (Y105), CPT1, PPARγ, and β-actin were determined by western blot. Date are represented as mean ± SEM of three different experiments; #P < 0.05 vs. normal group; *P < 0.05, **P < 0.01 vs. M1 group; ns, P > 0.05 vs M1 group.
4. Discussion
Owing to the essential role of macrophages in NASH, core events in their function might be suitable for therapeutic intervention. Here we firstly identified Anx A5 exhibits not only blockade in M1 macrophages but also activation in M2 macrophages, which is more potential for balancing the hepatic inflammation and improving NASH (Fig. 1, Fig. 7, Fig. 8). While, CVC, a subject in phase III clinical trial of NASH, only shows mild suppression in M1 macrophages at higher concentration compared with Anx A5, without influencing M2 characteristics, suggesting its restriction in NASH treatment (Fig. S9).
Mechanism about macrophage polarization is diversity, and classic NFκB, STAT1 and STAT6 signals are involved in it. Although Anx A5 displays regulation in these typical pathways (Fig. 2), this is not enough to reveal the mechanism of Anx A5 in modulating macrophage phenotype shift. As well known, Anx A5 is a phospholipid binding protein, however, in macrophage polarization we have confirmed Anx A5 plays a leading role in metabolic reprogramming from glycolysis to oxidative phosphorylation, and the related indexes are almost completely reversed (Fig. 3). Hence, we focus on macrophage metabolic events under Anx A5 treatment.
The production of ATP provide energy for cellular function, and generally glucose is as the main source for ATP production through glycolysis and tricarboxylic cycle (TCA)-dependent oxidative phosphorylation (OXPHOS) [38]. Although glycolysis produces less ATP than OXPHOS, the speed of ATP production is more quicker than the later, which is critical for maintaining energy requirements [39]. When macrophages are stimulated by foreign invaders, they need quick energy consumption to proliferation and fight infection [16]. Therefore, M1 macrophages increase metabolism of glucose to lactate and decrease OXPHOS, which is required for proinflammatory activation. While M2 macrophages are associated with tissue repairment and anti-inflammation, needing constant supply of energy and exhibiting absence of glycolysis and presence of OXPHOS [40].
During the process of macrophage polarization, Anx A5 can enter the cytoplasm, suggesting that A5 not only binds to phospholipid of cell membrane, but also induces intracellular signals. Furthermore, we have confirmed with various methods that Anx A5 highly integrates with PKM2 in macrophage cytoplasm (Fig. 4). Numerous studies have reported a key function for PKM2 in the switch of glycolysis and OXPHOS. We have revealed that Anx A5 directly binds with PKM2 at ASP101, LEU104, ARG106 (Fig. 4G–I). These binding sites are surrounding with Y105 of PKM2, probably leading to the inhibition of Anx A5 on Y105 phosphorylation. Y105 phosphorylation of PKM2 is the main characteristic of monomer/dimer form, which prevents tetramer form assembling and subsequent nuclear translocating. In nucleus, monomer/dimer form of PKM2 interacts with Hif1α and directly binds with IL-1β promoter, resulting in IL-1β transcription, which dominantly occurs in glycolysis of M1 macrophage [41,42]. While, tetramer form mainly stays in cytoplasm and is responsible for OXPHOS in anti-inflammation [23]. Our following results demonstrate Anx A5 obviously blocks PKM2 translocation in nucleus, induces tetramer formation in cytoplasm, activates pyruvate kinase enzymatic activity and prevents the interaction of Hif1α and PKM2, all of these are concurrent with Y105 phosphorylation inhibition.
While, effect of Anx A5 on PKM2 monomer/dimer form is not remarkable (Fig. 5D), which may be due to different timepoint for assay. And in some previous study, PKM2 dimer form was also very low to monitor [20]. Still, the increase of tetramer form induced by Anx A5 triggers notable change in ratio of monomer/dimer and tetramer form, which is the main reason for metabolic reprogramming and macrophage phenotype shift under Anx A5 treatment. What's more, we confirm this effect greatly dependent on PKM2 by using PKM2 enzymic inhibitor, compound 3k, and PKM2 (Y105E) mutation (Fig. 11).
NASH is recognized as a kind of chronic inflammatory disease. Hepatic macrophages, neutrophils, DC cells and NK cells are all involved in the development of NASH, among these, macrophages are likely to be more important for steatosis and fibrosis of NASH [43]. Mice subjected to macrophage depletion are protected from the development of steatosis [44]. And it has been verified that portal infiltration of macrophages seems to occur at the early stage before inflammation or fibrosis develops, but predicting progressive disease [45]. Upon liver injury, Kupffer cells and the recruited monocytes differentiate to M1 macrophages, secrete inflammatory cytokines and ROS, and incite HSC to release extracellular matrix, resulting in progressive fibrosis [46]. All these literatures reveal the essential role of macrophages in steatosis, inflammation and fibrosis. Here, we have proved that Anx A5 obviously controls hepatic macrophage functions, without significant influence in other non-parenchymal liver cells (Fig. 8) and hepatocytes (Fig. S10). Therefore, it is concluded that improvement of Anx A5 on NASH in vivo is mainly due to regulating hepatic macrophage phenotype shift via targeting PKM2 and consequent metabolic reprogramming, then initiating cell-cell interactions in liver.
Besides, PKM2 expresses and functions in steatosis of hepatocyte, while in this study Anx A5 does not obviously influence the lipid accumulation induced by PA in primary hepatocytes (Fig. S10), owing to many factors involved in lipid metabolism of hepatocyte and the effect of Anx A5 is covered. Instead, Anx A5 displays the amelioration in lipotoxicity, inflammation and fibrosis because of the major target cell is hepatic macrophages.
5. Conclusion
In conclusion, we have revealed a novel function of Anx A5 in macrophage polarization and HFD-induced NASH, providing important insights into the metabolic reprogramming changes in hepatic macrophage, which is important for NASH progression and therapy. Anx A5 targeting to PKM2 results in glycolysis inhibition and mitochondrial oxidative metabolism activation, which triggers macrophage phenotype shift and offers a novel therapeutic approach for NASH.
Declaration of competing interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
This work was supported by National Key R&D Program of China (No. 2017YFA0506000), National Natural Science Foundation of China (Nos. 81872916, 81673487, 81730100), National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program”, China (No. 2018ZX09201002), “The Drug Innovation Major Project” of National Science & Technology Ministry (No. 2018ZX09711001-003-007), The Open Project of State Key Laboratory of Natural Medicines,(No. SKLNMKF202003), and the Fundamental Research Funds for the Central Universities (020814380118).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101634.
Contributor Information
Xudong Wu, Email: xudongwu@nju.edu.cn.
Qiang Xu, Email: molpharm@163.com.
Author contributions
(1) Study conception, design and manuscript writing: Xudong Wu, and Fang Xu;(2) Acquisition, analysis and/or interpretation of data: Fang Xu, Mengmeng Guo, Wei Huang, Lili Feng, Kangkang Luo, Jiazhen Zhu, Jian Gao, Bingfeng Zheng, Ling-Dong Kong, Tao Pang; (3) Final approval and overall responsibility for the published work: Xudong Wu and Qiang Xu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Younossi Z. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Am. J. Gastroenterol. 2012;107:811–826. doi: 10.1038/ajg.2012.128. [DOI] [PubMed] [Google Scholar]
- 3.Dulai P.S. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. doi: 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotman Y., Sanyal A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66:180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 5.Spengler E.K., Loomba R. Recommendations for diagnosis, referral for liver biopsy, and treatment of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mayo Clin. Proc. 2015;90:1233–1246. doi: 10.1016/j.mayocp.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tacke F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017;66:1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 7.Tacke F., Zimmermann H.W. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 2014;60:1090–1096. doi: 10.1016/j.jhep.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Li P.Z., He K., Li J.Z., Liu Z.J., Gong J.P. The role of Kupffer cells in hepatic diseases. Mol. Immunol. 2017;85:222–229. doi: 10.1016/j.molimm.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Duarte N., Coelho I.C., Patarrão R.S., Almeida J.I., Penha-Gonçalves C., Macedo M.P. How inflammation impinges on NAFLD: a role for kupffer cells. BioMed Res. Int. 2015;2015:984578. doi: 10.1155/2015/984578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazankov K. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019;16:145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 11.Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y.C., Zou X.B., Chai Y.F., Yao Y.M. Macrophage polarization in inflammatory diseases. Int. J. Biol. Sci. 2014;10:520–529. doi: 10.7150/ijbs.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapouri-Moghaddam A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 14.Feng L. Seselin ameliorates inflammation via targeting Jak2 to suppress the proinflammatory phenotype of macrophages. Br. J. Pharmacol. 2019;176:317–333. doi: 10.1111/bph.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng L.L. 5, 7, 2 ', 4 ', 5 '-Pentamethoxyflavanone regulates M1/M2 macrophage phenotype and protects the septic mice. Chin. J. Nat. Med. 2019;17:363–371. doi: 10.1016/S1875-5364(19)30042-1. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths H.R., Gao D., Pararasa C. Redox regulation in metabolic programming and inflammation. Redox Biol. 2017;12:50–57. doi: 10.1016/j.redox.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Kasmi K.C., Stenmark K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin. Immunol. 2015;27:267–275. doi: 10.1016/j.smim.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Prados J.C. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 19.Vats D. Oxidative metabolism and PGC-1 beta attenuate macrophage-mediated inflammation. Cell Metabol. 2006;4 doi: 10.1016/j.cmet.2006.05.011. 255-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palsson-McDermott E.M. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metabol. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalaiarasan P., Subbarao N., Bamezai R.N. Molecular simulation of Tyr105 phosphorylated pyruvate kinase M2 to understand its structure and dynamics. J. Mol. Model. 2014;20:2447. doi: 10.1007/s00894-014-2447-6. [DOI] [PubMed] [Google Scholar]
- 22.Alquraishi M. Pyruvate kinase M2: a simple molecule with complex functions. Free Radic. Biol. Med. 2019;143:176–192. doi: 10.1016/j.freeradbiomed.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi W. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat. Med. 2017;23:753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaneton B. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mudaliar S. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology. 2013;145:574–582. doi: 10.1053/j.gastro.2013.05.042. e571. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Park H., Bae E.J. Efficacy of evogliptin and cenicriviroc against nonalcoholic steatohepatitis in mice: a comparative study. KOREAN J. PHYSIOL. PHARMACOL. 2019;23:459–466. doi: 10.4196/kjpp.2019.23.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouter A. Review: annexin-A5 and cell membrane repair. Placenta. 2015;36(Suppl 1):S43–S49. doi: 10.1016/j.placenta.2015.01.193. [DOI] [PubMed] [Google Scholar]
- 28.Wolgast L.R., Arslan A.A., Wu X.X., Beyda J.N., Pengo V., Rand J.H. Reduction of annexin A5 anticoagulant ratio identifies antiphospholipid antibody-positive patients with adverse clinical outcomes. J. Thromb. Haemostasis. 2017;15:1412–1421. doi: 10.1111/jth.13699. [DOI] [PubMed] [Google Scholar]
- 29.Peng B., Guo C., Guan H., Liu S., Sun M.Z. Annexin A5 as a potential marker in tumors. Clin. Chim. Acta. 2014;427:42–48. doi: 10.1016/j.cca.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Ewing M.M. Annexin A5 therapy attenuates vascular inflammation and remodeling and improves endothelial function in mice. Arterioscler. Thromb. Vasc. Biol. 2011;31:95–101. doi: 10.1161/ATVBAHA.110.216747. [DOI] [PubMed] [Google Scholar]
- 31.Pineda-Torra I., Gage M., de Juan A., Pello O.M. Isolation, culture, and polarization of murine bone marrow-derived and peritoneal macrophages. Methods Mol. Biol. 2015;1339:101–109. doi: 10.1007/978-1-4939-2929-0_6. [DOI] [PubMed] [Google Scholar]
- 32.Bale S.S., Geerts S., Jindal R., Yarmush M.L. Isolation and co-culture of rat parenchymal and non-parenchymal liver cells to evaluate cellular interactions and response. Sci. Rep. 2016;6:25329. doi: 10.1038/srep25329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim B. Western blot techniques. Methods Mol. Biol. 2017;1606:133–139. doi: 10.1007/978-1-4939-6990-6_9. [DOI] [PubMed] [Google Scholar]
- 34.Kilkenny C., Browne W., Cuthill I.C., Emerson M., Altman D.G. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br. J. Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath J.C., Lilley E. Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br. J. Pharmacol. 2015;172:3189–3193. doi: 10.1111/bph.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitosugi T. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000431. ra73-ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He X. PKM2 in carcinogenesis and oncotherapy. Oncotarget. 2017;8:110656–110670. doi: 10.18632/oncotarget.22529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rui L.Y. Energy metabolism in the liver. Compreh. Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha S., Shalova I.N., Biswas S.K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 2017;280:102–111. doi: 10.1111/imr.12603. [DOI] [PubMed] [Google Scholar]
- 40.Verdeguer F., Aouadi M. Macrophage heterogeneity and energy metabolism. Exp. Cell Res. 2017;360:35–40. doi: 10.1016/j.yexcr.2017.03.043. [DOI] [PubMed] [Google Scholar]
- 41.Corcoran S.E., O'Neill L.A. HIF1alpha and metabolic reprogramming in inflammation. J. Clin. Invest. 2016;126:3699–3707. doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1beta production and to prevent DSS-induced colitis in mice. Cell Rep. 2017;19:2331–2344. doi: 10.1016/j.celrep.2017.05.065. [DOI] [PubMed] [Google Scholar]
- 43.Nati M., Haddad D., Birkenfeld A.L., Koch C.A., Chavakis T., Chatzigeorgiou A. The role of immune cells in metabolism-related liver inflammation and development of non-alcoholic steatohepatitis (NASH) Rev. Endocr. Metab. Disord. 2016;17:29–39. doi: 10.1007/s11154-016-9339-2. [DOI] [PubMed] [Google Scholar]
- 44.Huang W. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gadd V.L. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology. 2014;59:1393–1405. doi: 10.1002/hep.26937. [DOI] [PubMed] [Google Scholar]
- 46.Schuppan D., Surabattula R., Wang X.Y. Determinants of fibrosis progression and regression in NASH. J. Hepatol. 2018;68:238–250. doi: 10.1016/j.jhep.2017.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.