Abstract
Objective
This study aimed to investigate the clinical significance of circular RNA 0016788 (circ_0016788) regarding its correlation with tumor progression and survival in hepatocellular carcinoma (HCC).
Methods
Two hundred and seventy‐eight HCC patients who received tumor resection were retrospectively analyzed. Circ_0016788 expression in 278 tumor tissues and 116 adjacent tissues was detected by reverse transcription‐quantitative polymerase chain reaction.
Results
Circ_0016788 was elevated in tumor tissue compared with adjacent tissue, and it had an excellent value in distinguishing tumor tissue from adjacent tissue (area under the curve: 0.913; 95% confidence interval: 0.885‐0.941). Furthermore, circ_0016788 correlated to higher performance status score, larger tumor size, increased Barcelona clinic liver cancer (BCLC) stage, abnormal aspartate aminotransferase, abnormal alpha‐fetoprotein, and abnormal carbohydrate antigen 199 levels. Moreover, by Kaplan‐Meier curve and log‐rank test, circ_0016788 high expression correlated with shorter overall survival (OS). Importantly, multivariate Cox's regression analysis displayed that circ_0016788 high expression, older age (≥60 years), history of hepatitis B, history of liver cirrhosis, raised Child‐Pugh stage (B vs A), multifocal disease, largest tumor size (≥5 cm), and increased BCLC stage (B vs A) were independent predictors for poorer OS.
Conclusion
Circ_0016788 displays the potential as a biomarker for assisting personalized treatment, tumor management, and prognosis surveillance in surgical HCC patients.
Keywords: biomarker, circular RNA 0 016 788, hepatocellular carcinoma, overall survival, progression
1. INTRODUCTION
Hepatocellular carcinoma (HCC), the most predominant histological subtype of primary liver cancers, typically arises on the background of chronic hepatitis B and hepatitis C, heavy alcohol intake, obesity, and exposure to aflatoxin‐contaminated foodstuffs. 1 , 2 HCC is the third leading cause of cancer deaths worldwide, with a rapidly increasing incidence, especially in sub‐Saharan Africa and eastern Asia. 3 Owning to atypical symptoms in the early stage, only 30%‐40% of HCC patients are eligible for curative resection, while these patients often suffer from HCC recurrence in a short time, 4 meaning that the majority of HCC patients are diagnosed in the late stages of malignancy with limited treatment approaches. 5 , 6 These above situations lead to dismal survival profiles of HCC. Therefore, understanding the genetic basis of HCC will provide insights into its pathogenesis and help identify novel biomarkers for facilitating treatment decisions making and aiding prognostication in HCC patients.
Circular RNAs (circRNAs) are a class of covalently closed RNA molecules that lack 5′ end caps and 3′ end poly (A) tails. 7 Along with deeper researches, multiple circRNAs have been posited to function as sponges of microRNAs (miRNAs), regulators of splicing and transcription, and modifiers of parental gene expressions, which are all essential in mediating important cellular processes and tumor pathogenesis (including HCC). 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 For instance, circ‐ASAP1 facilitates HCC cell proliferation, colony formation, migration, and invasion by modulating miR‐326/miR‐532‐5p‐MAPK1 in vitro; and HCC patients with increased tumor circ‐ASAP1 level exhibits lower OS as well as higher recurrence than these without increased tumor circ‐ASAP1 level. 8 Another study illustrates that circ‐FOXP1 accelerates HCC progression by sponging miR‐875‐3p and miR‐421, and clinically, circ‐FOXP1 high expression associates with larger tumor size, microvascular invasion, advanced tumor‐node‐metastasis (TNM) stage, and poor prognosis in HCC patients. 9 In addition, circ_0068669 exhibits the potency as a biomarker for HCC metastasis. 14 Previously, our collaborate institution uncovers a novel circRNA (circ_0016788) by microarray that is greatly increased in HCC tissues compared with adjacent normal tissues, and its knockdown inhibits HCC proliferation, migration, invasion in vitro as well as represses HCC tumor growth in vivo. 10 Considering the above data, we hypothesized that circ_0016788 might have clinical significance regarding the potential as a biomarker for progression and prognosis in HCC patients. However, there is currently no report about the clinical implication of circ_0016788 in HCC patients. Therefore, the present study retrospectively detected tumor circ_0016788 expression in 278 surgical HCC patients and investigated the correlation of circ_0016788 with clinical features, liver function indexes, tumor markers, and overall survival (OS) in these HCC patients, aiming to provide insights for tumor management and prognosis improvement of HCC in clinical practice.
2. MATERIALS AND METHODS
2.1. Patients
From January 2014 to December 2015, the data of 278 HCC patients who received tumor resection in our hospital were reviewed in this retrospective study. The screening criteria mainly included the following: (a) pathologically diagnosed as primary HCC; (b) Barcelona clinic liver cancer (BCLC) stage A or B; (c) Child‐pugh stage A or B; (d) about to receive resection; (e) snap‐frozen and well‐preserved tumor tissues; (f) aged 18‐80 years; (g) no history of neoadjuvant therapy; (h) complete preoperative clinical data and follow‐up records; (i) no relapse of HCC before surgery; (j) no other complicated malignancies or history of malignancies. This study was approved by the Institutional Review Board of our hospital. All patients or their guardians provided the informed consents before the inclusion.
2.2. Clinical data collection
The electronic medical records of each patient were reviewed for collecting the preoperative clinical characteristics including age, gender, history of hepatitis B (HB), history of liver cirrhosis, Child‐pugh stage, performance status (PS) score, tumor nodule number, largest tumor size, BCLC stage, liver function index (such as alanine transaminase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP) and total bilirubin (TBIL)), and tumor markers (such as alpha‐fetoprotein (AFP), carcinoembryonic antigen (CEA), and carbohydrate antigen 199 (CA199)). Besides, by reviewing the follow‐up records of each patient, survival data were collected for OS assessment, with the last follow‐up date of December 31, 2018. OS was defined as the duration from the date of resection to the date of death.
2.3. Circ_0016788 determination
The reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) was performed to detect the circ_0016788 expression in the tumor tissues. Moreover, there were 116 paired adjacent tissues available for RT‐qPCR among the total 278 HCC patients; therefore, circ_0016788 expression in these 116 paired adjacent tissues was detected by RT‐qPCR as well. First, total RNA was isolated from tumor and paired adjacent tissues with TRIzol™ Reagent (Thermo Fisher Scientific). Then, the isolated RNA was treated with RNase R (Epicentre) to digest linear RNAs. After that, the RNA underwent reverse transcription by PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara), followed by amplification using KOD SYBR® qPCR Mix (Toyobo)). GAPDH was applied as the internal reference for circ_0016788, and the relative expression of circ_0016788 was calculated by 2−△△Ct method. Primers were designed according to the previous study 10 : circ_0016788, forward, 5′‐CAGTTTATGATTGCCGTCCTCC‐3′, reverse, 5′‐GTGATCGTCTAGGTGGTAACC‐3′; GAPDH, forward, 5′‐TCGACAGTCAGCCGCATCTTCTTT‐3′, reverse, 5′‐ACCAAATCCGTTGACTCCGACCTT‐3′.
2.4. Statistical analysis
Kolmogorov‐Smirnov test was used for the determination of the normality of continuous variables. The normally distributed continuous variables were described as mean ± standard deviation (SD), and the non‐normally distributed continuous variables were displayed as median and interquartile range (IQR). Categorical variables were presented as count (percentage). According to the percentile of circ_0016788 expression in tumor tissues, patients were classified as circ_0016788 low expression (in the 0th to 50th percentile) and circ_0016788 high expression (in the 50th to 100th percentile). And as for circ_0016788 high expression patients, they were further classified as circ_0016788 high + expression (in the 50th to 75th percentile), circ_0016788 high++ expression (in the 75th to 90th percentile), and circ_0016788 high+++ expression (in the 90th to 100th percentile). Comparison of circ_0016788 expression between tumor and adjacent tissue was determined by Wilcoxon signed rank sum test. Comparison of preoperative clinical characteristics between circ_0016788 high expression patients and circ_0016788 low expression patients was determined by chi‐square test. Receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) were plotted to assess the performance of circ_0016788 in discriminating tumor tissue from adjacent tissue. The OS was illustrated by Kaplan‐Meier curve, and the difference of OS between/among groups was analyzed by log‐rank test. Univariate and multivariate Cox's proportional hazard regression model analyses were used for determining the factors related to OS P value <.05 was considered as significant. SPSS 22.0 software (IBM) was used for statistical analyses, and GraphPad Prism 7.00 (GraphPad Software) was applied for figures plotting.
3. RESULTS
3.1. Patients’ characteristics
Regarding the demographic features, the mean age of 55 (19.8%) female and 223 (80.2%) male HCC patients was 58.4 ± 9.3 years (Table 1). The history of HB was found in 238 (85.6%) patients, and the history of liver cirrhosis was found in 197 (70.9%) HCC patients, respectively. In terms of clinical features, 231 (83.1%) and 47 (16.9%) HCC patients presented with Child‐Pugh stage A and B, respectively; 218 (78.4%) and 60 (21.6%) HCC patients had PS score 0 and 1, respectively; 152 (54.7%) and 126 (45.3%) HCC patients presented with unifocal and multifocal nodule, respectively; 154 (55.4%) and 124 (44.6%) HCC patients were of largest tumor size <5.0 cm and largest tumor size ≥5.0 cm, respectively; 128 (46.0%) and 150 (54.0%) HCC patients were categorized as BCLC stage A and B, respectively. With respect to liver function indexes, the median ALT, AST, ALP, and TBIL levels were 27.1 (21.2‐39.4) U/L, 36.2 (26.9‐48.5) U/L, 105.1 (81.3‐144.7) U/L, and 15.1 (10.8‐25.7) μmol/L, respectively. As for tumor markers, the median AFP, median CEA, and median CA199 levels were 32.7 (5.2‐1079.7) ng/mL, 2.7 (2.0‐4.4) ng/mL, and 13.1 (5.3‐29.1) U/mL, respectively.
Table 1.
Baseline characteristics of HCC patients
| Items | HCC patients (N = 278) |
|---|---|
| Demographic features | |
| Age (years), mean ± SD | 58.4 ± 9.3 |
| Gender, No. (%) | |
| Female | 55 (19.8) |
| Male | 223 (80.2) |
| History of HB, No. (%) | |
| No | 40 (14.4) |
| Yes | 238 (85.6) |
| History of liver cirrhosis, No. (%) | |
| No | 81 (29.1) |
| Yes | 197 (70.9) |
| Clinical features | |
| Child‐Pugh stage, No. (%) | |
| A | 231 (83.1) |
| B | 47 (16.9) |
| PS score, No. (%) | |
| 0 | 218 (78.4) |
| 1 | 60 (21.6) |
| Tumor nodule number, No. (%) | |
| Unifocal | 152 (54.7) |
| Multifocal | 126 (45.3) |
| Largest tumor size, No. (%) | |
| <5.0 cm | 154 (55.4) |
| ≥5.0 cm | 124 (44.6) |
| BCLC stage, No. (%) | |
| A | 128 (46.0) |
| B | 150 (54.0) |
| Liver function indexes | |
| ALT (U/L), median (IQR) | 27.1 (21.2‐39.4) |
| AST (U/L), median (IQR) | 36.2 (26.9‐48.5) |
| ALP (U/L), median (IQR) | 105.1 (81.3‐144.7) |
| TBIL (μmol/L), median (IQR) | 15.1 (10.8‐25.7) |
| Tumor markers | |
| AFP (ng/mL), median (IQR) | 32.7 (5.2‐1079.7) |
| CEA (ng/mL), median (IQR) | 2.7 (2.0‐4.4) |
| CA199 (U/mL), median (IQR) | 13.1 (5.3‐29.1) |
Abbreviations: AFP, alpha‐fetoprotein; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; BCLC, Barcelona clinic liver cancer; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; HB, hepatitis B; HCC, hepatocellular carcinoma; IQR, interquartile range; PS score, performance status score; SD, standard deviation; TBIL, total bilirubin.
3.2. The value of circ_0016788 in differentiating tumor tissue from adjacent tissue
Circ_0016788 expression was elevated in tumor tissue compared with adjacent tissue (P < .001) (Figure 1A). Furthermore, ROC curve analysis revealed that circ_0016788 had an excellent value in distinguishing tumor tissue from adjacent tissue (AUC: 0.913; 95%CI: 0.885‐0.941) (Figure 1B).
Figure 1.
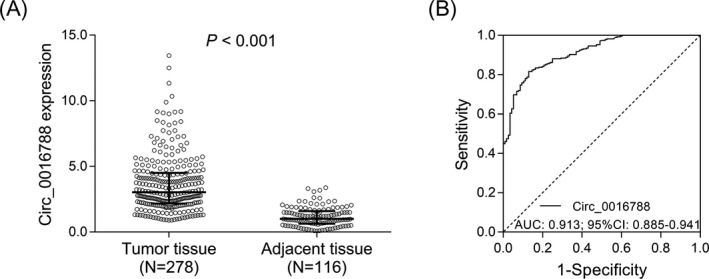
Circ_0016788 discriminated tumor tissue from adjacent tissue. Comparison of circ_0016788 expression between tumor tissue and adjacent tissue (A). ROC curve analysis of circ_0016788 in differentiating tumor tissue from adjacent tissue (B). Circ_0016788, circular RNA 0016788; ROC, receiver operating characteristic; AUC, area under the curve; CI, confidence interval
3.3. Correlation of tumor circ_0016788 with demographic features
No correlation of tumor circ_0016788 with age (P = .186) (Figure 2A), gender (P = .452) (Figure 2B), history of HB (P = .305) (Figure 2C), or history of liver cirrhosis (P = .692) (Figure 2D) was observed in HCC patients.
Figure 2.
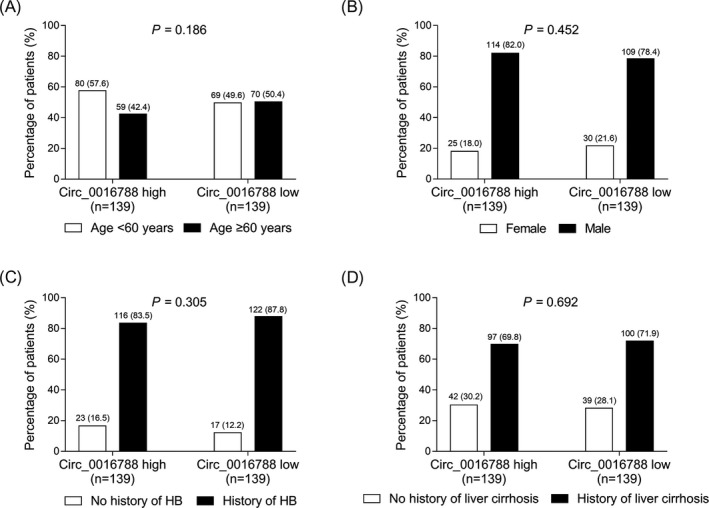
Comparison of demographic features between tumor circ_0016788 high expression patients and low expression patients. Comparisons of age (A), gender (B), history of HB (C), and history of liver cirrhosis (D) between tumor circ_0016788 high expression patients and tumor circ_0016788 low expression patients. Circ_0016788, circular RNA 0016788; HB, hepatitis B
3.4. Correlation of tumor circ_0016788 with clinical features
Tumor circ_0016788 correlated with higher PS score (1 vs 0) (P = .041) (Figure 3B), larger tumor size (largest tumor size ≥5.0 cm vs largest tumor size <5 cm) (P < .001) (Figure 3D), and more advanced BCLC stage (B vs A) (P = .016) (Figure 3E), while, there was no correlation of tumor circ_0016788 with Child‐Pugh stage (P = .078) (Figure 3A) or tumor nodule number (P = .810) (Figure 3C) in HCC patients.
Figure 3.
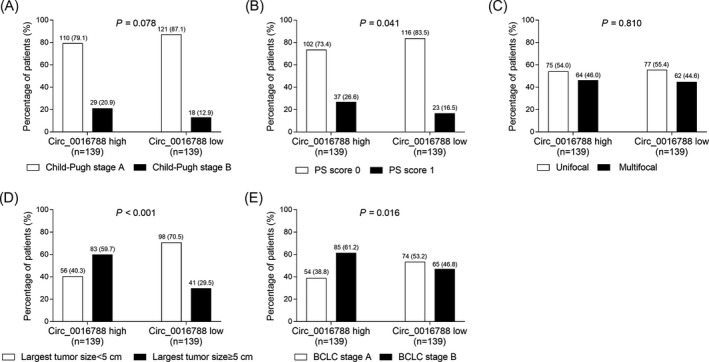
Comparison of clinical features between tumor circ_0016788 high expression patients and low expression patients. Comparisons of Child‐Pugh stage (A), PS score (B), tumor nodule number (C), largest tumor size (D), and BCLC stage (E) between tumor circ_0016788 high expression patients and tumor circ_0016788 low expression patients. Circ_0016788, circular RNA 0016788; PS score, performance status score; BCLC, Barcelona clinic liver cancer
3.5. Correlation of tumor circ_0016788 with liver function indexes and tumor markers
Tumor circ_0016788 correlated with abnormal AST (P = .004) (Figure 4B), abnormal AFP (P = .016) (Figure 4E) and abnormal CA199 (P = .047) (Figure 4G) levels; however, no correlation of tumor circ_0016788 with ALT (P = .486) (Figure 4A), ALP (P = .544) (Figure 4C), TBIL (P = .803) (Figure 4D), or CEA (P = .879) (Figure 4F) levels was revealed in HCC patients.
Figure 4.
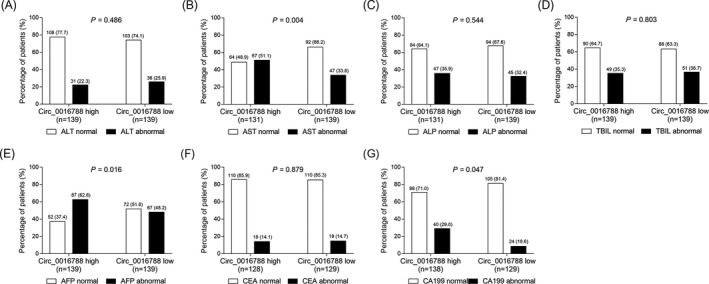
Comparison of liver function indexes and tumor markers between tumor circ_0016788 high expression patients and low expression patients. Comparisons of ALT (A), AST (B), ALP (C), TBIL (D), AFP (E), CEA (F), and CA199 (G) levels between tumor circ_0016788 high expression patients and tumor circ_0016788 low expression patients. Circ_0016788, circular RNA 0016788; ALT, alanine transaminase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; TBIL, total bilirubin; AFP, alpha‐fetoprotein; CEA, carcinoembryonic antigen; CA199, carbohydrate antigen 199
3.6. Correlation of tumor circ_0016788 with OS
Based on the percentile of circ_0016788 expression in tumor tissues, patients were divided into circ_0016788 low expression (0th‐50th percentile) and circ_0016788 high expression (50th‐100th percentile), and the latter was further classified as circ_0016788 high + expression (50th‐75th percentile), circ_0016788 high++ expression (75th‐90th percentile), and circ_0016788 high+++ expression (90th‐100th percentile). The OS was less satisfactory in tumor circ_0016788 high expression patients than that in tumor circ_0016788 low expression patients (P < .001) (Figure 5A). Furthermore, the OS was the lowest in tumor circ_0016788 high+++ expression patients, followed by tumor circ_0016788 high++ expression patients and circ_0016788 high + expression patients, and then the highest in tumor circ_0016788 low expression patients (P < .001) (Figure 5B).
Figure 5.
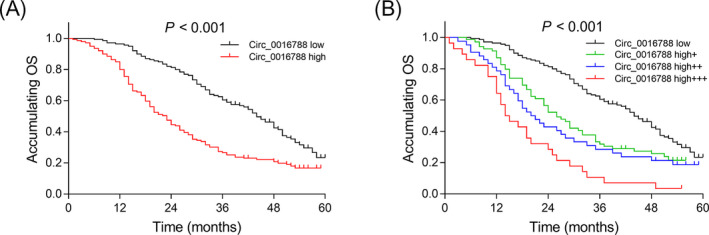
The predictive value of tumor circ_0016788 for OS. Comparison of OS between patients with tumor circ_0016788 high expression and patients with tumor circ_0016788 low expression (A). Comparison of OS among patients with circ_0016788 low expression, circ_0016788 high + expression, circ_0016788 high++ expression and circ_0016788 high+++ expression (B). Circ_0016788, circular RNA 0016788; OS, overall survival
3.7. Prognostic factors for OS
Univariate Cox's regression analysis disclosed that circ_0016788 high expression (P < .001; HR = 2.077), age (≥60 years) (P = .010; HR = 1.440), history of HB (P = .001; HR = 2.154), history of liver cirrhosis (P = .002; HR = 1.664), Child‐Pugh stage (B vs A) (P < .001; HR = 2.304), largest tumor size (≥5 cm) (P < .001; HR = 1.940), BCLC stage (B vs A) (P < .001; HR = 2.093), AST (abnormal) (P = .010; HR = 1.449), and TBIL (abnormal) (P = .002, HR = 1.554) were predictors for worse OS in HCC patients (Table 2). Subsequent multivariate Cox's regression analysis revealed that circ_0016788 high expression (P = .013; HR = 1.635), age (≥60 years) (P < .010; HR = 2.301), history of HB (P = .005; HR = 8.744), history of liver cirrhosis (P = .046; HR = 1.619), Child‐Pugh stage (B vs A) (P < .001; HR = 7.521), multifocal (P < .001; HR = 2.500), largest tumor size (≥5 cm) (P < .001; HR = 5.900), and BCLC stage (B vs A) (P < .001; HR = 1.906) were independent predictors for worse OS in HCC patients.
Table 2.
Analysis of factors correlated with OS
| Items | Univariate Cox's regression | Multivariate Cox's regression | ||
|---|---|---|---|---|
| P value | HR (95%CI) | P value | HR (95%CI) | |
| Circ_0016788 high | <.001 | 2.077 (1.572‐2.746) | .013 | 1.635 (1.111‐2.407) |
| Age (≥60 y) | .010 | 1.440 (1.092‐1.899) | <.001 | 2.301 (1.610‐3.288) |
| Male | .060 | 1.426 (0.986‐2.064) | .376 | 1.259 (0.756‐2.097) |
| History of HB | .001 | 2.154 (1.354‐3.428) | .005 | 8.744 (1.946‐39.301) |
| History of liver cirrhosis | .002 | 1.664 (1.201‐2.305) | .046 | 1.619 (1.010‐2.597) |
| Child‐Pugh stage (B vs A) | <.001 | 2.304 (1.632‐3.254) | <.001 | 7.521 (4.295‐13.170) |
| PS score (1 vs 0) | .415 | 1.148 (0.824‐1.598) | .406 | 1.188 (0.791‐1.784) |
| Multifocal | .272 | 1.168 (0.886‐1.539) | <.001 | 2.500 (1.642‐3.806) |
| Largest tumor size (≥5 cm) | <.001 | 1.940 (1.471‐2.558) | <.001 | 5.900 (3.689‐9.436) |
| BCLC stage (B vs A) | <.001 | 2.093 (1.562‐2.804) | <.001 | 1.906 (1.341‐2.709) |
| ALT (abnormal) a | .607 | 0.918 (0.662‐1.273) | .612 | 0.888 (0.562‐1.404) |
| AST (abnormal) a | .010 | 1.449 (1.093‐1.921) | .536 | 1.143 (0.750‐1.742) |
| ALP (abnormal) a | .092 | 1.282 (0.960‐1.712) | .805 | 0.952 (0.647‐1.402) |
| TBIL (abnormal) a | .002 | 1.554 (1.170‐2.064) | .725 | 1.074 (0.722‐1.598) |
| AFP (abnormal) a | .096 | 1.266 (0.959‐1.671) | .695 | 1.079 (0.737‐1.582) |
| CEA (abnormal) a | .398 | 0.825 (0.529‐1.288) | .237 | 0.740 (0.449‐1.218) |
| CA199 (abnormal) a | .221 | 1.224 (0.885‐1.693) | .973 | 0.992 (0.605‐1.625) |
Abbreviations: AFP, alpha‐fetoprotein; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; BCLC, Barcelona clinic liver cancer; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; CI, confidence interval; HB, hepatitis B; HR, hazard ratio; OS, overall survival; PS score, performance status score; TBIL, total bilirubin.
ALT abnormal ≥ 40.0 U/L, AST abnormal ≥ 40.0 U/L, ALP abnormal < 53.0 U/L or > 128.0 U/L, TBIL abnormal < 5.1 μmol/L, or > 19.0 μmol/L, AFP abnormal ≥ 25.0 ng/mL, CEA abnormal ≥ 5 ng/mL, CA199 abnormal ≥ 37.0 U/mL. Factors correlated with OS were analyzed by univariate and multivariate Cox's proportional hazard regression model.
4. DISCUSSION
The present study highlighted that in HCC patients, (a) Circ_0016788 was overexpressed in tumor tissue compared with adjacent tissue, and it had an excellent value in distinguishing tumor tissue from adjacent tissue. (b) Tumor circ_0016788 was associated with higher PS score, larger tumor size, BCLC stage B, abnormal AST, AFP, and CA199 levels. (c) Tumor circ_0016788 high expression was an independent predictive factor for worse OS.
Hepatocellular carcinoma is an aggressive and highly heterogeneous malignancy that derives from complex genetic and epigenetic alternations in the liver. 1 Genomic instability and the interactions between the tumor microenvironment and cancer cells drive tumor evolution, which are possible contributory factors for ineffective treatment and poor prognosis. 1 Further screening biomarkers for assisting in prognosis prediction of HCC are crucial for malignancy management and surveillance. As an emerging field of research, the role of several circRNAs in the initiation and progression of HCC has been uncovered by increasing studies. 10 , 11 , 12 For instance, circ‐PVT1 promotes HCC cell proliferation and migration by mediating miR‐203/homebox D3 pathway; in the clinical setting, circ‐PVT1 correlates with lymph node metastasis and advanced TNM stages in HCC patients. 11 Another study illuminates that circ‐ZNF652 enhances HCC metastasis by inducing snail‐mediated epithelial‐mesenchymal transition, and HCC patients with high circ‐ZNF652 expression are more vulnerable to vascular invasion, intrahepatic metastasis, and distal metastasis. 12 A previous exploration by our collaborative institution discloses that knockdown of circ_0016788 suppresses HCC cell proliferation, invasion and promotes the apoptosis in vitro, and inhibits tumor growth in vivo. 10 However, the clinical application of circ_0016788 in the tumor development and progression of HCC remains to be fully elucidated. In the present study, our findings elucidated that circ_0016788 was increased in tumor tissue compared with adjacent tissue, and it could effectively discriminate tumor tissue from adjacent tissue in HCC patients. This might be explained that (a) circ_0016788 might induce a series of somatic genomic alternations (eg, TP53 mutations and CTNNB1 mutations), which accelerated the malignant transformation of dysplastic nodules in patients with cirrhotic liver or hepatic adenomas in patients without underlying liver disease. 1 Thereby, circ_0016788 correlated with higher risk of HCC. (b) Circ_0016788 might promote cell proliferation, invasion and inhibit cell apoptosis by downregulating miR‐486/cyclin‐dependent kinase 4 expression (CDK4), which induced the formation of tumors. 10 Herein, circ_0016788 could distinguish tumor tissue from adjacent tissue in HCC. Additionally, the present study found that circ_0016788 correlated with higher PS score (1 vs 0), larger tumor size (largest tumor size ≥5 cm vs largest tumor size < 5 cm), more advanced BCLC stage (B vs A), abnormal AST, AFP, and CA199 levels in HCC patients. Several explanations have been suggested: (a) circ_0016788 might promote CDK4 expression by sponging miR‐486, which enhanced HCC cell proliferation, invasion, and tumor growth while repressed the apoptosis. 10 Thus, circ_0016788 high expression was associated with larger tumor size and more advanced BCLC stage in HCC patients. (b) Circ_0016788 might stimulate the level of pyruvate kinase isozymes M2 that encoded a key enzyme in the process of glycolysis, resulting in the enhanced glycolysis and malignancy of HCC cells. 17
Existing researches have proved the prognostic value of circRNAs in HCC patients. 9 , 18 For instance, HCC patients who have circ‐RHOT1 high expression show shorter OS and recurrence‐free survival compared to patients with circ‐RHOT1 low expression. 18 Another study discloses that high circ‐FOXP1 expression is associated with poorer OS in HCC patients. 9 As for circ_0016788, its correlation with HCC prognosis is still unclear. The present study assessed the impact of circ_0016788 on prognosis in HCC patients by Kaplan‐Meier analysis, and we observed that circ_0016788 correlated with shorter OS in HCC patients. Furthermore, the subsequent multivariate Cox's regression analysis exhibited that circ_0016788 high expression was an independent predictive factor for shorter OS in HCC patients. The possible explanations might include that (a) based on our findings, circ_0016788 was associated with abnormal AST, AFP, CA199 levels, larger tumor size, and more advanced BCLC stage, which might enlarge liver damage and accelerate the malignancy of HCC cells. Therefore, circ_0016788 high expression correlated with poorer prognosis. (b) Circ_0016788 might facilitate the HCC cell stemness, growth, and dissemination by contributing to the aberrant activation of Wnt/β signaling pathway, which led to chemoresistance, lower treatment responses, tumor recurrence, and metastasis. 19 , 20 Thereby, circ_0016788 high expression correlated with unfavorable OS in HCC patients.
The present study was the first to explore the clinical implications of circ_0016788 as a biomarker for tumor management and prognosis surveillance, which might provide a valuable reference for exploring the role of circ_0016788 in the development and progression of other hematological malignancies and solid tumors. However, some limitations needed to be noticed in the present study. First, HCC patients with advanced BCLC stage were not included since they were not suitable for resection; therefore, the clinical implication of circ_0016788 in these patients was not explored. Second, this is a single‐center study with relatively small sample size, which might cause selection bias and reduce statistic power. Thus, future study involving large sample size and multiple centers should be warranted to verify our findings. Thirdly, the present study was a retrospective study, and the screening criteria included snap‐frozen/well‐preserved tumor tissues. However, not all the included HCC patients were with archived tumor tissues and paired adjacent tissues. Therefore, only 116 paired adjacent tissues available among the total 278 HCC patients, which might reduce the statistic power. Lastly, a prospective study regarding the clinical significance of circ_0016788 in HCC patients would be desirable in future.
In conclusion, circ_0016788 is associated with the deteriorated liver function and tumor features of HCC, and it independently predicts worse survival in HCC patients. These findings provide additional information for personalized treatment, management, and surveillance of HCC from bench to clinic.
ACKNOWLEDGMENTS
This study supported by Research Foundation of Wuhan Municipal Healthy Commission (WX14D07) and Research Foundation of The Central Hospital of Wuhan (YB13B01).
Cheng F, Wang L, Zhang J. Circular RNA 0016788 displays as a biomarker for tumor progression and poor prognosis in surgical hepatocellular carcinoma patients. J Clin Lab Anal. 2020;34:e23300 10.1002/jcla.23300
Cheng and Wang contributed equally to this work.
REFERENCES
- 1. Craig AJ, von Felden J, Garcia‐Lezana T, et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139‐152. [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301‐1314. [DOI] [PubMed] [Google Scholar]
- 4. Chan AWH, Zhong J, Berhane S, et al. Development of pre and post‐operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69(6):1284‐1293. [DOI] [PubMed] [Google Scholar]
- 5. Xiong DD, Feng ZB, Lai ZF, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 2019;10(9):658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450‐1462. [DOI] [PubMed] [Google Scholar]
- 7. Salzman J, Circular RNA. Expression: its potential regulation and function. Trends Genet. 2016;32(5):309‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu ZQ, Zhou SL, Li J, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2019. 10.1002/hep.31068 [DOI] [PubMed] [Google Scholar]
- 9. Wang W, Li Y, Li X, et al. Circular RNA circ‐FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR‐875‐3p and miR‐421. Biomed Pharmacother. 2020;121:109517. [DOI] [PubMed] [Google Scholar]
- 10. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR‐486/CDK4 pathway. J Cell Physiol. 2018;234(1):500‐508. [DOI] [PubMed] [Google Scholar]
- 11. Zhu Y, Liu Y, Xiao B, et al. The circular RNA PVT1/miR‐203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol Open. 2019;8(9):bio043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo J, Duan H, Li Y, et al. A novel circular RNA circ‐ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail‐mediated epithelial‐mesenchymal transition by sponging miR‐203/miR‐502‐5p. Biochem Biophys Res Commun. 2019;513(4):812‐819. [DOI] [PubMed] [Google Scholar]
- 13. Sun X, Ge X, Xu Z, et al. Identification of circular RNA‐microRNA‐messenger RNA regulatory network in hepatocellular carcinoma by integrated analysis. J Gastroenterol Hepatol. 2020;35(1):157‐164. [DOI] [PubMed] [Google Scholar]
- 14. Yao T, Chen Q, Shao Z, et al. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;32(8):e22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang YG, Wang T, Ding M, et al. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa‐miR‐1307. Cancer Lett. 2019;460:128‐138. [DOI] [PubMed] [Google Scholar]
- 16. Fu L, Wu S, Yao T, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018;32(2):e22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298‐1316. [DOI] [PubMed] [Google Scholar]
- 18. Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR‐1324/FZD5/Wnt/beta‐catenin axis. Biochem Biophys Res Commun. 2018;497(2):626‐632. [DOI] [PubMed] [Google Scholar]
- 20. Perugorria MJ, Olaizola P, Labiano I, et al. Wnt‐beta‐catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121‐136. [DOI] [PubMed] [Google Scholar]


