Abstract
KIAA1429 (also known as vir-like m6A methyltransferase-associated protein (VIRMA)), a newly identified component of the RNA m6A methyltransferase complex, plays critical roles in guiding region-selective m6A deposition. However, in mammals, whether KIAA1429 mediates RNA m6A regulatory pathway functions in vivo remains unknown. Here, we show that the Kiaa1429-specific deficiency in oocytes resulted in female infertility with defective follicular development and fully grown germinal vesicle (GV) oocytes failing to undergo germinal vesicle breakdown (GVBD) and consequently losing the ability to resume meiosis. The oocyte growth is accompanied by the accumulation of abundant RNAs and posttranscriptional regulation. We found that the loss of Kiaa1429 could also lead to abnormal RNA metabolism in GV oocytes. RNA-seq profiling revealed that Kiaa1429 deletion altered the expression pattern of the oocyte-derived factors essential for follicular development. In addition, our data show that the conditional depletion of Kiaa1429 decreased the m6A levels in oocytes and mainly affected the alternative splicing of genes associated with oogenesis. In summary, the m6A methyltransferase KIAA1429-mediated RNA metabolism plays critical roles in folliculogenesis and the maintenance of oocyte competence.
Subject terms: RNA, RNA, RNA, RNA, Development
Introduction
RNA N6-methyladenosine (m6A) is the most prevalent RNA modification and mainly occurs at the consensus RNA motif of RRACH (R = A or G; H = A, U, or C), catalyzed by a large m6A methyltransferase complex (“m6A writers”) composed by METTL3 and METTL14 heterodimer, WTAP, KIAA1429, RBM15, ZC3H13, and other undefined proteins [1]. The m6A modification is reversible and can be removed by the demethylase enzymes ALKBH5 [2] and FTO (“m6A erasers”) [3]. With the recognition of m6A in RNA by its binding proteins (“m6A readers”), the biological functions of the m6A modification are carried out [4–7]. KIAA1429, a recently identified component of the m6A writers, has been proven to mediate preferential m6A deposition in the 3′UTR and near stop codon and is associated with alternative polyadenylation (APA) in HeLa cells [5]. However, the in vivo biological functions of KIAA1429 in mammalian development remain unknown. m6A modification-related factors are usually ubiquitously expressed and are involved in regulating diverse biological processes in various tissues. For example, METTL3 has been reported to modulate spermatogenesis, T cell homeostasis and long-term memory formation [8–10]; YTHDF2 is a m6A reader that is required for early zygotic development and maintenance of female fertility, yet in HeLa cells YTHDF2 is essential for heat shock response [11, 12]. While some factors involved in m6A dynamic modification appear to have tissue-specific functions, for example, ALKBH5 and YTHDC2 are particularly abundant in the testes where their deletion caused spermatogenic defects and impaired male fertility [13, 14]. Kiaa1429 is a highly expressed gene in oocytes compared with most mouse tissues/cells according to the BioGPS database [15], which indicates that it might be a critical factor that specifically functions in mouse oocytes. However, the precise role of KIAA1429 in oocytes remains unclear.
Dynamic m6A modification functions in various aspects of RNA metabolism, such as alternative splicing (AS), APA, nuclear export, mRNA stability, and translation [1]. During oogenesis, the oocytes synthesize and accumulate maternal RNAs for further oocyte meiotic maturation with the change of RNA metabolic activities, which are necessary to determine oocyte quality and consequently affect ovarian function [16, 17]. Although m6A methylation contributing to RNA regulation during oocyte maturation have been identified [11, 18], the pathways and unknown factors involved in this process still remain to be explored. The data on Kiaa1429 from the DBTMEE database [19] classify it as a maternal gene that is transcribed during oocyte growth and required for oocyte developmental competence in mice [20]. In addition, KIAA1429-mediated m6A regulates APA and affects cellular functions in HeLa cells. The possible mechanisms underlying the involvement of KIAA1429-mediated RNA metabolism in mouse oocytes is much worth being studied.
Oocyte growth is always accompanied by dynamic epigenetic modifications. In the mammalian ovary, each oocyte is enclosed within a follicle, and its growth is accompanied by follicular development [21]. At the terminal of folliculogenesis, the chromatin of GV oocyte in the antral follicle gradually transforms from a non-surrounded nucleolus (NSN) to a surrounded nucleolus (SN) chromatin configuration. Compared with NSN oocytes, SN oocytes usually have higher competence to resume meiosis and consequently develop to metaphase II (MII) oocytes [20, 22]. Thus, the NSN–SN transition is closely associated with oocyte meiotic competence. However, the regulatory mechanism involved in the NSN–SN transition is unclear. Classical epigenetic regulations, such as histone modification and DNA methylation, are tightly associated with the chromatin remodeling during meiotic maturation [23–25]. However, it is unclear whether RNA methylation, as another layer of epigenetic regulation at the RNA level, also participates in chromatin condensation from the NSN to SN configuration. Whether the KIAA1429-mediated accumulation of RNA methylation marks in the oocytes is necessary for meiotic resumption needs to be explored.
In this study, we constructed an oocyte-specific Kiaa1429 conditional knockout mouse model and found that the females were infertile. We confirmed that the female infertility was attributed to defective follicular development, which arose from abnormal oogenesis due to the deletion of Kiaa1429. We further identified KIAA1429-regulated splicing events during oogenesis that were crucial for sustaining the developmental competence of oocytes.
Material and methods
Mice
The Kiaa1429fl/fl mice in a C57BL/6N genomic background were constructed by the Bin Shen Laboratory, and Zp3-Cre mice were maintained with a C57BL/6J genomic background. Kiaa1429fl/fl mice were crossed with the Zp3-Cre mice to produce oocyte-specific Kiaa1429 knockout in Kiaa1429fl/flZp3-Cre mice (also referred as Kiaa1429Zp3cKO mice); while Kiaa1429fl/+Zp3-Cre mice were used as controls in the following experiments. The floxed Kiaa1429 gene was identified via PCR using the forward primer 5′-GCTTATATCATAGGCACTGG-3′ and the reverse primer 5′-CACAATTAGGAAGGAACTTC-3′, and the PCR products of the WT and floxed alleles were 160 and 194 bp, respectively. For Zp3-Cre, the forward primer was 5′-GCGGTCTGGCAGTAAAAACTATC-3′, the reverse primer was 5′-GTGAAACAGCATTGCTGTCACTT-3′. For fertility tests, 8-week-old Kiaa1429Zp3cKO and control females were housed with wild-type males for 6 months. The mice were housed under specific pathogen-free environmental conditions with free access to water and food, a temperature of 20–22 °C, 50–70% humidity, and a 12-h light/dark cycle at the Animal Core Facility of Nanjing Medical University. The animal care and experimental procedures were conducted according to the guidelines of the Animal Research Committee of Nanjing Medical University.
Oocyte collection and culture
Three-week-old female mice were intraperitoneally injected with 5 IU of pregnant mare serum gonadotropin (PMSG) (Ningbo Sansheng Pharmaceutical Corporation, Zhejiang, China) to stimulate antral follicular development. After 48 h, fully grown GV oocytes (at least 70 μm in diameter) were isolated by gently puncturing the antral follicles with a sterile insulin syringe needle under the visual field of a dissecting microscope and released into M2 medium (Sigma, M7167) with 2.5 μM milrinone (Sigma, M4659). Eight-week-old mouse ovaries were first digested in M2 medium supplemented with 4 mg/ml collagenase, and the granulosa cell-oocyte complexes were selected and digested with collagenase again to release the oocytes. Growing oocytes with a diameter of ~60 µm were from preantral (small antral) follicles, those with a diameter of ~40 µm were from secondary follicles, and those with a diameter of ~25 µm were from primary follicles. The follicles were collected with mouth-controlled pipettes. The ovarian remnants were then collected by centrifugation, and the supernatants were aspirated. The ovarian fragments were further digested with 0.25% trypsin-EDTA and 0.01% DNase and dispersed into single cells, and the smallest oocytes from primordial follicles with a diameter of ~10–20 µm were manually collected with pipettes [26, 27]. For in vitro maturation, the fully grown GV oocytes were cultured in M12 medium for 4 h (to evaluate the ratio of germinal vesicle breakdown (GVBD)) or 14 h (to evaluate the ratio of PB extrusions) in vitro at 37 °C under 5% CO2. For the superovulation assay, the mice were injected with 5 IU of human chorionic gonadotropin (hCG) (Ningbo Sansheng Pharmaceutical Corporation, Zhejiang, China) followed by 5 IU of PMSG after 48 h. MII oocytes were harvested from the oviducts 16 h post hCG injection.
Immunofluorescence
Oocytes were fixed in phosphate-buffered saline (PBS) supplemented with 4% paraformaldehyde (PFA) (Sigma, P6148) for 30 min at room temperature, followed by permeabilization with 0.5% Triton X-100 in PBS for 45 min. Then, the oocytes were blocked in blocking buffer (2.5% bovine serum albumin (BSA) and 0.2% Triton X-100 in PBS) at 4 °C for 8–12 h. After blocking, the oocytes were incubated with primary antibodies at 4 °C for 12–24 h. After three washes with PBS, the oocytes were incubated with secondary antibodies for 2 h at room temperature, washed three times with PBS, and stained with Hoechst 33342 (KeyGen BioTECH, KGA212-10) for 10 min. The oocytes were mounted on glass slides and detected under a confocal laser scanning microscope (LSM 700, Carl Zeiss, Germany). The antibodies used are listed in Supplementary Table S1.
Western blot analysis
Western blot was carried out as previously described [24, 28]. Briefly, a total of 150 oocytes were collected in RIPA lysis buffer (CWBIO, CW2333) supplemented with 1% protease inhibitor cocktail (CWBIO, CW2200) and 1% phosphatase inhibitor cocktail (CWBIO, CW2383), incubated on ice for 30 min and then mixed with 20% sodium lauryl sulfate (SDS) sample buffer (FDBIO, FD002). After boiling for 5 min, the total lysate was separated by 4–20% SDS-PAGE and transferred onto PVDF membranes. For blocking, the membranes were immersed in TBS with 0.1% Tween-20 (TBST) and 5% low-fat dry milk for 2 h at room temperature and then incubated overnight with the primary antibody at 4 °C. After three washes with TBST, the membranes were then incubated with the appropriate secondary antibody for 2 h at room temperature. Following three washes with TBST. The protein bands were then visualized using an enhanced chemiluminescence kit with HRP (FDBIO, FD8020) and detected using a Bio-Rad gel imaging system. The antibodies used are listed in Supplementary Table S1.
Construction of the gene targeting vector
The mouse Kiaa1429 coding sequence (CDS) corresponding to full-length Kiaa1429 (isoform 2), p1 (residues 1–2705), and p2 (residues 2645–5436) was PCR-amplified from a mouse ovarian cDNA pool, and HA tag sequences (TACCCATACGACGTCCCAGACTACGCT) were inserted after ATG and before TAG using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505). The PCR products were purified using the TIANgel Midi Purification Kit (DP209-02) and then digested by EcoRI and HindIII restriction enzymes (New England Biolabs, Inc.). N-terminal HA-tagged and C-terminal HA-tagged mouse full-length Kiaa1429 was cloned into pcDNA3.1(−) vectors by the ClonExpress MultiS One Step Cloning Kit (Vazyme, C113). The full-length mouse SRSF3 CDS was also amplified from a mouse ovarian cDNA pool by PCR, and the FLAG-tagged plasmid of SRSF3 was constructed in the same manner (the HA tag was replaced by the FLAG tag (GACTACAAGGATGACGACGATAAG)). The primers used are listed in Supplementary Table S2.
Cell culture, plasmid transfection, and immunoprecipitation
HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Cat. No. 11995065) supplemented with 10% fetal bovine serum (FBS), 1% penicillin–streptomycin solution, and 1% GlutaMAX (Gibco, Cat. No. 35050061) at 37 °C in a humidified 5% CO2 incubator. The 293T cells were transfected using Lipofectamine 3000 (Invitrogen) and cultured in Opti-MEM media for 48 h. The transfected cells were lysed in RIPA lysis buffer (Beyotime, P0013K) supplemented with 1 mM protease inhibitor cocktail (Bimake, B14001) and then rocked gently at 4 °C for 30 min. Following centrifugation at 16,000 × g for 30 min, the supernatant was subjected to immunoprecipitation with Protein A/G Magnetic Beads (Bimake, B23202) at 4 °C for 1 h after preclearing the beads. Tag antibody was incubated with the cell lysates with gentle rotation overnight at 4 °C. The samples were then washed three times with lysis buffer. SDS sample buffer was added to the beads, which were boiled for 5 min, and the respective input lysates were analyzed by western blot.
Histological analysis and immunofluorescence of ovaries
Ovaries were fixed in 4% PFA overnight at 4 °C, and then dehydrated through a series of graded ethanol solutions and xylene before paraffin embedding. Serial sections (5 μm) were attached to slides, heated at 60 °C, followed by washing in xylene and rehydration through a graded series of ethanol and double-distilled water and then staining with Hematoxylin and Eosin (HE) reagents. Follicles at various stages of development were counted in every third section throughout the whole ovary by double blind method. To avoid repeated counting of the same follicle, only follicles containing an oocyte with visible GV nuclei stained by hematoxylin were counted, and the total number of primordial, primary, preantral and antral follicles was characterized as described previously [27]. Immunofluorescence was also performed on 5-μm sections. Dewaxed and rehydrated tissue sections were placed on slides in a plastic jar containing 200 ml of 0.1 M citrate buffer, pH 6.0 under 350 W microwave irradiation for 5 min. The slides were then incubated in blocking buffer (PBS containing 5% BSA) for 2 h. Next, the slides were treated with primary antibodies at 4 °C for 12–24 h. After washing three times with PBS, the slides were then incubated with secondary antibodies for 2 h at room temperature and washed three times in PBS. Positive immunostaining was visualized using a DAB kit (ZSGB-BIO, ZLI9018). The images were analyzed under a microscope (Nikon Eclipse E200) equipped with a digital camera (MSHOT, MS60). The antibodies used are listed in Supplementary Table S1.
TUNEL assay
For the analysis of apoptosis during follicular development, a terminal deoxynucleotidyl transferase dUTP Nick-End Labeling (TUNEL) assay was performed on ovary paraffin sections from 4-week-old females using the In Situ Cell Death Detection Kit (Roche, 11684795910). The procedures were carried out according to the instructions provided by the manufacturer. Fluorescence was detected using a confocal laser scanning microscope (LSM 700, Carl Zeiss, Germany).
BrdU incorporation
To analyze proliferation during follicular development, 4-week-old mice were intraperitoneally injected with bromodeoxyuridine (BrdU) (50 mg/kg; Sigma, B5002) and sacrificed 2 h post injection. Ovarian paraffin sections were then incubated with anti-BrdU antibody, proceeding for immunohistochemistry as described with the previously procedure [29], and analyzed under a microscope (Nikon Eclipse E200) equipped with a digital camera (MSHOT, MS60). The antibody used is listed in Supplementary Table S1.
EU incorporation assay
Control or Kiaa1429Zp3cKO oocytes were cultured in M2 medium with 2.5 µM milrinone and 1 µM 5-ethynyl uridine (EU) for 2 h. Fixation, permeabilization, and staining were performed by the Click-iT RNA Alexa Fluor 594 Imaging Kit (Invitrogen, C10330) according to the manufacturer’s instructions. The oocytes were detected and imaged under a confocal laser scanning microscope (LSM 700, Carl Zeiss, Germany).
RNA isolation, cDNA synthesis, RT-PCR, and qPCR analysis
Total RNA was extracted from oocytes of 3-week-old mice using the TRIzol Reagent (Invitrogen, 15596026). The RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1621) was used for cDNA synthesis. Semiquantitative RT-PCR was performed using Premix Taq (Takara, RR901). qPCR was performed using AceQ qPCR SYBR Green Master Mix (Vazyme, Q141-02) according to the manufacturer’s instructions with an ABI Q5 real-time PCR System (Applied Biosystems, Thermo Fisher Scientific). Quantification of the fold change in gene expression was calculated using the comparative threshold cycle (Ct) method with 18 s as the internal control based on the following equation: fold change (cKO vs. control) = 2−▵▵Ct, where ▵▵Ct = (CtcKO/gene − Ctcontrol/gene) − (CtcKO/18s − Ctcontrol/18 s). The primers used are listed Supplementary Table S2.
Oocyte RNA methylation assessment
The RNA m6A levels in oocytes were quantified using the EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (EpiGentek). Approximately 800 control or Kiaa1429Zp3cKO oocytes were collected for the m6A analysis. ELISA-based assays were performed according to the manufacturer’s instructions (EpiGentek).
Oocyte library construction and sequencing
Oocyte library construction and sequencing were carried out using a total of eight samples from 20-day-old control or Kiaa1429Zp3cKO mice in four separate experiments; each sample contained 20 GV oocytes. The libraries for the isolated GV oocytes were generated based on Smart-Seq2 [30]. The KAPA HyperPlus Prep Kit (KK8514) was used to generate sequence libraries according to the manufacturer’s instructions. All samples were sequenced using a HiSeq X Ten (Illumina) with paired-end 150-bp sequencing.
RNA-seq analysis
The reads of the RNA-seq data were aligned to the mouse genome mm10 using STAR 2-pass mode with the annotations from Ensembl version 90 and the parameter ‘–outFilterMismatchNmax 6’. SAMtools were used to extract uniquely mapped reads with a mapping quality ≥20. The number of reads that mapped to each gene was counted using HTSeq-count tool with the ‘union’ overlap resolution mode and –stranded = no. The expression of genes was quantified as reads per kilobase million (RPKMs) by edgeR. The genes with RPKM >1 in at least three control samples were considered expressed. Only expressed genes were kept for the subsequent analysis. Differentially expressed genes were identified by DESeq2 using the following two criteria: the fold change in expression should be >2 and the adjusted p value should be lower than 0.05. Enriched Gene Ontology (GO) terms with a p value < 0.05 were obtained from the Database for Annotation, Visualization and Integrated Discovery.
m6A peak enrichment analysis
m6A peak data and their enrichment scores were downloaded from the MethylTranscriptome DataBase (MeT-DB) (http://compgenomics.utsa.edu/methylation/). The overlapping peaks were merged with bedtools. BedGraphToBigWig was used to convert the BED files into bigwig files. ComputeMatrix and PlotProfiles from DeepTools were used to compute the peak enrichments in downregulated, upregulated, and unchanged genes. The search region was 400 bp upstream of the start codon and 400 bp downstream of the stop codon in genes. Gene bodies were scaled to the same length (1 kb) with the parameter ‘scale-regions’.
Differential alternative polyadenylation identification
RNA-seq reads were mapped to the mm9 genome using STAR 2-pass mode with annotations from Ensembl version 65 and the parameter ‘–outFilterMismatchNmax 6’. Roar was performed to analyze APA using an annotation database of polyadenylation sites from PolyADB version 2. Transcript PRE and POST portions were defined by multiple APA annotation files. Significant events were obtained by a threshold p value < 0.05 and FPKMpre > 1. The m/M ratio was computed for each sample using the counts of mapped reads and the lengths of the transcript’s PRE and POST portions. A larger m/M ratio indicates a shorter isoform and vice versa. The roar value is the ratio of m/M in Kiaa1429Zp3cKO samples to m/M in control samples. Roar values > 1 indicate shorter isoforms in the Kiaa1429Zp3cKO samples and vice versa. The Integrative Genomic Viewer was used for data visualization. The changes of APA were validated by qPCR using primers (listed in Supplementary Table S2) flanking the PRE (representing both short and long transcripts) and POST (exclusively representing the long transcripts) portions of these genes.
Identification of differential alternative splicing events
RNA-seq data were aligned to the mouse genome mm10 using STAR with the following parameters: ‘–outSAMstrandField intronMotif –chimSegmentMin 2 –outFilterMismatchNmax 3 –alignEndsType EndToEnd –outSAMtype BAM SortedByCoordinate –alignIntronMax 299999 –alignSJDBoverhangMin 6’. For splicing analysis between the Kiaa1429Zp3cKO and control samples, we performed a replicate multivariate analysis of transcript splicing (rMATS) with the default parameters to identify AS events. rMATS identified five types of AS events (skipped exon (SE), mutually exclusive exon, retained intron (RI), alternative 5′ splice site, and alternative 3′ splice site). Significant AS events were obtained using a false discovery rate threshold <0.05. The validation of AS events was performed by RT-PCR analysis using primers (listed in Supplementary Table S2) flanking the SEs of these target genes.
YTHDC1 and SRSF3 motif enrichment analysis
YTHDC1 and SRSF3 motifs were obtained from previous literature [4]. scanMotifGenomeWide.pl from the HOMER program was used to find motifs around the splicing sites of skipped or inclusion exon events. The search regions were within 100 nt of exonic adjacent regions and 300 nt of intronic adjacent sites. YTHDC1 and SRSF3 motifs were assigned to the search regions. The number of motifs that fell into each search region was counted. The density of motifs was plotted using window 20 nt.
Morpholino microinjection
Before morpholino microinjection, the GV oocytes were collected and mixed together, then randomly allocated to different treatment group. Approximately 10 pL of morpholino (Gene Tools, USA) was injected at a concentration of 1 mM into GV oocytes from 3-week-old mice in M2 medium with 2.5 µM milrinone. The oocytes were then cultured for 8 h in M2 medium with 2.5 µM milrinone, and after culture, some of these oocytes were collected for assessment of the changes in AS levels by RT-PCR. Followed by the three washes were carried out in M2 medium alone to remove the milrinone, the remaining oocytes were cultured in M12 medium for an additional 4 h (to evaluate the ratio of GVBD) in vitro at 37 °C under 5% CO2. The sequences of morpholinos are listed in Supplementary Table S2.
Statistical analysis
All experiments were repeated at least three times for statistical analysis, and data from one representative experiment are shown unless otherwise stated. Statistical analyses were performed with GraphPad Prism 7.00 (GraphPad Software, Inc.). Quantifications are presented as mean values ± s.e.m (standard error of mean). Statistical comparison was performed using two-tailed Student’s t tests or chi-square test. Statistical significance was determined as indicated in the figure legends. P values < 0.05 were regarded as significant (*p < 0.05; **p < 0.01; ***p < 0.001). Sample size was chosen accordingly to the studies using similar methods and is comparable with what is routinely employed in the field. More details of experimental statistics (such as sample size (n), description of sample collection, and the number of replicates) are indicated in the corresponding figure legends. There was no statistical method used to predetermine sample size.
Results
Kiaa1429 is expressed during oocyte growth and is required for female fertility
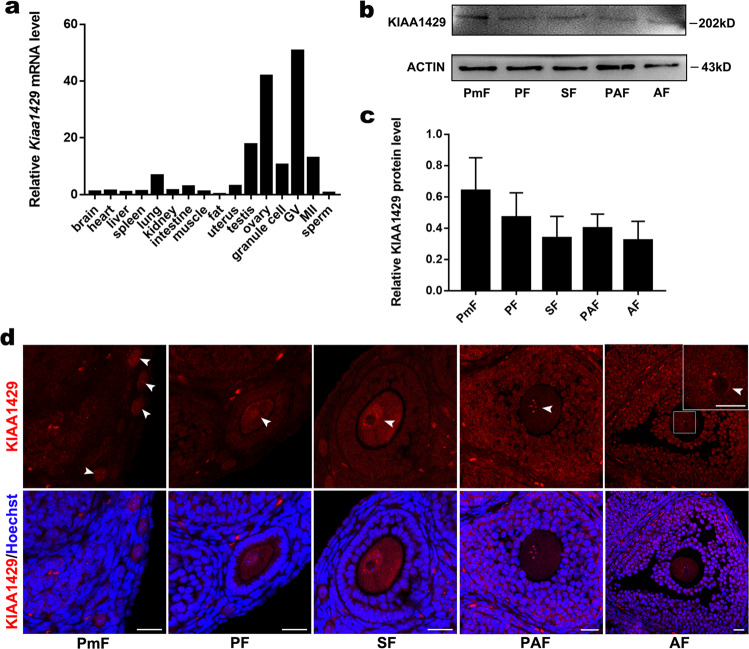
We first used qPCR to characterize the expression pattern of Kiaa1429 in mouse tissues and cells and noticed that Kiaa1429 mRNA expression was dominant in GV oocytes (Fig. 1a). KIAA1429 protein was examined to be expressed in the oocytes at different developmental stages of follicles collected from 8-week-old mice by western blot (Fig. 1b, c), and the immunofluorescence results showed its signal was mainly in the oocyte nuclei and granulosa cells during folliculogenesis (Fig. 1d, Supplementary Fig. S1a). These findings indicate that KIAA1429 may be an essential factor that specifically functions in mouse oocytes. Thus, we generated Kiaa1429fl/fl mice, and crossed them with Zp3-cre mice (the Cre recombinase is expressed in the oocytes at primary follicle stage and onward [31]) to obtain mice with oocyte-specific deletion of Kiaa1429 (Supplementary Fig. S1b). Kiaa1429 exon 2 was successfully deleted in Kiaa1429fl/flZp3-Cre (Kiaa1429Zp3cKO) mice, which results in premature termination of KIAA1429, while Kiaa1429fl/+Zp3-Cre mice were used as controls in the following experiments (Supplementary Fig. S1c). The Kiaa1429 mRNA was almost undetectable in the GV oocytes of Kiaa1429Zp3cKO mice, and the knockout efficiency was also validated at protein level (Supplementary Fig. S1d, e). Next, we carried out a breeding assay for up to 6 months, female Kiaa1429Zp3cKO mice were found completely infertile (Supplementary Fig. S1f). Together, these results demonstrate that KIAA1429 expression in oocytes is essential for female fertility.
Fig. 1. Kiaa1429 is a gene specifically expressed during oocyte development.
a mRNA profiles of various mouse tissues and cells by qPCR. The relative mRNA level in the brain was set to 1.0, and the fold changes in different tissues and cells are shown. GV germinal vesicle-stage oocytes, MII metaphase II (MII)-stage oocytes. b Western blot analysis of the expression of KIAA1429 protein and the internal control ACTIN in oocytes at different follicle stages. A representative western blot gel image is shown. Number of oocytes used for each blot: 200 primordial, 80 primary, 80 secondary, 80 preantral, and 50 antral follicle-stage oocytes. This experiment was repeated four times. c Quantification of the western blot data. Data are presented as the mean ± s.e.m. from four independent experiments, t test (two-tailed). There is no statistical difference in the expression levels of KIAA1429 between any two groups of the oocytes. d KIAA1429 Immunofluorescence location in oocytes and granulosa cells at different follicle stages. IgG was used as a negative control. PmF primordial follicle, PF primary follicle, SF secondary follicle, PAF preantral follicle, AF antral follicle. Scale bar: 20 μm.
Kiaa1429 deficiency causes a severe defect in follicular development
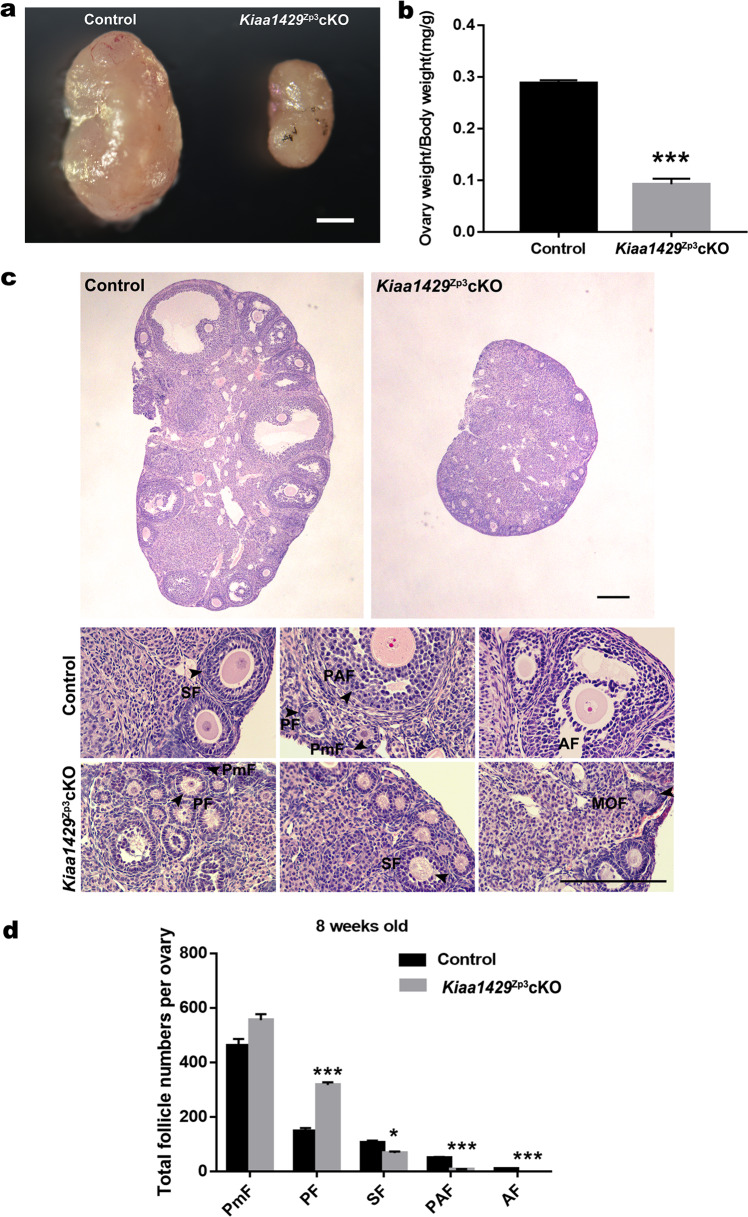
We further analyzed the phenotype of the Kiaa1429Zp3cKO and control mice to uncover the role of KIAA1429 in female fertility. The viable Kiaa1429Zp3cKO mice were indistinguishable from their control littermates in growth but had substantially smaller ovaries, with a 68% reduction in the ovary-to-body weight ratio at 8 weeks (Fig. 2a, b). Then, we performed histological sections with HE staining of the ovaries from 8-week-old females and quantified the number of follicles at each stage. The results showed there were fewer secondary and more primary follicles in the 8-week-old Kiaa1429Zp3cKO ovaries (Fig. 2c, d), indicating defective follicular development beyond the primary stage. In addition, multiple oocyte follicles (MOFs) were seen in Kiaa1429Zp3cKO ovaries (Fig. 2c). Large antral follicles first appear from the 20th–30th days in mice [32]. To investigate the potential developmental defect in preovulatory follicles more closely, we repeated the HE staining and quantification of follicles in 4-week-old ovaries and found 4-week-old Kiaa1429Zp3cKO ovaries contained fewer preantral follicles relative to the control ovaries and almost no antral follicles (Supplementary Fig. S2a, b). Furthermore, the TUNEL assay of 4-week-old Kiaa1429Zp3cKO ovaries showed significantly increased apoptosis signaling in the secondary follicles and preantral follicles (Supplementary Fig. S2c, d). Moreover, a decreased proliferation in the granulosa cells of Kiaa1429Zp3cKO ovaries was detected by BrdU incorporation (Supplementary Fig. S2e, f). In summary, KIAA1429 is required for folliculogenesis and its deletion in oocytes leads to follicular growth defect along with abnormal apoptosis and proliferation of granulosa cells.
Fig. 2. KIAA1429 expression in oocytes is essential for follicular development.
a Morphological analysis of 8-week-old Kiaa1429Zp3cKO and control ovaries. Scale bar: 0.5 mm. b Ratio of ovary weight to body weight in 8-week-old Kiaa1429Zp3cKO and control mice. Data are presented as the mean ± s.e.m. from three independent experiments. ***P < 0.001, t test (two-tailed). c HE-stained sections of 8-week-old Kiaa1429Zp3cKO and control ovaries. Representative images of the regions are shown below, Black arrows indicate follicles at different stages. MOF multiple oocyte follicle. Scale bar: 200 μm. d Quantitative analysis of follicles at different stages in 8-week-old Kiaa1429Zp3cKO and control ovaries. Unilateral ovaries were isolated from control and Kiaa1429Zp3cKO mice and counted in each experiment. Total three control ovaries and three Kiaa1429Zp3cKO ovaries were used for counting follicles. Data are presented as the mean ± s.e.m. *P < 0.05, ***P < 0.001, t test (two-tailed).
Kiaa1429 deficiency influences oocyte destiny during folliculogenesis
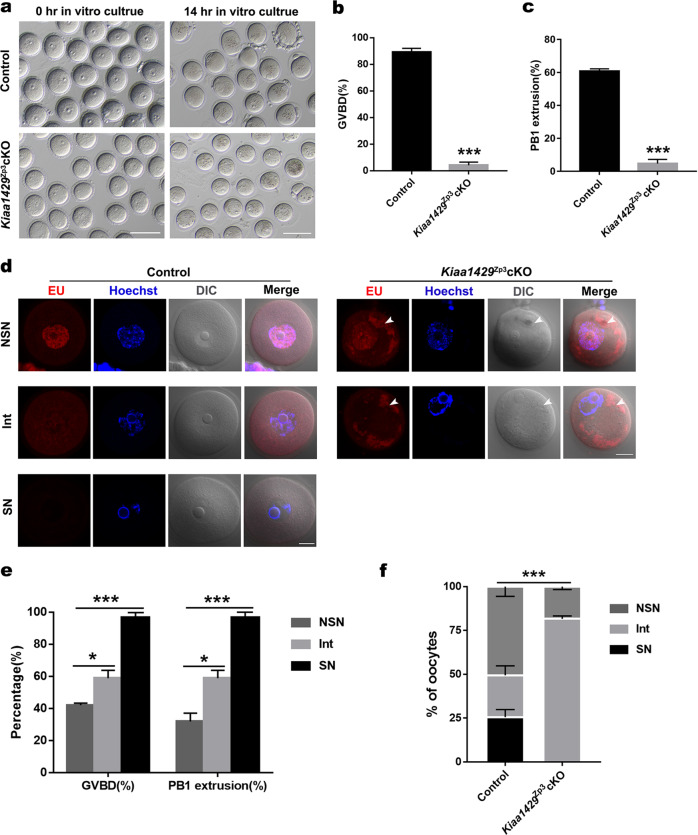
To further investigate the cause of follicular development defect after Kiaa1429 deletion, we analyzed follicular development in 3-week-old Kiaa1429Zp3cKO females and found that there was no significant difference in the number of follicles at different stages between the Kiaa1429Zp3cKO and control mice, except for the appearance of MOFs (Supplementary Fig. S3a, b). However, following PMSG treatment, the Kiaa1429Zp3cKO females had abnormal follicular morphology and smaller GV oocytes in the preovulatory follicles than the control ones (Supplementary Fig. S3c, d). To determine whether the developmental defect in Kiaa1429Zp3cKO oocytes occurred, we isolated GV-stage oocytes from Kiaa1429Zp3cKO and control ovaries. The Kiaa1429Zp3cKO oocytes exhibited significantly lower rates of GVBD and first polar body (PB1) extrusion than the control oocytes in in vitro culture (Fig. 3a–c), which is consistent with the results of superovulation in vivo, wherein no MII-stage oocytes ovulated in the Kiaa1429Zp3cKO mice (Supplementary Fig. S3e). Because the developmental competence of the resulting MII oocyte is associated with chromatin remodeling and transcriptional activity changes during oocyte maturation [22, 33], we next analyzed the global RNA transcription in the control and Kiaa1429Zp3cKO oocytes through the detection of EU incorporated into nascent RNA and found that transcription was active in the NSN-type configuration of both the Kiaa1429Zp3cKO and control oocytes. However, abnormal RNA granules were detected in the cytoplasm of Kiaa1429Zp3cKO oocytes (Fig. 3d). Given that the potential role of KIAA1429 in the modulation of RNA metabolism, we infer that incorrectly processed nascent RNAs are transported to the cytoplasm and form RNA granules due to the loss of m6A modification. Meanwhile, to validate the essential relationship between chromatin configuration and meiotic progression, we cultured GV oocytes with different nuclear configurations in vitro and found that SN oocytes had a higher GVBD rate than NSN and Intermediate (Int) oocytes remarkably (Fig. 3e); however, none of the Kiaa1429Zp3cKO oocytes reached the SN-type configuration, and the majority of the Kiaa1429Zp3cKO oocytes were at the stage of Int-type configuration (Fig. 3d, f). In summary, KIAA1429 is required for oocyte competence in mediating chromatin configuration and RNA metabolism in GV oocytes.
Fig. 3. KIAA1429 is essential for oocyte developmental competence and Kiaa1429Zp3cKO oocytes exhibit abnormal chromatin organization and RNA granules.
a GV oocytes were isolated from 3-week-old Kiaa1429Zp3cKO and control ovaries following superovulation in vivo and cultured for 14 h in vitro. A large number of Kiaa1429Zp3cKO oocytes were arrested at the GV stage, whereas the oocytes from control mice developed into MII oocytes. Scale bars: 100 μm. b Proportions of GVBD oocytes in Kiaa1429Zp3cKO and control GV oocytes cultured in vitro. Data are presented as the mean ± s.e.m. from three independent experiments (total number of oocytes analyzed: n = 83 for control; n = 84 for Kiaa1429Zp3cKO). ***P < 0.001, t test (two-tailed). c Proportions of oocytes with extruded first polar bodies (PBE) in Kiaa1429Zp3cKO and control GV oocytes cultured in vitro. Data are presented as the mean ± s.e.m. from three independent experiments (total number of oocytes analyzed: n = 83 for control; n = 84 for Kiaa1429Zp3cKO). ***P < 0.001, t test (two-tailed). d Kiaa1429Zp3cKO and control GV oocytes cultured with 5-ethynyl uridine (EU). The signals of nascent RNA are detected as red fluorescence, whereas the nuclei are blue (Hoechst). The oocyte morphology was observed by differential interference contrast (DIC) microscopy. Arrowheads indicate abnormal cytoplasmic granules. The images are representative of three independent experiments. Scale bars: 20 μm. e Percentage of GVBD oocytes and oocytes with extruded PBEs respectively in NSN, Int and SN oocytes cultured in vitro. Data are presented as the mean ± s.e.m. from three independent experiments (total number of oocytes analyzed: n = 70). *P < 0.05, ***P < 0.001, t test (two-tailed). f The proportion of the NSN, Int and SN nucleolus types in Kiaa1429Zp3cKO and control GV oocytes (total number of oocytes analyzed: n = 66 for control; n = 53 for Kiaa1429Zp3cKO). ***P < 0.001, chi-square test.
Kiaa1429 deficiency influences the expression pattern of oocyte-derived factors with a decrease of m6A levels in oocytes
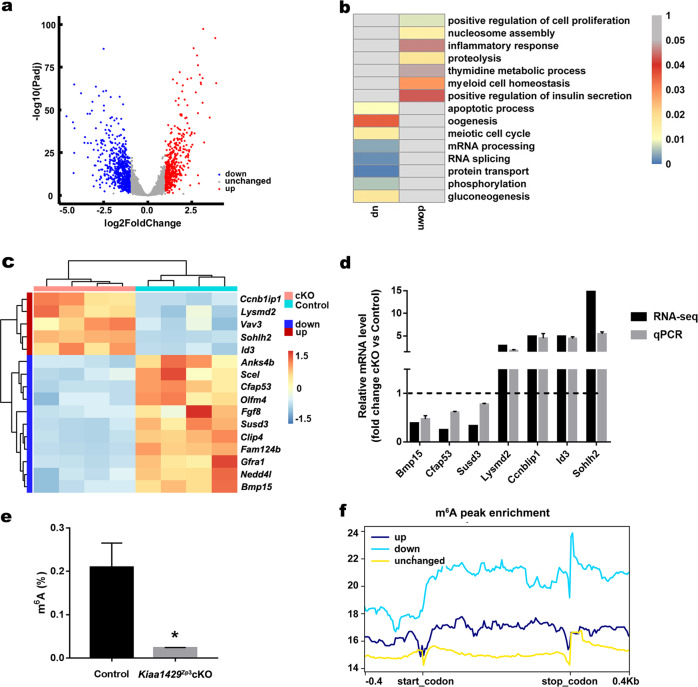
As given, it is possible that Kiaa1429Zp3cKO oocytes exhibit RNA granules caused by defect in RNA processing, we next performed RNA-seq, and the data revealed that a total of 1081 genes were significantly differentially expressed. A total of 448 genes were upregulated and 633 were downregulated in Kiaa1429Zp3cKO oocytes (Fig. 4a, Supplementary Table S3). GO enrichment analysis revealed that some changes in the transcriptome could induce the observed defect in follicular development and oocyte maturation after Kiaa1429 deletion. These include genes that control key processes such as cell proliferation, the apoptotic process, and oogenesis (Fig. 4b). Furthermore, we found that Kiaa1429 deletion altered the expression pattern of many oocyte-derived factors (Fig. 4c), which have been reported to be involved in the signaling pathways of the oocyte-granulosa cell dialog and play important roles in oocyte survival and folliculogenesis [34–39]. These oocyte-derived factors were randomly selected for validation of their expressional alterations by qPCR (Fig. 4d).
Fig. 4. Transcriptional profiling by RNA-seq and relevance to m6A modification.
a Volcano plots of the significantly differentially expressed genes (an adjusted p value < 0.05 and Fold change > 2; downregulated (blue), upregulated (Red), and unchanged genes (Gray) in Kiaa1429Zp3cKO oocytes are shown). b Enriched GO terms in downregulated and upregulated genes after Kiaa1429 deletion. The heatmap shows the −log10(p value) of the GO terms. c Heatmap of oocyte-derived factor genes. The color indicates the expression level after z-scoring. d Validation of RNA-seq data in oocytes from 20-day-old mice by qPCR. The tested genes were randomly selected from oocyte-derived factor genes. The RNA levels are shown relative to those of the control oocytes and were normalized to 1. Data of qPCR are presented as the mean ± s.e.m. from three independent experiments. e The m6A levels in total RNA from 20-day-old GV oocytes were measured using the EpiQuik m6A RNA Methylation Quantification Kit (EpiGentek). Data are presented as the mean ± s.e.m. from three independent experiments. *P < 0.05, t test (two-tailed). f The enrichment of m6A peaks in downregulated, upregulated, and unchanged genes after Kiaa1429 deletion.
We next sought to understand whether KIAA1429-mediated RNA m6A modification could regulate changes in the transcriptome. By an ELISA-based assay, we detected a decrease of m6A levels in the Kiaa1429Zp3cKO oocytes (Fig. 4e). Due to the sample limitations for m6A-seq, we analyzed the m6A peak enrichment and distribution in the transcripts from our RNA-seq data using the MeT-DB database [40] and found that the downregulated transcripts in Kiaa1429Zp3cKO oocytes had a higher m6A peak and a prominent m6A peak enrichment in the 3′UTR and near the stop codon (Fig. 4f), indicating that transcripts with more m6A modifications tend to have lower expression. To our knowledge, KIAA1429 participates in the regulation of APA in HeLa cells [5]. We therefore analyzed APA in the control and Kiaa1429Zp3cKO transcriptomes using Roar [41]. The results revealed 262 events with 3′UTR lengthening versus 221 events with 3′UTR shortening in Kiaa1429Zp3cKO oocytes (Supplementary Fig. S4a, Supplementary Table S4). The changes in APA events were not as dominant as previously reported in the HeLa cells [5]. Nevertheless, we examined the 3′UTR changes of some oocyte maturation- and follicular development-related genes [42–49] (Supplementary Fig. S4b–i) and validated these changes by qPCR (Supplementary Fig. S4j). In summary, Kiaa1429 deletion alters the expression of oocyte-derived factors at the RNA level and causes a significant decrease in the RNA m6A level. The APA analysis suggested that KIAA1429 mostly regulates other pathways in oocytes except for APA.
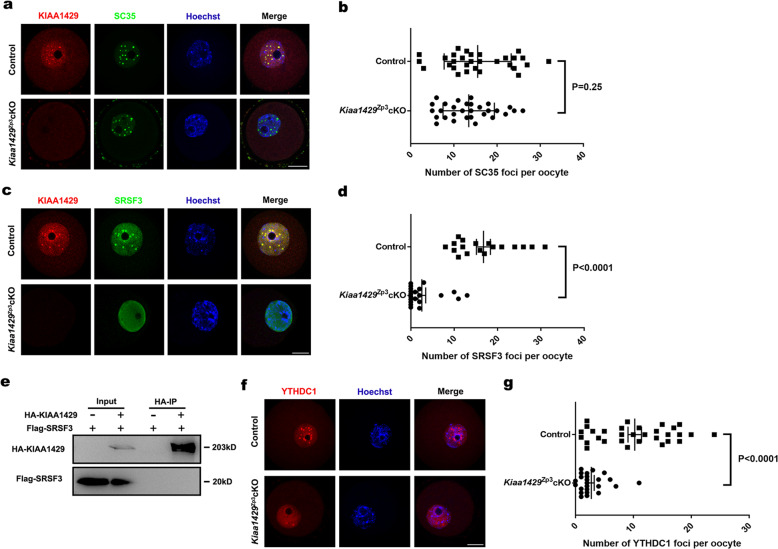
KIAA1429 colocalizes with SRSF3 in nuclear speckles and influences the localization of SRSF3 and YTHDC1
To further explore how KIAA1429 participates in nuclear RNA processing in oocytes, we performed immunofluorescence to characterize the localization of KIAA1429 in the GV oocytes. KIAA1429 was shown to be located in nuclear speckles by costaining with the nuclear speckle marker SC35 (Fig. 5a). A quantitative foci analysis of SC35 revealed that the deletion of Kiaa1429 had almost no effect on the number of nuclear speckles (Fig. 5b). To our knowledge, nuclear speckles are enriched with pre-mRNA processing factors, including splicing factors involved in AS [50]. SRSF3 is a splicing factor that belongs to the serine/arginine-rich protein family, and its deletion in mouse oocytes also caused GVBD defect [51]. The immunofluorescence revealed that KIAA1429 colocalized with SRSF3 in the nuclear speckles, and SRSF3 staining decreased or even disappeared completely in the nuclear speckles of Kiaa1429Zp3cKO oocytes (Fig. 5c, d), suggesting that KIAA1429 may participate in AS via SRSF3. To determine whether SRSF3 is required for interaction with KIAA1429, coimmunoprecipitation was performed and revealed that KIAA1429 did not interact with SRSF3 directly (Fig. 5e). Given that the m6A reader YTHDC1 regulates mRNA splicing by recruiting SRSF3 to its binding regions of targeted mRNAs [4], we next examined YTHDC1 nuclear staining in the Kiaa1429Zp3cKO and control GV oocytes. Intriguingly, we found that the YTHDC1 foci number per oocyte was significantly decreased with the deletion of Kiaa1429 (Fig. 5f, g). These findings demonstrate that in oocytes, KIAA1429 is important for the proper localization of certain mRNA processing factors in nuclear speckles.
Fig. 5. KIAA1429 colocalizes with SRSF3 in nuclear speckles and regulates YTHDC1 and SRSF3 localization.
a Immunofluorescence of Kiaa1429Zp3cKO and control GV oocytes with KIAA1429 and SC35 costaining. The images are representative of three independent experiments. Scale bars: 20 μm. b Quantitative foci analysis of SC35. GV oocytes from Kiaa1429Zp3cKO and control females in three experiments were randomly selected for foci analysis of SC35 (n = 30 biologically independent oocytes for control; n = 30 biologically independent oocytes for Kiaa1429Zp3cKO). P = 0.25, t test (two-tailed). c Immunofluorescence of Kiaa1429Zp3cKO and control GV oocytes with KIAA1429 and SRSF3 costaining. Images are representative of three independent experiments. Scale bars: 20 μm. d Quantitative foci analysis of SRSF3. GV oocytes from Kiaa1429Zp3cKO and control females in three experiments were randomly selected for foci analysis of SRSF3 (n = 24 biologically independent oocytes for control; n = 27 biologically independent oocytes for Kiaa1429Zp3cKO). P < 0.0001, t test (two-tailed). e Coimmunoprecipitation analysis of KIAA1429 with SRSF3. f YTHDC1 immunofluorescence of Kiaa1429Zp3cKO and control GV oocytes. The images are representative of three independent experiments. Scale bars: 20 μm. g Quantitative foci analysis of YTHDC1. GV oocytes from Kiaa1429Zp3cKO and control females in three experiments were randomly selected for foci analysis of YTHDC1 (n = 31 biologically independent oocytes for control; n = 24 biologically independent oocytes for Kiaa1429Zp3cKO). P < 0.0001, t test (two-tailed).
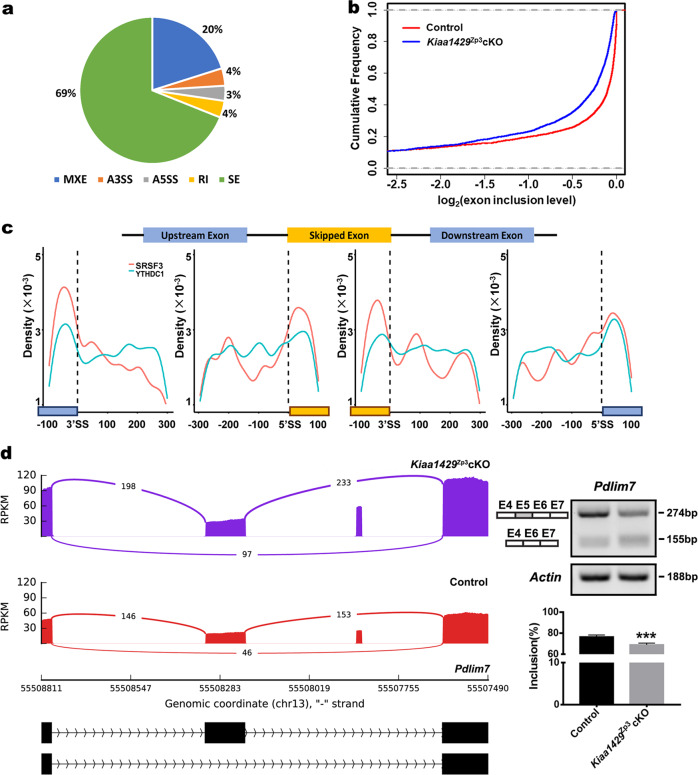
KIAA1429 regulates the alternative splicing of oogenesis-related transcripts
Our discovery of the nuclear speckle localization of KIAA1429 together with the splicing factor SRSF3 in oocytes raised the possibility that KIAA1429 plays a role in regulating AS. To confirm this, we analyzed the differential AS events in the RNA-seq data using rMATS [52]. We identified 4214 different AS events between the control and Kiaa1429Zp3cKO oocytes, with a dominant enrichment of SEs (Fig. 6a, Supplementary Table S5). In addition, we found that the genes in the Kiaa1429Zp3cKO oocytes had a lower exon inclusion level compared with the control oocytes (Fig. 6b). Based on our finding that KIAA1429 regulated the localization of SRSF3 and YTHDC1, we further analyzed AS events in Srsf3-knockout oocytes [51] and Ythdc1-deficient oocytes [53]. The results revealed that KIAA1429, consistent with SRSF3 and YTHDC1, tended to promote exon inclusion (Supplementary Fig. S5a, Supplementary Table S6), and there was an overlap of 280 SE genes among the three samples (Supplementary Fig. S5b), suggesting that these SE genes may be regulated by KIAA1429 along with SRSF3 and YTHDC1. Further analysis revealed a significant enrichment of the SRSF3 and YTHDC1-binding motifs [4] in the exon regions near splicing sites of the Kiaa1429Zp3cKO oocytes (Fig. 6c). We also noticed that 56% of the SE genes in the SRSF3 mutant oocytes overlapped with KIAA1429-regulated ones (Supplementary Fig. S5b, Supplementary Table S7). GO analysis revealed that these overlapping genes were associated with the phenotype of oocyte maturation arrest, abnormal nuclear programming, and RNA granules and included genes controlling processes such as cell division, mRNA processing, and chromatin remodeling (Supplementary Fig. S5c). Next, we screened several potential candidates with respect to oogenesis: Aurkc [54], Pdlim7 [51], Slbp [55], and Uchl3 [56]. Notably, SRSF3-mediated Pdlim7 exon inclusion has been proven to be essential for maintaining proper GVBD in mouse oocyte meiosis [51]. We performed a RT-PCR analysis of the control and Kiaa1429Zp3cKO oocytes using primers flanking the SEs of these target genes and verified that the exon inclusion levels of these four genes were all significantly decreased in the Kiaa1429Zp3cKO oocytes (Fig. 6d, Supplementary Fig. S5d–f). Furthermore, we microinjected morpholinos into wild-type oocytes to induce specific skipping events of Pdlim7 exon 5, Aurkc exon 3, Uchl3 exon 4, and Slbp exon 3, and found the change of AS level of Pdlim7, Uchl3, and Slbp by RT-PCR with the decrease of GVBD rate (Supplementary Fig. S5g–i). In summary, the m6A methyltransferase KIAA1429 participates in posttranscriptional regulation by mostly regulating AS in oocytes.
Fig. 6. KIAA1429 regulates the alternative splicing of transcripts that function in oogenesis.
a Five types of alternative splicing events were identified in Kiaa1429Zp3cKO samples (SE skipped exon, MXE mutually exclusive exon, RI retained intron, A5SS alternative 5′ splice site, A3SS alternative 3′ splice site). b The cumulative frequency plot for the alternative exon inclusion levels in control and Kiaa1429Zp3cKO oocytes. The exon inclusion levels were transformed by log2. P < 0.01, Wilcoxon signed-rank paired test. c Distribution of YTHDC1 and SRSF3 motifs near splice sites of skipped exon events (100 nt upstream and 300 nt downstream from 50 splice sites, 50 SS, and 300 nt upstream and 100 nt downstream from 30 splice sites, 30 SS). d Sashimi plot and RT-PCR validation of Pdlim7 gene in control and Kiaa1429Zp3cKO oocytes. The alternative exon (Exon 5 (E5)) is marked in gray.
Discussion
KIAA1429 is a recently identified component of m6A methyltransferase complex [5, 57] and has a homologous gene, Virilizer, in Drosophila, which is required for the AS of the sex determination factor Sex-lethal [58]. However, whether KIAA1429 plays in mammalian development remains unknown. Our study indicated that Kiaa1429 is highly expressed in GV oocytes during follicle growth and Kiaa1429Zp3cKO females were infertile. Further functional analysis demonstrated that the oocyte-specific deletion of Kiaa1429 resulted in defective folliculogenesis and serious damage on oocyte developmental competence. Kiaa1429 deletion altered the transcriptome expression profile of oocyte-derived factors those are necessary for the differentiation of granulosa cells and for these cells to support oocyte growth. Notably, Kiaa1429Zp3cKO oocytes exhibited an obvious enrichment of exon skipping events. Thus, our study revealed that KIAA1429, a novel m6A writer, plays a critical role in female fertility and participates in follicular development and oocyte meiotic maturation by primarily regulating AS.
Our results indicated that 3′UTR changes in oocytes were not as obvious as previously reported in HeLa cells after Kiaa1429 deletion [5]. Nevertheless, the nuclear speckle localization of KIAA1429 led us to analyze AS in oocytes. rMATS analysis demonstrated that KIAA1429 was involved in AS regulation in oocytes. The changes in the AS events after Kiaa1429 deletion were more distinct than the changes in the 3′UTR lengths. In oocytes, SE events accounted for ~69% of total AS events caused by Kiaa1429 deletion, while in HeLa cells, the ratio of SE events was about 52% by analyzing the published data [5]. In addition, Kiaa1429 deletion in oocytes led to a lower exon inclusion level, whereas in HeLa cells, this tendency was not observed (78% of SE events in oocytes were exon skipping events, but only 48% in HeLa cells), suggesting that the function of KIAA1429 is unique in different cells. Thus, the KIAA1429-mediated AS regulatory mechanism likely plays a major role in oocytes other than APA.
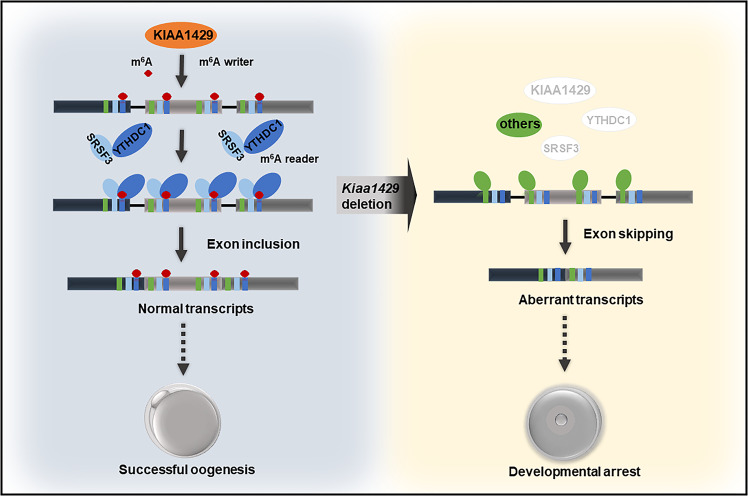
We observed that KIAA1429 colocalized with SRSF3 in oocytes and KIAA1429 deletion caused the decreased localization of SRSF3 and YTHDC1 in the nucleus of oocytes, with enrichment observed for the SRSF3-binding consensus and YTHDC1-binding consensus in the exon regions near the splicing sites. In addition, by integrating RNA-seq data with published SRSF3 and YTHDC1 RNA-seq data, we found that 56% of SRSF3-regulated SE events overlapped with SE events in Kiaa1429Zp3cKO oocytes. Pdlim7 exon 5 was skipped in the Kiaa1429Zp3cKO oocytes, as the same exon skipped in Srsf3-knockout oocytes. And the phenotype induced by the exon skipping in several oogenesis-related factors was also consistent with the GVBD defect after Kiaa1429 deletion. Thus, we suggest a potential mechanism: YTHDC1 recognizes the m6A modification on KIAA1429-mediated pre-mRNA and promotes exon inclusion by recruiting SRSF3 to their binding region. Upon Kiaa1429 deletion, SRSF3 and YTHDC1 fail to bind pre-mRNA. Then, exons are skipped with the binding of other factors, such as SRSF10, which was reported to bind and mask the region near the m6A sites, resulting in exon skipping upon the failure of m6A modification [59]. Aberrant transcripts appear, and consequently, GVBD defect in oocytes occurs, as illustrated in Fig. 7.
Fig. 7. Schematic diagram for the role of the m6A writer KIAA1429 in oocytes.
KIAA1429 mediates the m6A modification on pre-mRNA, then YTHDC1 recognizes the m6A signal and recruits SRSF3 to the binding regions on the KIAA1429-mediated pre-mRNA, resulting in exon inclusion together with the spliceosome. With the deletion of Kiaa1429, SRSF3, and YTHDC1 fail to bind pre-mRNA without KIAA1429-mediated m6A modification and instead, other factors such as SRSF10 bind to promote exon skipping. These skipped exons lead to the accumulation of aberrant transcripts in oocytes. Consequently, GVBD fails and oocyte growth arrests.
With a high proportion of follicles reached the antral stage, approximately half of the NSN oocytes converted their chromatin configuration to Int oocytes and SN oocytes [22, 25], which was confirmed in our study. During the NSN to SN transition, oocytes change their metabolic activities and accumulate maternal factors for further oocyte maturation [20]. Strikingly, the Kiaa1429Zp3cKO oocytes had abnormal chromatin configurations and failed to resume meiosis. And we observed the disordered spatiotemporal regulation of gene expression in the Kiaa1429Zp3cKO oocytes that are reflected in RNA-seq data. Thus, KIAA1429 may be involved in nuclear reprogramming to organize the genome to attend oocyte maturation by maintaining dynamic RNA modifications during oogenesis.
In summary, we first demonstrated that the recently identified m6A methyltransferase KIAA1429 is required for folliculogenesis and the oocyte quality control reflected in many processes including meiosis resumption, chromatin remodeling and RNA metabolism. With the decrease in the m6A levels in oocytes, Kiaa1429 deficiency alters the expression pattern of the oocyte-derived factors that orchestrate follicular development, providing a solid understanding about the role of KIAA1429 in mammalian reproduction and enriching the content of which RNA epigenetic modification contributes to oocyte meiotic competence and follicular development. Remarkably, our finding that KIAA1429 is the only known m6A writer in oocyte nuclear speckles implies an essential role of KIAA1429 in AS, and the phenotype induced by the change of the exon inclusion level in several oogenesis-related factors was consistent with the GVBD defect after Kiaa1429 deletion. Of importance, most of the differences between oocytes and HeLa cells suggest that the role of KIAA1429 in RNA metabolism was cell-specific. Nevertheless, the deletion of the m6A writer KIAA1429 might exert a much broader effect on the physiological function of cells and various biological processes. These still need to be discussed in further studies. In addition, due to the amount of RNA from oocyte samples required for m6A-seq, we could not determine whether the transcripts of target genes have m6A modifications, and a technological breakthrough of m6A-seq in oocytes needs to be achieved in the future.
Supplementary information
Supplementary information, supplementary table and figure legends
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFC1004002 and 2017YFC1001301), the Nature Science Foundation of China (31871505, 31622039 and 61873276) and the Science Foundation for Distinguished Young Scholars of Jiangsu Province (BK20160045).
Data availability
The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus under accession ID GSE129714.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yue Hu, Zhangyi Ouyang
Contributor Information
Bin Shen, Email: binshen@njmu.edu.cn.
Wenjie Shu, Email: shuwj@bmi.ac.cn.
Ran Huo, Email: huoran@njmu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-020-0516-1) contains supplementary material, which is available to authorized users.
References
- 1.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–19.. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–14.. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–42.. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Wang M, Xie D, Huang Z, Zhang L, Yang Y, et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018;28:1050–61.. doi: 10.1038/s41422-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, et al. The RNA m6A reader YTHDF2 Is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–67. e4. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–27.. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016;44:D313–6. [DOI] [PMC free article] [PubMed]
- 16.Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14:159–77. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137:859–70. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi ST, Ma JY, Wang ZB, Guo L, Hou Y, Sun QY. N6-methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J Biol Chem. 2016;291:23020–6. doi: 10.1074/jbc.M116.748889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SJ, Shirahige K, Ohsugi M, Nakai K. DBTMEE: a database of transcriptome in mouse early embryos. Nucleic Acids Res. 2015;43:D771–6. doi: 10.1093/nar/gku1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma JY, Li M, Luo YB, Song S, Tian D, Yang J, et al. Maternal factors required for oocyte developmental competence in mice: transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes. Cell Cycle. 2013;12:1928–38. doi: 10.4161/cc.24991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monniaux D. Driving folliculogenesis by the oocyte-somatic cell dialog: lessons from genetic models. Theriogenology. 2016;86:41–53. doi: 10.1016/j.theriogenology.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Zuccotti M, Garagna S, Merico V, Monti M, Alberto Redi C. Chromatin organisation and nuclear architecture in growing mouse oocytes. Mol Cell Endocrinol. 2005;234:11–7. doi: 10.1016/j.mce.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 23.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Liu XM, Yan MQ, Ji SY, Sha QQ, Huang T, Zhao H, et al. Loss of oocyte Rps26 in mice arrests oocyte growth and causes premature ovarian failure. Cell Death Dis. 2018;9:1144. doi: 10.1038/s41419-018-1196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnet-Garnier A, Feuerstein P, Chebrout M, Fleurot R, Jan HU, Debey P, et al. Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int J Dev Biol. 2012;56:877–87. doi: 10.1387/ijdb.120149ab. [DOI] [PubMed] [Google Scholar]
- 26.Schultz RM, Stein P, Svoboda P. The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod. 2018;99:160–74. doi: 10.1093/biolre/ioy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rankin TL, O’Brien M, Lee E, Wigglesworth K, Eppig J, Dean J. Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development. 2001;128:1119–26. doi: 10.1242/dev.128.7.1119. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YL, Zhao LW, Zhang J, Le R, Ji SY, Chen C, et al. DCAF13 promotes pluripotency by negatively regulating SUV39H1 stability during early embryonic development. EMBO J. 2018;37:e98981. doi: 10.15252/embj.201898981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Shi L, Gong X, Jiang M, Yin Y, Zhang X, et al. Oocyte-dependent activation of MTOR in cumulus cells controls the development and survival of cumulus-oocyte complexes. J Cell Sci. 2016;129:3091–103. doi: 10.1242/jcs.182642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 31.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–2. [PubMed] [Google Scholar]
- 32.Jones EC, Krohn PL. The relationships between age, numbers of ocytes and fertility in virgin and multiparous mice. J Endocrinol. 1961;21:469–95. doi: 10.1677/joe.0.0210469. [DOI] [PubMed] [Google Scholar]
- 33.Bouniol-Baly C, Hamraoui L, Guibert J, Beaujean N, Szollosi MS, Debey P. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod. 1999;60:580–7. doi: 10.1095/biolreprod60.3.580. [DOI] [PubMed] [Google Scholar]
- 34.Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–66.. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 35.Biase FH, Kimble KM. Functional signaling and gene regulatory networks between the oocyte and the surrounding cumulus cells. BMC Genomics. 2018;19:351. doi: 10.1186/s12864-018-4738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estienne A, Price CA. The fibroblast growth factor 8 family in the female reproductive tract. Reproduction. 2018;155:R53–62. [DOI] [PubMed]
- 37.Shin YH, Ren Y, Suzuki H, Golnoski KJ, Ahn HW, Mico V, et al. Transcription factors SOHLH1 and SOHLH2 coordinate oocyte differentiation without affecting meiosis I. J Clin Investig. 2017;127:2106–17.. doi: 10.1172/JCI90281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Yan Z, Qin Q, Nisenblat V, Chang HM, Yu Y, et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell. 2018;72:1021–34.e4. doi: 10.1016/j.molcel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 39.Monti M, Redi C. Oogenesis specific genes (Nobox, Oct4, Bmp15, Gdf9, Oogenesin1 and Oogenesin2) are differentially expressed during natural and gonadotropin-induced mouse follicular development. Mol Reprod Dev. 2009;76:994–1003. doi: 10.1002/mrd.21059. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Flores MA, Meng J, Zhang L, Zhao X, Rao MK, et al. MeT-DB: a database of transcriptome methylation in mammalian cells. Nucleic Acids Res. 2015;43:D197–203. [DOI] [PMC free article] [PubMed]
- 41.Grassi E, Mariella E, Lembo A, Molineris I, Provero P. Roar: detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinforma. 2016;17:423. doi: 10.1186/s12859-016-1254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livera G, Uzbekov R, Jarrier P, Fouchecourt S, Duquenne C, Parent AS, et al. Loss of oocytes due to conditional ablation of Murine double minute 2 (Mdm2) gene is p53-dependent and results in female sterility. FEBS Lett. 2016;590:2566–74. doi: 10.1002/1873-3468.12275. [DOI] [PubMed] [Google Scholar]
- 43.Freimer JW, Krishnakumar R, Cook MS, Blelloch R. Expression of alternative Ago2 isoform associated with loss of microRNA-driven translational repression in mouse oocytes. Curr Biol. 2018;28:296–302.e3. doi: 10.1016/j.cub.2017.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kugler JM, Lem C, Lasko P. Reduced cul-5 activity causes aberrant follicular morphogenesis and germ cell loss in Drosophila oogenesis. PLoS ONE. 2010;5:e9048. doi: 10.1371/journal.pone.0009048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachut M, Sood P, Levin Y, Moallem U. Proteomic analysis of preovulatory follicular fluid reveals differentially abundant proteins in less fertile dairy cows. J Proteom. 2016;139:122–9. doi: 10.1016/j.jprot.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Gurbuz F, Desai S, Diao F, Turkkahraman D, Wranitz F, Wood-Trageser M, et al. Novel inactivating mutations of the DCAF17 gene in American and Turkish families cause male infertility and female subfertility in the mouse model. Clin Genet. 2018;93:853–9. doi: 10.1111/cge.13183. [DOI] [PubMed] [Google Scholar]
- 47.Gache V, Waridel P, Winter C, Juhem A, Schroeder M, Shevchenko A, et al. Xenopus meiotic microtubule-associated interactome. PLoS ONE. 2010;5:e9248. doi: 10.1371/journal.pone.0009248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandormael-Pournin S, Guigon CJ, Ishaq M, Coudouel N, Ave P, Huerre M, et al. Oocyte-specific inactivation of Omcg1 leads to DNA damage and c-Abl/TAp63-dependent oocyte death associated with dramatic remodeling of ovarian somatic cells. Cell Death Differ. 2015;22:108–17. doi: 10.1038/cdd.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craig J, Orisaka M, Wang H, Orisaka S, Thompson W, Zhu C, et al. Gonadotropin and intra-ovarian signals regulating follicle development and atresia: the delicate balance between life and death. Front Biosci. 2007;12:3628–39. doi: 10.2741/2339. [DOI] [PubMed] [Google Scholar]
- 50.Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, et al. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell. 2012;23:3694–706. doi: 10.1091/mbc.E12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Do DV, Strauss B, Cukuroglu E, Macaulay I, Wee KB, Hu TX, et al. SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements. Cell Discov. 2018;4:33. doi: 10.1038/s41421-018-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601. [DOI] [PMC free article] [PubMed]
- 53.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci USA. 2012;109:E2215–22. [DOI] [PMC free article] [PubMed]
- 55.Allard P, Yang Q, Marzluff WF, Clarke HJ. The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Developmental Biol. 2005;286:195–206. doi: 10.1016/j.ydbio.2005.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mtango NR, Sutovsky M, Vandevoort CA, Latham KE, Sutovsky P. Essential role of ubiquitin C-terminal hydrolases UCHL1 and UCHL3 in mammalian oocyte maturation. J Cell Physiol. 2012;227:2022–9. doi: 10.1002/jcp.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balacco DL, Soller M. The m6A writer: rise of a machine for growing tasks. Biochemistry. 2019;58:363–78. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 58.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–4. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 59.Roundtree IA, He C. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Trends Genet. 2016;32:320–1. doi: 10.1016/j.tig.2016.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, supplementary table and figure legends
Data Availability Statement
The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus under accession ID GSE129714.