Abstract
Background
CircRNA plays an important role in the development of tumors, but its mechanism of action in ovarian cancer is still unclear.
Methods
The expression level of hsa_circ_0013958 in 45 pairs of ovarian cancer tissues and cells was quantified by qRT‐PCR, further revealing whether it is related to clinicopathological features and diagnostic value. Next, the effects of hsa_circ_0013958 on the proliferation, migration, invasion, and apoptosis of A2780 and OVCAR‐3 cells were detected by CCK‐8 assay, Transwell assay, and flow cytometry, respectively. Last, the expression levels of epithelial‐mesenchymal transition‐related proteins (E‐cadherin and Vimentin) and apoptosis‐related proteins (Bcl‐2 and Bax) were detected by Western blotting.
Results
Hsa_circ_0013958 was highly expressed in ovarian cancer tissues and cells, and its expression was closely related to patient FIGO stage and lymph node metastasis. Further, in vitro studies showed that knockdown of hsa_circ_0013958 suppressed proliferation, migration, and invasion of ovarian cancer cells but elevated the cell apoptotic rate. The expression levels of both epithelial‐mesenchymal transition‐related proteins and apoptosis‐related proteins were also changed.
Conclusions
Hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial‐mesenchymal transition and apoptotic signaling pathways.
Keywords: apoptosis, hsa_circ_0013958, invasion, ovarian cancer, proliferation
1. INTRODUCTION
Ovarian cancer is one of the common malignant tumors in women worldwide, with a high morbidity and mortality rate. In 2018, in the United States, there were 22 240 new cases of ovarian cancer and 14 070 deaths.1 Because ovarian cancer is concealed, it is difficult to diagnose early, and relapse and metastasis are common. Therefore, the 5‐year survival rate of patients with advanced ovarian cancer is only 25%.2 CA125, human epididymis protein 4 (HE4), and calculation of Risk of Ovarian Malignancy Algorithm (ROMA) are reported to be of great value in the diagnosis of ovarian cancer.3 Some studies have also shown that the derived neutrophil‐to‐lymphocyte ratio (dNLR) may be an important indicator for the diagnosis of ovarian cancer, and even the identification of early and advanced ovarian cancer.4 However, these markers may not be the best diagnostic markers. Exploring early diagnostic markers and treatment targets for ovarian cancer is of particular importance clinically.
CircRNA, a novel non‐coding RNA, was first discovered in RNA viruses. CircRNA is more stable than linear RNA because it does not include a 5′ end cap and a 3′ end poly (A) tail.5 CircRNAs are usually formed by exons or introns by reverse splicing or lasso introns. CircRNAs mainly include three types as follows: exon circRNA (EcircRNA), exon‐intron hybridization circRNA (EIciRNA), and intron that circulates itself to form circRNA (ciRNA).6 The most common type is EcircRNA. The function of circRNA is mainly related to its localization in the cell. EcircRNA is mainly located in the cytoplasm, while EIciRNA and ciRNA can be located in the nucleus or cytoplasm.7 Numerous studies have shown that circRNA is closely related to the occurrence and development of tumors. Hsa_circ_0058124 promoted the tumorigenesis and invasion of papillary thyroid carcinoma by making miR‐218‐5p sponge and upregulate its target gene NUMB and then inhibit the NOTCH3/GATAD2A axis.8 In lung squamous cell carcinoma, circTP63 competed with miR‐873‐3p to affect FOXM1.9 Circ‐ABCB10 is associated with advanced clinical pathological features and poor prognosis of patients and affects the biological function of ovarian cancer.10
Hsa_circ_0013958 is located on chromosome 1, and its genomic length is 816. It has been reported that hsa_circ_0013958 acts as a sponge of miRNA‐134 and upregulates the expression of cyclin D1 to affect the development of non‐small cell lung cancer.11 However, the function of hsa_circ_0013958 and its potential mechanism in ovarian cancer is still unclear.
In this study, we first demonstrated that hsa_circ_0013958 was upregulated in ovarian cancer tissues and cells and promoted the malignant biological behavior of ovarian cancer cells. The proliferation, migration, and invasion of ovarian cancer cells were inhibited when hsa_circ_0013958 was knocked down, but cell apoptosis was promoted. In addition, the expression levels of both epithelial‐mesenchymal transition‐related proteins and apoptosis‐related proteins were changed, correspondingly. The results may provide a new strategy for the treatment of ovarian cancer.
2. MATERIALS AND METHODS
2.1. Patient samples
A total of 90 patients who were admitted to Shengjing Hospital affiliated with China Medical University from March 2017 to October 2018 were selected. The postoperative pathology of all cases was confirmed by two chief physicians in the Department of Pathology of the hospital, and all clinical data regarding the cases were collected. Forty‐five cases of ovarian cancer were pathologically diagnosed as ovarian serous carcinoma. In the control group, another 45 patients were confirmed by postoperative pathology to have normal ovaries removed because of benign uterine lesions. All patients did not receive chemoradiotherapy before surgery. All patients had signed informed consent before the operation, and the study was approved by the Ethics Committee of Shengjing Hospital affiliated to China Medical University.
2.2. Cell culture
The human ovarian cancer cell lines including A2780 and OVCAR‐3 cells and human normal ovarian epithelial cell line HOSEpiC were obtained from the Tumor Cell Band Research Institute of the Chinese Academy of Medical Science. A2780 and OVCAR‐3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. HOSEpiC cells were cultured in 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All these cell lines were cultured in a humidified incubator at 37°C and 5% CO2.
2.3. RNA extraction and quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted from tumor tissues and cells using TRIzol reagent (Invitrogen), and 400 ng of total RNA in each sample was converted to cDNA using a PrimeScriptVR RT kit (Takara). qRT‐PCR was performed using SYBR Premix Ex Taq (Takara). The primers were designed for this experiment, and the sequences of hsa_circ_0013958 and GAPDH were as follows: hsa_circ_0013958 Forward primer: 5′‐GAACCCCCAGCTATTAGAGGTCCC‐3′, hsa_circ_0013958 Reverse primer: 5′‐GGAAAAGCCCAGCCAGCAAAC‐3′, GAPDH Forward primer: 5′‐TTCTTTTGCGTCGCCAGCCG‐3′, GAPDH Reverse primer: 5′‐GGTGACCAGGCGCCCAATAC‐3′. GAPDH was used as an internal control, and the relative expression level of hsa_circ_0013958 was measured using the 2−ΔΔ C t method.
2.4. Cell transfection
siRNA‐negative control (si‐NC) and small interfering RNA (siRNA) targeting hsa_circ_0013958 were purchased from GenePharma Co., Ltd. The sequences of si‐NC and siRNA were as follows: si‐NC sense 5′‐UUCUUCGAACGUGUCACGUTT‐3′; si‐NC antisense 5′‐ACGUGACACGUUCGGAGAATT‐3′; siRNA sense 5′‐CCGAAACCAUAUUCUCCUUTT‐3′; siRNA antisense 5′‐AAGGAGAAUAUGGUUUCGGTT‐3′. Ovarian cancer cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer instructions.
2.5. Cell counting kit‐8 assay (CCK‐8 assay)
The CCK‐8 assay was used to detect ovarian cancer cell proliferation. Transfected ovarian cancer cells were evenly spread on 96‐well plates at a density of 2000 cells/100 μL per well. Then, 10 μL CCK‐8 was added to each well at different time points (0, 24, 48, 72, and 96 hours) and incubated in the humidified incubator at 37°C at 5% CO2 for 1 hour. Absorbance was detected at 450 nm using a spectrophotometer (XFLUOR4, version: V 4.51), and the proliferation changes were calculated.
2.6. Cell transwell assay
The cell transwell assay was used to explore the ovarian cancer cell migration and invasion. After transfection, 30 000 cells/200 μL of serum‐free DMEM were put into the upper chamber (Corning), and 600 μL of DMEM was placed in the lower chamber, which contained 20% FBS. After 48 hours incubation, the chamber was removed, washed three times with PBS, fixed with 4% paraformaldehyde, and stained with crystal violet solution.
2.7. Cell apoptosis assay
Transfected cells were trypsinized without EDTA and washed twice with cold PBS. Cell apoptosis was assessed by flow cytometry using the Annexin V‐FITC Apoptosis Detection Kit (YuhengCo., Ltd.) according to the manufacturer's instructions.
2.8. Western blot analysis
RIPA Lysis Buffer (Beyotime) was used to harvest ovarian cancer cell proteins. The BCA protein assay kit (Beyotime) was used to measure the ovarian cancer cell protein concentrations. After processing by SDS‐PAGE electrophoretically, the protein was transferred to a PVDF membrane (US Microwells). After the PVDF membrane was blocked by tris‐buffered saline and tween 20 (TBST) containing 5% fat‐free milk for 2 hours at room temperature, the PVDF membranes were incubated overnight at 4°C with primary antibodies from Wanleibio against Bcl‐2 (1:500, Rabbit), Bax (1:500, Rabbit), E‐cadherin (1:1000, Rabbit), Vimentin (1:1000, Rabbit), and GAPDH (1:1000, Mouse) as a visual loading control. On the second day, after three TBST washes, the membrane was incubated with the secondary antibody for 2 hours at room temperature, and then the protein bands on the membranes were exposed using an enhanced chemiluminescence system (Santa Cruz Biotechnology).
2.9. Statistical analysis
The statistical analysis was performed using the data adopted software IBM SPSS Statistics 25.0. The data came from three independent experiments and are expressed by the mean ± SD. Student's t test was used to analyze the differences between two independent samples. The correlation between hsa_circ_0013958 and clinicopathological characteristics was evaluated by the chi‐square test and Fisher's exact test (frequency < 5). P < .05 indicates statistical significance.
3. RESULTS
3.1. Hsa_circ_0013958 was highly expressed in ovarian cancer tissues and cells
To investigate the expression of hsa_circ_0013958 in ovarian cancer tissues and cells, the level of hsa_circ_0013958 expression in 45 ovarian cancer tissues and normal ovarian tissues was detected by qRT‐PCR. The results showed that hsa_circ_0013958 was significantly upregulated in ovarian cancer tissues compared with normal ovarian tissues (Figure 1A). We further analyzed the relationship between hsa_circ_0013958 and clinicopathological features of patients. The results showed that the expression of hsa_circ_0013958 was closely related to patient FIGO stage and lymph node metastasis (P < .05), and the difference was statistically significant. However, there was no correlation between the age, menopausal status, differentiation, and expression of hsa_circ_0013958 (P > .05; Table 1).
Figure 1.
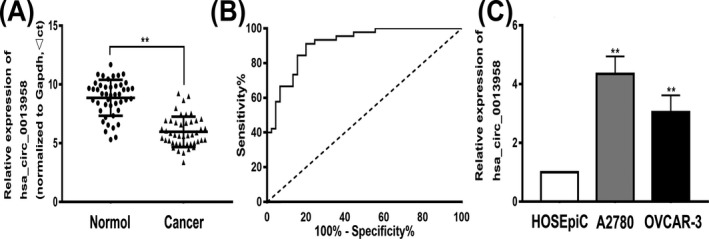
Hsa_circ_0013958 endogenous expression and ROC curve in ovarian cancer. A, Hsa_circ_0013958 was highly expressed in ovarian cancer tissues compared with normal tissues. B, The ROC curves of hsa_circ_0013958. C, The hsa_circ_0013958 expression level was significantly higher in ovarian cancer cell lines (A2780 and OVCAR‐3) than in the normal ovarian cell line HOSEpiC. The data are presented as the mean ± SD. *P < .05, **P < .01
Table 1.
Correlation between hsa_circ_0013958 expression and clinical pathological characteristics
| Clinicopathological features | Numbers | Hsa_circ_0013958 | P value | |
|---|---|---|---|---|
| Low expression | High expression | |||
| Age(y) | ||||
| <50 | 14 | 10 | 4 | .057 |
| ≥50 | 31 | 12 | 19 | |
| Menopause | ||||
| Yes | 29 | 12 | 17 | .175 |
| No | 16 | 10 | 6 | |
| Differentiation | ||||
| Well and moderate | 11 | 3 | 8 | .165 |
| Poor | 34 | 19 | 15 | |
| FIGO stage | ||||
| I‐II | 15 | 11 | 4 | .029 |
| III‐IV | 30 | 11 | 19 | |
| Lymph node metastasis | ||||
| Yes | 30 | 10 | 20 | .005 |
| No | 15 | 12 | 3 | |
We further analyzed the accuracy of using hsa_circ_0013958 in the diagnosis of ovarian cancer using receiver operating characteristic (ROC) curves (Figure 1B). The area under the curve (AUC) was 0.912 (95% CI = 0.854‐0.969, P < .01). When the cut‐off value was 7.6435, the Youden index, sensitivity, and specificity were 0.711, 80.0%, and 91.1%, respectively. These results indicate that hsa_circ_0013958 can be used as a diagnostic marker for ovarian cancer.
In addition to high expression of hsa_circ_0013958 in ovarian cancer cells compared with normal ovarian cells (Figure 1C), the data indicated that hsa_circ_0013958 may play a role as a cancer‐promoting gene in ovarian cancer.
3.2. Knockdown of hsa_circ_0013958 inhibited the proliferation of ovarian cancer cells
In the present study, siRNA inhibiting hsa_circ_0013958 was transfected into ovarian cancer cell lines A2780 and OVCAR‐3, and the expression of hsa_circ_0013958 in ovarian cancer cells was detected by qRT‐PCR. The results showed that the expression level of hsa_circ_0013958 in the si‐circRNA group was lower than that of the blank control group (Figure 2A‐B).
Figure 2.
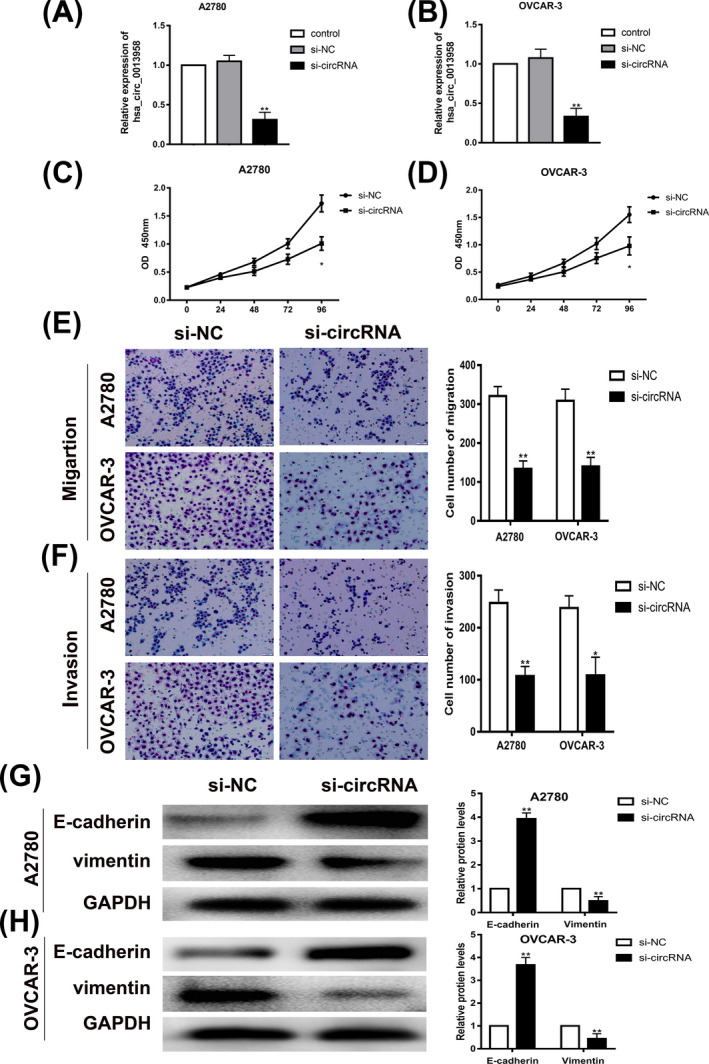
Knockdown of hsa_circ_0013958 significantly inhibited proliferation, migration, and invasion of ovarian cancer cells. A, Transfection with siRNA against hsa_circ_0013958 reduced the expression of hsa_circ_0013958 in A2780. B, Transfection with siRNA against hsa_circ_0013958 reduced the expression of hsa_circ_0013958 in OVCAR‐3. C, Silencing of hsa_circ_0013958 in A2780 slowed cell proliferation. D, Silencing of hsa_circ_0013958 in OVCAR‐3 slowed cell proliferation. E, Silencing of hsa_circ_0013958 in A2780 and OVCAR‐3 slowed cell migration. F, Silencing of hsa_circ_0013958 in A2780 and OVCAR‐3 slowed cell invasion. G, Silencing of hsa_circ_0013958 increased the protein expression level of E‐cadherin but reduced theVimentin protein expression level in A2780. H, Silencing of hsa_circ_0013958 increased the protein expression level of E‐cadherin but reduced the Vimentin protein expression level in OVCAR‐3. The data are presented as the mean ± SD. *P < .05, **P < .01
To further investigate the effect of hsa_circ_0013958 on the proliferation of ovarian cancer cells, we conducted CCK‐8 assays. The results showed that the proliferation of A2780 and OVCAR‐3 cells was significantly inhibited after hsa_circ_0013958 was knocked down (Figure 2C‐D).
3.3. Knockdown of hsa_circ_0013958 significantly inhibited the migration and invasion of ovarian cancer cells
To investigate the effect of hsa_circ_0013958 on the migration and invasion of ovarian cancer cells, Transwell assays were conducted in ovarian cancer cell lines A2780 and OVCAR‐3. Compared with the blank control group, the migration and invasion of ovarian cancer cells in the si‐circRNA group were significantly decreased (Figure 2E‐F).
Subsequently, Western blotting experiments were performed to detect the expression of the epithelial‐mesenchymal transition‐related proteins E‐cadherin and Vimentin. The protein level of E‐cadherin in ovarian cancer cells with knockdown of hsa_circ_0013958 was significantly increased, but the protein level of Vimentin was significantly decreased (Figure 2G‐H).
3.4. Hsa_circ_0013958 down‐regulation promoted apoptosis in ovarian cancer cells
Further, we studied the effect of hsa_circ_0013958 on the apoptosis rate of ovarian cancer cells. Using apoptosis experiments, the results showed that the apoptosis rate of ovarian cancer cells in the si‐circRNA group was significantly increased (Figure 3A‐B).
Figure 3.
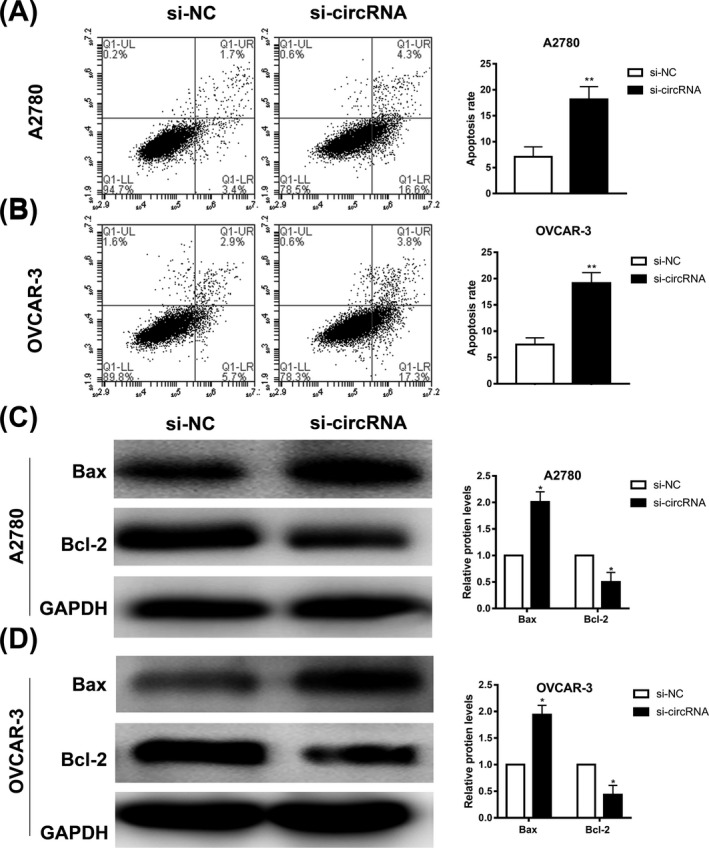
Knockdown of hsa_circ_0013958 significantly promoted apoptosis of ovarian cancer cells. A, Silencing of hsa_circ_0013958 in A2780 promoted apoptosis. B, Silencing of hsa_circ_0013958 in OVCAR‐3 promoted apoptosis. C, Silencing of hsa_circ_0013958 increased the protein expression level of Bax but reduced Bcl‐2 protein expression level in A2780. D, Silencing of hsa_circ_0013958 increased the protein expression level of Bax but reduced Bcl‐2 protein expression level in OVCAR‐3. The data are presented as the mean ± SD. *P < .05, **P < .01
Western blotting assays were used to detect the expression of apoptosis‐related proteins Bcl‐2 and Bax. Compared with the blank control group, the protein level of Bax was significantly increased in the si‐circRNA group, whereas the protein level of Bcl‐2 was significantly decreased (Figure 3C‐D).
4. DISCUSSION
CircRNA is mainly generated after the process of precursor RNA being variably spliced.12 Current studies show that circRNA is closely related to the occurrence and development of disease. CircRNA exerts biological functions through sponging miRNA, binding to RNA binding proteins, translating protein, and regulating transcription.13 The role of circRNA in various types of cancer has also been considered. Recent studies have shown that hsa_circ_0072309 inhibited breast cancer cell proliferation and migration by negatively regulating miR‐492.14 Tang et al15 found that circKIF4A can promote the occurrence and development of triple‐negative breast cancer by competing with miR‐375 to bind with the KIF4A mRNA 3′UTR region and can be used as a prognostic marker and therapeutic target for triple‐negative breast cancer. Circ‐HuR affects the progression of gastric cancer through interacting with CNBP protein.16 While there are few studies about circRNA in ovarian cancer, current studies have found that circRNA UBAP2, circ‐SMAD7, circ‐CSPP1, circ‐ABCB10, and hsa_circ_0061140 are highly expressed in ovarian cancer and promote the development of ovarian cancer.10, 17, 18, 19, 20 Circ‐ITCH was down‐expressed in ovarian cancer; interestingly, Luo et al21 reported that circ‐ITCH negatively regulates miR‐10a and inhibits ovarian cancer cell proliferation but promotes apoptosis. Hu et al22 found that circ‐ITCH acts as a sponge for miR‐145, increasing its target RASA1 expression level and then affects the malignant biological behavior of ovarian cancer cells.
In our study, the expression of hsa_circ_0013958 in ovarian cancer tissues and cells was upregulated, and the expression level of hsa_circ_0013958 was closely related to FIGO stage and lymph node metastasis. The area under the curve (AUC) was 0.912, indicating that it has good diagnostic value. Therefore, it is speculated that it may act as an oncogene in ovarian cancer by promoting the proliferation, migration, and invasion of ovarian cancer cells, while inhibiting the apoptosis.
Epithelial‐mesenchymal transition is widely involved in tumorigenesis, developments, and angiogenesis. E‐cadherin is a transmembrane protein, and its loss is an important characteristic of epithelial‐mesenchymal transition, indicating that it may be closely related to tumor metastasis and recurrence.23, 24 Vimentin is a type III intermediate silk protein expressed in mesenchymal cells and is the main skeleton component of mesenchymal cells. Therefore, Vimentin is often used as a marker for mesenchymal‐derived cells or cells undergoing epithelial‐mesenchymal transition.25, 26 Increased E‐cadherin expression and decreased Vimentin expression often indicate decreased epithelial‐mesenchymal transition function. In the present study, the protein level of E‐cadherin was increased, but the protein level of Vimentin was decreased when hsa_circ_0013958 was knocked down. This indicates that hsa_circ_0013958 may affect the migration and invasion of ovarian cancer cells by affecting epithelial‐mesenchymal transition. Bax/Bcl‐2 are apoptosis‐related proteins that participate in and regulate the apoptosis of many tumors.27 Activated Bcl‐2 family proteins affect the permeability of the outer membrane of the mitochondria and depolarize the outer membrane to release some pro‐apoptotic proteins, such as cytochrome C, apoptosis‐inducing factors, etc.28 Bcl‐2 is located on the nuclear membrane, endoplasmic reticulum, and mitochondrial outer membrane, and its expression decreases during apoptosis.29 Bax is mainly located in the cytoplasm and can form a heterodimer with Bcl‐2 or form a homodimer by itself, with imbalanced expression in apoptosis.30 The increase in Bax expression and decrease in Bcl‐2 expression often indicates increased apoptosis. In the present study, the protein expression level of Bax was increased and the protein expression level of Bcl‐2 was decreased after hsa_circ_0013958 knockdown, further confirming that hsa_circ_0013958 acted as an oncogene to inhibit apoptosis of ovarian cancer cells.
In summary, hsa_circ_0013958 was highly expressed and played a role as an oncogene in ovarian cancer. Knockdown of hsa_circ_0013958 inhibited the malignant biological behavior of ovarian cancer cells. This may provide a new target for the diagnosis and treatment of ovarian cancer.
ACKNOWLEDGMENTS
This study was supported by grants from the Natural Science Foundation of Liaoning Province (Grant No. 201800832) and The Outstanding Scientific Fund of Shengjing Hospital (Grant No. 201705).
Pei C, Wang H, Shi C, Zhang C, Wang M. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial‐mesenchymal transition and apoptotic signaling pathways. J Clin Lab Anal. 2020;34:e23292 10.1002/jcla.23292
REFERENCES
- 1. Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Timmermans M, Sonke GS, Van de Vijver KK, van der Aa MA, Kruitwagen RFPM. No improvement in long‐term survival for epithelial ovarian cancer patients: A population‐based study between 1989 and 2014 in the Netherlands. Eur J Cancer. 2018;88:31‐37. [DOI] [PubMed] [Google Scholar]
- 3. Kim B, Park Y, Kim B, et al. Diagnostic performance of CA 125, HE4, and risk of Ovarian Malignancy Algorithm for ovarian cancer. J Clin Lab Anal. 2019;33:e22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Y‐Y, Qin Y‐Y, Qin J‐Q, Zhang X, Lin F‐Q. Diagnostic value of derived neutrophil‐to‐lymphocyte ratio in patients with ovarian cancer. J Clin Lab Anal. 2019;33:e22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235:718‐724. [DOI] [PubMed] [Google Scholar]
- 6. Zhang H, Shen Y, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34:e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019;38(1):318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Ye X, Xia X, Lin X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26:151‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170‐2182. [DOI] [PubMed] [Google Scholar]
- 12. Santer L, Bar C, Thum T. Circular RNAs: a novel class of functional rna molecules with a therapeutic perspective. Mol Ther. 2019;27:1350‐1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675‐691. [DOI] [PubMed] [Google Scholar]
- 14. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR‐492. Cancer Manag Res. 2019;11:1033‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang H, Huang X, Wang J, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple‐negative breast cancer. Mol Cancer. 2019;18:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang F, Hu A, Li D, et al. Circ‐HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheng M, Wei N, Yang HY, Yan M, Zhao QX, Jing LJ. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA‐144. Eur Rev Med Pharmacol Sci. 2019;23:7283‐7294. [DOI] [PubMed] [Google Scholar]
- 18. Zhao Y, Qin XP, Lang YP, Kou D, Shao ZW. Circular RNA circ‐SMAD7 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur Rev Med Pharmacol Sci. 2019;23:5603‐5610. [DOI] [PubMed] [Google Scholar]
- 19. Li QH, Liu Y, Chen S, et al. circ‐CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR‐1236‐3p sponge. Biomed Pharmacother. 2019;114:108832. [DOI] [PubMed] [Google Scholar]
- 20. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1‐mediated cell growth and metastasis in ovarian cancer through miR‐370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55‐63. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Luo L, Gao YQ, Sun XF. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR‐10a‐alpha. Eur Rev Med Pharmacol Sci. 2018;22:8119‐8126. [DOI] [PubMed] [Google Scholar]
- 22. Hu J, Wang L, Chen J, et al. The circular RNA circ‐ITCH suppresses ovarian carcinoma progression through targeting miR‐145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505:222‐228. [DOI] [PubMed] [Google Scholar]
- 23. Yu W, Yang L, Li T, Zhang Y. Cadherin signaling in cancer: its functions and role as a therapeutic target. Front Oncol. 2019;9:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venhuizen JH, Jacobs FJC, Span PN, Zegers MM. P120 and E‐cadherin: double‐edged swords in tumor metastasis. Semin Cancer Biol. 2019. 10.1016/j.semcancer.2019.07.020 [DOI] [PubMed] [Google Scholar]
- 25. Cheng F, Eriksson JE. Intermediate filaments and the regulation of cell motility during regeneration and wound healing. Cold Spring Harb Perspect Biol. 2017;9(9):a022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pluciennik E, Nowakowska M, Pospiech K, et al. The role of WWOX tumor suppressor gene in the regulation of EMT process via regulation of CDH1‐ZEB1‐VIM expression in endometrial cancer. Int J Oncol. 2015;46:2639‐2648. [DOI] [PubMed] [Google Scholar]
- 27. Adams JM, Cory S. The BCL‐2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chandrasekar AP, Cummins NW, Badley AD. The role of the BCL‐2 family of proteins in HIV‐1 pathogenesis and persistence. Clin Microbiol Rev. 2019;33:e00107‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salam AAA, Nayek U, Sunil D. Homology modeling and docking studies of Bcl‐2 and Bcl‐xL with small molecule inhibitors: identification and functional studies. Curr Top Med Chem. 2018;18:2633‐2663. [DOI] [PubMed] [Google Scholar]
- 30. Garner TP, Reyna DE, Priyadarshi A, et al. An autoinhibited dimeric form of BAX regulates the BAX activation pathway. Mol Cell. 2016;63:485‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]


