Abstract
High-grade serous carcinoma (HGSC) is the most common and deadliest ovarian cancer (OC) type, accounting for 70–80% of OC deaths. This high mortality is largely due to late diagnosis. Early detection is thus crucial to reduce mortality, yet the tumor pathogenesis of HGSC remains poorly understood, making early detection exceedingly difficult. Faithfully and reliably representing the clinical nature of human HGSC, a recently developed triple-knockout (TKO) mouse model offers a unique opportunity to examine the entire disease spectrum of HGSC. Metabolic alterations were investigated by applying ultra-performance liquid chromatography–mass spectrometry (UPLC–MS) to serum samples collected from these mice at premalignant, early, and advanced stages of HGSC. This comprehensive analysis revealed a panel of 29 serum metabolites that distinguished mice with HGSC from controls and mice with uterine tumors with over 95% accuracy. Meanwhile, our panel could further distinguish early-stage HGSC from controls with 100% accuracy and from advanced-stage HGSC with over 90% accuracy. Important identified metabolites included phospholipids, sphingomyelins, sterols, N-acyltaurine, oligopeptides, bilirubin, 2(3)-hydroxysebacic acids, uridine, N-acetylneuraminic acid, and pyrazine derivatives. Overall, our study provides insights into dysregulated metabolism associated with HGSC development and progression, and serves as a useful guide toward early detection.
Keywords: high-grade serous ovarian cancer, carcinoma, biomarkers, mouse model, untargeted metabolomics, ultra-performance liquid chromatography–mass spectrometry
Graphical Abstract
INTRODUCTION
Ovarian cancer (OC) is the most lethal gynecologic malignancy and the seventh most commonly diagnosed cancer among women in the world.1 The overall 5-year survival rate of OC is 47%.2 Due to the lack of specific symptoms, however, ovarian cancer is frequently diagnosed in late stages,3,4 where 5-year survival rates are only 29%.2 Among all the OC subtypes, high-grade serous carcinoma (HGSC), also known as high-grade serous ovarian cancer, is the most common and deadliest type, accounting for 70–80% of OC deaths.2,5,6 The high mortality of HGSC is largely attributable to advanced-stage diagnosis. Approximately 80% of HGSC cases are diagnosed at advanced stage (stage III or IV), in which the 5-year and 10-year survival rates are 32.1% and 15%, respectively. Though only about 20% of HGSCs are detected at early stage (stage I or II), the early-stage diagnosis of HGSC dramatically raises patient survival to 71.4% (5 year) and 53% (10 year).6,7 Hence, early detection of HGSC would be crucial to improving OC patients’ survival. Unfortunately, the cellular origin and tumor pathogenesis of HGSC still remain poorly understood, making early detection difficult.5,8,9
Presently, no diagnostic tests are available for detecting HGSC at an early stage among at-risk patients, let alone screening tests for asymptomatic women in the general population.6,10–13 The conventional strategy for OC risk evaluation includes trans-vaginal ultrasound and measurement of the serum tumor biomarker cancer antigen 125 (CA125) levels. However, this biomarker is of limited use, as serum elevations of CA125 can be observed in a number of different conditions unrelated to OC.14 In addition to CA125, there have been several biomarker-based tests approved by the FDA in ovarian cancer.15 However, these FDA-approved tests are neither diagnostic tests nor screening tests for OC. They are merely for referral purposes after ovarian tumor diagnosis is established. For example, the multivariate index assay OVA1 test is applied mainly to evaluate the likelihood of malignancy in women presenting an ovarian adnexal mass prior to surgery.15,16
Metabolomics has emerged as a promising tool for biomarker discovery leading to enhanced diagnostics as well as providing insight into the molecular underpinnings of disease biology.17 Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) are the two techniques that have been most used for profiling metabolic alterations associated with OC in serum,18–23 plasma,24–27 urine,28–31 tissue,32–34 and ovarian cyst fluid.35 Dysregulation in nucleotide, histidine, tryptophan, mucin,28 phospholipid, and piperidine metabolic pathways,24 fatty acid β-oxidation, and glycolysis33 have been associated with OC development and progression. The majority of these new metabolic markers, however, are non-HGSC related.
One major impediment to HGSC early detection is the lack of knowledge of early tumor development and progression. To better understand the pathogenesis of ovarian cancer, we developed two mouse models of HGSC: (1) a double-knockout (DKO) mouse (Dicer1flox/flox Ptenflox/flox Amhr2cre/+) by inactivating the Dicer1 and Pten genes and (2) a triple mutant (TKO) mouse (p53LSL‑R172H/+ Dicer1flox/flox Ptenflox/flox Amhr2cre/+) by adding a p53 mutation (R172H), which is equivalent to human p53-R175H mutant, one of the frequent p53 mutations found in human OC.36,37 As p53 mutations are observed in almost all human HGSC cases (96%),38,39 the TKO mouse model would be genetically closer to human HGSC than the DKO model we previously studied.20 These mice develop metastatic HGSC that faithfully model human HGSC phenotypically, histopathologically, and at the molecular level.37 Remarkably, HGSC from these mice reproduces the clinical metastasis of human HGSC in 100% of cases. In these models, HGSC originates and develops progressively in the fallopian tube, envelops the ovaries, and then aggressively metastasizes throughout the peritoneal cavity, including the omentum, diaphragm, mesentery, and peritoneal membrane. These extensive metastases invariably induce massive ascites and inevitably kill the mice (100%). These mouse models therefore present a rare and unique opportunity to study the entire disease spectrum of HGSC—from inception to early progression to metastasis—particularly early-stage progression.
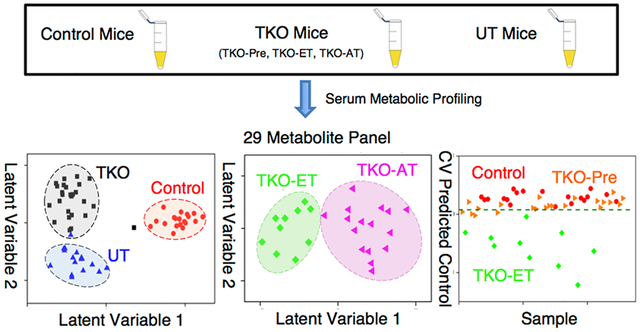
Here, using a “deep” ultra-performance liquid chromatography–mass spectrometry (UPLC–MS) approach, we characterize serum metabolic profiles of p53-Dicer1-Pten TKO mice at the premalignant stage, early stage, and advanced stage of HGSC. Among the identified are metabolites closely associated with HGSC, which discriminate HGSC from controls and from uterine tumors (UT) with over 95% accuracy, sensitivity, and specificity. The same metabolite panel is able to further distinguish early-stage HGSC from advanced-stage HGSC with over 90% accuracy, sensitivity, and specificity and distinguish early-stage HGSC against control samples with 100% sensitivity, specificity, and accuracy, respectively. The biological relevance of the differential metabolites is discussed, gaining insights into disease development and progression. This 29-feature panel enables effective detection of early stage as well as advanced-stage HGSCs, offering potential to diagnose HGSC at early stages.
MATERIALS AND METHODS
Chemicals
LC–MS-grade methanol, LC–MS-grade 2-propanol, LC–MS-grade acetonitrile, LC–MS-grade water, formic acid (99.5+%), ammonium acetate, and ammonium hydroxide were purchased from Fisher Chemical (Fisher Scientific International, Inc. Pittsburgh, PA) and used to prepare mobile phases and solutions.
Mice and Serum Sampling
In this study, triple mutant (TKO) mice (p53LSL‑R172H/+ Dicer1flox/flox Ptenflox/flox Amhr2cre/+) were generated by mating p53LSL‑R172H/+ Dicer1flox/flox Ptenflox/flox female mice with Dicer1flox/flox Ptenflox/flox Amhr2cre/+ male mice. Serum samples were collected from TKO mice at different stages of tumor progression: from premalignant stage to early stage to advanced stage. Precursor lesions are in the fallopian tubes at the premalignant stage, when no tumors are yet present. TKO mice with precursor (i.e., premalignant stage) lesions in the fallopian tubes are referred to as “TKO-Pre”, TKO-ET (early tumor) are TKO mice with early stage tumors in the fallopian tubes without evidence of metastasis, TKO-AT (advanced stage tumor) are TKO mice with ovarian and peritoneal metastases accompanied by ascites, and TKO-Ctrl are control mice (p53LSL‑R172H/+ Dicer1flox/flox Ptenflox/flox) that have the same genetic background as TKO mice but develop no tumors. All TKO mice developed high-grade serous carcinoma (HGSC, i.e., high-grade serous ovarian cancer). To enhance the selectivity for HGSC-specific metabolite markers, a tumor control group was also included. These were uterine tumor (UT) mice (p53LSL‑R172H/+ Ptenflox/flox Amhr2cre/+) that developed uterine tumors, but not HGSC.
Blood samples were collected via retro-orbital bleeding after anesthesia, from 22 TKO-Pre mice (average age, 2.0 months; age range, 1.3–3.1 months), 10 TKO-ET mice (4.8 months, 2.5–5.9 months), 16 TKO-AT mice (6.4 months, 4.3–10.1 months), 19 TKO-Ctrl mice (3.0 months, 3.0–8.2 months), and 17 UT mice (5.4 months, 3.7–6.8 months). Blood samples were centrifuged at 14 000 rpm for 5 min at room temperature, and serum was collected and stored at −80 °C until UPLC–MS analysis.
Sample Preparation
Serum samples were thawed on ice and subject to two different sample preparation protocols to obtain profiles of both nonpolar and polar submetabolomes. Reverse phase (RP) and hydrophilic interaction liquid chromatography (HILIC) UPLC–MS analysis in both positive and negative ion modes were combined for these different polarity metabolite fractions to obtain complementary and “deeper” metabolome information. Methanol (for polar) or 2-propanol (for non-polar) was added to a 50 μL serum sample in a 3:1 ratio to precipitate proteins. Samples were vortex-mixed for 10 s and centrifuged at 13 000 rpm for 7 min. Then, 150 μL of supernatant was frozen at −80 °C for UPLC–MS analysis. A sample blank was prepared with 50 μL of LC–MS-grade water, and a pooled quality control (QC) sample was created by mixing a 10 μL aliquot of each serum sample. Both the sample blank and the pooled sample were processed with the same procedure as the murine serum samples. Samples were run in randomized order on consecutive days. Solvent blanks and sample blanks were analyzed together with murine serum samples. QC samples were analyzed every 12 runs to assess UPLC–MS system stability and were used to compensate for time-dependent batch effects with a QC-based regression curve for each detected compound.
UPLC–MS Analysis
Chromatography was performed with an Ultimate 3000 UPLC (Thermo Fisher Scientific, Inc., Waltham, MA) system equipped with a Waters ACQUITY UPLC BEH C18, 2.1 × 50 mm, 1.7 μm particle column or a Waters ACQUITY UPLC BEH HILIC, 2.1 × 75 mm, 1.7 μm particle column. A Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) was used in all cases. For reverse-phase (RP) separations, mobile phase A was water/ acetonitrile (40:60 v/v), and mobile phase B was acetonitrile/ 2-propanol (10:90 v/v). Both mobile phases included 10 mM ammonium formate and 0.1% formic acid additives to improve peak shape and ionization efficiency. For hydrophilic interaction chromatography (HILIC) separations, mobile phase A was water/acetonitrile (95:5 v/v), 10 mM ammonium acetate, and 0.05% ammonium hydroxide. Mobile phase B was acetonitrile with 0.05% ammonium hydroxide. Chromatographic gradients are described in Table S1 of the Supporting Information (SI). The column temperature was 55 °C, while samples were maintained at 5 °C in the autosampler. Injection volumes of 5 and 2 μL were used in RP and HILIC methods, respectively. RP and HILIC chromatographies were performed both in positive and negative ion modes, and MS parameters can be found in the Supporting Information (Table S2).
For metabolite identification purposes, the top five data-dependent acquisition (DDA) experiments were used to collect MS/MS spectra using a stepped normalized collision energy (NCE) of 10, 30, and 50 V. For compounds missed by DDA, parallel reaction monitoring (PRM) experiments were performed at collision energies ranging from 10 to 40 V to obtain fragmentation data for identification purposes.
Data Processing
Spectral features were extracted from the raw data using Compound Discoverer v2.1 software (Thermo Fisher Scientific, Inc., Waltham, MA). This procedure included chromatographic alignment, peak picking, peak area integration, and QC-based compound area normalization. Features that eluted with the chromatographic solvent front with retention times <0.5 min in RP data sets and <0.9 min in HILIC data sets were considered unreliable due to potential ion suppression effects40 and were thus removed. Further filtering was carried out by removing features that were not present in 50% of at least one of the serum sample groups at 10 times the baseline abundance, defined as the peak area of the sample blank run. Welch’s t test with a Benjamini Hochberg correction was applied to TKO vs TKO-Ctrl mouse groups and to TKO vs UT mouse groups. The union of differential features from both control and UT comparisons was chosen for down-selecting HGSC-specific features. The most useful set of features for classification was produced with genetic algorithms41 (GAs, MATLAB R2016a, The MathWorks, Natick, MA, with PLS_Toolbox v.8.1.1, Eigenvector Research, Inc., Wenatchee, WA). GAs are evolutionary algorithms that generate solutions to optimization problems. They have been widely used in variable selection due to their high performance in large-scale variable selection rate and classification accuracy.41 Parameters for GA variable selection are provided in the Supporting Information (Table S3). After the GA variable selection process, features were chosen on the basis of frequency criteria. Principal component analysis (PCA) and orthogonal partial least square discriminant analysis (oPLS-DA)42 were performed to assess the discriminating power of metabolite data sets in nonsupervised and supervised manners, respectively. Data were preprocessed by autoscaling prior to PCA and oPLS-DA analysis and cross-validated using 10 iterations of random sample subsets. Data generated in this work are available through the NIH Metabolomics Workbench (http://www.metabolomicsworkbench.org/) with project ID PR000784 (doi 10.21228/M8BH6F, study ID ST001172).
Discriminant Feature Identification
Metabolite identification was attempted for the panel of best discriminant features. Mass spectral ion adduct analysis was first performed to ensure the unambiguous assignment of the feature of interest in each mass spectrum. Elemental formulas were then generated on the basis of exact masses with a maximum mass error of 10 mDa and isotopic patterns using Compound Discoverer v2.1. The elements included in the elemental formula search were constrained to C, H, N, O, P, and S. Tentative identities were searched against the human metabolome database (HMDB),43 Lipid Maps database,44 and Metlin database45 using both the accurate mass and generated elemental formulas with a mass error of 10 mDa. Tandem MS databases such as Metlin, mzCloud,46 and MassBank47 were used together with literature searches to further confirm the identity of the metabolite candidates for which tandem MS/ MS data were successfully acquired. Fragmentation patterns were also manually analyzed in a few cases to distinguish between different isobaric species.
RESULTS AND DISCUSSION
Multivariate Classification Performance
A data set comprising a total of 5937 spectral features that were above background and chromatographically retained was produced by combining the RP ESI(+), RP ESI(−), HILIC ESI(+), and HILIC ESI(−) data sets. Initially, the extent by which UPLC–MS metabolic profiling could differentiate serum samples of 19 TKO-Pre, 10 TKO-ET, and 16 TKO-AT mice, against 22 TKO-ctrl and 17 UT mice was investigated. An unsupervised PCA exploration of these data sets showed large overlap between TKO-Pre and TKO-Ctrl mice (Figure 1), indicating that, as expected, only subtle differences existed between the two groups because TKO-Pre mice had not yet developed any tumors and were phenotypically closer to the healthy controls. Thus, for the following data analysis, the TKO mouse group is a combination of TKO-ET and TKO-AT mice.
Figure 1.
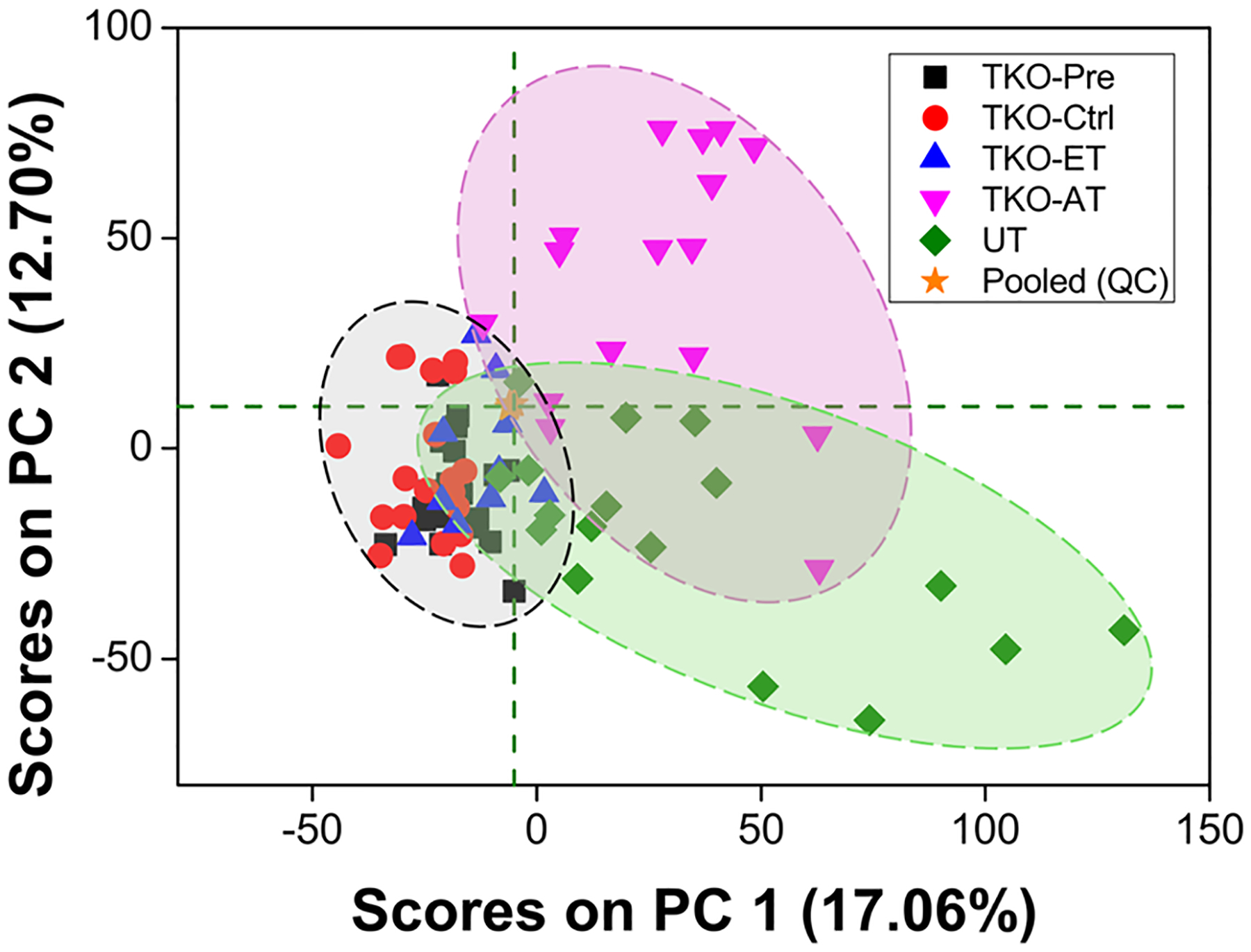
PCA score plot including all sample groups using the total initial set of 5937 spectral features. The clustering of samples in this plot reveals clear separation between TKO-ET mice from TKO-AT mice and UT mice, a moderate separation between TKO-ET mice and TKO-Ctrl mice with some overlap, and complete overlap of TKO-Pre and TKO-Ctrl mice. Pooled QC samples, represented by orange stars, clustered toward the center of the plot, indicating that the technical variance was minimal.
The set of 5937 features was utilized to build an oPLS-DA model that distinguished serum samples from TKO mice against TKO-Ctrl and UT mice. Performance characteristics of this model (Figure 2A,B) were 88.1, 92.8, and 90.4% for the cross-validated sensitivity, specificity, and accuracy, respectively. Five serum samples were systematically misclassified, including two TKO-ET mice with ID No. 42 and 51, one TKO-AT mouse labeled No. 62, and two UT mice labeled No. 69 and 78. Sample cohort information can be found in the Supporting Information. The model employed a total of four latent variables and interpreted 42.1% and 54.48% variance from the X (feature peak area) and Y (sample class membership) blocks, respectively. Although this model’s performance was acceptable, GA feature selection was used to obtain a smaller, more robust, metabolic feature set that could better discriminate TKO mice from TKO-Ctrl and UT mice. GA variable selection led to a 29-feature panel (Figure 2C,D) with 96.2, 97.2, and 96.7% cross-validated sensitivity, specificity, and accuracy, respectively. In this case, only two serum samples were systematically misclassified. The two misclassified samples were collected from a TKO-ET mouse labeled No. 42 and a UT mouse labeled No. 68. The TKO-ET mouse was repeatedly misclassified as a TKO-Ctrl mouse. This could be due to the age of the mouse, 2.5 months, when the sample was collected, which was much younger than the rest of the TKO-ET mice (average age 4.8 months) and close to the average age of the TKO-Pre mice (average age 2.0 months). As TKO-Pre mice were phenotypically closer to the TKO-Ctrl mice, this may explain why the young TKO-ET mouse was classified as a TKO-Ctrl mouse in the models. The three latent variable model interpreted 58.7% and 56.1% variance from the X and Y blocks, respectively. Binary comparisons (TKO vs TKO-Ctrl, TKO vs UT) were also performed (Figure 3A,B), indicating that the 29-feature panel was strongly HGSC-specific and could differentiate TKO mice from control and UT mice with high performance at the same time. In these cases, the classification accuracy ranged from 95.3 to 98.1%, with a cross-validated sensitivity of 96.2–96.5% and specificity of 94.1–100%. PCA was utilized to further evaluate the discriminant performance of the 29-feature panel in an unsupervised manner and rule out any overfitting. Score plots were generated for both the initial 5937-feature set and the selected 29-feature panel (Figure S1, SI). Good group clustering and increased variance captured were observed with the 29-feature panel, even in an unsupervised fashion.
Figure 2.
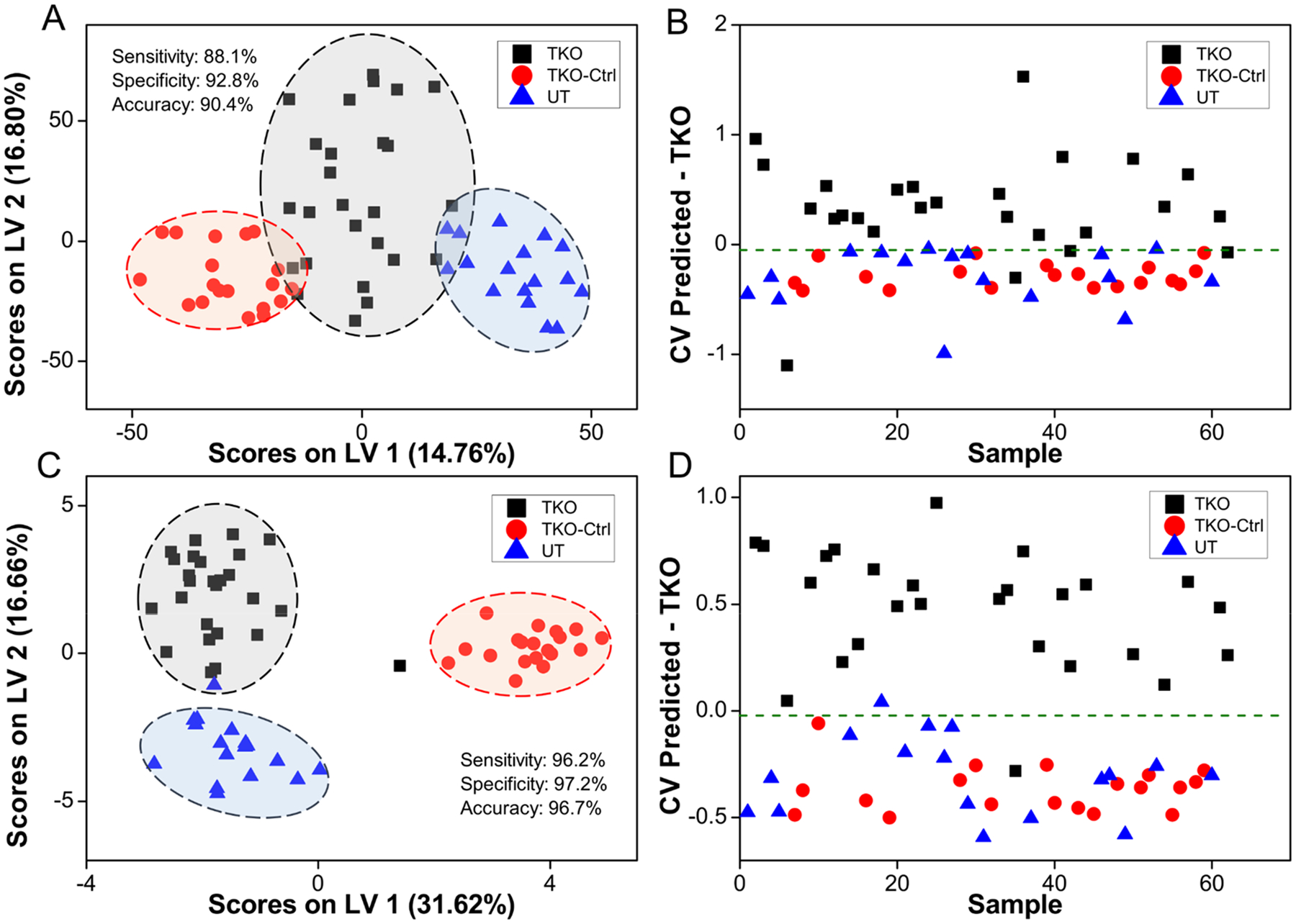
(A) oPLS-DA score plot depicting clustering of samples using the initial set of 5937 spectral features. (B) oPLS-DA cross-validated classification plot using 5937 spectral features. The x-axis represents randomized sample number, and the y-axis represents the cross-validated predicted scores of the oPLS-DA classification model. (C) oPLS-DA scores plot using the GA-selected 29-feature panel. (D) oPLS-DA cross-validated classification plot using the 29-feature panel. TKO, TKO-Ctrl, and UT samples are represented by black squares, red circles, and blue triangles, respectively. The threshold for sample classification is represented by the green dashed line. Cross-validated sensitivity, specificity, and accuracy values are given for each model. TKO samples are a combination of TKO-ET and TKO-AT samples.
Figure 3.
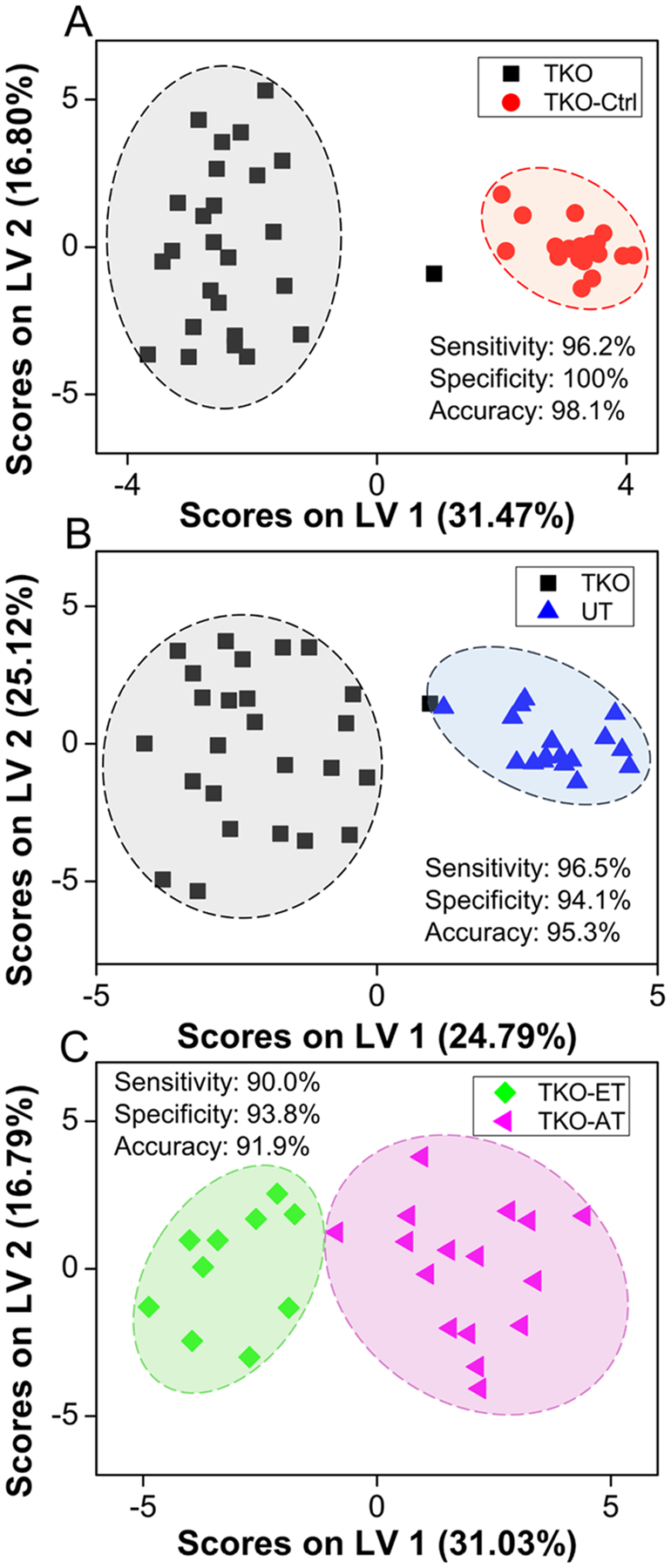
oPLS-DA scores plot depicting clustering of samples between (A) TKO and TKO-Ctrl samples, (B) TKO and UT samples, and (C) TKO-ET and TKO-AT samples using the 29-feature panel. Variance between classes is captured across the x-axis. TKO, TKO-Ctrl, UT, TKO-ET, and TKO-AT samples are represented by black squares, red circles, blue triangles, green diamonds, and pink triangles, respectively. Cross-validated sensitivity, specificity, and accuracy values are given for each model. TKO samples are TKO-ET and TKO-AT samples combined.
As accurate staging and early-stage-disease detection greatly improve clinical outcome, we investigated the performance of the 29-feature panel in discriminating early and late stages of HGSC. This panel of 29 metabolites distinguished TKO-ET from TKO-AT mice with 90.0%, 93.8%, and 91.9% for the cross-validated sensitivity, specificity, and accuracy, respectively (Figure 3C). Permutation tests with 2000 iterations returned a p-value of 0.017 measured using group separation distance,48 avoided overfitting, and further validated the model. We further explored the potential of the 29-feature panel in disease early detection. The oPLS-DA model successfully discriminated TKO-ET against TKO-Ctrl mice with 100% cross-validated sensitivity, specificity, and accuracy, respectively. This panel, however, was indistinguishable between precursors and controls. We used the 22 precursor (TKO-Pre) samples as an unknown sample set, and input into this classification model, 18 out of 22 TKO-Pre samples were predicted as being similar to TKO-Ctrl samples (Figure 4), echoing our previous statement that the TKO-Pre mice were phenotypically closer to the TKO-Ctrl mice. Nevertheless, this 29-feature panel enabled effective detection of early stage as well as advanced-stage HGSCs, offering potential to diagnose HGSC at early stage.
Figure 4.
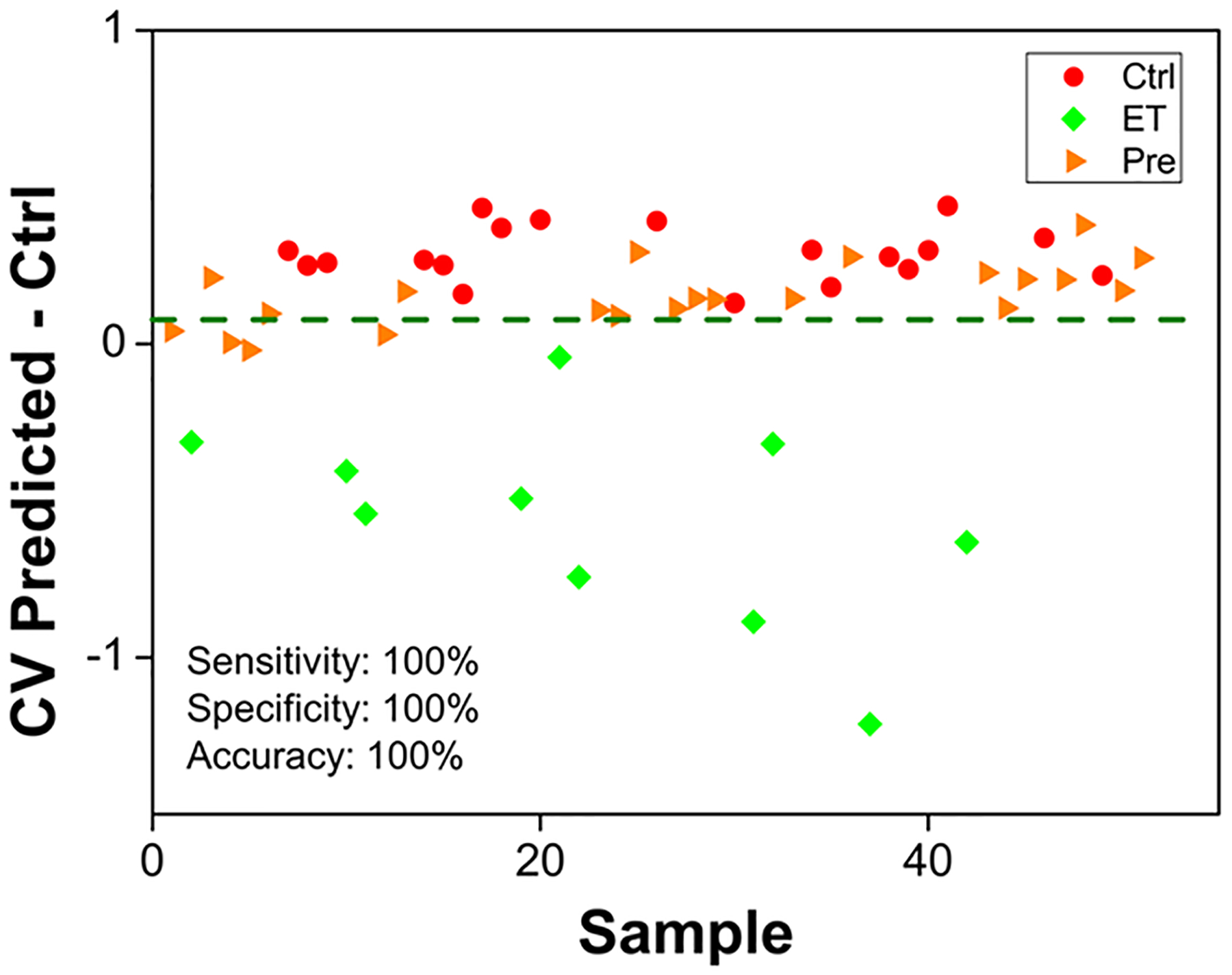
oPLS-DA cross-validated classification plot between ET and Ctrl samples using the 29-feature panel. The x-axis represents the randomized sample number, and the y-axis represents the cross-validated predicted scores of the binary oPLS-DA classification model. ET and Ctrl samples are represented by green diamonds and red circles, respectively. Precursor (Pre) samples projected into the model are represented by orange triangles. The threshold for sample classification is represented by the green dashed line. Cross-validated sensitivity, specificity, and accuracy values are given for the model.
Discriminant Metabolite Identification
Following multivariate analysis, metabolite identification was carried out for the species in the 29-feature panel. Twenty-four of the 29 metabolic features were identified by both high-resolution MS and MS/MS (Table 1). Table S4 (SI) provides detailed MS/MS fragmentation information and confidence level for each identified species. Metabolites in the panel included lipids, oligopeptides, and other small molecules, such as bilirubin, uridine, hydroxysebacic acids, and N-acetylneuraminic acid (NeuAc). For features that did not yield informative MS/MS information, tentative assignments were made on the basis of high-resolution MS and isotopic relative ion abundances only (Table 1, italicized) and should be considered tentative until further research is pursued.
Table 1.
Annotation of Metabolites in the 29-Feature Panel, Including the Acquisition Mode, Retention Time, Observed Exact Mass and Mass Error, Theoretical Mass, Predicted Elemental Formula, Observed Adduct, p-Value of Abundances between TKO and TKO-Ctrl Samples and between TKO and UT Samples, and Fold Changes
| m/z | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feature no. | Acquisition mode | retention time (min) | expl | theor | Elemental formula | adduct type | mass error (ppm) | Fold changea | p-valueb HGSC vs control/HGSC vs UT | metabolite identityc |
| 208 | HILIC ESI(+) | 4.96 | 109.0765 | 109.0760 | C6H8N2 | [M + H]+ | 4.58 | −0.19 | 0.003 47/8.45 × 10−7 | dimethypyrazine, ethylpyrazine, 2-picolylamine |
| 202 | HILIC ESI(+) | 2.67 | 271.1151 | 271.1149 | C9H14N6O4 | [M + H]+ | 0.74 | 1.08 | 0.000 261/1.52 × 10−6 | – |
| 385 | HILIC ESI(−) | 1.65 | 516.7524 | – | – | [M + Br]− | – | 0.68 | 0.001 16/2.86 × 10−6 | – |
| 277 | RP ESI(−) | 1.02 | 583.2576 | 583.2562 | C33H36N4O6 | [M − H]− | 2.40 | −0.88 | 1.10 × 10−7/5.57 × 10−6 | bilirubin |
| 494 | HILIC ESI(−) | 1.58 | 514.7545 | – | – | [M + Br]− | – | 1.33 | 0.000 762/2.51 × 10−5 | – |
| 363 | HILIC ESI(+) | 2.21 | 398.2022 | 398.2034 | C17H27N5O6 | [M + H]+ | −3.01 | 0.80 | 0.001 48/3.13 × 10−5 | Glu-His-Leu Glu-His-He |
| 427 | RP ESI(+) | 8.15 | 693.5567 | 693.5581 | C47H74O2 | [M + Na]+ | −2.02 | 1.26 | 0.001 04/0.000 112 | CE(20:5) |
| 303 | RP ESI(+) | 1.42 | 552.4015 | 552.4024 | C28H58NO7P | [M + H]+ | −1.63 | 0.93 | 0.005 46/0.000 114 | LysoPC(20:0) |
| 751 | HILIC ESI(+) | 1.28 | 696.4993 | 696.5012 | C76H144O17P2 | [M + 2H]2+ | −2.73 | 0.57 | 0.0116/0.000 124 | CL(67:2) |
| 37 | HILIC ESI(−) | 2.20 | 217.1075 | 217.1081 | C10H18O5 | [M − H]− | −2.76 | −0.42 | 0.006 97/0.000 137 | 2(3)-hydroxysebacic acid |
| 572 | HILIC ESI(+) | 4.56 | 454.8905 | – | – | [M + H]+ | – | −1.50 | 9.44 × 10−7/0.000 149 | – |
| 651 | HILIC ESI(−) | 1.27 | 281.0358 | 281.0360 | C9H12N2O6 | [M + Cl]− | −0.71 | −0.22 | 0.009 34/0.000 351 | uridine (isotopic peak) |
| 557 | HILIC ESI(−) | 3.22 | 308.0988 | 308.0987 | C11H19NO9 | [M − H]− | 0.32 | 0.94 | 0.003 16/0.000 360 | NeuAc |
| 88 | RP ESI(+) | 1.22 | 607.2522 | 607.2527 | C33H36N4O6 | [M + Na]+ | −0.82 | −1.10 | 1.34 × 10−6/0.000 491 | bilirubin |
| 274 | RP ESI(+) | 1.25 | 540.4465 | 540.4445 | C32H63NO4S | [M + H − H20]+ | 3.70 | −0.84 | 3.19 × 10−6/0.000 718 | NAT(30:1) |
| 696 | HILIC ESI(−) | 1.27 | 279.0387 | 279.0389 | C9H12N2O6 | [M + Cl]− | −0.72 | −2.21 | 9.20 × 10−14/0.000 865 | uridine |
| 449 | RP ESI(−) | 6.88 | 673.5272 | 673.5290 | C37H75N2O6P | [M − H]− | −2.67 | −1.75 | 0.000 209/0.001 21 | SM(d32:1) |
| 189 | RP ESI(+) | 1.20 | 585.2700 | 585.2708 | C33H36N4O6 | [M + H]+ | −1.37 | −0.21 | 0.0161/0.001 52 | bilirubin |
| 210 | HILIC ESI(+) | 4.31 | 619.1976 | – | – | [M + H]+ | – | 0.18 | 0.009 58/0.002 14 | – |
| 98 | HILIC ESI(−) | 2.68 | 247.1184 | 247.1187 | C11H20O6 | [M − H]− | −1.21 | −1.98 | 3.57 × 10−10/0.002 37 | fatty acyl glucoside |
| 191 | RP ESI(+) | 4.47 | 818.6049 | 818.6058 | C48H84NO7P | [M + H]+ | −1.10 | −0.36 | 0.0145/0.003 84 | PC(P-40:6) |
| 485 | RP ESI(+) | 6.27 | 978.7103 | 978.7158 | C56H100NO10P | [M + H]+ | −5.62 | 0.16 | 0.000 306/0.004 13 | PS(50:5), PS(O-50:6(OH)), PS(P-50:5(OH)) |
| 700 | RP ESI(−) | 1.06 | 472.1559 | 472.1586 | C20H23N7O7 | [M − H]− | −5.72 | 0.75 | 0.001 16/0.006 80 | folinic acid, 10-formyltetrahydrofolate, pteroyl-d-glutamic acid |
| 22 | RP ESI(−) | 3.70 | 491.3419 | 491.3412 | C26H52O6S | [M − H]− | 1.42 | 3.03 | 0.002 25/0.007 11 | fatty acid ester derivatives |
| 644 | HILIC ESI(+) | 3.93 | 276.1190 | 276.1190 | C10H17N3O6 | [M + H]+ | 0.00 | 2.93 | 0.002 93/0.008 67 | norophthalmic acid, γ-glutamyl glutamine |
| 725 | HILIC ESI(−) | 1.25 | 219.0187 | 219.0193 | C5H8N4O4S | [M − H]− | −2.74 | −0.29 | 0.0120/0.008 90 | taurine derivatives |
| 48 | RP ESI(−) | 0.93 | 462.0594 | 462.0616 | C14H25NO11 | [M + Br]− | −4.76 | 2.78 | 0.002 69/0.009 63 | N-acetyllactosamine, β-1,4-mannose-N-acetylglucosamine, lacto-N-biose I |
| 772 | HILIC ESI(−) | 3.61 | 217.1075 | 217.1081 | C10H18O5 | [M − H]− | −2.76 | −0.90 | 0.001 14/0.0117 | 2(3)-hydroxysebacic acid |
| 29 | RP ESI(+) | 3.27 | 383.3306 | 383.3308 | C27H42O | [M + H]+ | −0.52 | −1.06 | 1.25 × 10−9/0.0127 | 7-dehydrodesmosterol, 5a-cholesta-8,24-dien-3-one, cholesta-4,6-dien-3-one |
Fold change (FC) was calculated as the base 2 logarithm of the average abundance ratios between TKO and TKO-Ctrl samples). Positive FC values indicate increased abundance in TKO samples, while negative values indicate higher abundance in TKO-Ctrl samples.
All p-values are calculated using an FDR-corrected t-test. For additional fragmentation information and level of confidence for each identification, please refer to Table S4 (SI).
Abbreviations: Glu, glutamic acid; His, histidine; Leu, leucine; Ile, isoleucine; CE, cholesteryl ester; LysoPC, lysophosphatidylcholine; PC, phosphatidylcholine; CL, cardiolipin; NAT, N-acyltaurine; SM, sphingomyelin; PS, phosphatidylserine; NeuAc, N-acetylneuraminic acid.
HGSC-Related Metabolic Alterations
Understanding the biological role of specific metabolites is also crucial to enhance our understanding of HGSC metabolism. Discussed below is the potential role of the differential metabolites that were altered in different HGSC stages (Ctrl, Pre, ET, and AT). The biological roles of the remaining identified metabolites can be found in the Supporting Information.
Alterations in metabolism of phosphatidylcholine (PC) and its ester-bond hydrolysis product, lysophosphatidylcholine (LysoPC), have been reported in several gynecological cancers, including ovarian cancer studies,49–53 cervical cancer,54 and breast cancer.55 Our findings of decreased PC(P-40:6) (Table 1; fold change −0.36) and elevated LysoPC(20:0) (Table 1; fold change +0.93) serum levels in TKO-AT mice are in agreement with published ovarian cancer studies.51,53 The alterations are likely due to the activation of PC-cycle enzymes, including choline kinase (ChoK) and PC-specific phospholipase C (PC-plc).50,56 Moreover, the significantly decreased PC and elevated LysoPC alterations were found in advanced-stage cancer (Figure 5), suggesting that these changes were related to tumor metastasis and progression. This observation awaits further validation and investigation.
Figure 5.
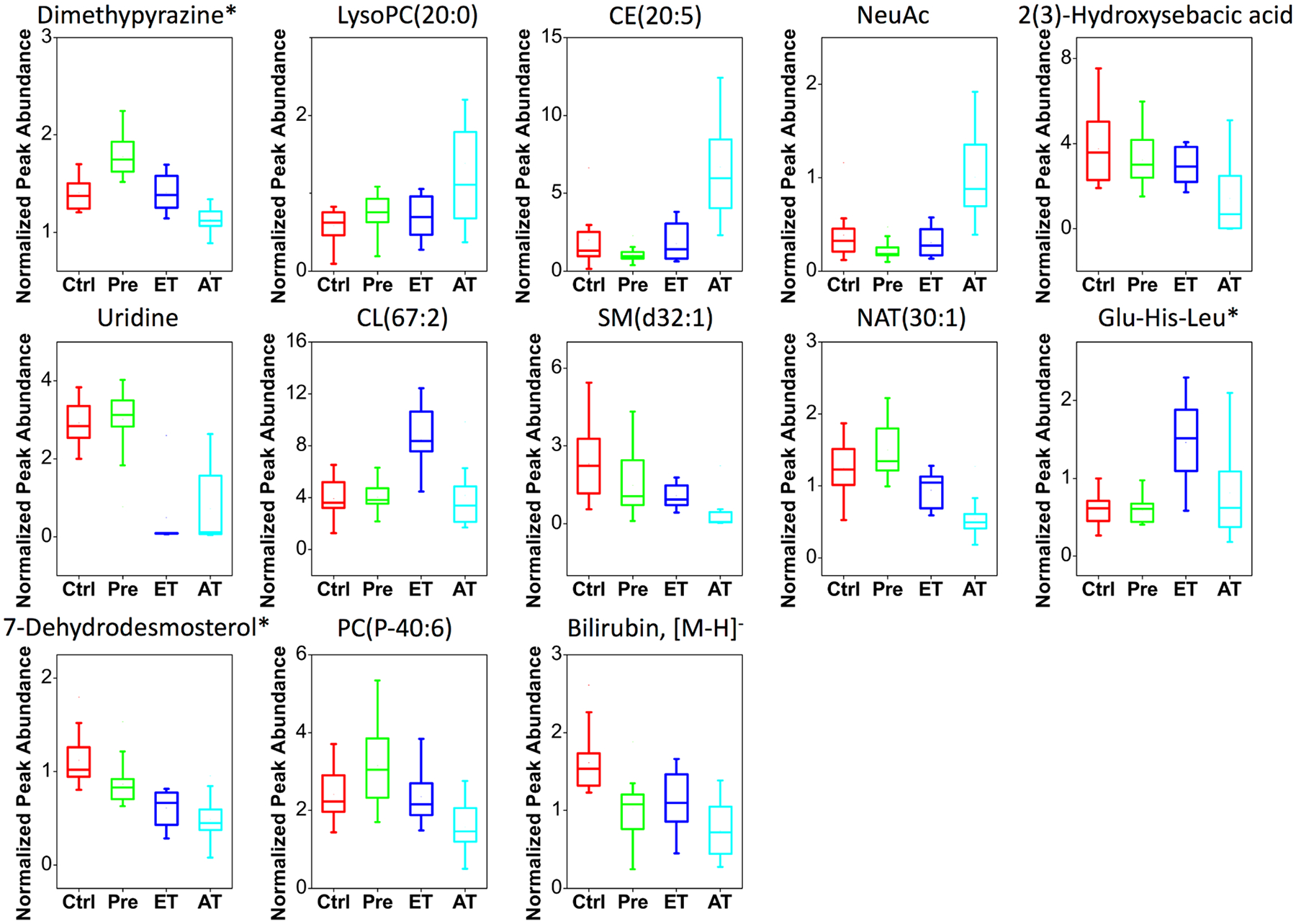
Box plots showing changes of the selected identified metabolites in Ctrl (n = 19), Pre (n = 22), ET (n = 10), and AT (n = 16) samples. The mean, median, upper and lower quartiles, outliers, and minimum and maximum (whiskers) values are displayed. *Features have isomers.
Cardiolipins (CLs) are a unique mitochondrial phospholipid class that regulates bioenergetic processes and signaling events related to apoptosis and aging.57,58 An increase of CL(67:2) serum levels in TKO-ET mice was observed (Figure 5). However, further studies are still needed to understand the role of CL in cellular function and signaling pathways as they relate to ovarian cancer.
One of the identified features in the 29-metabolite panel was the sphingomyelin (SM) SM(d32:1) (Table 1). SM is an essential element of plasma membrane structure and plays important roles in cancer biology, with the hydrolysis by sphingomyelinases resulting in the formation of ceramides, key players in cellular proliferation, growth, and apoptosis.59,60 Consequently, changes in SM levels can have a profound effect on the biophysical properties of cellular membranes and signaling.61 We observed decreased levels of SM(d32:1) in HGSC mice, especially in the advanced stage (Table 1, Figure 5; fold change −1.75). Along the same lines, Braicu et al. have detected a decrease of SM levels in serum samples from ovarian cancer patients,62 strengthening the findings of our study.
Sterol metabolites derived from the cholesterol biosynthetic pathway are important structural components of cell membranes.63 They regulate membrane fluidity and permeability as well as biological activities, including lipid synthesis, cell growth, and apoptosis.64 One discriminant feature was identified as a sterol metabolite, cholesteryl ester [CE(20:5)]. CEs are formed by the esterification of cholesterol with long-chain fatty acids and are transported through the blood by lipoproteins. Overexpression of CEs has been reported in multiple malignant tumors,65 including ovarian cancer.66 We observed a significant increase in serum levels of CE(20:5) in TKO-AT mice (Table 1, Figure 5; fold change +1.26), which is in line with these findings. Another cholesterol derivative, C27H43O [7-dehydrodesmosterol, 5a-cholesta-8,24-dien-3-one, or cholesta-4,6-dien-3-one], was identified in our 29-feature panel. Gradually decreased serum levels of 7-dehydrodesmosterol (and/or its isomers) were detected in TKO with different stages (Table 1, Figure 5; fold change −1.06). 7-Dehydrodesmosterol (and/or its isomers) is an intermediate in the cholesterol biosynthesis pathway. The decrease can be attributed to the enhanced biosynthesis activity of cholesterol, as an increased serum cholesterol level is associated with ovarian cancer.67
N-Acyltaurines (NATs) are molecules with fatty acids conjugated to taurine. We observed decreased serum levels of NAT(30:1) in HGSC mice, especially in the advanced stage (Table 1, Figure 5; fold change −0.84). Limited research has been reported on these species, and their biological role remains largely unexplored. In support of our findings, Chatzakos et al. found that NAT reduced proliferation in human prostate cancer cells.68 Therefore, one likely explanation is the diminished antiproliferative effect with decreased levels of NAT, resulting in further disease progression.
A tripeptide (Glu-His-Leu or Glu-His-Ile) was found to be higher in TKO-ET mice (Table 1, Figure 5; fold change +0.80). Literature searches revealed no known roles for small peptides in ovarian cancer. Some proteases such as protease M, however, have been reported to be overexpressed in serum of ovarian cancer patients69–72 and could, therefore, increase the levels of small circulating peptides in HGSC.
Bilirubin, the end product of heme catabolism in mammals, exhibits antioxidant properties by scavenging peroxyl radicals.73,74 Increased serum bilirubin concentration has been found to be associated with decreased risk for cancer mortality.75 We observed decreased serum levels of bilirubin in HGSC mice, with further decrease in the advanced stage (Table 1, Figure 5; fold change [M − H]− −0.88). This finding agrees with previous work by our group, showing lowered concentration of bilirubin for early-stage ovarian cancer in Dicer-Pten double-knockout (DKO) mice compared with controls.20 One likely explanation of this finding is the diminished protection of bilirubin against oxidative stress in ovarian cancer, leading to further disease progression.
2(3)-Hydroxysebacic acids have been found to be important diagnostic markers for impaired peroxisomal fatty acid oxidation disorders.76 Their levels are regulated by medium-chain acyl-CoA dehydrogenase in the fatty acid β-oxidation pathway. Increased excretion of 2(3)-hydroxysebacic acids is associated with acyl-CoA dehydrogenase deficiency and decreased β-oxidation of fatty acids.76–78 Our experiments reported lower serum levels of 2(3)-hydroxysebacic acids in TKO-AT mice (Table 1, Figure 5; fold change −0.90), suggesting increased activity of acyl-CoA dehydrogenase and fatty acid β-oxidation. In support of this hypothesis, both Fong et al. and Ke et al. have observed increased fatty acid β-oxidation in ovarian cancer patients.24,33
We observed significantly decreased serum levels of uridine in HGSC mice, especially in the early stage (Table 1, Figure 5; fold change −2.21). The level of circulating uridine is a reflection of de novo pyrimidine biosynthesis and the utilization of uridine by tissue via the salvage pathway.79 As cell proliferation in tumors requires a large pool of pyrimidines for rapid DNA and RNA synthesis, the observed relative decrease in serum uridine levels could be due to either tissue uridine uptake or rapid uridine catabolism in ovarian cancer.
N-Acetylneuraminic acid (NeuAc), also known as sialic acid (SA), is a major constituent of glycoproteins and glycolipids.80 We observed significantly increased serum NeuAc levels in TKO-AT mice (Table 1, Figure 5; fold change 0.94). In agreement with this finding, increased SA serum levels have been reported in ovarian cancer patients compared to healthy controls.81,82 This elevation of NeuAc serum levels could be attributed to high levels of glycoprotein and glycolipid release due to the high turnover of malignant tumor cells.80,83 Moreover, the detected elevation of NeuAc levels in TKO-AT mice suggests its strong association with disease progression, confirming its potential application as a diagnostic indicator for HGSC malignancy.81,84,85
One feature in the discriminant metabolite panel was identified as C6H8N2, likely a pyrazine derivative, with lower abundance in TKO-AT mice (Table 1, Figure 5; fold change −0.19). Many pyrazines are exogenous metabolites that originate from food sources, such as cereals, soybeans, potatoes, cocoa products, and other natural products,86 and have been reported to exhibit anticancer activities.87 Although all mice in this study were fed the same diet, it is plausible that altered metabolism in HGSC mice led to differences in abundance of dietary metabolites following disease progression.
CONCLUSIONS
Serum from TKO p53-Dicer1-Pten mice with both early- and advanced-stage HGSC tumors was successfully profiled using a UPLC–MS-based, nontargeted metabolomics strategy. Identified are a panel of 29 metabolites that distinguishes TKO mice from TKO-Ctrl and UT mice and further distinguishes TKO-ET from TKO-AT and TKO-Ctrl. Metabolic alterations in TKO mice—mainly characterized by aggressive fatty acid β-oxidation; abnormal metabolism of phospholipids, glycoproteins, and glycolipids; heme catabolism; cholesterol biosynthesis; and pyrimidine biosynthesis—were found to be associated with HGSC development and progression. Identified biomarkers that differentially expressed in early and advanced stages of HGSC were phospholipids, sphingomyelin, sterols, N-acyltaurine, oligopeptide, and other small molecules, including bilirubin, 2(3)-hydroxysebacic acids, uridine, N-acetylneuraminic acid, and pyrazine derivatives. Taken together, our deep metabolomics study provided insights into dysregulated metabolism in HGSC, which could aid in disease diagnosis, as well as support our understanding of disease development and progression. In particular, this panel of 29 metabolites will serve as a useful guide toward early detection of high-grade serous ovarian cancer.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate Dr. Yong-Hyun Shin and Solji Hyeon for their technical assistance. We also thank the staff from the Georgia Institute of Technology’s Systems Mass Spectrometry Core Facility for technical assistance. F.M.F. and J.K. acknowledge support from NIH/NCI award number 1R01CA218664.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteo-me.9b00263.
Multigroup and binary PCA score plots of TKO, TKO-Ctrl, and UT samples using the initial 5937-feature set and the 29-feature panel; box plots showing changes of the 29 selected spectral features in TKO-Ctrl, TKO-ET, TKO-AT, and UT samples; liquid chromatography gradients for RP and HILIC separation methods; MS acquisition parameters; GA variable selection parameters; detailed MS/MS annotation of the 29-feature panel; sample cohort information; and biological roles of identified features not discussed in the paper (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Reid BM; Permuth JB; Sellers TA Epidemiology of ovarian cancer: a review. Cancer Biol. Med 2017, 14 (1), 9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Siegel R; Miller K; Jemal A Cancer Statistics, 2018. Ca-Cancer J. Clin 2018, 68 (1), 7–30. [DOI] [PubMed] [Google Scholar]
- (3).Seidman JD; Horkayne-Szakaly I; Haiba M; Boice CR; Kurman RJ; Ronnett BM The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int. J. Gynecol. Pathol 2004, 23 (1), 41–44. [DOI] [PubMed] [Google Scholar]
- (4).Seidman JD; Zhao P; Yemelyanova A “Primary peritoneal” high-grade serous carcinoma is very likely metastatic from serous tubal intraepithelial carcinoma: assessing the new paradigm of ovarian and pelvic serous carcinogenesis and its implications for screening for ovarian cancer. Gynecol. Oncol 2011, 120 (3), 470–473. [DOI] [PubMed] [Google Scholar]
- (5).Bowtell DD; Bohm S; Ahmed AA; Aspuria PJ; Bast RC Jr.; Beral V; Berek JS; Birrer MJ; Blagden S; Bookman MA; Brenton JD; Chiappinelli KB; Martins FC; Coukos G; Drapkin R; Edmondson R; Fotopoulou C; Gabra H; Galon J; Gourley C; Heong V; Huntsman DG; Iwanicki M; Karlan BY; Kaye A; Lengyel E; Levine DA; Lu KH; McNeish IA; Menon U; Narod SA; Nelson BH; Nephew KP; Pharoah P; Powell DJ Jr.; Ramos P; Romero IL; Scott CL; Sood AK; Stronach EA; Balkwill FR Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15 (11), 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kim J; Park E; Kim O; Schilder J; Coffey D; Cho C; Bast R Cell Origins of High-Grade Serous Ovarian Cancer. Cancers 2018, 10 (11), No. 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Peres L; Cushing-Haugen K; Kobel M; Harris H; Berchuck A; Rossing M; Schildkraut J; Doherty J Invasive Epithelial Ovarian Cancer Survival by Histotype and Disease Stage. J. Natl. Cancer Inst 2019, 111 (1), 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bowtell DDL The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer 2010, 10 (11), 803–808. [DOI] [PubMed] [Google Scholar]
- (9).Bast RCJ; Hennessy B; Mills GB The biology of ovarian cancer: new opportunities for translation. Nat. Rev. Cancer 2009, 9 (6), 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mai PL; Wentzensen N; Greene MH Challenges related to developing serum-based biomarkers for early ovarian cancer detection. Cancer Prev. Res 2011, 4 (3), 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Buys SS; Partridge E; Black A; Johnson CC; Lamerato L; Isaacs C; Reding DJ; Greenlee RT; Yokochi LA; Kessel B; Crawford ED; Church TR; Andriole GL; Weissfeld JL; Fouad MN; Chia D; O’Brien B; Ragard LR; Clapp JD; Rathmell JM; Riley TL; Hartge P; Pinsky PF; Zhu CS; Izmirlian G; Kramer BS; Miller AB; Xu JL; Prorok PC; Gohagan JK; Berg CD Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011, 305 (22), 2295–2303. [DOI] [PubMed] [Google Scholar]
- (12).Moyer VA Screening for ovarian cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann. Intern. Med 2012, 157 (12), 900–904. [DOI] [PubMed] [Google Scholar]
- (13).Jacobs IJ; Menon U; Ryan A; Gentry-Maharaj A; Burnell M; Kalsi JK; Amso NN; Apostolidou S; Benjamin E; Cruickshank D; Crump DN; Davies SK; Dawnay A; Dobbs S; Fletcher G; Ford J; Godfrey K; Gunu R; Habib M; Hallett R; Herod J; Jenkins H; Karpinskyj C; Leeson S; Lewis SJ; Liston WR; Lopes A; Mould T; Murdoch J; Oram D; Rabideau DJ; Reynolds K; Scott I; Seif MW; Sharma A; Singh N; Taylor J; Warburton F; Widschwendter M; Williamson K; Woolas R; Fallowfield L; McGuire AJ; Campbell S; Parmar M; Skates SJ Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 2016, 387 (10022), 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kobayashi E; Ueda Y; Matsuzaki S; Yokoyama T; Kimura T; Yoshino K; Fujita M; Kimura T; Enomoto T Biomarkers for Screening, Diagnosis, and Monitoring of Ovarian Cancer. Cancer Epidemiol Biomarkers Prev 2012, 21 (11), 1902–1912. [DOI] [PubMed] [Google Scholar]
- (15).Ueland F A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics 2017, 7 (1), No. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bristow R; Smith A; Zhang Z; Chan D; Crutcher G; Fung E; Munroe D Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol. Oncol 2013, 128 (2), 252–259. [DOI] [PubMed] [Google Scholar]
- (17).Zhang A; Sun H; Yan G; Wang P; Wang X Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed Res. Int 2015, 2015, 354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Odunsi K; Wollman RM; Ambrosone CB; Hutson A; McCann SE; Tammela J; Geisler JP; Miller G; Sellers T; Cliby W; Qian F; Keitz B; Intengan M; Lele S; Alderfer JL Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int. J. Cancer 2005, 113 (5), 782–788. [DOI] [PubMed] [Google Scholar]
- (19).Guan W; Zhou M; Hampton CY; Benigno BB; Walker LD; Gray A; McDonald JF; Fernandez FM Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinf 2009, 10, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jones CM; Monge ME; Kim J; Matzuk MM; Fernández FM Metabolomic Serum Profiling Detects Early-Stage High-Grade Serous Ovarian Cancer in a Mouse Model. J. Proteome Res 2015, 14 (2), 917–927. [DOI] [PubMed] [Google Scholar]
- (21).Gaul DA; Mezencev R; Long TQ; Jones CM; Benigno BB; Gray A; Fernández FM; McDonald JF Highly-accurate metabolomic detection of early-stage ovarian cancer. Sci. Rep 2015, 5, 16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chen J; Zhang X; Cao R; Lu X; Zhao S; Fekete A; Huang Q; Schmitt-Kopplin P; Wang Y; Xu Z; Wan X; Wu X; Zhao N; Xu C; Xu G Serum 27-nor-5beta-cholestane-3,7,12,24,25 pentol glucuronide discovered by metabolomics as potential diagnostic biomarker for epithelium ovarian cancer. J. Proteome Res 2011, 10 (5), 2625–2632. [DOI] [PubMed] [Google Scholar]
- (23).Zhou M; Guan W; Walker LD; Mezencev R; Benigno BB; Gray A; Fernandez FM; McDonald JF Rapid mass spectrometric metabolic profiling of blood sera detects ovarian cancer with high accuracy. Cancer Epidemiol., Biomarkers Prev 2010, 19 (9), 2262–2271. [DOI] [PubMed] [Google Scholar]
- (24).Ke C; Hou Y; Zhang H; Fan L; Ge T; Guo B; Zhang F; Yang K; Wang J; Lou G; Li K Large-scale profiling of metabolic dysregulation in ovarian cancer. Int. J. Cancer 2015, 136 (3), 516–526. [DOI] [PubMed] [Google Scholar]
- (25).Ke C; Li A; Hou Y; Sun M; Yang K; Cheng J; Wang J; Ge T; Zhang F; Li Q; Li J; Wu Y; Lou G; Li K Metabolic phenotyping for monitoring ovarian cancer patients. Sci. Rep 2016, 6, 23334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang T; Wu X; Yin M; Fan L; Zhang H; Zhao F; Zhang W; Ke C; Zhang G; Hou Y; Zhou X; Lou G; Li K Discrimination between malignant and benign ovarian tumors by plasma metabolomic profiling using ultra performance liquid chromatography/mass spectrometry. Clin. Chim. Acta 2012, 413 (9–10), 861–868. [DOI] [PubMed] [Google Scholar]
- (27).Fan L; Zhang W; Yin M; Zhang T; Wu X; Zhang H; Sun M; Li Z; Hou Y; Zhou X; Lou G; Li K Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol 2012, 51 (4), 473–479. [DOI] [PubMed] [Google Scholar]
- (28).Zhang T; Wu X; Ke C; Yin M; Li Z; Fan L; Zhang W; Zhang H; Zhao F; Zhou X; Lou G; Li K Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res 2013, 12 (1), 505–512. [DOI] [PubMed] [Google Scholar]
- (29).Chen J; Zhou L; Zhang X; Lu X; Cao R; Xu C; Xu G Urinary hydrophilic and hydrophobic metabolic profiling based on liquid chromatography-mass spectrometry methods: Differential metabolite discovery specific to ovarian cancer. Electrophoresis 2012, 33 (22), 3361–3369. [DOI] [PubMed] [Google Scholar]
- (30).Slupsky CM; Steed H; Wells TH; Dabbs K; Schepansky A; Capstick V; Faught W; Sawyer MB Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res 2010, 16 (23), 5835–5841. [DOI] [PubMed] [Google Scholar]
- (31).Woo HM; Kim KM; Choi MH; Jung BH; Lee J; Kong G; Nam SJ; Kim S; Bai SW; Chung BC Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta 2009, 400 (1–2), 63–69. [DOI] [PubMed] [Google Scholar]
- (32).Denkert C; Budczies J; Kind T; Weichert W; Tablack P; Sehouli J; Niesporek S; Konsgen D; Dietel M; Fiehn O Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res 2006, 66 (22), 10795–10804. [DOI] [PubMed] [Google Scholar]
- (33).Fong MY; McDunn J; Kakar SS Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One 2011, 6 (5), No. e19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ben Sellem D; Elbayed K; Neuville A; Moussallieh FM; Lang-Averous G; Piotto M; Bellocq JP; Namer IJ Metabolomic Characterization of Ovarian Epithelial Carcinomas by HRMAS-NMR Spectroscopy. J. Oncol 2011, 2011, 174019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Boss EA; Moolenaar SH; Massuger LF; Boonstra H; Engelke UF; de Jong JG; Wevers RA High-resolution proton nuclear magnetic resonance spectroscopy of ovarian cyst fluid. NMR Biomed 2000, 13 (5), 297–305. [DOI] [PubMed] [Google Scholar]
- (36).Kim J; Coffey DM; Creighton CJ; Yu Z; Hawkins SM; Matzuk MM High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (10), 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kim J; Coffey DM; Ma L; Matzuk MM The ovary is an alternative site of origin for high-grade serous ovarian cancer in mice. Endocrinology 2015, 156 (6), 1975–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ahmed AA; Etemadmoghadam D; Temple J; Lynch AG; Riad M; Sharma R; Stewart C; Fereday S; Caldas C; Defazio A; Bowtell D; Brenton JD Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol 2010, 221 (1), 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).The Cancer Genome Atlas Research Network.. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474 (7353), 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Annesley T Ion suppression in mass spectrometry. Clin. Chem 2003, 49 (7), 1041–1044. [DOI] [PubMed] [Google Scholar]
- (41).Tsai C; Eberle W; Chu C Genetic algorithms in feature and instance selection. Knowl Based Syst 2013, 39, 240–247. [Google Scholar]
- (42).Worley B; Powers R Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1 (1), 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wishart D; Feunang Y; Marcu A; Guo A; Liang K; Vazquez-Fresno R; Sajed T; Johnson D; Li C; Karu N; Sayeeda Z; Lo E; Assempour N; Berjanskii M; Singhal S; Arndt D; Liang Y; Badran H; Grant J; Serra-Cayuela A; Liu Y; Mandal R; Neveu V; Pon A; Knox C; Wilson M; Manach C; Scalbert A HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018, 46 (D1), D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Fahy E; Sud M; Cotter D; Subramaniam S LIPID MAPS online tools for lipid research. Nucleic Acids Res 2007, 35, W606–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Guijas C; Montenegro-Burke J; Domingo-Almenara X; Palermo A; Warth B; Hermann G; Koellensperger G; Huan T; Uritboonthai W; Aisporna A; Wolan D; Spilker M; Benton H; Siuzdak G METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem 2018, 90 (5), 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mistrik R mzCLOUD: A spectral tree library for the identification of “unknown unknowns”. Abstracts of Papers of the 255th American Chemical Society National Meeting; 2018. [Google Scholar]
- (47).Horai H; Arita M; Kanaya S; Nihei Y; Ikeda T; Suwa K; Ojima Y; Tanaka K; Tanaka S; Aoshima K; Oda Y; Kakazu Y; Kusano M; Tohge T; Matsuda F; Sawada Y; Hirai M; Nakanishi H; Ikeda K; Akimoto N; Maoka T; Takahashi H; Ara T; Sakurai N; Suzuki H; Shibata D; Neumann S; Iida T; Funatsu K; Matsuura F; Soga T; Taguchi R; Saito K; Nishioka T; Tanaka K MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom 2010, 45 (7), 703–714. [DOI] [PubMed] [Google Scholar]
- (48).Bijlsma S; Bobeldijk L; Verheij E; Ramaker R; Kochhar S; Macdonald I; van Ommen B; Smilde A Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem 2006, 78 (2), 567–574. [DOI] [PubMed] [Google Scholar]
- (49).Iorio E; Mezzanzanica D; Alberti P; Spadaro F; Ramoni C; D’Ascenzo S; Millimaggi D; Pavan A; Dolo V; Canevari S; Podo F Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res 2005, 65 (20), 9369–9376. [DOI] [PubMed] [Google Scholar]
- (50).Podo F; Sardanelli F; Iorio E; Canese R; Carpinelli G; Fausto A; Canevari S Abnormal choline phospholipid metabolism in breast and ovary cancer: Molecular bases for noninvasive imaging approaches. Curr. Med. Imaging Rev 2007, 3 (2), 123–137. [Google Scholar]
- (51).Okita M; Gaudette D; Mills G; Holub B Elevated levels and altered fatty acid composition of plasma lysophosphatidylcholine (lysoPC) in ovarian cancer patients. Int. J. Cancer 1997, 71 (1), 31–34. [DOI] [PubMed] [Google Scholar]
- (52).Sutphen R; Xu Y; Wilbanks G; Fiorica J; Grendys E; LaPolla J; Arango H; Hoffman M; Martino M; Wakeley K; Griffin D; Blanco R; Cantor A; Xiao Y; Krischer J Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol. Biomarkers Prev 2004, 13 (7), 1185–1191. [PubMed] [Google Scholar]
- (53).Xiao Y; Schwartz B; Washington M; Kennedy A; Webster K; Belinson J; Xu Y Electrospray ionization mass spectrometry analysis of lysophospholipids in human ascitic fluids: Comparison of the lysophospholipid contents in malignant vs nonmalignant ascitic fluids. Anal. Biochem 2001, 290 (2), 302–313. [DOI] [PubMed] [Google Scholar]
- (54).Yin M; Tan S; Li X; Hou Y; Cao G; Li K; Kou J; Lou G Identification of phosphatidylcholine and lysophosphatidylcholine as novel biomarkers for cervical cancers in a prospective cohort study. Tumor Biol 2016, 37 (4), 5485–5492. [DOI] [PubMed] [Google Scholar]
- (55).Qiu Y; Zhou B; Su M; Baxter S; Zheng X; Zhao X; Yen Y; Jia W Mass Spectrometry-Based Quantitative Metabolomics Revealed a Distinct Lipid Profile in Breast Cancer Patients. Int. J. Mol. Sci 2013, 14 (4), 8047–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Iorio E; Ricci A; Bagnoli M; Pisanu ME; Castellano G; Di Vito M; Venturini E; Glunde K; Bhujwalla ZM; Mezzanzanica D; Canevari S; Podo F Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res 2010, 70 (5), 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Patil V; Greenberg M Cardiolipin-Mediated Cellular Signaling. Adv. Exp. Med. Biol 2013, 991, 195–213. [DOI] [PubMed] [Google Scholar]
- (58).Paradies G; Paradies V; De Benedictis V; Ruggiero F; Petrosillo G Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta, Bioenerg 2014, 1837 (4), 408–417. [DOI] [PubMed] [Google Scholar]
- (59).Bienias K; Fiedorowicz A; Sadowska A; Prokopiuk S; Car H Regulation of sphingomyelin metabolism. Pharmacol. Rep 2016, 68 (3), 570–581. [DOI] [PubMed] [Google Scholar]
- (60).Hannun Y The sphingomyelin cycle and the 2nd messenger function of ceramide. J. Biol. Chem 1994, 269 (5), 3125–3128. [PubMed] [Google Scholar]
- (61).Milhas D; Clarke C; Hannun Y Sphingomyelin metabolism at the plasma membrane: Implications for bioactive sphingolipids. FEBS Lett 2010, 584 (9), 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Braicu E; Darb-Esfahani S; Schmitt W; Koistinen K; Heiskanen L; Poho P; Budczies J; Kuhberg M; Dietel M; Frezza C; Denkert C; Sehouli J; Hilvo M High-grade ovarian serous carcinoma patients exhibit profound alterations in lipid metabolism. Oncotarget 2017, 8 (61), 102912–102922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Barenholz Y Cholesterol and other membrane active sterols: from membrane evolution to “rafts. Prog. Lipid Res 2002, 41 (1), 1–5. [DOI] [PubMed] [Google Scholar]
- (64).Edwards P; Ericsson J Sterols and isoprenoids: Signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem 1999, 68, 157–185. [DOI] [PubMed] [Google Scholar]
- (65).Tosi M; Tugnoli V Cholesteryl esters in malignancy. Clin. Chim. Acta 2005, 359 (1–2), 27–45. [DOI] [PubMed] [Google Scholar]
- (66).Ades A; Carvalho J; Graziani S; Amancio R; Souen J; Pinotti J; Maranhao R Uptake of a cholesterol-rich emulsion by neoplastic ovarian tissues. Gynecol. Oncol 2001, 82 (1), 84–87. [DOI] [PubMed] [Google Scholar]
- (67).Tania M; Khan M; Song Y Association of lipid metabolism with ovarian cancer. Curr. Oncol 2010, 17 (5), 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Chatzakos V; Slatis K; Djureinovic T; Helleday T; Hunt M N-Acyl Taurines are Anti-Proliferative in Prostate Cancer Cells. Lipids 2012, 47 (4), 355–361. [DOI] [PubMed] [Google Scholar]
- (69).Anisowicz A; Sotiropoulou G; Stenman G; Mok S; Sager R A novel protease homolog differentially expressed in breast and ovarian cancer. Mol. Med 1996, 2 (5), 624–636. [PMC free article] [PubMed] [Google Scholar]
- (70).Diamandis E; Yousef G; Soosaipillai A; Bunting P Human kallikrein 6 (zyme/protease M/neurosin): A new serum biomarker of ovarian carcinoma. Clin. Biochem 2000, 33 (7), 579–583. [DOI] [PubMed] [Google Scholar]
- (71).Luo L; Katsaros D; Scorilas A; Fracchioli S; Bellino R; van Gramberen M; de Bruijn H; Henrik A; Stenman U; Massobrio M; van der Zee A; Vergote I; Diamandis E The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res 2003, 63 (4), 807–811. [PubMed] [Google Scholar]
- (72).Yousef G; Polymeris M; Grass L; Soosaipillai A; Chan P; Scorilas A; Borgono C; Harbeck N; Schmalfeldt B; Dorn J; Schmitt M; Diamandis E Human kallikrein 5: A potential novel serum biomarker for breast and ovarian cancer. Cancer Res 2003, 63 (14), 3958–3965. [PubMed] [Google Scholar]
- (73).Stocker R; Yamamoto Y; Mcdonagh A; Glazer A; Ames B Bilirubin is an antioxidant of possible physiological importance. Science 1987, 235 (4792), 1043–1046. [DOI] [PubMed] [Google Scholar]
- (74).Stocker R; Glazer A; Ames B Antioxidant activity of albumin-bound bilirubin. Proc. Natl. Acad. Sci. U. S. A 1987, 84 (16), 5918–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Temme E; Zhang J; Schouten E; Kesteloot H Serum bilirubin and 10-year mortality risk in a Belgian population. Cancer Causes Control 2001, 12 (10), 887–894. [DOI] [PubMed] [Google Scholar]
- (76).Muth A; Mosandl A; Wanders R; Nowaczyk M; Baric I; Bohles H; Sewell A Stereoselective analysis of 2-hydroxysebacic acid in urine of patients with Zellweger syndrome and of premature infants fed with medium-chain triglycerides. J. Inherited Metab. Dis 2003, 26 (6), 583–592. [DOI] [PubMed] [Google Scholar]
- (77).Pollitt R; Losty H; Westwood A 3-Hydroxydicarboxylic aciduria: a distinctive type of intermittent dicarboxylic aciduria of possible diagnostic-significance. J. Inherited Metab. Dis 1987, 10, 266–269. [Google Scholar]
- (78).Duran M; Deklerk J; Vanpelt J; Wadman S; Scholte H; Beekman R; Jennekens F The analysis of plasma and urinary organic-acids during prolonged fasting differentiates between systematic carnitine deficiency and a defect of fatty-acid oxidation. J. Inherited Metab. Dis 1983, 6, 121–122.6321854 [Google Scholar]
- (79).Karle J; Anderson L; Erlichman C; Cysyk R Serum uridine levels in patients receiving N-(Phosphonacetyl)-L-Aspartate. Cancer Res 1980, 40 (8), 2938–2940. [PubMed] [Google Scholar]
- (80).Plucinsky M; Riley WM; Prorok J; Alhadeff J Total and lipid-associated serum sialic-acid levels in cancer-patients with different primary sites and differing degrees of metastatic involvement. Cancer 1986, 58 (12), 2680–2685. [DOI] [PubMed] [Google Scholar]
- (81).Berbec H; Paszkowska A; Siwek B; Gradziel K; Cybulski M Total serum sialic acid concentration as a supporting marker of malignancy in ovarian neoplasia. Eur. J. Gynaecol. Oncol 1999, 20 (5–6), 389–392. [PubMed] [Google Scholar]
- (82).Aranganathan S; Senthil K; Nalini N A case control study of glycoprotein status in ovarian carcinoma. Clin. Biochem 2005, 38 (6), 535–539. [DOI] [PubMed] [Google Scholar]
- (83).Stefenelli N; Klotz H; Engel A; Bauer P Serum sialic-acid in malignant-tumors, bacterial-infections, and chronic liver-diseases. J. Cancer Res. Clin. Oncol 1985, 109 (1), 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Silver H; Karim K; Salinas F; Swenerton K Significance of sialic-acid and carcinoembryonic antigens as monitors of tumor burden among patients with carcinoma of the ovary. Surg. Gynecol. Obstet 1981, 153 (2), 209–213. [PubMed] [Google Scholar]
- (85).Vardi J; Tadros G; Foemmel R; Shebes M Plasma lipid-associated sialic-acid and serum CA-125 as indicators of disease status with advanced ovarian-cancer. Obstet. Gynecol 1989, 74 (3), 379–383. [PubMed] [Google Scholar]
- (86).Maga J; Sizer C Pyrazines in foods - review. J. Agric. Food Chem 1973, 21 (1), 22–30. [Google Scholar]
- (87).Miniyar P; Murumkar P; Patil P; Barmade M; Bothara K Unequivocal Role of Pyrazine Ring in Medicinally Important Compounds: A Review. Mini-Rev. Med. Chem 2013, 13 (11), 1607–1625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


