Abstract
Muscle disuse impairs muscle quality and is associated with increased mortality. Little is known regarding additive effects of multiple bouts of disuse, which is a common occurrence in patients experiencing multiple surgeries. Mitochondrial quality is vital to muscle health and quality; however, to date mitochondrial quality control has not been investigated following multiple bouts of disuse. Therefore, the purpose of this study was to investigate mitochondrial quality controllers during multiple bouts of disuse by hindlimb unloading. Male rats (n ~ 8/group) were assigned to the following groups: hindlimb unloading for 28 days, hindlimb unloading with 56 days of reloading, 2 bouts of hindlimb unloading separated by a recovery phase of 56 days of reloading, 2 bouts of hindlimb unloading and recovery after each disuse, or control animals with no unloading. At designated time points, tissues were collected for messenger RNA and protein analysis of mitochondrial quality. Measures of mitochondrial biogenesis, such as proliferator-activated receptor gamma coactivator 1 alpha, decreased 30%–40% with unloading with no differences noted between unloading conditions. Measures of mitochondrial translation were 40%–50% lower in unloading conditions, with no differences noted between bouts of unloading. Measures of mitophagy were 40%–50% lower with reloading, with no differences noted between reloading conditions. In conclusion, disuse causes alterations in measures of mitochondrial quality; however, multiple bouts of disuse does not appear to have additive effects.
Keywords: muscle, disuse, bed rest, mitochondrial biogenesis, mitochondrial dynamics, mitophagy
Résumé:
L’inactivité des muscles nuit à leurs qualités et est associée à une mortalité accrue. On sait peu de choses sur les effets additifs de plusieurs épisodes d’inactivité, un phénomène fréquent chez les patients subissant plusieurs chirurgies. La qualité mitochondriale est essentielle à la santé et à la qualité des muscles; cependant, à ce jour, aucun contrôle de la qualité mitochondriale n’est documenté après plusieurs épisodes d’inactivité. Ainsi, le but de cette étude est d’examiner les contrôleurs de qualité mitochondriale au cours de plusieurs épisodes d’inactivité, soit par le déchargement des membres postérieurs. Des rats mâles (n ~ 8/groupe) sont affectés aux groupes suivants : déchargement des membres postérieurs pendant 28 jours, déchargement des membres postérieurs et 56 jours de rechargement, 2 périodes de déchargement des membres postérieurs séparées par une phase de récupération de 56 jours de rechargement, 2 périodes de déchargement et de récupération des membres postérieurs après chaque inactivité, animaux témoins sans déchargement. À des moments donnés, des tissus sont prélevés pour l’analyse de l’ARN messager et des protéines de la qualité mitochondriale. Les mesures de la biogenèse mitochondriale, telles que PGC-1α, diminuent de 30 à 40 % avec le déchargement, et ce, sans différence observée entre les conditions de déchargement. Les mesures de la traduction mitochondriale sont réduites de 40 à 50 % dans les conditions de déchargement, sans différence observée entre les épisodes de déchargement. Les mesures de la mitophagie sont inférieures de 40 à 50 % avec le rechargement et aucune différence n’est observée entre les conditions de rechargement. En conclusion, l’inactivité suscite des altérations dans les mesures de la qualité mitochondriale; cependant, les multiples épisodes d’inactivité ne semblent pas avoir d’effets additifs. [Traduit par la Rédaction]
Mots-clés: muscle, inactivité, repos au lit, biogenèse mitochondriale, dynamique mitochondriale, mitophagie
Introduction
Skeletal muscle is one of the most metabolically active tissues in the human body (Kanehisa et al. 1994,1995) and alterations in skeletal muscle quality and function have significant implications for whole body health (Bell et al. 2016; Bye et al. 2017; Chacon-Cabrera et al. 2016; Kenny et al. 2017; Leitner et al. 2017; Peng et al. 2012; Phillips et al. 2017). Treatment of chronic diseases often requires multiple extended hospital visits, which leads to health consequences independent of the disease itself, such as skeletal muscle wasting (Bell et al. 2016; Bye et al. 2017; Kenny et al. 2017; Leitner et al. 2017; Peng et al. 2012). For example, traumatic wounds such as burn injuries may involve multiple hospital stays over an extended period of time (Sahin et al. 2011). Furthermore, astronauts often experience multiple extended sessions of unloading because of space flight. While the effects of mechanical unloading on skeletal muscle have been well-studied, to our knowledge, few studies have investigated if multiple occurrences of disuse exhibit additive detriments on muscle quality and function, which may have significant implications for patients with chronic diseases or traumatic injuries. Shimkus et al. (2018) recently reported that repeated bouts of hindlimb unloading does not change muscle mass and skeletal muscle protein synthesis; however, collagen content increased with subsequent bouts of disuse when compared with a single bout of disuse. However, to our knowledge, this model has not been utilized to evaluate other potential mediators of muscle quality and size, such as mitochondrial quality. Mitochondria are critical for muscle mass maintenance (Koves et al. 2008; Sandri et al. 2006), as such understanding alterations in mitochondrial quality control mechanisms during sequential bouts of disuse may be critical for effective patient management in those undergoing multiple surgical bouts.
Previous work has found mitochondrial dysregulation with disuse-induced muscle loss (Cannavino et al. 2015; Liu et al. 2014). As the primary energy and peroxide producers of the cell, mitochondrial aberrations can cause significant pathologies in skeletal muscle (Hyatt et al. 2019; Powers 2014; Powers et al. 2012). Mitochondria have multiple quality control processes to maintain a functioning network, including: mitochondrial biogenesis (the creation of new mitochondrial components), translation of mitochondrial encoded proteins (Lee et al. 2016b), mitochondrial dynamics (the fusion and fission of the mitochondrial network), and mitophagy (the removal of damaged mitochondria) (Yan et al. 2012). In many cases, dysregulated mitochondrial quality control can induce skeletal muscle atrophy (Oishi et al. 2008; Romanello and Sandri 2015; Sandri et al. 2006).
Skeletal muscle disuse causes disruptions to all of these mitochondrial quality control processes (Calvani et al. 2013). Proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α), the master regulator of mitochondrial biogenesis (Puigserver et al. 1998), has been reported to be downregulated during hindlimb immobilization (Oishi et al. 2008), which impairs the synthesis of new mitochondrial components and directly leads to muscle loss (Sandri et al. 2006). Mitochondrial dynamics are also disrupted during inactivity (Cannavino et al. 2015). Fission, mediated by mitochondrial regulatory signals (mitochondrial fission 1 protein (FIS1), dynamin-related protein 1 (DRP1), and mitochondrial fission factor (MFF)), is the removal of mitochondrial components from the mitochondrial network (typically for subsequent autophagic removal); whereas fusion of mitochondrial networks (mediated by mitofusion 1 (MFN1), mitofusion 2 (MFN2), and mitochondrial dynamin-like GTPase (OPA1)) allows for efficient sharing of mitochondrial components (Chan 2012; Twig et al. 2008). However, discoordination of the balance between fission/fusion events can lead to impaired muscle function. Specifically, prior work has found increased fission proteins (FIS1, DRP1, and MFF) (Romanello et al. 2010; Westermann 2010) and decreased fusion proteins (MFN1/2 and OPA1) (Westermann 2010) during disuse atrophy, which may directly contribute to muscle loss (Cannavino et al. 2015; Chen et al. 2010; Standley et al. 2017). Moreover, key proteins involved in autophagy and mitophagy (BCL2 interacting protein 3 (BNIP3), PTEN-induced putative kinase 1 (PINK1), and Parkin) are often dysregulated during skeletal muscle atrophy (Brown et al. 2017a; Kang et al. 2016), often leading to muscle dysfunction and reduced force output (Zhu et al. 2015). Finally, translation of mitochondrially encoded proteins (commonly referred to as mitochondrial translation) may have significant impact on mitochondrial adenosine triphosphate generation. The content of many of these regulators (mitochondrial translational initiation factor 2 (mtIF2), mitochondrial translational initiation factor 3 (mtIF3), Tu translation elongation factor mitochondrial (TuFM), and translational activator of cytochrome c oxidase I (Taco1)) are known to be disrupted during muscle and mitochondrial pathologies (Boczonadi and Horvath 2014; Lee et al. 2016b). However, to our knowledge the relative influence of multiple bouts of disuse atrophy on mitochondrial translation has yet to be investigated.
It is clear that mitochondrial quality regulators are imperative for overall muscle health and function. More so, these processes may largely influence the maintenance of skeletal muscle mass; however, if these processes are differentially altered during multiple bouts of unloading remains unknown and could largely influence patient outcomes. As such, understanding alterations in mitochondrial quality during multiple bouts of disuse may be critical for the development of effective therapeutics for muscle pathologies. Therefore, the purpose of this study was to examine the effects of multiple bouts of disuse and reuse on regulators of mitochondrial quality control using gastrocnemius muscle from our previous works (Shimkus et al. 2018; Shirazi-Fard et al. 2014).
Materials and methods
Animals and interventions
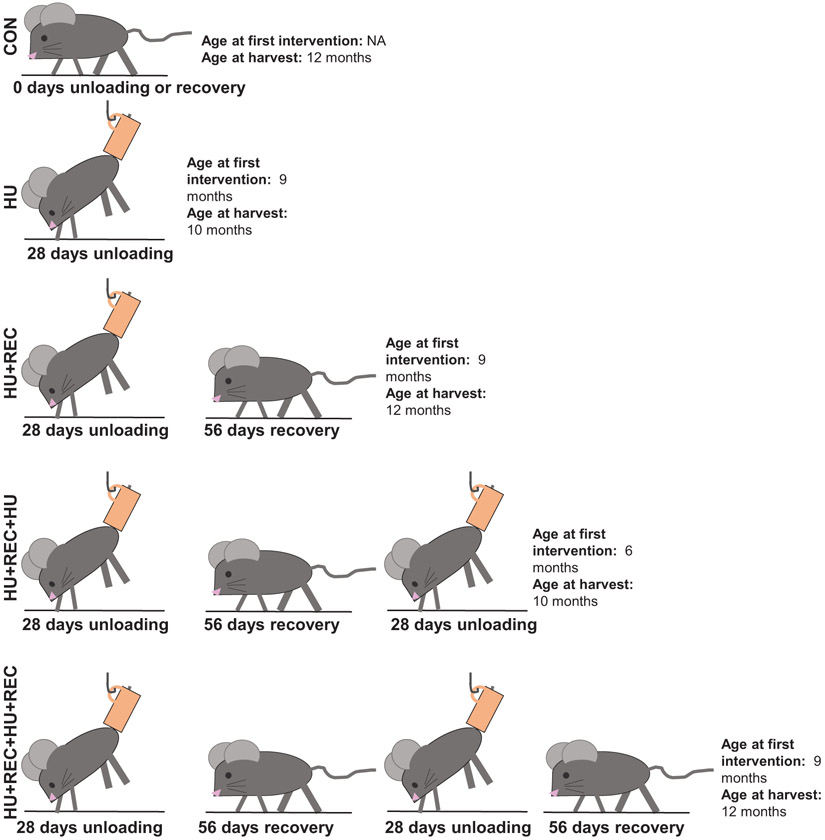
Experimental design has previously been described (Shimkus et al. 2018; Shirazi-Fard et al. 2013, 2014). Briefly, male Sprague–Dawley rats aged 5.5 months were assigned to the following groups: (i) cage control (CON), (ii) 28 days of hindlimb unloading (HU), (iii) 28 days HU followed by 56 days of recovery (HU+REC), (iv) 2 HU cycles of 28 days with 56 days of recovery in between unloading (HU+REC+HU), and (v) 2 alternating cycles of 28 days HU and 56 days recovery (HU+REC+HU+REC). All groups contained ~8 animals/group. A pictorial description of the experimental time course can be found in Fig. 1. Animal interventions were implemented to allow for animals to be harvested at approximately the same age (10 or 12 months as detailed in Fig. 1). While these animals were not all the same age at harvest, prior works using rats measuring similar outcomes have found no age-related differences in related measures in rats aged 10–12 month (Kimball et al. 2004). Animals were pair-fed during the first week of each HU period. At designated timepoints, animals were administered ketamine/xylazine via intraperitoneal injection, euthanized via exsanguination followed by cervical dislocation. At designated timepoints, hindlimb muscles were removed and snap frozen in liquid nitrogen for later biochemical analyses. All procedures and animal work described herein were approved by and conducted in accordance with the Institutional Animal Care and Use Committee of Texas A&M University (AUP no. 2008-93), and all animal experiments were performed at Texas A&M University.
Fig. 1.
Pictorial description of the experimental design. HU and HU+REC animals-initiated interventions at 9 months of age. HU+REC+HU- and HU+REC+HU+REC-initiated interventions at 6 months of age. HU and HU+REC+HU animals were harvested at 10 months of age. HU+REC, HU+REC+HU+REC, and CON animals were harvested at 12 months of age. NA, not applicable. See Materials and methods section for explanations of groups.
RNA isolation, complimentary DNA (cDNA) synthesis, and quantitative real-time polymerase chain reaction (PCR)
RNA isolation from gastrocnemius muscle, cDNA synthesis, and quantitative real-time PCR were performed as previously reported (Brown et al. 2017b; Greene et al. 2015; Lee et al. 2016b). Gastrocnemius muscles were chosen because of their mixed fiber-type phenotype, in this strain of male rats (Armstrong and Phelps 1984). More so, because disuse-atrophy appears to differently affect fiber types (Gao et al. 2018; Thomason et al. 1987), the gastrocnemius muscle provides a mixture of both atrophy-resistant and atrophyprone fibers. Additionally, because a majority of human skeletal muscle is composed of mixed fibers (Dahmane et al. 2005; Suter et al. 1993), as such, the gastrocnemius muscle allows for greater external validity for this model in relation to the human pathologies. SYBR Green primers were designed using Primer-BLAST through PubMed (Table 1). Custom primers were designed to produce amplicons between (Kimball et al. 2004) 70 and 150 bp with melting temperatures at 60 °C, forced to span exon–exon junctions and produced pairs with potential binding on unintended targets were eliminated from consideration. Amplicon products were separated on agarose gel and visualized using ethidium bromide under ultraviolet light to confirm the existence of a single band at the intended size; images were acquired using a customized gel documentation system with a Nikon Coolpix L820 digital camera.
Table 1.
Polymerase chain reaction (PCR) primer sequences for PCR analysis—primer pairs used for SYBR-green-based quantitative real-time reverse-transcription PCR.
| Target | Forward primer | Reverse primer | Tm (°C) | Amplicon length (bp) |
RefSeq ID no. |
|---|---|---|---|---|---|
| Nrf2 | TTTGTAGATGACCATGAGTCGC | CTCCATGTCCTGCTGTATGCT | 60 | 147 | NM_031789.2 |
| Tfam | TGCTAAGAACACTGGGGCG | TACAGATAAGGCTGACAGGCG | 60 | 84 | NM_031326.1 |
| Pparα | TGCGACATCATGGAACCCAA | GGCCGATCTCCACAGCAAAT | 60 | 113 | NM_013196.1 |
| Opa1 | CAGTTCAGAAGACCTCGCCA | GGTGTACCCGCAGTGAAGAA | 60 | 93 | NM_133585.3 |
| Mfn1 | ACATACAGGAACCCGGAACT | TTCTGTAGCCCTGTATCTCCAC | 59 | 70 | NM_138976.1 |
| Mfn2 | AGGCGATTTGAGGAGTGCAT | ACAATAAACCCGCTGCTCCT | 60 | 144 | NM_130894.4 |
| Fis1 p6 | ACGAAGCTGCAAGGAATTTTGA | AACCAGGCACCAGGCATATT | 60 | 98 | NM_001163243.1 |
| Drp1 | TCACCCGGAGACCTCTCATT | TGCTTCAACTCCATTTTCTTCTCC | 60 | 89 | NM_001025947.2 |
| Mff | AAGCGAAGAGATCCGAGCAG | CCTCTGACGCTCCTTCAACA | 60 | 146 | NM_001039015.2 |
| Bnip3 | AAGCGCACAGCTACTCTCAG | ACGCCTTCCAATGTAGATCCC | 60 | 147 | NM_053420.3 |
| mtIF2 | GCCAAGGTCCCCAGTTGTTA | CTGCCACTTGAGTTTCCCGA | 60 | 89 | NM_001004254.1 |
| mtIF3 | GGACAGCATGATTTGGACACC | TCTCATCCATCTCACTCCCG | 59 | 124 | NM_001115041.2 |
| Tufm | GCCTCCAGGGAAGGAACTTG | GTGCCAATGGTCTTGTTGCC | 60 | 126 | NM_001106295.1 |
| Taco1 | CTTAGAGGTGTGTCGCAGCA | CCACGACCCTCATACAGCAA | 60 | 102 | NM_001108302.1 |
Isolation of protein and immunoblotting
Immunoblotting was performed as we have previously described (Brown et al. 2018, 2015; Lee et al. 2016a; Perry et al. 2016; Rosa-Caldwell et al. 2017). The following antibodies were used to probe Western blot membranes: cytochrome c oxidase subunit IV (COXIV) (Cell Signaling Technology, Danvers, Mass., USA; 4844), voltage-dependent anion channel (VDAC) (Cell Signaling; 4661), PGC-1α (Santa Cruz Biotechnologies, Santa Cruz, Calif., USA; sc-13067), Parkin (Cell Signaling; 4211), phosphorylated (p)-Parkinser65 (Abcam, Cambridge, Mass., USA; ab154995), Pink1 (Santa Cruz; sc-517353), MFN1 (Santa Cruz; sc-50330), MFN2 (Santa Cruz; sc-50331), Fis1 (Santa Cruz; sc-48865), Opa1 (Santa Cruz; sc-367890), microtubule-associated proteins 1A/1B light chain 3A and 3B (LC3A/B) (also referred to as LC3II/I; Cell Signaling, 4108), BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3) (Cell Signaling; 3769), p62/sequestosome-1 (SQSTM1) (MilliporeSigma, St. Louis, Mo., USA; p0067).
Statistical analysis
To delineate differences between CON, HU, HU+REC, HU+REC+HU, and HU+REC+HU+REC, a 1-way ANOVA with preplanned contrasts was utilized (PROC GLM). These preplanned contrasts were chosen to investigate the specific research question regarding differences between multiple bouts of unloading and recovered conditions while maximizing power over a traditional 1-way ANOVA. More so, these contrast statements are orthogonal as previously described (Myers et al. 2010) and thereby providing the type I error control of a 1-way ANOVA while partitioning the comparisons. Specific comparisons included CON versus hindlimb unloading (HU combined; combining both the single (HU) and repeat (HU+REC+HU) bouts of hindlimb unloading) to compare how unloading overall compares to control (in the results section delineated as HU combined); 1 HU versus HU+REC+HU, to compare how multiple bouts of unloading compare to a singular unloading; CON versus recovery (recovery combined: combining both single (HU+REC) and repeat (HU+REC+HU+REC) bouts of recovery to investigate how recovery after unloading overall compares to CON (in the results section delineated as recovery combined); HU+REC versus HU+REC+HU+REC to investigate differences between bouts of recovery; and finally HU (combining both HU groups) versus recovery (combining both reloading groups) to investigate how unloading compares with recovery. Differences were considered statistically significant at p ≤ 0.05. All messenger RNA (mRNA) and protein data are presented relative to CON animals. For clarification, all mRNA targets are italicized and lowercase, while protein targets are not italicized and uppercase. All data were analyzed using the Statistical Analysis System (SAS; version 9.3, Cary, N.C., USA) and expressed as means ± SEM.
Results
Both a single bout of 28-day hindlimb unloading and multiple bouts of hindlimb unloading (HU+REC+HU) impair mitochondrial biogenesis, and PGC-1α protein content does recover with reloading
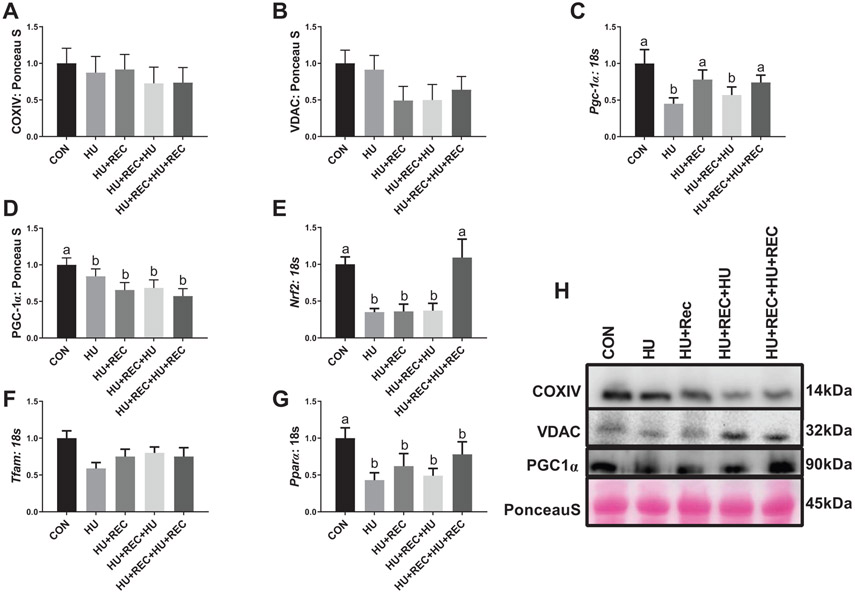
Phenotypic characteristics of these animals have been previously reported (Shimkus et al. 2018; Shirazi-Fard et al. 2014). In short, tissue weight of the gastrocnemius muscle was significantly and equally reduced by ~25% following either a single bout of hindlimb unloading or a repeat disuse period (HU+REC+HU) (HU combined) without differences between bouts (Shimkus et al. 2018; Shirazi-Fard et al. 2014). There were no differences noted among any groups on protein content of COXIV (Figs. 2A and 2H; p value range 0.4982–0.935). There were also no differences noted among groups on protein content of VDAC (Figs. 2B and 2H; p value range 0.153–0.970). Pgc-1α mRNA content was approximately 50% lower in HU combined compared with CON (Fig. 2C; p = 0.002), though no differences were noted between unloaded groups (Fig. 2C; p = 0.559). There was a nonsignificant mean decrease in Pgc-1α mRNA (p = 0.085) with recovery combined (HU+REC and HU+REC+HU+REC) compared with CON animals (Fig. 2C). There was no difference between recovery conditions (HU+REC and HU+REC+HU+REC) (Fig. 2C; p = 0.544), although HU combined had ~30% lower Pgc-1α mRNA content compared with recovery combined (Fig. 2C; p = 0.050). PGC-1α protein content was ~50% lower in HU combined compared with CON (Figs. 2D and 2H; p = 0.0418), and there were no differences between HU and HU+REC+HU (Fig. 2D; p = 0.151). More so recovery combined had ~40% lower PGC-1α content compared with CON (p = 0.030), though no differences were noted between these groups and HU groups, respectively (Fig. 2D; p = 0.950). Nrf2 mRNA content was reduced ~70% with HU combined (Fig. 2E; p = 0.0002), with no differences between these conditions (Fig. 2E; p = 0.913). Additionally, recovery combined were ~50% lower overall compared with CON (Fig. 2E; p = 0.0009); however, this appeared to be largely driven by differences between recovery conditions, with HU+REC having 70% less Nrf2 mRNA compared with HU+REC+HU+REC (Fig. 2E; p = 0.0231). Mitochondrial transcription factor A mRNA content was not different between any groups (Fig. 2F; p values ranging from 0.0922–0.696). Furthermore, Pparα mRNA content was ~50% lower in HU combined compared with CON (Fig. 2G; p = 0.0015), with no differences between recovery combined groups compared with CON (Fig. 2G; p = 0.602).
Fig. 2.
Immunoblot and rt-qPCR analysis for regulators of mitochondrial biogenesis and markers for mitochondrial content during bouts of hindlimb unloading or recovery. (A–G) rt-qPCR or immunoblot quantification. (H) Representative immunoblots. n of ~8 per group was utilized. Lettering denotes statistical significance at an α set at p < 0.05. See text for definitions of terms.
Hindlimb unloading dysregulates mitochondrial genome-specific translation machinery
There were no significant differences in mRNA content noted on mitochondrial translation initiation factor mtIf2 between conditions (Fig. 3A; p values range from 0.116–0.969). There was 50% lower mtIF3 mRNA content in HU animals compared with control (Fig. 3B; p = 0.0156). HU animals also had ~40% lower mtIF3 mRNA content compared with recovery animals; however, this did not reach statistical significance (p = 0.054; Fig. 3B) without any further differences between groups noted (p value range 0.418–0.667). HU combined rats had ~50% lower Tufm mRNA content compared with CON (Fig. 3C; p = 0.0085) with no differences noted between HU conditions (Fig. 2C; p = 0.7196). CON did not differ from recovery combined (Fig. 3C; p = 0.6032) and there were no differences between HU+REC and HU+REC+HU+REC (Fig. 3C; p = 0.6585). HU combined had ~40% lower mRNA content of Tufm compared with recovery conditions (Fig. 3C; p = 0.0067). Similarly, Taco1 mRNA content was ~50% lower in HU combined groups compared with CON (Fig. 3D; p = 0.0099) with no differences between HU and HU+REC+HU (Fig. 3D; p = 0.477), between CON and recovery combined (Fig. 3D; p = 0.228), between either HU+REC and HU+REC+HU+REC (Fig. 3D; p = 0.281) or recovery combined and HU combined conditions (Fig. 3D; p = 0.0692).
Fig. 3.
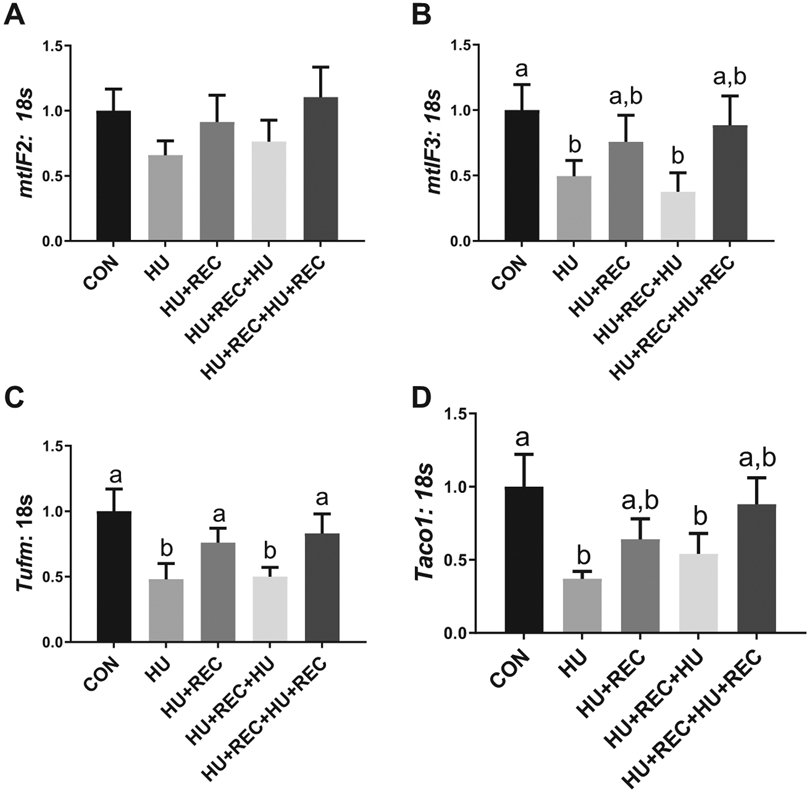
rt-qPCR analysis for translation initiation and elongation factors for proteins encoded in the mitochondrial genome. (A–D) rt-qPCR quantification. n of ~8 per group was utilized. Lettering denotes statistical significance at an alpha set at p < 0.05. See text for definitions of terms.
Mitochondrial fission is dysregulated following multiple bouts of disuse, though mitochondrial fusion does not appear affected by single or multiple bouts of disuse
There were no significant changes in the mRNA or protein content between groups of the regulators of mitochondrial fusion regulator Opa1 (Fig. 4A). For Mfn1, CON was not different from HU combined conditions (Fig. 4B; p = 0.989); however, this appears to be largely driven by differences in Mfn1 content between HU conditions, with HU+REC+HU having ~8.5-fold greater Mfn1 content compared with HU (Fig. 4B; p = 0.026). No further differences for Mfn1 were noted (Fig. 4B; p values ranging from 0.113–0.459). However, there was a nonsignificant mean increase in Mfn2 mRNA content in HU combined compared with recovery combined (p = 0.052; Fig. 4C). There were no other differences noted between groups (p values ranging from 0.087–0.776; Fig. 3C). For Drp1 mRNA content, CON compared with HU combined conditions approached statistical significance (p = 0.069; Fig. 4D), with an overall mean decrease of ~30% in HU combined conditions. There were no differences between HU versus HU+REC+HU conditions (p = 0.735; Fig. 4D), or CON versus recovery combined (p = 0.724; Fig. 4D) for Drp1 mRNA content. However, HU+REC+HU+REC had approximately 200% greater Drp1 mRNA content compared with HU+REC (p = 0.007; Fig. 4D). Additionally, HU combined and recovery combined conditions were different, with recovery combined having ~2-fold more Drp1 content compared with HU combined (p = 0.013; Fig. 4D), which appeared to be driven by HU+REC+HU+REC. Fis1 mRNA content was ~60% lower in HU groups compared with CON (Fig. 4E; p = 0.012), with no differences between HU and HU+REC+HU (Fig. 4E; p = 0.343). No other differences for Fis1 mRNA were noted (Fig. 4E; p value range 0.863–0.720). mRNA content of Mff was not altered between experimental groups (p values ranging from 0.0941–0.974; Figs. 4E and 4F).
Fig. 4.
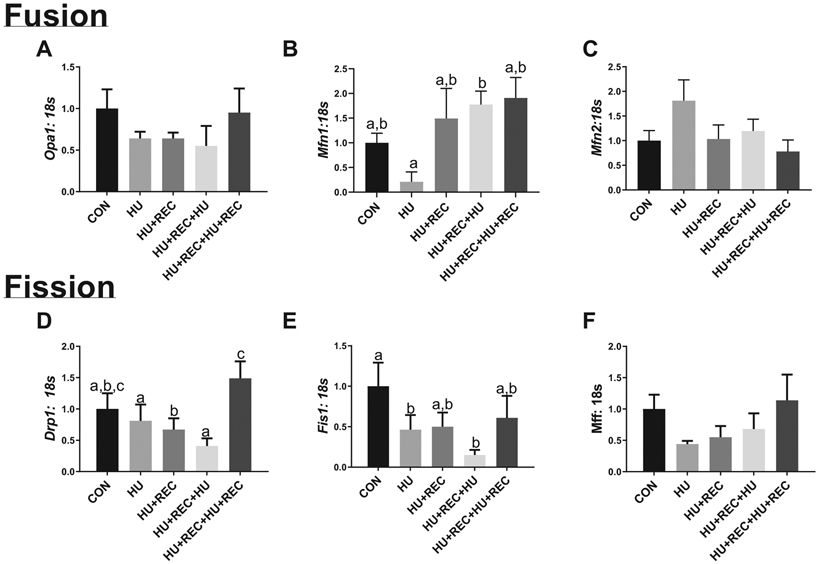
rt-qPCR analysis for regulators of the fission and fusion of mitochondrial membranes. (A–C) Regulators for fusion of mitochondrial membranes. (D–F) Regulators for fission of mitochondrial membranes. n of ~8 per group was utilized. Lettering denotes statistical significance at an α set at p < 0.05. See text for definitions of terms.
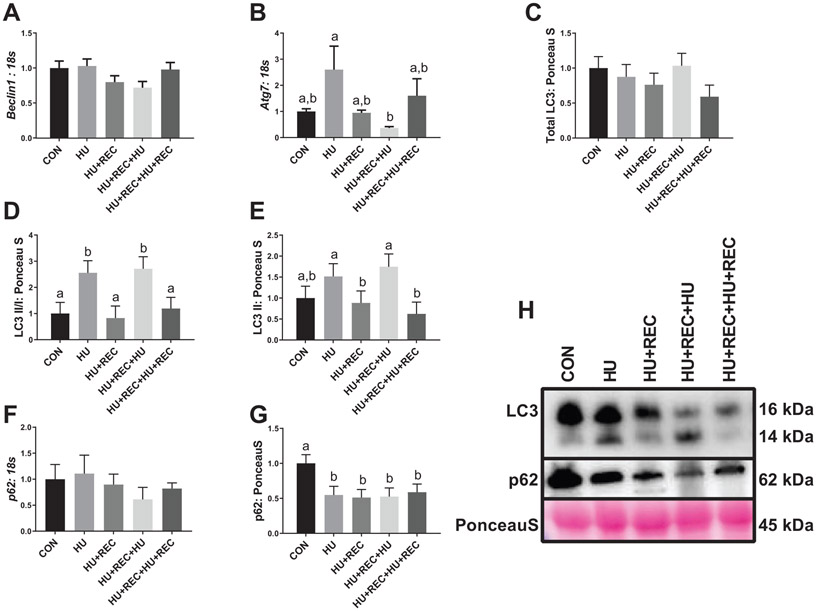
Disuse caused an increase in machinery for autophagosome initiation and clearance
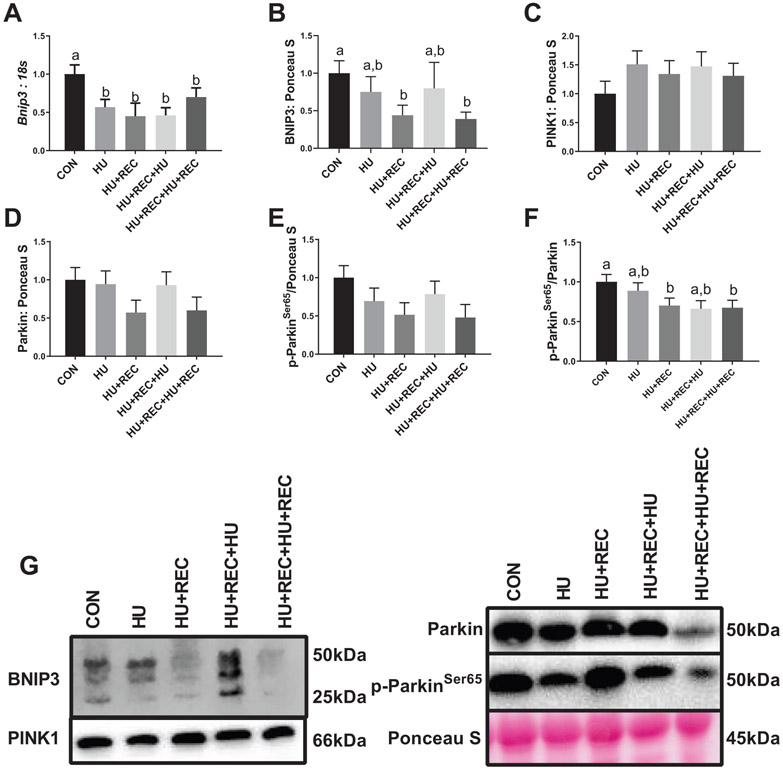
There was no difference between groups with regard to mRNA content of Beclin (p values ranging from 0.392–0.842; Fig. 5A). Regarding autophagy related 7 mRNA content, while HU combined conditions did not differ from CON (Fig. 5B; p = 0.264), HU was ~4-fold greater compared with HU+REC+HU (Fig. 5B; p = 0.0038), suggesting a divergent response between multiple bouts of unloading. Total LC3 protein content did not change across conditions (p values ranging from 0.077–0.458, Figs. 5C and 5H). LC3II/I ratio was approximately 2-fold greater in HU combined conditions compared with CON (Figs. 5D and 5H; p = 0.008) as well as approximately 2-fold greater compared with recovery combined conditions (Figs. 5D and 5H; p = 0.004), with no other differences noted between HU conditions (Figs. 5D and 5H; p = 0.561), recovery conditions (Figs. 5D and 5H; p = 0.871,) or recovery conditions compared with CON (Figs. 5D and 5H; p = 0.819). More so, LC3II itself demonstrated a similar pattern, though HU combined conditions compared with CON did not reach statistical significance (Figs. 5E and 5H; p = 0.052). However, recovery combined compared with HU combined conditions did reach statistical significance (Figs. 5E and 5H; p = 0.003), with no further differences noted (p values ranging 0.318–0.513; Figs. 5E and 5H). mRNA content of p62 was not different across any comparisons (p values ranging from 0.154–0.999, Fig. 5F). p62 protein content was ~50% lower in HU combined and recovery combined conditions compared with CON (Figs. 5G and 5H; p = 0.007 and p = 0.004, respectively). Regarding mitophagy, HU combined and recovery combined conditions had ~40% lower mRNA content of Bnip3 compared with CON (Fig. 6A; p = 0.047 and p = 0.046, respectively), with no other differences noted (Fig. 6A; p values ranging from 0.486–0.900). BNIP3 protein content was also ~50% lower in recovery combined conditions compared with CON (Figs. 6B and 6G; p = 0.028); however, no further differences were noted (p values ranging from 0.094–0.863; Figs. 6B and 6G). There were no differences in PINK1 protein content between any comparisons (p values ranging from 0.221–0.844; Figs. 6C and 6G). Also, no differences were noted for Parkin protein content between comparisons (p values ranging from 0.176–0.806; Figs. 6D and 6G). However, for phosphorylated-ParkinSer65, a mean decrease of ~40% was noted between recovery combined compared with CON; however, this did not reach statistical significance (p = 0.051; Figs. 6E and 6G). More so, a similar pattern was noted in p-ParkinSer65/Parkin ratios, and this difference between recovery combined and CON conditions reached statistical significance (Figs. 6F and 6G; p = 0.027). No other differences between p-Parkin or p-Parkin/Parkin ratios were noted (p values ranging from 0.113–0.860; Figs. 6E-6G).
Fig. 5.
Immunoblot and rt-qPCR analysis for regulators of autophagy. (A–G) rt-qPCR or immunoblot quantification. (H) Representative immunoblots. n of ~8 per group was utilized. Lettering denotes statistical significance at an α set at p < 0.05. See text for definitions of terms.
Fig. 6.
Immunoblot and rt-qPCR analysis for regulators of mitophagy. (A–F) rt-qPCR or immunoblot quantification. (G) Representative immunoblots. n of ~8 per group was utilized. Lettering denotes statistical significance at an α set at p < 0.05. See text for definitions of terms.
Discussion
Prior literature has reported that single bouts of disuse negatively affect mitochondrial quality control mechanisms (Cannavino et al. 2015; Chacon-Cabrera et al. 2016; Sandri et al. 2006); however, to our knowledge we are the first to investigate the effects of multiple bouts of unloading on these parameters. These data are clinically relevant for pathologies requiring multiple extended hospital visits or countless sedentary populations. The current study provides important initial data on the cellular signaling mechanisms occurring within skeletal muscle mitochondria exposed to multiple bouts of disuse. Our data demonstrate that disuse induced few differences between single and repeated bouts of unloading on many measures of mitochondrial quality; however, some markers of mitochondrial biogenesis appear to never fully recover following unloading and many markers of mitophagy appear altered during recovery following hindlimb unloading.
Mitochondrial content (measured by both COXIV and VDAC protein) did not change throughout multiple bouts of hindlimb unloading. Intriguingly, while mitochondrial content may not be affected from repeated bouts of disuse, PGC-1α (the master regulator of mitochondrial biogenesis (Puigserver et al. 1998) protein and mRNA along with other markers of mitochondrial biogenesis are disrupted after 28 days of unloading and do not appear to recover from 56 days of reloading. Overall, this discoordination between biogenesis and total mitochondrial content may suggest a reduction in mitochondrial turnover. Reduced mitochondrial turnover may then influence overall mitochondrial respiration; however, it is important to note we did not specifically measure mitochondrial respiration and future research would be needed to substantiate these claims. Additionally, disuse downregulated the mRNA content of mitochondrial genome specific translation activators and elongation factors Taco1 and Tufm, respectively. These enzymes are crucial for synthesizing protein subunits encoded in the mitochondrial genome that are vital for electron transport (Perez-Martinez et al. 2008). The influence of Taco1 and Tufm may be interesting future research avenues for disuse atrophy, as disruptions in electron transport appear to influence peroxide production in mitochondria (Alleman et al. 2014; Battersby and Richter 2013; Souto Padron de Figueiredo et al. 2015). Increased reactive oxygen species (ROS) production is currently one of the leading hypothesizes for the development of atrophy (Powers 2014; Powers et al. 2012); therefore, further investigations into upstream regulators of ROS production such as mitochondrial translation may be fruitful research avenues. Taken together, our data suggest multiple bouts of unloading decrease measures of mitochondrial biogenesis, without affecting content, and these alterations appear to persist even after other indices of skeletal muscle recovery (such as muscle mass (Shimkus et al. 2018)) have stabilized with reloading.
Overall, mitochondrial fusion s did not appear to be differentially altered by single or multiple bouts of hind limb unloading. However, mitochondrial fission, measured by Drp1 and Fis1 mRNA content, which showed a nonsignificant mean decrease with multiple bouts of unloading. DRP1 and FIS1 proteins are critical for the fission of mitochondria, so a reduction in the expression of these markers may lead to mitochondrial structural and functional dysregulation (Kornfeld et al. 2018). Dysregulated Drp1 content influences protein turnover by disrupting protein synthesis (Touvier et al. 2015), which is altered during unloading in these rats (Shimkus et al. 2018). More so, mitochondrial fission is also imperative for effective mitophagy, allowing for fission of dysfunctional mitochondrial components for mitophagy (Chan 2012). The pathological stimuli of HU may be sufficient to induce decreases in mitochondrial fission, which then may precipitate a decrease in overall mitophagy that we found in recovery conditions. However, we cannot delineate the specific role of altered fission expression on hindlimb unloading-mediated muscle loss at this time, as such, future investigations are warranted on the mechanistic influence of mitochondria fission on multiple bouts of disuse.
Our mRNA and protein data suggest autophagosomal formation and clearance are increased as a response to hindlimb unloading. We observed marked increases in LC3II/I ratio in HU and HU+REC+HU animals as well as an increase in LC3II protein content without any changes in total LC3 content, suggesting that disuse atrophy results in enhanced autophagosomal formation. Interestingly, this process does not appear to have any additive effects with multiple bouts of disuse, provided that sufficient recovery is given in between disuse bouts. More so, autophagosomal clearance, as measured by p62 protein (Klionsky et al. 2016), was increased in all experimental groups without any differences in p62 mRNA content. Our combined mRNA and protein data for p62 suggest the muscle cell is not altering the manufacture of p62 protein, but in fact degrading p62 at a higher rate through the autophagic process. Interestingly, this alteration affected all experimental groups and may suggest rates of autophagic clearance are still elevated during disuse recovery. It is entirely possible that the induction of autophagy machinery provides a protective effect such as the removal of damaged or misfolded proteins (Mammadova et al. 2019), which are elevated with hindlimb immobilization (Baehr et al. 2016). However, excessive autophagy induction can also play a large role in disuse-associated muscle wasting (Zhao et al. 2018), and excessive autophagy resolution may have unknown impacts on musculoskeletal health across the lifespan. More research is needed to fully understand the implications of autophagy resolution on long term muscular health.
We did not note any differences between multiple bouts of unloading on measures of mitophagy; however, we do note decreases in multiple markers of mitophagy, specifically BNIP3 (both mRNA and protein) and p-Parkinser65/Parkin ratio during recovery from hindlimb unloading. This finding contradicts prior research indicating increases in mitophagy machinery with hindlimb unloading (Kang and Ji 2016). These differences may be partially explained by differences of experimental design, where the prior study utilized much shorter time points (14 days of immobilization followed by 5 days of remobilization). As such it is plausible that mitophagy is increased shortly following reloading, potentially as a protective mechanism to remove swollen and/or dysfunctional mitochondria that are commonly noted in disuse atrophy (Grumati et al. 2010; Masiero et al. 2009). However, after sufficient reloading, mitophagy may then undergo a compensatory decrease to align with the reduced mitochondrial biogenesis that occurred during the disuse atrophy. This hypothesis would agree with prior work from our laboratory (Greene et al. 2015) establishing that PGC-1α regulates many aspects of mitochondrial quality control mechanisms, including mitophagy, and may suggest a temporal component for mitochondrial alterations during disuse atrophy and reloading. Importantly, this decrease in mitophagy may influence contractile function, despite mass being restored (Gouspillou et al. 2018; Zhang et al. 2013) and potentially be related to the increased collagen deposition noted in these animals (Shimkus et al. 2018).
In summary, the current study utilizes the hindlimb unloading model as a preclinical model to examine the effects of multiple bouts of extended disuse that may occur with imposed bedrest or hospital visits. This is the first study to investigate mitochondrial quality control regulators during disuse and after recovery from disuse, both with single and multiple bouts. We observed altered signaling of the mitochondrial regulatory processes (biogenesis, dynamics, and mitophagy), which provides important insight into the potential mechanisms behind mitochondrial degeneration that are observed in hindlimb unloading. Intriguingly, these data suggest that mitochondrial content is not altered during these stimuli; whereas, markers of mitochondrial biogenesis and mitochondrial breakdown are decreased with unloading and recovery, respectively, suggesting a possible temporal response of mitochondrial turnover during disuse and reloading and warranting further study. Regardless, alterations in mitochondrial turnover processes may influence the development of muscle pathologies and subsequent muscle quality (Kang and Li Ji 2012). Additionally, as prior research has clearly demonstrated differential responses of different fiber types to disuse atrophy (Gao et al. 2018; Thomason et al. 1987), more studies on repeated bouts of disuse using muscles with different fiber type compositions are likely warranted to determine if these responses are conserved across different muscle fibers. In conclusion, this is the first study to provide valuable data on mitochondrial alterations after multiple bouts of disuse and is an important first step in examining muscle pathologies present with repeated bouts of disuse.
Novelty.
Disuse atrophy causes multiple alterations to mitochondrial quality control.
With sufficient recovery most detriments to mitochondrial quality control are fixed.
In general, multiple bouts of disuse do not produce additive effects.
Les nouveautés.
L’atrophie d’inactivité entraîne de multiples altérations du contrôle de la qualité des mitochondries.
Une récupération suffisante permet une correction de la plupart des inconvénients du contrôle de la qualité des mitochondries.
En général, de multiples épisodes d’inactivité ne produisent pas d’effets additifs.
Acknowledgements
The authors would like to thank the Cell and Molecular Biology program at the University of Arkansas, Fayetteville, for supporting Jacob L. Brown’s graduate education and National Institutes of Health (NIH) Training Grant T32AG052363 for supporting Jacob L. Brown’s postdoctoral training. We would also like to extend our gratitude to the numerous other faculty, staff, and students of the Exercise Science Research Center at the University of Arkansas. Funded was provided by National Aeronautics and Space Administration (NASA) Grant Number NNX08AQ35G and NIH R15AR069913/AR/NIAMS.
Footnotes
Conflict of interest statement
The authors have no conflicts to disclose.
Contributor Information
Megan E. Rosa-Caldwell, Integrative Muscle Metabolism Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA..
Jacob L. Brown, Integrative Muscle Metabolism Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA..
Richard A. Perry, Jr., Exercise Muscle Biology Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA.
Kevin L. Shimkus, Muscle Biology Laboratory, Department of Health & Kinesiology, Texas A&M University, College Station, TX 77843, USA.
Yasaman Shirazi-Fard, Bone Biomechanics Laboratory, Department of Mechanical Engineering, Texas A&M University, College Station, TX 77843, USA..
Lemuel A. Brown, Exercise Muscle Biology Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA.
Harry A. Hogan, Bone Biomechanics Laboratory, Department of Mechanical Engineering, Texas A&M University, College Station, TX 77843, USA.
James D. Fluckey, Muscle Biology Laboratory, Department of Health & Kinesiology, Texas A&M University, College Station, TX 77843, USA.
Tyrone A. Washington, Exercise Muscle Biology Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA.
Michael P. Wiggs, Integrated Physiology and Nutrition Laboratory, Department of Health and Kinesiology, University of Texas at Tyler, Tyler, TX 75799, USA.
Nicholas P. Greene, Integrative Muscle Metabolism Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA; Exercise Muscle Biology Laboratory, Exercise Science Research Center, Department of Health, Human Performance and Recreation, University of Arkansas, Fayetteville, AR 72701, USA.
References
- Alleman RJ, Katunga LA, Nelson MA, Brown DA, and Anderson EJ 2014. The “Goldilocks Zone” from a redox perspective-Adaptive vs. deleterious responses to oxidative stress in striated muscle. Front. Physiol 5:358. doi: 10.3389/fphys.2014.00358. PMID:25278906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RB, and Phelps RO 1984. Muscle fiber type composition of the rat hindlimb. Am. J. Anat 171(3): 259–272. doi: 10.1002/aja.1001710303. PMID:6517030. [DOI] [PubMed] [Google Scholar]
- Baehr LM, West DWD, Marcotte G, Marshall AG, De Sousa LG, Baar K, and Bodine SC 2016. Age-related deficits in skeletal muscle recovery following disuse are associated with neuromuscular junction instability and ER stress, not impaired protein synthesis. Aging, 8(1): 127–146. doi: 10.18632/aging.100879. PMID:26826670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby BJ, and Richter U 2013. Why translation counts for mitochondria—retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J. Cell. Sci 126(Pt19): 4331–4338. doi: 10.1242/jcs.131888. PMID:24013545. [DOI] [PubMed] [Google Scholar]
- Bell KE, von Allmen MT, Devries MC, and Phillips SM 2016. Muscle disuse as a pivotal problem in sarcopenia-related muscle loss and dysfunction. J. Frailty Aging 5(1): 33–41. doi: 10.14283/jfa.2016.78. PMID:26980367. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, and Horvath R 2014. Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell Biol 48:77–84. doi: 10.1016/j.biocel.2013.12.011. PMID:24412566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Rosa-Caldwell ME, Lee DE, Blackwell TA, Brown LA, Perry RA, et al. 2017a. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J. Cachexia Sarcopenia Muscle, 8(6): 926–938. doi: 10.1002/jcsm.12232. PMID:28845591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Rosa-Caldwell ME, Lee DE, Brown LA, Perry RA, Shimkus KL, et al. 2017b. PGC-1alpha4 gene expression is suppressed by the IL-6-MEK-ERK 1/2 MAPK signalling axis and altered by resistance exercise, obesity and muscle injury. Acta Physiol. 220(2): 275–288. doi: 10.1111/apha.12826. [DOI] [PubMed] [Google Scholar]
- Brown JL, Lee DE, Rosa-Caldwell ME, Brown LA, Perry RA, Haynie WS, et al. 2018. Protein imbalance in the development of skeletal muscle wasting in tumour-bearing mice. J. Cachexia Sarcopenia Muscle, 9(5): 987–1002. doi: 10.1002/jcsm.12354. PMID:30328290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LA, Lee DE, Patton JF, Perry RA Jr., Brown JL, Baum JI, et al. 2015. Diet-induced obesity alters anabolic signalling in mice at the onset of skeletal muscle regeneration. Acta Physiol. 215(1): 46–57. doi: 10.1111/apha.12537. [DOI] [PubMed] [Google Scholar]
- Bye A, Sjøblom B, Wentzel-Larsen T, Gronberg BH, Baracos VE, Hjermstad MJ, et al. 2017. Muscle mass and association to quality of life in non-small cell lung cancer patients. J. Cachexia Sarcopenia Muscle, 8(5): 759–767. doi: 10.1002/jcsm.12206. PMID:28493418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvani R, Joseph AM, Adhihetty PJ, Miccheli A, Bossola M, Leeuwenburgh C, et al. 2013. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biol. Chem 394(3): 393–414. doi: 10.1515/hsz-2012-0247. PMID:23154422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavino J, Brocca L, Sandri M, Grassi B, Bottinelli R, and Pellegrino MA. 2015. The role of alterations in mitochondrial dynamics and PGC-1alpha overexpression in fast muscle atrophy following hindlimb unloading. J. Physiol 593(8): 1981–1995. doi: 10.1113/jphysiol.2014.286740. PMID:25565653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Cabrera A, Lund-Palau H, Gea J, and Barreiro E 2016. Time-course of muscle mass loss, damage, and proteolysis in gastrocnemius following unloading and reloading: implications in chronic diseases. PLoS One, 11(10): e0164951. doi: 10.1371/journal.pone.0164951. PMID:27792730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet 46:265–287. doi: 10.1146/annurev-genet-110410-132529. PMID:22934639. [DOI] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, and Chan DC 2010. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell, 141(2): 280–289. doi: 10.1016/j.cell.2010.02.026. PMID:20403324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane R, Djordjevič S, Simunic B, and Valenčič V 2005. Spatial fiber type distribution in normal human muscle histochemical and tensiomyographical evaluation. J. Biomech 38(12): 2451–2459. doi: 10.1016/j.jbiomech.2004.10.020. PMID:16214493. [DOI] [PubMed] [Google Scholar]
- Gao Y, Arfat Y, Wang H, and Goswami N 2018. Muscle atrophy induced by mechanical unloading: mechanisms and potential countermeasures. Front. Physiol 9: doi: 10.3389/fphys.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouspillou G, Godin R, Piquereau J, Picard M, Mofarrahi M, Mathew J, et al. 2018. Protective role of Parkin in skeletal muscle contractile and mitochondrial function. J. Physiol 596(13): 2565–2579. doi: 10.1113/JP275604. PMID:29682760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene NP, Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA, et al. 2015. Mitochondrial quality control, promoted by PGC-1alpha, is dysregulated by Western diet-induced obesity and partially restored by moderate physical activity in mice. Physiol. Rep 3(7): doi: 10.14814/phy2.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, et al. 2010. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med 16(11): 1313–1320. doi: 10.1038/nm.2247. PMID:21037586. [DOI] [PubMed] [Google Scholar]
- Hyatt H, Deminice R, Yoshihara T, and Powers SK 2019. Mitochondrial dysfunction induces muscle atrophy during prolonged inactivity: a review of the causes and effects. Arch. Biochem. Biophys 662: 49–60. doi: 10.1016/j.abb.2018.11.005. PMID:30452895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa H, Ikegawa S, Tsunoda N, and Fukunaga T 1994. Cross-sectional areas of fat and muscle in limbs during growth and middle age. Int. J. Sports Med 15(7): 420–425. doi: 10.1055/s-2007-1021081. PMID:8002122. [DOI] [PubMed] [Google Scholar]
- Kanehisa H, Yata H, Ikegawa S, and Fukunaga T 1995. A cross-sectional study of the size and strength of the lower leg muscles during growth. Eur. J. Appl. Physiol 72(1–2): 150–156. doi: 10.1007/BF00964130. PMID:8789586. [DOI] [PubMed] [Google Scholar]
- Kang C, and Ji LL 2016. PGC-1alpha overexpression via local transfection attenuates mitophagy pathway in muscle disuse atrophy. Free Radic. Biol. Med 93: 32–40. doi: 10.1016/j.freeradbiomed.2015.12.032. PMID:26746585. [DOI] [PubMed] [Google Scholar]
- Kang C, and Li, Ji L 2012. Role of PGC-1α signaling in skeletal muscle health and disease. Ann. NY. Acad. Sci 1271(1): 110–117. doi: 10.1111/j.1749-6632.2012.06738.x. PMID:23050972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Yeo D, and Ji LL 2016. Muscle immobilization activates mitophagy and disrupts mitochondrial dynamics in mice. Acta Physiol. 218(3): 188–197. doi: 10.1111/apha.12690. [DOI] [PubMed] [Google Scholar]
- Kenny HC, Rudwill F, Breen L, Salanova M, Blottner D, Heise T, et al. 2017. Bed rest and resistive vibration exercise unveil novel links between skeletal muscle mitochondrial function and insulin resistance. Diabetologia, 60(8): 1491–1501. doi: 10.1007/s00125-017-4298-z. PMID:28500394. [DOI] [PubMed] [Google Scholar]
- Kimball SR, O’Malley JP, Anthony JC, Crozier SJ, and Jefferson LS 2004. Assessment of biomarkers of protein anabolism in skeletal muscle during the life span of the rat: sarcopenia despite elevated protein synthesis. Am. J. Physiol. Endocrinol. Metab 287(4): E772–E780. doi: 10.1152/ajpendo.00535.2003. PMID:15187001. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy, 12(1): 1–222. doi: 10.1080/15548627.2015.1100356. PMID:26799652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld OS, Qvit N, Haileselassie B, Shamloo M, Bernardi P, and Mochly-Rosen D 2018. Interaction of mitochondrial fission factor with dynamin related protein 1 governs physiological mitochondrial function in vivo. Sci. Rep 8(1): 1062. doi: 10.1038/s41598-018-32228-1. PMID:30232469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. 2008. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7(1): 45–56. doi: 10.1016/j.cmet.2007.10.013. PMID:18177724. [DOI] [PubMed] [Google Scholar]
- Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA Jr., Wiggs MP, et al. 2016a. microRNA-16 is downregulated during insulin resistance and controls skeletal muscle protein accretion. J. Cell. Biochem 117(8): 1775–1787. doi: 10.1002/jcb.25476. PMID:26683117. [DOI] [PubMed] [Google Scholar]
- Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA, Washington TA, and Greene NP 2016b. Translational machinery of mitochondrial mRNA is promoted by physical activity in Western diet-induced obese mice. Acta Physiol. 218(3): 167–177. doi: 10.1111/apha.12687. [DOI] [PubMed] [Google Scholar]
- Leitner LM, Wilson RJ, Yan Z, and Gödecke A 2017. reactive oxygen species/nitric oxide mediated inter-organ communication in skeletal muscle wasting diseases. Antioxid. Redox. Signal 26(13): 700–717. doi: 10.1089/ars.2016.6942. PMID:27835923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Peng Y, Feng Z, Shi W, Qu L, Li Y, et al. 2014. Reloading functionally ameliorates disuse-induced muscle atrophy by reversing mitochondrial dysfunction, and similar benefits are gained by administering a combination of mitochondrial nutrients. Free Radic. Biol. Med 69: 116–128. doi: 10.1016/j.freeradbiomed.2014.01.003. PMID:24418157. [DOI] [PubMed] [Google Scholar]
- Mammadova N, Summers CM, Kokemuller RD, He Q, Ding S, Baron T, et al. 2019. Accelerated accumulation of retinal alpha-synuclein (pSer129) and tau, neuroinflammation and autophagic dysregulation in a seeded mouse model ofParkinson’s disease. Neurobiol. Dis 121:1–16. doi: 10.1016/j.nbd.2018.09.013. PMID:30218757. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. 2009. Autophagy is required to maintain muscle mass. Cell Metab. 10(6): 507–515. doi: 10.1016/j.cmet.2009.10.008. PMID:19945408. [DOI] [PubMed] [Google Scholar]
- Myers JL, Well AD, and Lorch RFJ 2010. Research Design and Statistical Analysis (3 ed.). New York, NY: Routledge. [Google Scholar]
- Oishi Y, Ogata T, Yamamoto KI, Terada M, Ohira T, Ohira Y, et al. 2008. Cellular adaptations in soleus muscle during recovery after hindlimb unloading. Acta Physiol. 192(3): 381–395. doi: 10.1111/j.1748-1716.2007.01747.x. [DOI] [PubMed] [Google Scholar]
- Peng P, Hyder O, Firoozmand A, Kneuertz P, Schulick RD, Huang D, et al. 2012. Impact of sarcopenia on outcomes following resection of pancreatic adenocarcinoma. J. Gastrointest. Surg 16(8): 1478–1486. doi: 10.1007/s11605-012-1923-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Funes S, Camacho-Villasana Y, Marjavaara S, Tavares-Carreon F, and Shingu-Vazquez M 2008. Protein synthesis and assemblyin mitochondrial disorders. Curr. Top. Med. Chem 8(15): 1335–1350. doi: 10.2174/156802608786141124. PMID:18991722. [DOI] [PubMed] [Google Scholar]
- Perry RA Jr., Brown LA, Lee DE, Brown JL, Baum JI, Greene NP, and Washington TA 2016. Differential effects of leucine supplementation in young and aged mice at the onset of skeletal muscle regeneration. Mech. Ageing Dev 157: 7–16. doi: 10.1016/j.mad.2016.05.007. PMID:27327351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SM, Dickerson RN, Moore FA, Paddon-Jones D, and Weijs PJ 2017. Protein turnover and metabolism in the elderly intensive care unit patient. Nutr. Clin. Pract 32(1suppl): 112s–120s. doi: 10.1177/0884533616686719. PMID:28388378. [DOI] [PubMed] [Google Scholar]
- Powers SK 2014. Can antioxidants protect against disuse muscle atrophy? Sports Med. 44(Suppl2): S155–S165. doi: 10.1007/s40279-014-0255-x. PMID:25355189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Smuder A, and Judge A 2012. Oxidative stress and disuse muscle atrophy: cause or consequence? Curr. Opin. Clin. Nutr. Metab. Care, 15(3): 240–245. doi: 10.1097/MCO.0b013e328352b4c2. PMID:22466926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell, 92(6): 829–839. doi: 10.1016/S0092-8674(00)81410-5. PMID:9529258. [DOI] [PubMed] [Google Scholar]
- Romanello V, and Sandri M 2015. Mitochondrial quality control and muscle mass maintenance. Front. Physiol. 6: 422. doi: 10.3389/fphys.2015.00422. PMID:26793123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, et al. 2010. Mitochondrial fission and remodelling contributes to muscle atrophy. Embo. J 29(10): 1774–1785. doi: 10.1038/emboj.2010.60. PMID:20400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Caldwell ME, Lee DE, Brown JL, Brown LA, Perry RA Jr., Greene E, et al. 2017. Moderate physical activity promotes basal hepatic autophagy in diet-induced obese mice. Appl. Physiol. Nutr. Metab 42(2): 148–156. doi: 10.1139/apnm-2016-0280. PMID:28084795. [DOI] [PubMed] [Google Scholar]
- Sahin I, Ozturk S, Alhan D, Açikel C, and Isik S 2011. Cost analysis of acute burn patients treated in a burn centre: the Gulhane experience. Ann. Burns Fire Disasters, 24(1): 9–13. PMID:21991233. [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, et al. 2006. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA, 103(44): 16260–16265. doi: 10.1073/pnas.0607795103. PMID:17053067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkus KL, Shirazi-Fard Y, Wiggs MP, Ullah ST, Pohlenz C, Gatlin DM, et al. 2018. Responses of skeletal muscle size and anabolism are reproducible with multiple periods of unloading/reloading. J. Appl. Physiol. (1985), 125(5): 1456–1467. doi: 10.1152/japplphysiol.00736.2017. [DOI] [PubMed] [Google Scholar]
- Shirazi-Fard Y, Anthony RA, Kwaczala AT, Judex S, Bloomfield SA, and Hogan HA 2013. Previous exposure to simulated microgravity does not exacerbate bone loss during subsequent exposure in the proximal tibia of adult rats. Bone, 56(2): 461–473. doi: 10.1016/j.bone.2013.07.004. PMID:23871849. [DOI] [PubMed] [Google Scholar]
- Shirazi-Fard Y, Metzger CE, Kwaczala AT, Judex S, Bloomfield SA, and Hogan HA. 2014. Moderate intensity resistive exercise improves metaphyseal cancellous bone recovery following an initial disuse period, but does not mitigate decrements during a subsequent disuse period in adult rats. Bone, 66: 296–305. doi: 10.1016/j.bone.2014.06.005. PMID:24929241. [DOI] [PubMed] [Google Scholar]
- Souto Padron de Figueiredo A, Salmon AB, Bruno F, Jimenez F, Martinez HG, Halade GV, et al. 2015. Nox2 mediates skeletal muscle insulin resistance induced by a high-fat diet. J. Biol. Chem 290(21): 13427–13439. doi: 10.1074/jbc.M114.626077. PMID:25825489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley RA, Distefano G, Pereira SL, Tian M, Kelly OJ, Coen PM, et al. 2017. Effects of beta-hydroxy-beta-methylbutyrate (HMB) on skeletal muscle mitochondrial content and dynamics, and lipids after 10 days of bed rest in older adults. J. Appl. Physiol. (1985), 123(5): 1092–1100. doi: 10.1152/japplphysiol.00192.2017. [DOI] [PubMed] [Google Scholar]
- Suter E, Herzog W, Sokolosky J, Wiley JP, and Macintosh BR 1993. Muscle fiber type distribution as estimated by Cybex testing and by muscle biopsy. Med. Sci. Sports. Exerc 25(3): 363–370. PMID:8455452. [PubMed] [Google Scholar]
- Thomason DB, Herrick RE, Surdyka D, and Baldwin KM 1987. Time course of soleus muscle myosin expression during hindlimb suspension and recovery. J. Appl. Physiol. (1985), 63(1): 130–137. doi: 10.1152/jappl.1987.63.1.130. [DOI] [PubMed] [Google Scholar]
- Touvier T, De Palma C, Rigamonti E, Scagliola A, Incerti E, Mazelin L, et al. 2015. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 6: e1663. doi: 10.1038/cddis.2014.595. PMID:25719247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo. J 27(2): 433–446. doi: 10.1038/sj.emboj.7601963. PMID:18200046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol 11(12): 872–884. doi: 10.1038/nrm3013. PMID:21102612. [DOI] [PubMed] [Google Scholar]
- Yan Z, Lira VA, and Greene NP 2012. Exercise training-induced regulation of mitochondrial quality. Exerc. Sport Sci. Rev 40(3): 159–164. doi: 10.1097/JES.0b013e3182575599. PMID:22732425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Davis C, Sakellariou GK, Shi Y, Kayani AC, Pulliam D, et al. 2013. CuZnSOD gene deletion targeted to skeletal muscle leads to loss of contractile force but does not cause muscle atrophy in adult mice. Faseb J. 27(9): 3536–3548. doi: 10.1096/fj.13-228130. PMID:23729587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YN, Li ZY, Kan K, and Su HT 2018. Mechanism of cell autophagy for regulating skeletal muscle wasting of rats after severe burns. Zhonghua Shao Shang Za Zhi, 34(2): 102–106. doi: 10.3760/cma.j.issn.1009-2587.2018.02.008. PMID:29973028. [DOI] [PubMed] [Google Scholar]
- Zhu M, Li X, Tian X, and Wu C 2015. Mask loss-of-function rescues mitochondrial impairment and muscle degeneration of Drosophila pink1 and parkin mutants. Hum. Mol. Genet 24(11): 3272–3285. doi: 10.1093/hmg/ddv081. PMID:25743185. [DOI] [PMC free article] [PubMed] [Google Scholar]