Abstract
Background
Circular RNAs (circRNAs) are structural ubiquitous RNA molecules. Accumulating evidences have elucidated that circRNAs play essential roles in the pathogenesis of diseases including cancers. Exosomal circRNAs are those circRNAs stably existing in exosomes and having high clinical values as novel potential diagnostic biomarkers of many diseases. Gastrointestinal (GI) malignancies, including pancreatic cancer, colorectal cancer, hepatocellular carcinoma (HCC), and gastric cancer, are leading causes of mortality worldwide and a major global health burden. However, no ideal tumor biomarkers of screening early GI cancers are currently available.
Methods
We collected data through Web of Science. The search terms used were as follows: circular RNA, circRNA, exosomes, exosomal circRNAs, biomarkers, gastrointestinal malignancies, pancreatic cancer, hepatocellular carcinoma, HCC, gastric cancer, colorectal cancer, physiological functions, biogenesis, molecular mechanism. Only articles published in English were included.
Results
We found that several circRNAs and exosomal circRNAs have been used as potential biomarkers to screen GI cancers including pancreatic cancer (hsa_circ_0001649, circ_0007534, circ_0030235, circRHOT1, circZMYM2, circ‐LDLRAD3, chr14:101402109‐101464448C, chr4:52729603‐52780244C, circ‐IARS, and circ‐PDE8A), HCC (circSETD3, circADAMTS13, hsa_circ_0007874, hsa_circ_104135, circFBLIM1, cSMARCA5, circRNA‐100338, and circPTGR1), colorectal cancer (hsa_circ_0001178, hsa_circ_0000826, hsa_circ_0004771, circDDX17, circITGA7, and circHIPK3), and gastric cancer (hsa_circ_0074362, circNRIP1, circAKT3, circ‐DONSON, circPSMC3, circ‐KIAA1244, circPVRL3, circPVT1, hsa_circ_0000096, ciRS‐133, hsa_circ_0001017, and hsa_circ_0061276).
Conclusion
CircRNAs and exosomal circRNAs have the potential high clinical diagnostic values for GI malignancies.
Keywords: biomarkers, circular RNAs, exosome, gastrointestinal malignancies, non‐invasive diagnosis
1. INTRODUCTION
In the recent few decades, with the accomplishment of Human Genome Project, approximately 20 000 genes encoding proteins have been identified. 1 However, only lower than 2% of genome actually codes proteins. The vast majority of genome, which accounts for almost 98%, is transcribed to non‐coding RNAs (ncRNAs). 2 Compared with microRNAs (miRNAs) and traditional linear RNAs, circular RNAs (circRNAs) resist to ribonuclease R. 3 Because of their covalently closed structure, circRNAs are stable and present in high abundance in blood. 4 , 5 CircRNAs are now considered as potential biomarkers in gastrointestinal (GI) malignancies. 6 , 7 , 8 Thanks to the advancement of high‐throughput functional genomic screening biotechnology, an increasing number of researches have confirmed tumor‐associated circRNAs. 6 , 9
Exosomes are vesicles generated and released by cells. Exosomes, carrying a cargo of lipids, genetic materials, proteins, and their derivatives, modulate cells' behaviors, influence the extracellular system, and may be a source of human disease biomarkers. 10 , 11 Exosomal circRNAs (exo‐circRNAs) have unique profiles reflecting the characteristics of tumors. 10 They would be new GI cancers biomarkers.
Gastrointestinal malignancies, which mainly include gastric cancer (GC), colorectal cancer (CRC), hepatocellular carcinoma (HCC), and pancreatic cancer, are main causes of mortality worldwide and a global burden. For example, pancreatic cancer with only approximately 5% patients surviving over 5 years is the deadliest cancer; CRC is the third most frequent tumor and the second major origin of deaths related to cancers in the USA; HCC is the fourth major origin of cancer‐associated death worldwide; GC is the fifth most frequent tumor and the third major burden associated with cancers in modern society. 12 Nevertheless, most of patients with GI malignancies miss the therapeutic window. 12 When found early, GI malignancies are highly curable. However, the early diagnosis of GI malignancies continues to be a major challenge.
Recent years, researchers have gradually realized that the developments of new technology for non‐invasive early detection are likely to be the most effective method for cutting down mortality of GI malignancies.
Commonly used biomarkers such as carbohydrate antigen 125 (CA125), CA 19‐9, squamous cell carcinoma antigen (SCCA), α fetoprotein (AFP), tissue inhibitor of metalloproteinases 1 (TIMP‐1), cytokeratin 19 fragments (CyFra21‐1), and carcinoembryonic antigen (CEA) could be found not only in serum of patients with GI malignancies but also in serum of healthy individuals. 13 For example, the specificity and sensitivity of CEA to detect GC were 0.686 and 0.593, respectively, and those for CA19‐9 were 0.605 and 0.559, respectively. 14 Obviously, these biomarkers interfere the diagnostic accuracy and sensitivity.
CircRNAs and exo‐circRNAs extracted from plasma are high‐efficient blood‐based biomarkers with great clinical significance. In recent years, with dramatic successes of considerable technological advances and the progression of techniques, such as real‐time and high‐sensitivity liquid biopsy assays, sensitive sequencing, high‐depth targeted next‐generation sequencing, sequencing somatic mutations, polymerase chain reaction (PCR), next‐generation sequencing, and droplet digital PCR (ddPCR) analysis, circRNAs and exo‐circRNAs have appeared potential high clinical values and could be used to detect various GI malignancies. 15 , 16 Compared with lower specificity and sensitivity of CEA and CA19‐9 to detect GC, we explored that the specificity and sensitivity of hsa_circ_0001017 in blood reached 0.794 and 0.811, respectively. 4 Therefore, there is a great hope and promising broad clinical applications in screening GI malignancies through circRNA‐based approaches.
This review summarizes GI cancer‐associated circRNAs and exo‐circRNAs and their possible application in the diagnosis of GI cancers.
2. circRNAs AND EXOSOMES
2.1. The history of circRNA research
CircRNAs were first discovered in viruses 41 years ago. 17 For decades, researchers considered circRNAs as products of splicing errors. Later, in 1996, circRNAs were described as self‐replicating molecules in rodent and human cancer cells. 18 Nonetheless, circRNAs have been re‐evaluated mainly owing to the great advances in deep sequencing and computational approaches. 19
2.2. The biogenesis and physiological functions of circRNAs
Nowadays, a major group of circRNAs have been reported to be generated by different mechanisms. 9 , 19 Most circRNAs are arisen from precursor mRNAs (pre‐mRNAs) through “out‐of‐order” splicing (a process called back‐splicing). 9 , 20 Multiple known circRNAs are from exons of protein‐encoding genes. 19 circRNAs are crucial players involved in numerous biological processes, such as sponging of miRNAs, regulating parental gene transcription and tumorigenicity, protein kinase activation, angiogenic sprouting, generation of short proteins, and post‐transcriptional regulation in the pathogenesis of malignancies. 20 , 21 , 22 , 23 , 24 Additionally, recent functional studies have reported many tissue‐specific and cell‐specific circRNAs, which were functionally characterized as efficient biomarkers in human diseases. 25 For example, ciRS‐7 (also termed CDR1as) has been demonstrated to be a risk factor through sponging corresponding miRNAs. 26 , 27 CircSETD3 (hsa_circ_0000567) and circADAMTS13 also act as sponges of miRNAs and participate in HCC tumorigenesis. 28 , 29 Strikingly, hundreds of circRNAs including ciRS‐7 are highly abundant in the mammalian brains. 30 Some researchers believe that ciRS‐7 could be possibly used as therapeutic target agents against Alzheimer's neuronal injury. 31 Moreover, several studies showed that the expression of circRNAs elucidated various cardiovascular and cerebrovascular diseases. 32 , 33 Furthermore, some circRNAs may be potential therapeutic targets of gestational diabetes mellitus. 34 Similarly, circHIPK3 was identified as an applicable therapeutic intervention for diabetic proliferative retinopathy. 35
2.3. The functions of exosomes
Exosomes (diameter, 30‐160 nm) act on many biological processes and participate in substance delivery and signal transduction. 36 Exosome formation occurs spontaneously within the endosomal network via re‐routing of multivesicles and are released through exocytosis. 37 Trams et al described the concept of exosomes as microvesicles composed of plasma membrane in 1981. 38 In 1985, Pan et al confirmed the presence of exosomes inside multivesicular endocytic compartments using immunoelectron microscopy. 39 Nonetheless, the official name of these structures as “exosomes” was provided by Johnstone et al in 1987. 40 Several recent dynamic light scattering, transmission, and atomic force microscopy studies have helped us address the exosomes function sites. 41 , 42 Several groups have performed diverse studies on potential biological functions of exosomes in medicine. 43 , 44 For instance, an exosome‐based drug delivery system with high application potential in cancer targeted therapy was reported. 43 Additionally, cancer‐derived exosomes show a great potential for acting as biomarkers. 45 These studies have provided new ideas about how exosomes may be used to treat cancers efficiently and effectively. However, exosomes may present as two‐edged sword. For instance, researchers found that exosomes play critical roles in neurological disorders and could spread pathological proteins in these neuropsychiatric deficits, such as ischemic stroke, Parkinson neurodegeneration, brain infarction, and Alzheimer's neuronal injury. 46 Unfortunately, some tumor‐derived exosomes may promote cancer progression and accelerate metastasis by modulating the primary site of tumors and assisting metastatic cancer cells escape from immunologic surveillance. 47
3. circRNAs AS BIOMARKERS IN GI MALIGNANCIES
Most advanced GI malignancies and their metastases are difficult to be cured. In contrast, patients with early GI cancer can be treated effectively. However, the main clinical imaging equipments such as digital X‐ray radiography, magnetic resonance imaging (MRI), and positron emission tomography‐computed tomography (PET‐CT) are not only expensive but also often quite subjective. Therefore, circRNAs and exo‐circRNAs as potential effective biomarkers may provide opportunities to properly treat patients with GI malignancies (Figure 1). CircRNAs are not easily degraded by enzymes such as exonucleases and ribonucleases. They have a greater stability and a longer half‐life in body fluids. Additionally, the expression of circRNAs in fluids and tissues often provides relatively high sensitivity and specificity in different developmental stages of GI malignancy patients. Therefore, circRNAs and exo‐circRNAs may be applied as biomarkers for GI cancer patients (Figure 2).
FIGURE 1.
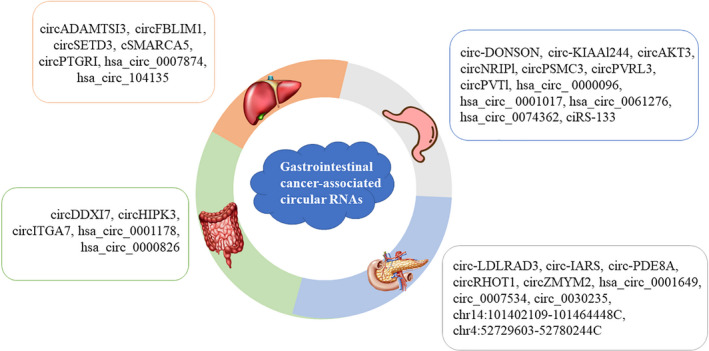
Gastrointestinal cancer‐associated circular RNAs
FIGURE 2.
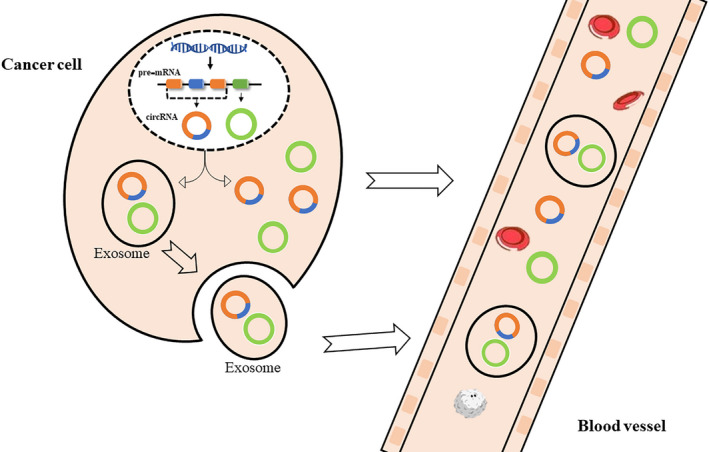
CircRNAs and exosomal circRNAs used as biomarkers. In the nuclei of cancer cells, circRNAs are produced by pre‐mRNA after transcription. The formation of circRNAs may involve one exon, several exons, several introns, or only one intron. CircRNAs may be released into the blood directly or with exosomes
3.1. circRNAs as biomarkers of gastric cancer
Gastric cancer is a main origin of death in the modern society. 48 Recent years, researchers have investigated circRNAs as biomarkers of GC. Based on RNA sequencing biotechnology, circNRIP1 and circAKT3 (hsa_circ_0000199) were explored to be increasingly expressed in human GC tissues; the increments of circNRIP1 and circAKT3 in human GC tissues were related to the metastasis potential and recurrence risk. 49 , 50 After purifying exosomes, Zhang et al found upregulated expression of circNRIP1 in GC patients. 49 Their findings suggested that high levels of circNRIP1 indicated reduced survival. 49 Huang et al found that the upregulation of circAKT3 in GC patients elucidated reduced five‐year disease‐free survival (DFS). 50 The area under the curve (AUC) is 0.91, which elucidated that circAKT3 could be biomarker to predict survival. 50 In addition, circ‐DONSON (hsa_circ_0004339) was markedly increased in GC tissues. 51 The expression of circ‐DONSON was related to DFS, confirming circ‐DONSON as a novel GC biomarker. 51
In contrast, the expression of circPSMC3 and circ‐KIAA1244 was significantly reduced in GC patients' plasmas. 52 , 53 Mechanism study elucidated that circPSMC3 might regulate the tumor progression through sponging miR‐296‐5p and might be applied as a potential therapeutic target of GC. 52 Sun et al found that the attenuated expression of circPVRL3 could boost GC cells growth. 54
Our group measured the expression profiles of circRNAs in cancer plasma from GC patients. 4 , 6 Aberrant expression of 308 circRNAs was demonstrated in GC tissues. 6 Total 343 circRNAs in plasma from GC patients were further found. 4 The above studies elucidated that circRNAs have advantages on the potential novel biomarkers of GC.
3.2. circRNAs as biomarkers of hepatocellular carcinoma
Currently, the incidence of HCC is supreme in Central America. 55 Thousands of patients with HCC were diagnosed at late stages and lost the best opportunity of a surgical cure. Reduced circMTO1 expression in HCC was related to irreversible miss of therapeutic window for HCC patients. 56 Bai et al discovered that silencing of circFBLIM1 could markedly inhibit HCC cells by directly sponging miR‐346. 57 Additionally, underlying mechanism experiments showed that circSETD3 suppresses tumor growth by activating the cascade pathways. 29 Furthermore, cSMARCA5 is known to act an essential role in the tumorigenesis of HCC. 58 Our group found that hsa_circ_0068669 expression elucidated stages of HCC patients. 8 These studies helped us uncover the great clinical value of circRNAs as biomarkers of HCC.
3.3. circRNAs as biomarkers of colorectal cancer
Colorectal cancer is common malignancies and the second cancer killer in the USA. 12 Increasing the screening rate of CRC among people and finding effective treatment targets of CRC is imperative. Recently, scholars have identified circRNAs as effective biomarkers of CRC. Known to be associated with unfavorable clinicopathological factors, hsa_circ_0001178 and hsa_circ_0000826 clearly upregulated in CRC mucosae compared with adjacent normal mucosae. 59 The AUCs were 0.816 and 0.945 for hsa_circ_0000826 and hsa_circ_0001178, respectively. 59 Moreover, circDDX17 (hsa_circ_0002211) expression was lower in the CRC mucosae; and knockdown of circDDX17 could accelerate CRC cell growth and differentiation as well as suppress apoptosis. 60 These results suggest that circDDX17 could serve as a tumor suppressor. Li et al further found that reduced circDDX17 expression was related to distant stage of CRC. 60
Additionally, circITGA7 markedly inhibited CRC tumorigenesis. 61 Interestingly, the level of circHIPK3 was notably higher. 62 Moreover, increased expression of circHIPK3 was related to CRC distant metastasis, T status of tumor, and advanced clinical stage. 62 These findings highlight circRNAs as biomarkers for CRC.
3.4. circRNAs as biomarkers of pancreatic cancer
Pancreatic cancer is largely incurable cancer with the lowest survival in the worldwide. 12 , 63 This challenge has attracted the interest of researchers, who have investigated circRNAs as new biomarkers of pancreatic cancer. Recently, two distinctly expressed circRNAs (chr14:101402109‐101464448C and chr4:52729603‐52780244C) were demonstrated to be novel potential biomarkers and drug targets of pancreatic ductal adenocarcinoma (PDAC). 64 Circ_0007534 and circ_0030235 were markedly upregulated in PDAC; the expression of these circRNAs elucidated the poor prognosis of PDAC patients. 65 , 66 Furthermore, the expression of hsa_circ_0007534 indicated lymph node invasion. 65 Xu et al found that positive lymph node invasion and higher tumor stage were associated with high expression of hsa_circ_0030235. 66
CircRHOT1 (hsa_circ_0005397) and circ‐LDLRAD3 were also overexpressed in pancreatic cancer cells. 67 , 68 The circ‐LDLRAD3 in plasmas of PDAC patients was associated with stages. 68 In addition, hsa_circ_0001649 was significantly downregulated in cells and tissue specimens of PDAC. 69 Jiang et al confirmed that the low expression of hsa_circ_0001649 elucidated irreversible differentiation grade and stage through Fisher's exact tests. 69 Jiang et al found that high hsa_circ_0001649 expression elucidated high overall survival (OS) rate. 69 These studies verified that hsa_circ_0001649 could be applied as a biomarker of PDAC.
4. EXOSOMAL circRNAs AS BIOMARKERS OF GI MALIGNANCIES
Exosomes have been the increasingly interesting particles in medicine and biotechnology, especially for using as biomarkers in clinical diagnosis. 37 , 70 Their molecular constituents such as proteins, miRNAs, and circRNAs are stable and promising as diagnostic biomarkers in GI cancers. 71 , 72 Multiple previous studies have made the point that numerous tumor‐derived or tumor‐associated exo‐circRNAs could be reliable tumor‐specific biomarkers of GI cancers. 73 , 74 , 75 , 76 , 77 , 78 , 79 For instance, circulating exosomal ciRS‐133 (hsa_circ_0010522) extracted from GC patients' serum and plasma samples was markedly higher compared to normal subjects. 73 Wang et al also investigated that the expression levels of circPTGR1 (hsa_circ_0008043) in HCC patients' serum exosomes were clearly upregulated. 74 Li et al discovered that the expression levels of exosomal circRNA IARS (circ‐IARS) in pancreatic cancer tissues and plasma specimens were significantly higher than healthy controls. 75 Additionally, circulating exosomal circRNAs PDE8A (circ‐PDE8A) derived from PDAC patients' plasma exosomes was also abnormally overexpressed in pancreatic cancer patients; and its levels were closely related to the tumor invasion, clinicopathological characteristics, and survival of PDAC patients. 76 Besides, Pan et al showed that hsa_circ_0004771 in serum exosomes from early CRC patients was significantly increased. 77 The fact that the AUC was reached to 0.90 elucidated that exosomal hsa_circ_0004771 could be underlying biomarker to predict early CRC. 77 In addition, circRNA‐100338 was markedly increased in HCC patients' exosomes. 78 Its expression was related to DFS and metastases, confirming circRNA‐100338 as a novel HCC biomarker. 78
5. CONCLUSIONS AND FUTURE DIRECTIONS
There is great hope that circRNAs and exo‐circRNAs may be used in medical oncology (Table 1). Our understanding of exosomes has rapidly advanced over the last few years. We look forward to the day of application of circRNAs and exo‐circRNAs in the diagnosis and treatment of GI malignancies.
TABLE 1.
Overview of identified gastrointestinal malignancy‐associated circRNAs
| Type of cancer | CircRNA | Expression | Host gene | Chromosome | Putative function | Potential clinical value | Type of biomarker | Target | Sample detected | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Gastric cancer | hsa_circ_0000096 | Down | HIAT1 | Chr1 | miRNA sponge | Biomarker | Diagnosis | Not investigated | Tissues | [79] |
| hsa_circ_0000190 | Down | CNIH4 | Chr1 | miRNA sponge | Biomarker | Diagnosis | Not investigated | Tissues and plasma | [17] | |
| hsa_circ_0005654 | Down | PRDM5 | Chr4 | miRNA sponge | Biomarker | Diagnosis | Not investigated | Tissues | [80] | |
| hsa_circ_0032627 (circDLST) | Up | DLST | Chr14 | miRNA sponge | Biomarker | Prognosis | miR‐502‐5p | Tissues | [81] | |
| hsa_circ_0004339 (circ‐DONSON) | Up | DONSON | Chr21 | via NURF complex | Biomarker | Prognosis | Not investigated | Tissues | [51] | |
| hsa_circ_0002320 (circYAP1) | Down | YAP1 | Chr11 | miRNA sponge | Biomarker | Prognosis | miR‐367‐5p/p27 Kip1 axis | Tissues | [82] | |
| hsa_circ_0000199 (circAKT3) | Up | AKT3 | Chr1 | miRNA sponge | Therapeutic target | Prognosis | miR‐198 | Tissues | [49] | |
| hsa_circ_0081143 | Up | COL1A2 | Chr7 | miRNA sponge | Biomarker | Prognosis | miR‐646 | Tissues | [83] | |
| hsa_circ_0000993 | Up | ATL2 | Chr2 | miRNA sponge | Biomarker | Prognosis | miR‐214‐5p | Tissues | [84] | |
| hsa_circ_0000140 | Down | KIAA0907 | Chr1 | Not investigated | Biomarker | Diagnosis | Not investigated | Tissues and plasma | [85] | |
| circLARP4 | Up | LARP4 | Chr12 | miRNA sponge | Biomarker | Prognosis | miR‐424‐5p | Tissues | [86] | |
| hsa_circ_0130810 (circ‐KIAA1244) | Down | KIAA1244 | Chr6 | Not investigated | Biomarker | Diagnosis and prognosis | Not investigated | Tissues, plasma and cells | [53] | |
| hsa_circ_0000745 | Down | SPECC1 | Chr17 | Not investigated | Biomarker | Diagnosis | Not investigated | Tissues, plasma and cells | [87] | |
| circ‐PSMC3 | Down | PSMC3 | Chr11 | miRNA sponge | Biomarker | Diagnosis and prognosis | miR‐296‐5p | Tissues and cells | [52] | |
| Hepatocellular cancer | hsa_circ_0016788 | Up | TRIM11 | Chr1 | Not investigated | Biomarker | Diagnosis | miR‐486/CDK4 pathway | Tissues and cells | [88] |
| hsa_circ_0007874 (circMTO1) | Down | MTO1 | Chr6 | miRNA sponge | Biomarker and therapeutic target | Diagnosis | miR‐9 | Tissues and cells | [56] | |
| hsa_circ_0001445 (cSMARCA5) | Down | SMARCA5 | Chr4 | miRNA sponge | Biomarker and therapeutic target | Prognosis | miR‐17‐3p and miR‐181b‐5p | Tissues | [58] | |
| hsa_circ_0010090 (circFBLIM1) | Up | FBLIM1 | Chr1 | miRNA sponge | Biomarker | Diagnosis | miR‐346 | Tissues and cells | [57] | |
| circZKSCAN1 | Down | ZKSCAN1 | Chr7 | ceRNA | Biomarker | Diagnosis | Not investigated | Tissues | [89] | |
| hsa_circ_0000567 (circSETD3) | Down | SETD3 | Chr14 | miRNA sponge | Biomarker | Prognosis | miR‐421 | Tissues and cells | [29] | |
| hsa_circRNA86 62‐12 (circTRIM33‐12) | Down | TRIM33‐12 | Chr1 | miRNA sponge | Biomarker | Prognosis | miR‐191 | Tissues and cells | [90] | |
| hsa_circ_0005075 | Up | EIF4G3 | Chr1 | miRNA sponge | Therapeutic target | Diagnosis | miR‐431 | Tissues and cells | [91] | |
| hsa_circ_0072088 (circZFR) | Down | ZFR | Chr5 | miRNA sponge | Biomarker | Not investigated | miR‐511 | Tissues and cells | [92] | |
| circSMARCA5 | Down | SMARCA5 | Chr4 | Not investigated | Biomarker | Diagnosis | Not investigated | Tissues, plasma and cells | [93] | |
| hsa_circ_0000284 (circHIPK3) | Up | HIPK3 | Chr11 | miRNA sponge | Not investigated | Not investigated | Not investigated | Not investigated | [94] | |
| Colorectal cancer | hsa_circ_0009361 | Down | GNB1 | Chr1 | miRNA sponge | Biomarker | Diagnosis | miR‐582 | Tissues and cells | [95] |
| hsa_circ_0136666 | Up | PRKDC | Chr8 | miRNA sponge | Biomarker and therapeutic target | Prognosis | miR‐136 | Tissues and cells | [96] | |
| hsa_circ_0006990 (circVAPA) | Up | VAPA | Chr18 | miRNA sponge | Biomarker and therapeutic target | Diagnosis | miR‐101 | Tissues and plasma | [60] | |
| hsa_circ_0055625 (circ_0055625) | Up | DUSP2 | Chr2 | miRNA sponge | Biomarker | Diagnosis | miR‐106b‐5p | Tissues | [97] | |
| hsa_circ_0000284 (circHIPK3) | Up | HIPK3 | Chr11 | miRNA sponge | Biomarker and therapeutic target | Prognosis | miR‐7 | Tissues and cells | [62] | |
| hsa_circ_0142527 | Down | PREX2 | Chr8 | miRNA sponge | Biomarker | Diagnosis | Not investigated | Tissues | [98] | |
| Pancreatic cancer | hsa_circ_0005397 (circRHOT1) | Up | RHOT1 | Chr17 | miRNA sponge | Therapeutic target | Not investigated | miR‐26b, miR‐125a, miR‐330, and miR‐382 | Tissues and cells | [99] |
Here, we speculate on two potential future directions for circRNAs and exo‐circRNAs. First, circRNAs and exo‐circRNAs as novel clinical biomarkers of GI malignancies will become the cornerstones of new clinical laboratory analysis methods. By detecting circRNAs and exo‐circRNAs, clinical doctors may diagnose early‐stage GI malignancies, monitor the recurrence and metastasis of GI malignancies, and predict the postoperative survival time. Second, circRNAs and exo‐circRNAs as potential therapeutic targets for GI malignancies may help clinicians extend advanced GI malignancies patients' life and improve their quality of life.
CONFLICT OF INTEREST
None.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (no. 81772279), the Scientific Innovation Team Project of Ningbo (no. 2017C110019), the National Undergraduate Training Program for Innovation and Entrepreneurship (no. 201811646025), the Student Research and Innovation Program of Ningbo University (no. 2017SRIP1918, no. 2018SRIP2507 and no. 2019SRIP1902), and the K. C. Wong Magna Found in Ningbo University.
Wang Y, Li Z, Xu S, Guo J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal. 2020;34:e23359 10.1002/jcla.23359
REFERENCES
- 1. Lupski JR. A human in human genetics. Cell. 2019;177(1):9‐15. [DOI] [PubMed] [Google Scholar]
- 2. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32(4):e22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT‐PCR detection. J Mol Med (Berl). 2018;96(1):85‐96. [DOI] [PubMed] [Google Scholar]
- 5. Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(1):9‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3):e22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yao T, Chen Q, Shao Z, Song Z, Fu L, Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;32(8):e22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018;7(7):3101‐3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 13. Phallen J, Sausen M, Adleff V, et al. Direct detection of early‐stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403);eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167‐171. [DOI] [PubMed] [Google Scholar]
- 15. Gkountela S, Castro‐Giner F, Szczerba BM, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1–2):98‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsu MT, Coca‐Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339‐340. [DOI] [PubMed] [Google Scholar]
- 18. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93(13):6536‐6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao T, Chen Q, Fu L, Guo J. Circular RNAs: biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47(6):497‐504. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y. Comment on “Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer,” Cancer Lett. 2020 Jan 2; 473(2020);139–147. Cancer Lett. 2020;425:1. [DOI] [PubMed] [Google Scholar]
- 21. Zhang JX, Lu J, Xie H, et al. circHIPK3 regulates lung fibroblast‐to‐myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10(3):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865‐880. [DOI] [PubMed] [Google Scholar]
- 23. Boeckel JN, Jaé N, Heumüller AW, et al. Identification and characterization of hypoxia‐regulated endothelial circular RNA. Circ Res. 2015;117(10):884‐890. [DOI] [PubMed] [Google Scholar]
- 24. Huang MS, Zhu T, Li L, et al. LncRNAs and CircRNAs from the same gene: masterpieces of RNA splicing. Cancer Lett. 2018;415:49‐57. [DOI] [PubMed] [Google Scholar]
- 25. Xia S, Feng J, Chen K, et al. CSCD: a database for cancer‐specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925‐D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu B, Yang T, Wang Z, Zhang Y, Liu S, Shen M. CircRNA CDR1as/miR‐7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871‐4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR‐484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA‐421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piwecka M, Glažar P, Hernandez‐Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357);eaam8526. [DOI] [PubMed] [Google Scholar]
- 31. Huang JL, Xu ZH, Yang SM, et al. Identification of differentially expressed profiles of Alzheimer's disease associated circular RNAs in a panax notoginseng saponins‐treated Alzheimer's disease mouse model. Comput Struct Biotechnol J. 2018;16:523‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR‐15b. J Mol Cell Cardiol. 2019;130:10‐22. [DOI] [PubMed] [Google Scholar]
- 33. Kong P, Yu Y, Wang L, et al. circ‐Sirt1 controls NF‐κB activation via sequence‐specific interaction and enhancement of SIRT1 expression by binding to miR‐132/212 in vascular smooth muscle cells. Nucleic Acids Res. 2019;47(7):3580‐3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bai C, Yang W, Lu Y, Wei W, Li Z, Zhang L. Identification of circular RNAs regulating islet β‐cell autophagy in type 2 diabetes mellitus. Biomed Res Int. 2019;2019:4128315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629‐1642. [DOI] [PubMed] [Google Scholar]
- 36. Karnati HK, Garcia JH, Tweedie D, et al. Neuronal enriched extracellular vesicle proteins as biomarkers for traumatic brain injury. J Neurotrauma. 2019;36(7):975‐987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177(2):428‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trams EG, Lauter CJ, Salem N Jr, Heine U. Exfoliation of membrane ecto‐enzymes in the form of micro‐vesicles. Biochim Biophys Acta. 1981;645(1):63‐70. [DOI] [PubMed] [Google Scholar]
- 39. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262(19):9412‐9420. [PubMed] [Google Scholar]
- 41. Genschmer KR, Russell DW, Lal C, et al. Activated PMN exosomes: Pathogenic entities causing matrix destruction and disease in the lung. Cell. 2019;176(1–2):113‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu H, Han H, Song S, et al. Exosome‐transmitted PSMA3 and PSMA3‐AS1 promote proteasome inhibitor resistance in multiple myeloma. Clin Cancer Res. 2019;25(6):1923‐1935. [DOI] [PubMed] [Google Scholar]
- 43. Kibria G, Ramos EK, Wan Y, Gius DR, Liu H. Exosomes as a drug delivery system in cancer therapy: potential and challenges. Mol Pharm. 2018;15(9):3625‐3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang B, Chen Y, Shi J. Exosome biochemistry and advanced nanotechnology for next‐generation theranostic platforms. Adv Mater. 2019;31(2):e1802896. [DOI] [PubMed] [Google Scholar]
- 45. Soung YH, Ford S, Zhang V, Chung J. Exosomes in cancer diagnostics. Cancers (Basel). 2017;9(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi M, Sheng L, Stewart T, Zabetian CP, Zhang J. New windows into the brain: central nervous system‐derived extracellular vesicles in blood. Prog Neurogibol. 2019;175:96‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li K, Chen Y, Li A, Tan C, Liu X. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144(7):1486‐1495. [DOI] [PubMed] [Google Scholar]
- 48. Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12(1):11‐20. [DOI] [PubMed] [Google Scholar]
- 49. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA‐149‐5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding L, Zhao Y, Dang S, et al. Circular RNA circ‐DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR‐296‐5p. Mol Cancer. 2019;18(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun HD, Xu ZP, Sun ZQ, et al. Down‐regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8(1):10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD‐3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population‐based registries in 71 countries. Lancet. 2018;391(10125):1023‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151‐1164. [DOI] [PubMed] [Google Scholar]
- 57. Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR‐346. J Exp Clin Cancer Res. 2018;37(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214‐1227. [DOI] [PubMed] [Google Scholar]
- 59. Xu H, Wang C, Song H, Xu Y, Ji G. RNA‐Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166‐179. [DOI] [PubMed] [Google Scholar]
- 62. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR‐7. Cell Death Dis. 2018;9(4):417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63(5):318‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shao F, Huang M, Meng F, Huang Q. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol. 2018;9:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hao L, Rong W, Bai L, et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR‐625 and miR‐892b. J Cell Biochem. 2019;120(3):3780‐3789. [DOI] [PubMed] [Google Scholar]
- 66. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR‐1253 and miR‐1294. Biochem Biophys Res Commun. 2019;509(1):138‐142. [DOI] [PubMed] [Google Scholar]
- 67. Qu S, Hao X, Song W, et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11(1):53‐63. [DOI] [PubMed] [Google Scholar]
- 68. Yang F, Liu DY, Guo JT, et al. Circular RNA circ‐LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345‐8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88‐93. [DOI] [PubMed] [Google Scholar]
- 70. Pluchino S, Smith JA. Explicating exosomes: reclassifying the rising stars of Intercellular Communication. Cell. 2019;177(2):225‐227. [DOI] [PubMed] [Google Scholar]
- 71. Baghaei K, Tokhanbigli S, Asadzadeh H, Nmaki S, Reza Zali M, Hashemi SM. Exosomes as a novel cell‐free therapeutic approach in gastrointestinal diseases. J Cell Physiol. 2019;234(7):9910‐9926. [DOI] [PubMed] [Google Scholar]
- 72. Nedaeinia R, Manian M, Jazayeri MH, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24(2):48‐56. [DOI] [PubMed] [Google Scholar]
- 73. Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR‐133/PRDM16 pathway. Int J Cancer. 2019;144(10):2501‐2515. [DOI] [PubMed] [Google Scholar]
- 74. Wang G, Liu W, Zou Y, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a‐MET pathway. EBioMedicine. 2019;40:432‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ‐IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li Z, Yanfang W, Li J, et al. Tumor‐released exosomal circular RNA PDE8A promotes invasive growth via the miR‐338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237‐250. [DOI] [PubMed] [Google Scholar]
- 77. Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa‐circ‐0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang XY, Huang ZL, Huang J, et al. Exosomal circRNA‐100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang Y, Xu S, Chen Y, Zheng X, Li T, Guo J. Identification of hsa_circ_0005654 as a new early biomarker of gastric cancer. Cancer Biomark. 2019;26(4):403‐410. [DOI] [PubMed] [Google Scholar]
- 81. Zhang J, Hou L, Liang R, et al. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR‐502‐5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 2019;18(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu H, Liu Y, Bian Z, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR‐367‐5p/p27 Kip1 axis [published correction appears in Mol Cancer. 2019 Jul 9;18(1):117]. Mol Cancer. 2018;17(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xue M, Li G, Fang X, Wang L, Jin Y, Zhou Q. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR‐646/CDK6 pathway. Cancer Cell Int. 2019;19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhong S, Wang J, Hou J, et al. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. 2018;10(10):1301‐1313. [DOI] [PubMed] [Google Scholar]
- 85. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. [DOI] [PubMed] [Google Scholar]
- 86. Zhang J, Hu H, Zhao Y, Zhao Y. CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour's progression via miR‐7 signals. Cell Prolif. 2018;51(6):e12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330‐6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR‐486/CDK4 pathway. J Cell Physiol. 2018;234(1):500‐508. [DOI] [PubMed] [Google Scholar]
- 89. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways [published correction appears in Mol Oncol. 2019 Nov; 13(11):2511]. Mol Oncol. 2017;11(4):422‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang PF, Wei CY, Huang XY, et al. Circular RNA circTRIM33‐12 acts as the sponge of MicroRNA‐191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li MF, Li YH, He YH, et al. Emerging roles of hsa_circ_0005075 targeting miR‐431 in the progress of HCC. Biomed Pharmacother. 2018;99:848‐858. [DOI] [PubMed] [Google Scholar]
- 92. Yang X, Liu L, Zou H, Zheng YW, Wang KP. circZFR promotes cell proliferation and migration by regulating miR‐511/AKT1 axis in hepatocellular carcinoma. Dig Liver Dis. 2019;51(10):1446‐1455. [DOI] [PubMed] [Google Scholar]
- 93. Li Z, Zhou Y, Yang G, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37‐44. [DOI] [PubMed] [Google Scholar]
- 94. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Geng Y, Zheng X, Hu W, et al. Hsa_circ_0009361 acts as the sponge of miR‐582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond). 2019;133(10):1197‐1213. [DOI] [PubMed] [Google Scholar]
- 96. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR‐136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247‐7256. [DOI] [PubMed] [Google Scholar]
- 97. Zhang J, Liu H, Zhao P, Zhou H, Mao T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR‐106b‐5p. J Cell Biochem. 2019;120(3):3027‐3037. [DOI] [PubMed] [Google Scholar]
- 98. Ge J, Jin Y, Lv X, et al. Expression profiles of circular RNAs in human colorectal cancer based on RNA deep sequencing. J Clin Lab Anal. 2019;33(7):e22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846‐2858. [DOI] [PMC free article] [PubMed] [Google Scholar]


