Abstract
Background
Genetic risk is thought to drive clinical variation on a spectrum of schizophrenia-like traits, but the underlying changes in brain structure that mechanistically link genomic variation to schizotypal experience and behavior are unclear.
Methods
We assessed schizotypy using a self-reported questionnaire and measured magnetization transfer as a putative microstructural magnetic resonance imaging marker of intracortical myelination in 68 brain regions in 248 healthy young people (14–25 years of age). We used normative adult brain gene expression data and partial least squares analysis to find the weighted gene expression pattern that was most colocated with the cortical map of schizotypy-related magnetization.
Results
Magnetization was significantly correlated with schizotypy in the bilateral posterior cingulate cortex and precuneus (and for disorganized schizotypy, also in medial prefrontal cortex; all false discovery rate–corrected ps < .05), which are regions of the default mode network specialized for social and memory functions. The genes most positively weighted on the whole-genome expression map colocated with schizotypy-related magnetization were enriched for genes that were significantly downregulated in two prior case-control histological studies of brain gene expression in schizophrenia. Conversely, the most negatively weighted genes were enriched for genes that were transcriptionally upregulated in schizophrenia. Positively weighted (downregulated) genes were enriched for neuronal, specifically interneuronal, affiliations and coded a network of proteins comprising a few highly interactive “hubs” such as parvalbumin and calmodulin.
Conclusions
Microstructural magnetic resonance imaging maps of intracortical magnetization can be linked to both the behavioral traits of schizotypy and prior histological data on dysregulated gene expression in schizophrenia.
Keywords: Adolescence, Allen Human Brain Atlas, Development, Fast-spiking GABAergic interneurons, Multiparameter MRI mapping, Myelination, Schizophrenia
SEE COMMENTARY ON PAGE 212
The genetic architecture of schizophrenia spectrum disorders assumes many independent allelic variations, each of small effect, contributing to the probability of diagnosis. Individuals with the greatest accumulation of genetic risk have the more severe psychotic disorder; individuals with a lower genetic risk may have less severe, nonpsychotic schizotypal personality disorder (1), characterized by social eccentricity and unusual beliefs (2). The genetic risk for schizophrenia has been resolved more clearly by recent genome-wide association studies (3,4) and postmortem human brain transcriptional studies (3,5). However, it remains unclear how expression of these schizophrenia-related genes might be related to neuroimaging markers of schizophrenia spectrum disorders.
Macrostructural magnetic resonance imaging (MRI) studies—which measure anatomical parameters like cortical thickness—have collectively provided robust evidence for reduced volume or thickness in a network of interconnected cortical areas in patients with schizophrenia (6). There have been fewer MRI studies of schizotypy, and the pattern of macrostructural results has not been consistent, perhaps reflecting their relatively small sample sizes (Table 1).
Table 1.
Previous Studies Relating Schizotypal Traits and Macrostructural Magnetic Resonance Imaging Metrics
| Study | Sample Size, n | Mean Age, Years | Experimental Design | Brain Coverage | Schizotypy Measure | Type I Error Control | Structural Index | Directionality | Brain Regions |
|---|---|---|---|---|---|---|---|---|---|
| Evans et al., 2016 (63) | 28 | 11 | VBM | Whole brain | PSI-C | Bonferroni | Volume | − | Caudate, amygdala, hippocampal gyrus, middle temporal |
| Nenadic et al., 2015 (64) | 59 | 31 | VBM / positive and negative factor of schizotypy | Whole brain | CAPE | FWE | Volume | − | R precuneus |
| Wang et al., 2015 (65) | 69 | 19 | VBM / dividing subjects with high/low schizotypy | Whole brain | SPQ | AlphaSim permutation | Volume | − | Dorsolateral prefrontal cortex, insula, posterior temporal, cerebellum |
| DeRosse et al., 2015 (66) | 138 | 36 | ANCOVA / dividing subjects with high/low schizotypy | ROIs | SPQ | None | Thickness/GM and WM volume | − | Frontal, temporal |
| Kühn et al., 2012 (67) | 34 | 36 | Vertexwise / positive and negative factor of schizotypy | Whole cortex/thalamus | SPQ | FDR | Thickness/thalamus volume | + | R dorsolateral prefrontal cortex, R dorsal premotor |
| Ettinger et al., 2012 (68) | 55 | 27 | VBM | Whole brain | RISC | FWE cluster | Volume | − | Medial prefrontal, orbitofrontal, temporal |
| Modinos et al., 2010 (69) | 38 | 20 | VBM / dividing subjects with high/low schizotypy | Whole brain | CAPE | FDR | Volume | + | Medial posterior cingulate, precuneus |
| Moorhead et al., 2009 (70) | 98 | 16 | VBM / longitudinal study | Whole brain | SIS | Unspecified multiple comparison correction | Volume | − | L medial temporal, L amygdala, L parahippocampal gyrus |
| Stanfield et al., 2008 (71) | 143 | 16 | ANCOVA | Whole brain | SIS | None | Folding | + | R prefrontal |
Directionality refers to whether the study reports a positive (+) or negative (−) association between a schizotypy measure and a structural metric.
ANCOVA, analysis of covariance; CAPE, Community Assessment of Psychic Experience; FDR, false discovery rate; FWE, familywise error; GM, gray matter; L, left; PSI-C: Psychiatric and Schizotypal Inventory for Children; R, right; RISC, Rust Inventory of Schizotypal Cognitions; ROI, region of interest; SIS, Structured Interview for Schizotypy; SPQ, Schizotypal Personality Questionnaire; VBM, voxel-based morphometry; WM, white matter.
Microstructural MRI provides information about the composition of tissue within a voxel (7). For example, magnetization transfer (MT) images (8) and “myelin maps” derived from the ratio of conventional T1- and T2-weighted images (9) are sensitive to the proportion of fatty brain tissue represented by each voxel, which, according to histological studies on animal models, is related to myelin content (10, 11, 12, 13). MT maps have been used as markers of myelination in white matter and the cortex (14) in healthy subjects (15) and in demyelinating disorders such as multiple sclerosis (16, 17, 18). Schizophrenia has been associated with reduced MT in the frontal, temporal, and insular cortices (19, 20, 21, 22, 23, 24, 25, 26), and the cortical expression of schizophrenia-related genes was (negatively) correlated with T1- and T2-weighted maps (27).
In this context, we measured schizotypy, using the Schizotypal Personality Questionnaire (SPQ), and MT, using a multiparameter MRI scanning procedure, in a sample of 248 healthy young people (14–25 years of age) (Table S1). We tested 3 key hypotheses in a logical sequence: 1) that intracortical MT was correlated with the SPQ total score (and subscale scores), 2) that the cortical pattern of schizotypy-related magnetization (SRM) was colocated with a cortical map of weighted whole-genome expression, and 3) that the gene transcripts most strongly coupled to SRM were enriched for genes that were transcriptionally dysregulated in histological case-control studies of schizophrenia.
Methods and Materials
Participants
A total of 2135 healthy young people, 14 to 25 years of age, were recruited from schools, colleges, National Health Service primary care services, and direct advertisement in north London and Cambridgeshire, United Kingdom. This primary cohort was stratified into 5 contiguous age-related strata, balanced for gender and ethnicity (28). A secondary cohort of 297 individuals was recruited by randomly subsampling the primary cohort so that ∼60 participants were assigned to each of the same age-related strata, balanced for gender and ethnicity, as in the primary cohort. Participants were excluded if they had a current or past history of clinical treatment for a psychiatric disorder, drug or alcohol dependence, neurological disorder including epilepsy, head injury causing loss of consciousness, or learning disability (see Supplement for details).
Written informed consent was provided by all participants as well as written parental assent for participants less than 16 years of age. The study was approved by the National Research Ethics Service and conducted in accordance with National Health Service research governance standards.
Schizotypy Assessment
The SPQ (29) is a self-report scale, comprising 74 dichotomous items that are grouped on 9 subscales, measuring the complex trait of schizotypy. Participants completed the SPQ on 2 assessments, separated by 6 to 18 months, so that traitlike scores on total and subscale SPQ metrics could be estimated by the number of questionnaire items positively endorsed by each participant on average over time.
MRI Data Acquisition
Structural MRI scans were acquired on 1 of 3 identical 3T MRI systems in London or Cambridge, United Kingdom (Magnetom TIM Trio [Siemens Healthcare, Erlangen, Germany], software version VB17). The multiparametric mapping protocol (8) yielded 3 multiecho fast low-angle shot scans with variable excitation flip angles. By appropriate choice of repetition time and flip angle α, acquisitions were predominantly weighted by T1 (repetition time = 18.7 ms, α = 20°), proton density, or MT (repetition time = 23.7 ms, α = 6°). Other acquisition parameters were 1-mm3 voxel resolution, 176 sagittal slices, and a field of view of 256 × 240 mm. A pilot study demonstrated satisfactory levels of between-site reliability in multiparametric mapping data acquisition (8). MT images (15) and T1 images (30,31) from this sample have been previously reported.
MRI Reconstruction, Cortical Parcellation, and Estimation of SRM
We used a standard automated processing pipeline for skull stripping, tissue classification, surface extraction, and cortical parcellation (FreeSurfer [http://surfer.nmr.mgh.harvard.edu]) applied to longitudinal relaxation rate (R1) maps (R1 = 1/T1). Expert visual quality control ensured accurate segmentation of pial and gray/white matter boundaries. Regional MT values were estimated at each of 68 cortical regions for each subject, resulting in a 248 × 68 regional MT data matrix. The Euler number for the R1 images was calculated as a proxy measure of image quality in the simultaneously acquired MT images (32).
A simple linear model of age-related change in MT was used to estimate two key parameters for each region: baseline MT at 14 years of age and age-related rate of change in the period from 14 to 24 years of age (15). For the principal analyses, effects of age on MT were controlled by regression before estimating the correlation of the age-corrected MT residuals with SPQ total score. The Kolmogorov-Smirnov normality test was used to determine the appropriate correlation estimator (Pearson’s or Spearman’s).
Estimation of Regional Gene Expression
We used the Allen Human Brain Atlas (AHBA), a whole-genome expression atlas of the adult human brain created by the Allen Institute for Brain Sciences using 6 donors 24 to 57 years of age (http://human.brain-map.org) (33). Probe-to-gene and sample-to-region mapping strategies can have a major impact on regional gene expression estimation (34). Here, we used the genome assembly hg19 (http://sourceforge.net/projects/reannotator/) (35) to reannotate the probe sequences into genes (36). When genes were mapped by multiple complementary RNA hybridization probes, the probe showing the highest average expression across samples was selected (37). MRI images of the AHBA donors were parcellated using the Desikan-Killiany atlas, and each cortical tissue sample was assigned to an anatomical structure. Regional expression levels were compiled to form a 68 × 20,647 regional transcription matrix (38) (see Supplement).
SRM and Human Brain Gene Expression
We used partial least squares (PLS) to analyze covariation between SRM and gene expression because it is technically well suited to the high collinearity of the gene expression data (39,40), and because PLS and the related multivariate method of canonical correlation analysis have been extensively developed and used for neuroimaging and transcriptional data analysis (41, 42, 43, 44). Specifically, we used PLS to analyze the relationship between the vector of 68 regional measures of SRM and the 68 × 20,647 matrix of 68 regional messenger RNA measurements for 20,647 genes (44). The first PLS component (PLS1) was defined as the weighted sum of whole-genome expression that was most strongly correlated, or most closely colocated, with the anatomical map of SRM. Permutation testing based on spherical rotations or “spins” of the spatially correlated SRM map was used to test the null hypothesis that PLS1 explained no more covariance between SRM and whole-genome expression than expected by chance (31). Bootstrapping was used to estimate the variability of each gene’s positive or negative weight on PLS1, and we tested the null hypothesis of zero weight for each gene with a false discovery rate (FDR) of 5% (42). The set of genes that were significantly (positively or negatively) weighted on PLS1 was called the SRM gene list or set.
Enrichment Analysis
We assigned a cellular affiliation score to each gene in the SRM gene list according to prior criteria for 4 cell types—neuron, astrocyte, microglia, or oligodendroglia (45)—and for a more fine-grained set of cell types (46) (Table S2). We used a data resampling procedure to test the null hypothesis that SRM genes were randomly assigned to different cell types.
We used 2 lists of genes that were differentially expressed, or transcriptionally dysregulated, in postmortem brain tissue measurements of messenger RNA from case-control studies of schizophrenia: 1) the list of genes reported by Gandal et al. (5) as upregulated (n = 845) or downregulated (n = 1175) in the prefrontal and parietal brain regions in schizophrenia (n = 159); and 2) the list of genes reported by Fromer et al. (47) as upregulated (n = 304) or downregulated (n = 345) in the dorsolateral prefrontal cortex in schizophrenia (n = 258). The 2 gene lists were partially overlapping (Table S3), and differential expression of all genes subsumed by the union of the 2 lists was strongly correlated between studies (ρ = .76, p < 10−129) (Figure S1).
We used repeated random relabeling of genes to test the null hypothesis that the SRM gene list included no more schizophrenia-related genes than expected by chance. We also applied the same resampling procedures to comparable prior data on differential gene expression from case-control studies of inflammatory bowel disease, bipolar disorder (BPD), major depressive disorder, and autism spectrum disorder (ASD) (5).
Data and Code Sharing
Data
Regional MT for 68 cortical regions, schizotypy scores, age, gender, socioeconomic status, scanning site, total brain volume, and Euler values (for N = 248) are available at https://github.com/RafaelRomeroGarcia/Schizotypy_MT_geneExp.
Code
Cortical parcellation of gene expression maps to estimate regional mean gene expression (48) can be found at https://github.com/RafaelRomeroGarcia/geneExpression_Repository. The rotate_parcellation code generates null models that preserve the spatial contiguity of cortical regions for permutation testing (31): https://github.com/frantisekvasa/rotate_parcellation. PLS analysis and bootstrapping to estimate PLS weights (15) can be found at https://github.com/KirstieJane/NSPN_WhitakerVertes_PNAS2016/tree/master/SCRIPTS. To generate Figure S1 from the raw Gandal et al. (5) and Fromer et al. (47) datasets, we used https://github.com/RafaelRomeroGarcia/Schizotypy_MT_geneExp. For mapping regional values to the cortical surface for visualization (BrainsForPublication v0.2.1), see https://doi.org/10.5281/zenodo.1069156.
Results
Sample Characteristics
After quality control checks, complete, evaluable MRI and behavioral data were available for analysis on 248 participants: mean age 19.11 ± 2.93 years; 123 (50%) female participants; 213 (86%) right-handed participants; IQ = 112.0 ± 10.5; and 214 (86%) white Caucasian, 10 Asian, 4 black/African American/Caribbean American, 17 mixed, and 3 other ethnic groups (see Table S1 for details).
Schizotypy and MT
Schizotypal personality scores in this healthy (nonpsychotic) sample followed a positively skewed distribution (mean = 0.23, median = 0.20) that was normalized by square root transformation prior to statistical analysis. There was no significant effect of age (R2 < .001, p = .69) (Figure S2), gender (R2 < .001, p = .77), or age-by-gender interaction (R2 < .001; p = .82) on SPQ total score or subscale scores.
SPQ total score was modestly positively correlated with global MT, estimated as the average MT over all 68 regions (R2 = .02, p = .015) (Figure S3). SPQ total score was significantly correlated with age-corrected regional MT in 4 of 68 regions individually tested (R2 > .04, p < .05, FDR corrected) (Figure 1A, B and Table S4): the left isthmus cingulate, left posterior cingulate, left precuneus, and right isthmus cingulate. These medial posterior cortical regions had high MT signals at 14 years of age (MT14) and relatively slow rates of increase in MT over the period of 14 to 25 years of age (ΔMT) (Figure 1C).
Figure 1.
Schizotypy-related magnetization (SRM): association between intracortical magnetization transfer (MT) and Schizotypal Personality Questionnaire (SPQ) score. (A) Cortical surface maps highlighting areas where SPQ total score was significantly positively correlated with regional MT after controlling for age by regression: pink regions had nominally significant SRM (2-tailed p < .05); red regions had significant SRM controlled for multiple comparisons over 68 cortical regions tested (false discovery rate < .05). (B) Scatterplot of SPQ total score for each participant vs. mean MT in regions of significant SRM (R2246 = .04, p = .002); each dot represents 1 of 248 healthy people 14 to 25 years of age. (C) Scatterplots of SRM vs. MT at 14 years of age (MT14) (left) (R267 = .34, permutation testing based on spherical rotations: p = .002) and SRM vs. change in magnetization 14 to 25 years of age (ΔMT) (right) (R267 = .28, permutation testing based on spherical rotations: p = .006). Each point represents a cortical region and colored points represent regions with significant SRM (pink: p < .05; red: false discovery rate < .05). (D) Word cloud representing ontological terms most frequently associated with functional activation of the medial posterior cortical areas of significant SRM. (E) Cortical surface maps highlighting areas where scores on the disorganized factor of schizotypy was significantly positively correlated with regional MT after controlling for age by regression (pink, 2-tailed p < .05; red, false discovery rate < .05).
We used prior functional MRI data to identify experimental task conditions that were most robustly associated with functional activation of these areas of significant SRM (Neurosynth [https://neurosynth.org/]) (49): memory, social cognition or theory of mind, and executive functions (Figure 1D). The posterior cingulate and medial parietal cortical areas of significant SRM were also enriched for default mode network–related terms in ontological analysis of prior functional MRI data (49).
Sensitivity Analyses of SRM
We used a linear model to control the association between SPQ and MT for the potentially confounding effects of age, gender, site, socioeconomic status, and total brain volume (Figure S4), and robust estimators to mitigate the influence of the small number of high SPQ scores on the estimation of SRM (Figure S5). In both cases, the key results were conserved: namely, significant SRM in default mode network areas and significant correlations between PLS1 weights and differential gene expression in schizophrenia. We also noted a negative correlation between Euler number and global MT, indicating reduced MT in a minority of poor-quality images (ρ = −.14, p = .03). When we excluded the 10% of participants with the poorest image quality, the correlation between Euler number and MT was no longer significant (ρ = .04, p = .11), but the key results were conserved (Figure S6).
Schizotypy is a complex trait comprising multiple dimensions of cognition, emotion, and behavior. In addition to the principal analysis of SPQ total score, we also considered 2 possible decompositions of the schizotypal trait. Nine subscales of the SPQ defined by Raine (29) were positively correlated with regional MT, but these associations were less robust than for SPQ total score (Figure S7). All 3 factors of schizotypy defined by Raine et al. (50), i.e., positive, negative, and disorganized dimensions, were positively correlated with MT. The correlation between disorganized schizotypy and MT was strongest and statistically significant when controlling for multiple comparisons (Figure 1E and Figure S8).
We also measured cortical thickness for each of the same 68 regions, using R1 images collected as part of the same MRI sequence used to measure MT. SPQ scores were negatively correlated with cortical thickness in some regions, but the associations between schizotypy and cortical thickness were not significant when corrected for multiple tests (Figure S9).
SRM and Gene Expression
PLS1 defined a weighted sum of whole-genome expression that accounted for ∼40% of the cortical patterning of SRM, significantly more than expected by chance (permutation testing based on spherical rotations: p = .027) (Figure 2A).
Figure 2.
Gene expression and schizotypy-related magnetization (SRM). (A) (Left panel) The first partial least squares component (PLS1) defined a linear combination of genes that had a similar cortical pattern of expression to the cortical map of SRM, representing the correlation between Schizotypal Personality Questionnaire and magnetization transfer at each of 68 cortical regions. (Center panel) Scatterplot of PLS1 scores versus SRM; each point is a cortical region. (Right panel) The combination of genes defined by PLS1 explains more of the variance in SRM (dotted line) than expected by chance (histogram of permutation distribution). (B) Illustrative example of the weights assigned to representative genes on PLS1. Genes with the highest positive weights are colored in pink, nonsignificantly weighted genes are shown in white, and the genes with the lowest negative weights are colored in blue. Tables summarize p values by permutation testing for enrichment analysis by 4 lists of genes affiliated to specific cell types and 4 lists of genes associated with schizophrenia: Gandal and Fromer up-reg (5, 47) are lists of genes transcriptionally upregulated postmortem in schizophrenia; Gandal and Fromer down-reg are lists of genes transcriptionally downregulated in schizophrenia. Scatterplots and cortical maps illustrate that positively weighted genes, like ANK1, are overexpressed in cortical regions with high levels of schizotypy-related myelination, whereas negatively weighted genes, like PTPRC, are underexpressed in regions with high levels of SRM. FDR, false discovery rate.
Multiple univariate Z tests were used to test the set of null hypotheses that the weight of each gene on PLS1 was zero. We found that this null hypothesis was refuted for 1932 positively weighted genes and 2153 negatively weighted genes (p < .05, FDR corrected for whole-genome testing at 20,647 genes) (Figure 2B). Positively weighted genes were normally overexpressed, and negatively weighted genes were normally underexpressed, in cortical areas with high SRM. These 4085 genes constituted the SRM gene list.
Functional and Schizophrenia-Related Enrichment of SRM Genes
The SRM gene list was tested for enrichment by genes characteristic of specific cell types using 2 sets of prior criteria (45,46). Positively weighted SRM genes were enriched for neuronal affiliation (permutation test, p < 10−4) (45) and, more specifically, for genes differentially expressed in fast-spiking parvalbumin (PVALB)-positive inhibitory interneurons (permutation test, FDR-corrected p < .01) (Table S2) (46). Negatively weighted SRM genes were enriched for astrocytes, microglia, and neuronal affiliation (permutation tests, all ps < 10−4) (Figure 2B) (45,46).
The positive or negative weighting of each SRM gene was strongly related to its differential expression in 2 postmortem studies of schizophrenia (5,47). Positively weighted SRM genes were enriched for genes that were significantly downregulated in both studies (Table S5) but not for significantly upregulated genes in either study. Additionally, positively weighted SRM genes were also enriched for genes previously associated with white matter connectivity differences in schizophrenia described by Romme et al. (51) (Figure S10). In contrast, negatively weighted SRM genes were enriched for genes that were significantly upregulated in both studies (permutation tests, all ps < 10−4) (Figure 2B and Table S6) but not for significantly downregulated genes.
Convergently, there was a significant negative correlation (Spearman’s ρ = −.16, p < 10−6) between the PLS1 weights of all genes in the genome and the differential expression values reported for all genes by Gandal et al. (5) and Fromer et al. (47) (Figure 3A). PLS1 gene weights were not correlated with differential expression in inflammatory bowel disease or major depressive disorder; however, they were negatively correlated with differential expression in BPD and ASD (Figure 3B).
Figure 3.
Weights of gene expression from partial least squares (PLS) analysis of schizotypy-related magnetization (SRM) were related to histological measures of differential gene expression from case-control studies of schizophrenia and other disorders. (A) The weight of each gene on the first PLS component was significantly negatively correlated with differential gene expression postmortem in schizophrenia according to prior data reported by Gandal et al. (5) (Spearman’s rank correlation, ρ11111 = −.16, Bonferroni-corrected adjusted p11111 < 10−65) and by Fromer et al. (47) [Spearman’s rank correlation, ρ586 = −.30, adjusted p586 < 10−12; for this dataset, only significantly different expression values have been reported (46)]. (B) Correlations between PLS weights and differential expression were also evaluated for other conditions (5): inflammatory bowel disease (ρ586 = −.02, adjusted p586 = .10), major depressive disorder (ρ15281 = .007, adjusted p15281 = .37), bipolar disorder (ρ16064 = −.09, adjusted p16064 < 10−19 ), and autism spectrum disorder (ρ11131 = .11, adjusted p11131 < 10−35). Red and blue points represent genes that are significantly up- and downregulated in postmortem data.
We analyzed the network of known protein-protein interactions [STRING (http://string-db.org) (52)] between proteins coded by the 213 genes that were significantly downregulated in schizophrenia (5) and significantly positively weighted in the PLS analysis of SRM (Table S5). There were significantly more interactions (edges) between proteins coded by these genes than expected by chance (permutation test, p < 10−5) (Figure 4 and Figure S11). Topologically, the network comprised several clusters of densely interconnected and functionally specialized proteins. The biggest cluster was enriched for synaptic terms and centered around highly connected “hub” proteins (Figure 4).
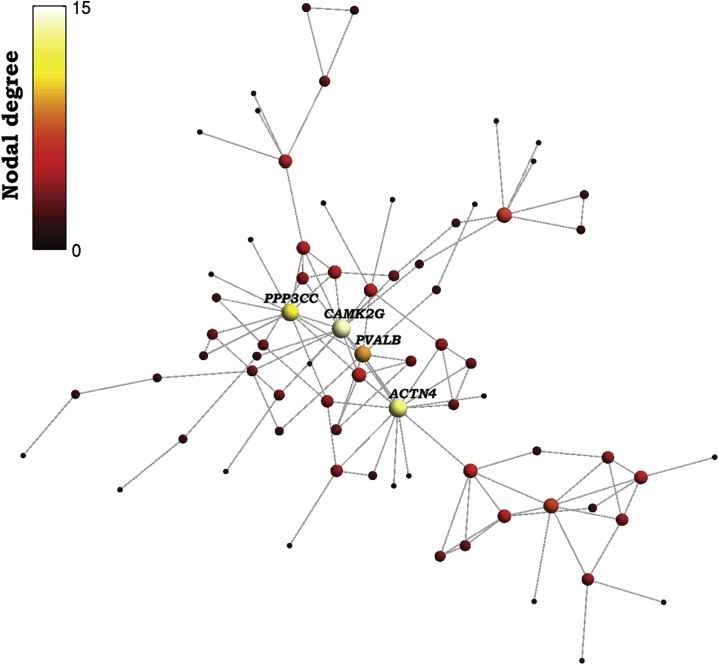
Figure 4.
Protein-protein interaction network for a set of 213 proteins coded by genes associated with both schizotypy-related magnetization and postmortem brain transcriptional dysregulation in schizophrenia. Nodes represent genes that were both 1) downregulated in brain tissue from 159 patients with schizophrenia and 2) positively weighted on the partial least squares component most strongly associated with schizotypy-related myelination in 248 healthy adolescents. Edges represent known protein-protein interactions. The color and size of each node represents its degree centrality or “hubness,” simply the number of interactions that protein has with the other proteins in the network. The top 4 most highly connected hubs are highlighted: PPP3CC is a calmodulin dependent phosphatase, calcineurin; CAMK2G is a calcium/calmodulin dependent kinase; PVALB is a calcium binding protein, parvalbumin; ACTN4 is a microfilamentous protein, actinin-alpha-4. This network is specialized for calcium-dependent processes that have been previously associated with interneurons and with pathogenesis of schizophrenia. For the complete list of gene names on the protein-protein interaction network, see Figure S11 and see https://version-10-5.string-db.org/cgi/network.pl?taskId=RMpA04wbWG8k for a full interactive version of the protein-protein interaction network.
Discussion
Schizotypy was associated with intracortical magnetization of posterior cortical regions of the default mode network and colocated with a normative cortical pattern of weighted whole-genome expression. The gene transcripts most strongly weighted in association with this MRI map of SRM were significantly enriched for genes specifically expressed by neuronal and glial cells, especially PVALB-positive interneurons, and for genes that were transcriptionally dysregulated in 2 prior postmortem studies of schizophrenia.
Magnetization, Intracortical Myelination, and Schizotypy
MT is a microstructural MRI measurement that is sensitive to the ratio of fatty and watery tissue represented by each voxel, and in the brain, most of the fat is myelin. Intracortical myelination, especially of the deeper layers of the cortex, has been recognized since seminal cytoarchitectonic and myeloarchitectonic studies in the early 20th century (14). MT images represent a stark contrast between the cortex and the central white matter as well as more nuanced variations across different cortical areas and layers (53). Histological measurements of myelin were positively correlated with MRI measurements of MT in postmortem brains (54). Intracortical measurements of MT in humans have been validated as microstructural MRI markers of myelination in healthy volunteers (55) and in patients with multiple sclerosis (56).
One plausible interpretation of SRM, therefore, is as a proxy marker for a biological state of schizotypy-related myelination. On this assumption, the results are open to further interpretation at a cellular level. For example, greater “myelination” could imply a greater density of myelinated neurons per voxel (a neuronal process), or a greater density of myelin per neuron (an oligodendroglial process), or some combination of these and other cellular parameters. The data available to us did not allow direct resolution of the relationships between SRM and myelination. Instead, we used open data on human brain gene expression (n = 6, mean age = 42.5 years) to explore these questions more indirectly.
The adult brain gene expression profile that was most closely colocated with the adolescent brain map of SRM (n = 248, mean age = 19 years) was enriched for neuronal, but not oligodendroglial, affiliations. This pattern of results arguably favors the interpretation that higher MT indicates a greater density of myelinated neurons per voxel, rather than a greater density of myelin per neuron, in people with higher schizotypy scores. However, the 20+-year age gap between the MRI measurements and the messenger RNA measurements precludes definitive resolution of these and other possible cellular interpretations of SRM. Although the SRM gene set is not known to demonstrate major developmental changes in expression after childhood (Figure S12), in the future it will be important to colocate MT phenotypes in children and young people (and animal models) with more precisely age-matched data on brain gene expression and histology.
The macroscopic medial posterior cortical areas where MT was most strongly correlated with schizotypy in general—SPQ total score—are key components of the default mode network [as defined by functional MRI studies (57)] and specialized for memory, social cognitive, and theory-of-mind functions that are known to be abnormal in patients with schizophrenia (58). Interestingly, the schizotypal factor of disorganization was also correlated with MT in the medial prefrontal cortical areas that also form part of the default mode network. All these regions had high levels of magnetization at 14 years of age and no significant subsequent change in magnetization over the period 14 to 25 years of age. This contrasts with areas of the lateral association cortex, which have a low level of MT at 14 years of age but show significant increase in MT over the course of adolescence (15). We can infer that these medial posterior cortical areas matured as part of a preadolescent wave of cortical development (59), which would be compatible with the stable, traitlike properties of schizotypal personality in these data and in other studies of adolescents and adults.
Cortical Gene Expression, Schizotypy-Related Myelination, and Schizophrenia
We wanted to identify which genes in the whole genome had a cortical expression pattern that was most similar to the cortical map of SRM. A large number (>20,000) of nonindependent statistical tests would be entailed in testing the association between each transcript’s spatially correlated cortical expression map and the cortical map of SRM. Therefore, we favored a multivariate approach and used PLS to identify a cortical pattern of weighted whole-genome expression that was significantly colocated with the SRM map and to identify which particular gene transcripts were most positively or negatively weighted.
We found that the positively weighted genes (n = 1932) were overexpressed in cortical areas with high levels of SRM, whereas the negatively weighted genes (n = 2153) were overexpressed in cortical areas with low levels of SRM. Both positive and negative genes were enriched for neuronal, but not for oligodendroglial, affiliations. Positive genes were specifically enriched for PVALB-positive inhibitory interneurons; negative genes were enriched for astrocytes and microglia.
We predicted hypothetically that the SRM gene set would be enriched for genes that are known to be transcriptionally dysregulated in schizophrenia. This prediction was supported by convergent results from enrichment analysis using two prior, independently discovered, and partially overlapping lists of genes differentially expressed in postmortem case-control studies of schizophrenia. In both cases, there were significantly more histologically downregulated genes in the list of positively weighted SRM genes, and significantly more upregulated genes in the list of negatively weighted SRM genes, than expected by chance. In other words, genes with reduced brain transcription postmortem in schizophrenia were normally more highly expressed in cortical areas with higher levels of SRM. A subset of the positively weighted SRM genes has been previously associated with white matter dysconnectivity in schizophrenia (51), suggesting that SRM of cortex and schizophrenia-related disruption of central white matter tracts may be different imaging phenotypes related to expression of genes in common.
The subset of 213 SRM-positive genes that were also significantly downregulated in one or both of the prior histological studies coded for a protein-protein interaction network comprising a small number of highly connected hub proteins (ACTN4 [alpha-actinin-4], CAMK2G [calcium/calmodulin-dependent protein kinase II gamma], PPP3CC [protein phosphatase 3 catalytic subunit gamma], and PVALB), each hub having up to 14 known biochemical interactions with other proteins in the network. CAMK2G is one of a family of serine/threonine kinases that mediate many of the second messenger effects of Ca2+ that are crucial for plasticity at glutamatergic synapses. PVALB is a calcium-binding albumin protein that is expressed particularly by the fast-spiking class of GABAergic (gamma-aminobutyric acidergic) interneurons that has been strongly implicated in the pathogenesis of schizophrenia (Figure 4) (60).
We found that the SRM gene list was also enriched by genes differentially expressed in ASD and BPD. These results are consistent with the postmortem evidence (5) that the differential gene expression profile of schizophrenia (compared with healthy controls) is strongly correlated with the case-control differences of transcription in BPD and ASD (ρ > .45, p < .001). They are also consistent with clinical evidence that both ASD and BPD are associated with increased schizotypal traits (61,62).
Methodological Issues
The brain tissue samples used for RNA sequencing in the AHBA were not homogeneously distributed across the cortex, so estimates of regional expression are based on different numbers of experimental measurements in each of the 68 regions. The case-control differences in frontal or parietal lobar gene transcription reported by Gandal et al. (5) and Fromer et al. (47), although based on a relatively large number of patients, are not as precisely localized or representative of the whole brain as the AHBA and MRI data. This study has a considerably larger sample size than any previously reported MRI study of schizotypy, and it is the first to evaluate a microstructural MRI marker, which was more strongly related to schizotypy than the more conventional macrostructural MRI marker of cortical thickness. Nonetheless, it is theoretically surprising that there was limited evidence for significant SRM of the frontal and lateral temporal cortex (although magnetization of the medial prefrontal cortex was significantly associated with the disorganized component of schizotypy) (Figure 1E), possibly reflecting limited statistical power. The SPQ is a self-report questionnaire measure of schizotypy; more refined and objective assessments of schizotypal traits would likely add value to future studies.
Conclusions
Overall, these correlational results do not unambiguously resolve questions of causality, but they are consistent with the interpretation that SRM, putatively an imaging marker of intracortical density of myelinated neurons, represents cellular processes determined in part by transcription of genes related to schizophrenia and other neuropsychiatric disorders.
Acknowledgments and Disclosures
This work was supported by a strategic award from the Wellcome Trust to the University of Cambridge and University College London: the Neuroscience in Psychiatry Network (NSPN), the National Institute for Health Research Cambridge Biomedical Research Centre, the NSPN (to RR-G), the Guarantors of Brain (to RR-G), MQ fellowship Grant No. MQF17_24 (to PEV), a fellowship of the Alan Turing Institute funded under the Engineering and Physical Sciences Research Council Grant No. EP/N510129/1 (to PEV); the National Institutes of Health Oxford-Cambridge Scholars’ Program (to JS). ETB is a National Institute for Health Research Senior Investigator.
We thank the Allen Brain Institute for access to human brain gene expression data, as well as all the members of the NSPN consortium for data collection, storage, and preprocessing: Edward Bullmore (Chief Investigator), Raymond Dolan, Ian Goodyer, Peter Fonagy, Peter Jones, Matilde Vaghi, Michael Moutoussis, Tobias Hauser, Sharon Neufeld, Michelle St Clair, Petra Vértes, Kirstie Whitaker, Rafael Romero-Garcia, Becky Inkster, Gita Prabhu, Cinly Ooi, Umar Toseeb, Barry Widmer, Junaid Bhatti, Laura Villis, Ayesha Alrumaithi, Sarah Birt, Aislinn Bowler, Kalia Cleridou, Hina Dadabhoy, Emma Davies, Ashlyn Firkins, Sian Granville, Elizabeth Harding, Alexandra Hopkins, Daniel Isaacs, Janchai King, Danae Kokorikou, Christina Maurice, Cleo McIntosh, Jessica Memarzia, Harriet Mills, Ciara O’Donnell, Sara Pantaleone, Jenny Scott, Pasco Fearon, Anne-Laura van Harmelen, and Rogier Kievit.
This article was published as a preprint on bioRxiv: doi: https://www.biorxiv.org/content/10.1101/487108v1.
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
NSPN Consortium member list: Principal investigators: Edward Bullmore (Chief Investigator from January 1, 2017), Raymond Dolan, Ian Goodyer (Chief Investigator until January 1, 2017), Peter Fonagy, Peter Jones; NSPN (funded) staff: Matilde Vaghi, Michael Moutoussis, Tobias Hauser, Sharon Neufeld, Rafael Romero-Garcia, Michelle St Clair, Kirstie Whitaker, Becky Inkster, Gita Prabhu, Cinly Ooi, Umar Toseeb, Barry Widmer, Junaid Bhatti, Laura Villis, Ayesha Alrumaithi, Sarah Birt, Aislinn Bowler, Kalia Cleridou, Hina Dadabhoy, Emma Davies, Ashlyn Firkins, Sian Granville, Elizabeth Harding, Alexandra Hopkins, Daniel Isaacs, Janchai King, Danae Kokorikou, Christina Maurice, Cleo McIntosh, Jessica Memarzia, Harriet Mills, Ciara O’Donnell, Sara Pantaleone, Jenny Scott; Affiliated scientists: Pasco Fearon, John Suckling, Anne-Laura van Harmelen, Rogier Kievit, Petra Vértes; full member list available in the Supplement.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2019.12.005.
Contributor Information
Rafael Romero-Garcia, Email: rr480@cam.ac.uk.
NSPN Consortium:
Edward Bullmore, Raymond Dolan, Ian Goodyer, Peter Fonagy, Peter Jones, Matilde Vaghi, Michael Moutoussis, Tobias Hauser, Sharon Neufeld, Rafael Romero-Garcia, Michelle St Clair, Kirstie Whitaker, Becky Inkster, Gita Prabhu, Cinly Ooi, Umar Toseeb, Barry Widmer, Junaid Bhatti, Laura Villis, Ayesha Alrumaithi, Sarah Birt, Aislinn Bowler, Kalia Cleridou, Hina Dadabhoy, Emma Davies, Ashlyn Firkins, Sian Granville, Elizabeth Harding, Alexandra Hopkins, Daniel Isaacs, Janchai King, Danae Kokorikou, Christina Maurice, Cleo McIntosh, Jessica Memarzia, Harriet Mills, Ciara O’Donnell, Sara Pantaleone, Jenny Scott, Pasco Fearon, John Suckling, Anne-Laura van Harmelen, Rogier Kievit, and Petra Vértes
Supplementary Material
References
- 1.Walter E.E., Fernandez F., Snelling M., Barkus E. Genetic consideration of schizotypal traits: A review. Front Psychol. 2016;7:1769. doi: 10.3389/fpsyg.2016.01769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calkins M.E., Curtis C.E., Grove W.M., Iacono W.G. Multiple dimensions of schizotypy in first degree biological relatives of schizophrenia patients. Schizophr Bull. 2004;30:317–325. doi: 10.1093/oxfordjournals.schbul.a007081. [DOI] [PubMed] [Google Scholar]
- 3.Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Schizophrenia Consortium. Purcell S.S.M., Wray N.N.R., Stone J.L.J., Visscher P.M.P., O’Donovan M.M.C. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandal M.J., Haney J.R., Parikshak N.N., Leppa V., Ramaswami G., Hartl C. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlich S., Brauns S., Yendiki A., Ho B.-C., Calhoun V., Schulz S.C. Associations of cortical thickness and cognition in patients with schizophrenia and healthy controls. Schizophr Bull. 2012;38:1050–1062. doi: 10.1093/schbul/sbr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasser M.F., Goyal M.S., Preuss T.M., Raichle M.E., Van Essen D.C. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. Neuroimage. 2014;93:165–175. doi: 10.1016/j.neuroimage.2013.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiskopf N., Suckling J., Williams G., Correia M.M., Inkster B., Tait R. Quantitative multi-parameter mapping of R1, PD(*), MT, and R2(*) at 3T: A multi-center validation. Front Neurosci. 2013;7:95. doi: 10.3389/fnins.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shafee R., Buckner R.L., Fischl B. Gray matter myelination of 1555 human brains using partial volume corrected MRI images. Neuroimage. 2015;105:473–485. doi: 10.1016/j.neuroimage.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janve V.A., Zu Z., Yao S.Y., Li K., Zhang F.L., Wilson K.J. The radial diffusivity and magnetization transfer pool size ratio are sensitive markers for demyelination in a rat model of type III multiple sclerosis (MS) lesions. Neuroimage. 2013;74:298–305. doi: 10.1016/j.neuroimage.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehto L.J., Sierra A., Gröhn O. Magnetization transfer SWIFT MRI consistently detects histologically verified myelin loss in the thalamocortical pathway after a traumatic brain injury in rat. NMR Biomed. 2017;30:3678. doi: 10.1002/nbm.3678. [DOI] [PubMed] [Google Scholar]
- 12.Turati L., Moscatelli M., Mastropietro A., Dowell N.G., Zucca I., Erbetta A. In vivo quantitative magnetization transfer imaging correlates with histology during de- and remyelination in cuprizone-treated mice. NMR Biomed. 2015;28:327–337. doi: 10.1002/nbm.3253. [DOI] [PubMed] [Google Scholar]
- 13.Zaaraoui W., Deloire M., Merle M., Girard C., Raffard G., Biran M. Monitoring demyelination and remyelination by magnetization transfer imaging in the mouse brain at 9.4 T. MAGMA. 2008;21:357–362. doi: 10.1007/s10334-008-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt O. Die myeloarchitektonische Felderung des menschlichen Stirnhirns. J Psychol Neurol. 1910;15:221–232. [Google Scholar]
- 15.Whitaker K.J., Vértes P.E., Romero-Garcia R., Váša F., Moutoussis M., Prabhu G. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci. 2016;113:9105–9110. doi: 10.1073/pnas.1601745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samson R.S., Cardoso M.J., Muhlert N., Sethi V., Wheeler-Kingshott C.A.M., Ron M. Investigation of outer cortical magnetisation transfer ratio abnormalities in multiple sclerosis clinical subgroups. Mult Scler. 2014;20:1322–1330. doi: 10.1177/1352458514522537. [DOI] [PubMed] [Google Scholar]
- 17.Pardini M., Sudre C.H., Prados F., Yaldizli Ö., Sethi V., Muhlert N. Relationship of grey and white matter abnormalities with distance from the surface of the brain in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2016;87:1212–1217. doi: 10.1136/jnnp-2016-313979. [DOI] [PubMed] [Google Scholar]
- 18.Derakhshan M., Caramanos Z., Narayanan S., Arnold D.L., Louis Collins D. Surface-based analysis reveals regions of reduced cortical magnetization transfer ratio in patients with multiple sclerosis: A proposed method for imaging subpial demyelination. Hum Brain Mapp. 2014;35:3402–3413. doi: 10.1002/hbm.22410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foong J., Symms M.R., Barker G.J., Maier M., Woermann F.G., Miller D.H., Ron M. Neuropathological abnormalities in schizophrenia: Evidence from magnetization transfer imaging. Brain. 2001;124:882–892. doi: 10.1093/brain/124.5.882. [DOI] [PubMed] [Google Scholar]
- 20.Bagary M.S., Symms M.R., Barker G.J., Mutsatsa S.H., Joyce E.M., Ron M.A. Gray and white matter brain abnormalities in first-episode schizophrenia inferred from magnetization transfer imaging. Arch Gen Psychiatry. 2003;60:779–788. doi: 10.1001/archpsyc.60.8.779. [DOI] [PubMed] [Google Scholar]
- 21.Du F., Cooper A.J., Thida T., Shinn A.K., Cohen B.M., Öngür D. Myelin and axon abnormalities in schizophrenia measured with magnetic resonance imaging techniques. Biol Psychiatry. 2013;74:451–457. doi: 10.1016/j.biopsych.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubicki M., Park H., Westin C.F., Nestor P.G., Mulkern R.V., Maier S.E. DTI and MTR abnormalities in schizophrenia: Analysis of white matter integrity. Neuroimage. 2005;26:1109–1118. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicki M., Westin C.F., Frumin M., Ersner-Hershfield H., Jolesz F.A., McCarley R.W., Shenton M.E. DTI and MTR abnormalities in schizophrenia — Voxel wise analysis of white matter integrity. Schizophr Res. 2003;60:199. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaniyappan L., Al-Radaideh A., Mougin O., Gowland P., Liddle P.F. Combined white matter imaging suggests myelination defects in visual processing regions in schizophrenia. Neuropsychopharmacology. 2013;38:1808–1815. doi: 10.1038/npp.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antosik-Biernacka A., Peuskens H., De Hert M., Peuskens J., Sunaert S., Van Hecke P., Goraj B. Magnetization transfer imaging in chronic schizophrenia. Med Sci Monit. 2006;12:MT17–MT21. [PubMed] [Google Scholar]
- 26.Wei Y., Collin G., Mandl R.C.W., Cahn W., Keunen K., Schmidt R. Cortical magnetization transfer abnormalities and connectome dysconnectivity in schizophrenia. Schizophr Res. 2018;192:172–178. doi: 10.1016/j.schres.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Burt J.B., Demirtaş M., Eckner W.J., Navejar N.M., Ji J.L., Martin W.J. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat Neurosci. 2018;21:1251–1259. doi: 10.1038/s41593-018-0195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiddle B., Inkster B., Prabhu G., Moutoussis M., Whitaker K.J., Bullmore E.T. Cohort profile: The NSPN 2400 Cohort: A developmental sample supporting the Wellcome Trust Neuro Science in Psychiatry Network. Int J Epidemiol. 2018;47:18–19. doi: 10.1093/ije/dyx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raine A. The SPQ: A scale for the assessment of schizotypal personality based on DSM-III-R criteria. Schizophr Bull. 1991;17:555–564. doi: 10.1093/schbul/17.4.555. [DOI] [PubMed] [Google Scholar]
- 30.Seidlitz J., Váša F., Shinn M., Romero-Garcia R., Whitaker K.J., Vértes P.E. Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron. 2018;97:231–247.e7. doi: 10.1016/j.neuron.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Váša F., Seidlitz J., Romero-Garcia R., Whitaker K.J., Rosenthal G., Vértes P.E. Adolescent tuning of association cortex in human structural brain networks. Cereb Cortex. 2017;28:281–294. doi: 10.1093/cercor/bhx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen A.F.G., Roalf D.R., Ruparel K., Blake J., Seelaus K., Villa L.P. Quantitative assessment of structural image quality. Neuroimage. 2018;169:407–418. doi: 10.1016/j.neuroimage.2017.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnatkevic̆iūtė A., Fulcher B.D., Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage. 2019;189:353–367. doi: 10.1016/j.neuroimage.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Arloth J., Bader D.M., Röh S., Altmann A. Re-Annotator: Annotation pipeline for microarray probe sequences. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richiardi J., Altmann A., Jonas R., Milazzo A.-C., Chang C., Chakravarty M.M. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J.A., Cai C., Langfelder P., Geschwind D.H., Kurian S.M., Salomon D.R., Horvath S. Strategies for aggregating gene expression data: The collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero-Garcia R., Whitaker K.J., Váša F., Seidlitz J., Shinn M., Fonagy P. Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage. 2018;171:256–267. doi: 10.1016/j.neuroimage.2017.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farahani H.A., Rahiminezhad A., Same L., Immannezhad K. A comparison of partial least squares (PLS) and ordinary least squares (OLS) regressions in predicting of couples mental health based on their communicational patterns. Procedia Soc Behav Sci. 2010;5:1459–1463. [Google Scholar]
- 40.Rosipal R., Krämer N. Overview and recent advances in partial least squares. In: Saunders C., Grobelnik M., Gunn S., Shawe-Taylor J., editors. LNCS 3940: Subspace, Latent Structure and Feature Selection. Springer; Berlin, Germany: 2006. pp. 34–51. [Google Scholar]
- 41.McColgan P., Gregory S., Seunarine K.K., Razi A., Papoutsi M., Johnson E. Brain regions showing white matter loss in Huntington’s disease are enriched for synaptic and metabolic genes. Biol Psychiatry. 2018;83:456–465. doi: 10.1016/j.biopsych.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero-Garcia R., Warrier V., Bullmore E.T., Baron-Cohen S., Bethlehem R.A.I. Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry. 2018;24:1053–1064. doi: 10.1038/s41380-018-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan S.E., Seidlitz J., Whitaker K.J., Romero-Garcia R., Clifton N.E., Scarpazza C. Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc Natl Acad Sci U S A. 2019;116:9604–9609. doi: 10.1073/pnas.1820754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vértes P.E., Rittman T., Whitaker K.J., Romero-Garcia R., Váša F., Kitzbichler M.G. Gene transcription profiles associated with inter-modular hubs and connection distance in human functional magnetic resonance imaging networks. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150362. doi: 10.1098/rstb.2015.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley K.W., Nakao-Inoue H., Molofsky A.V., Oldham M.C. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat Neurosci. 2018;21:1171–1184. doi: 10.1038/s41593-018-0216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toker L., Mancarci B.O., Tripathy S., Pavlidis P. Transcriptomic evidence for alterations in astrocytes and parvalbumin interneurons in subjects with bipolar disorder and schizophrenia. Biol Psychiatry. 2018;84:787–796. doi: 10.1016/j.biopsych.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero-Garcia R, Whitaker KJ, Vasa F, Seidlitz J, Shinn M, Fonagy P, et al. (2017): Data supporting NSPN publication: “Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex.” Neuroimage [Dataset]. Available at: 10.17863/CAM.21082. Accessed April 6, 2018. [DOI] [PMC free article] [PubMed]
- 49.Yarkoni T, Poldrack R, Nichols T, Van Essen D, Wager T (2011): NeuroSynth: A new platform for large-scale automated synthesis of human functional neuroimaging data. Presented at the 4th INCF Congress of Neuroinformatics, September 4–6, Boston, Massachusetts.
- 50.Raine A., Reynolds C., Lencz T., Scerbo A., Triphon N., Kim D. Cognitive-perceptual, interpersonal, and disorganized features of schizotypal personality. Schizophr Bull. 1994;20:191–201. doi: 10.1093/schbul/20.1.191. [DOI] [PubMed] [Google Scholar]
- 51.Romme I.A.C., de Reus M.A., Ophoff R.A., Kahn R.S., van den Heuvel M.P. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry. 2016;81:495–502. doi: 10.1016/j.biopsych.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lerch J.P., Van Der Kouwe A.J.W., Raznahan A., Paus T., Johansen-Berg H., Miller K.L. Studying neuroanatomy using MRI. Nat Neurosci. 2017;20:314–326. doi: 10.1038/nn.4501. [DOI] [PubMed] [Google Scholar]
- 54.Stüber C., Morawski M., Schäfer A., Labadie C., Wähnert M., Leuze C. Myelin and iron concentration in the human brain: A quantitative study of MRI contrast. Neuroimage. 2014;93:95–106. doi: 10.1016/j.neuroimage.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 55.Carey D., Caprini F., Allen M., Lutti A., Weiskopf N., Rees G. Quantitative MRI provides markers of intra-, inter-regional, and age-related differences in young adult cortical microstructure. Neuroimage. 2018;182:429–440. doi: 10.1016/j.neuroimage.2017.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmierer K., Scaravilli F., Altmann D.R., Barker G.J., Miller D.H. Magnetization transfer ratio and myelin in postmortem multiple sclerosis brain. Ann Neurol. 2004;56:407–415. doi: 10.1002/ana.20202. [DOI] [PubMed] [Google Scholar]
- 57.Fransson P., Marrelec G. The precuneus/posterior cingulate cortex plays a pivotal role in the default mode network: Evidence from a partial correlation network analysis. Neuroimage. 2008;42:1178–1184. doi: 10.1016/j.neuroimage.2008.05.059. [DOI] [PubMed] [Google Scholar]
- 58.Lener M.S., Wong E., Tang C.Y., Byne W., Goldstein K.E., Blair N.J. White matter abnormalities in schizophrenia and schizotypal personality disorder. Schizophr Bull. 2015;41:300–310. doi: 10.1093/schbul/sbu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grydeland H., Vértes P.E., Váša F., Romero-Garcia R., Whitaker K., Alexander-Bloch A.F. Waves of maturation and senescence in micro-structural MRI markers of human cortical myelination over the lifespan. Cereb Cortex. 2019;29:1369–1381. doi: 10.1093/cercor/bhy330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashimoto T., Volk D.W., Eggan S.M., Mirnics K., Pierri J.N., Sun Z. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barneveld P.S., Pieterse J., de Sonneville L., van Rijn S., Lahuis B., van Engeland H., Swaab H. Overlap of autistic and schizotypal traits in adolescents with Autism Spectrum Disorders. Schizophr Res. 2011;126:231–236. doi: 10.1016/j.schres.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Schürhoff F., Laguerre A., Szöke A., Méary A., Leboyer M. Schizotypal dimensions: Continuity between schizophrenia and bipolar disorders. Schizophr Res. 2005;80:235–242. doi: 10.1016/j.schres.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Evans D.W., Michael A.M., Ularevic M., Lusk L.G., Buirkle J.M., Moore G.J. Neural substrates of a schizotypal spectrum in typically developing children: Further evidence of a normal-pathological continuum. Behav Brain Res. 2016;315:141–146. doi: 10.1016/j.bbr.2016.08.034. [DOI] [PubMed] [Google Scholar]
- 64.Nenadic I., Lorenz C., Langbein K., Dietzek M., Smesny S., Schönfeld N. Brain structural correlates of schizotypy and psychosis proneness in a non-clinical healthy volunteer sample. Schizophr Res. 2015;168:37–43. doi: 10.1016/j.schres.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Yan C., Yin D.Z., Fan M.X., Cheung E.F.C., Pantelis C., Chan R.C.K. Neurobiological changes of schizotypy: Evidence from both volume-based morphometric analysis and resting-state functional connectivity. Schizophr Bull. 2015;41:444–454. doi: 10.1093/schbul/sbu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeRosse P., Nitzburg G.C., Ikuta T., Peters B.D., Malhotra A.K., Szeszko P.R. Evidence from structural and diffusion tensor imaging for frontotemporal deficits in psychometric schizotypy. Schizophr Bull. 2015;41:104–114. doi: 10.1093/schbul/sbu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kühn S., Schubert F., Gallinat J. Higher prefrontal cortical thickness in high schizotypal personality trait. J Psychiatr Res. 2012;46:960–965. doi: 10.1016/j.jpsychires.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 68.Ettinger U., Williams S.C.R., Meisenzahl E.M., Möller H.-J., Kumari V., Koutsouleris N. Association between brain structure and psychometric schizotypy in healthy individuals. World J Biol Psychiatry. 2012;13:544–549. doi: 10.3109/15622975.2011.559269. [DOI] [PubMed] [Google Scholar]
- 69.Modinos G., Mechelli A., Ormel J., Groenewold N.A., Aleman A., McGuire P.K. Schizotypy and brain structure: A voxel-based morphometry study. Psychol Med. 2010;40:1423–1431. doi: 10.1017/S0033291709991875. [DOI] [PubMed] [Google Scholar]
- 70.Moorhead T.W.J., Stanfield A., Spencer M., Hall J., McIntosh A., Owens D.C. Progressive temporal lobe grey matter loss in adolescents with schizotypal traits and mild intellectual impairment. Psychiatry Res. 2009;174:105–109. doi: 10.1016/j.pscychresns.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 71.Stanfield A.C., Moorhead T.W.J., Harris J.M., Owens D.G.C., Lawrie S.M., Johnstone E.C. Increased right prefrontal cortical folding in adolescents at risk of schizophrenia for cognitive reasons. Biol Psychiatry. 2008;63:80–85. doi: 10.1016/j.biopsych.2007.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.