Abstract
Objective
We aimed to evaluate the correlation of circular RNA La‐related RNA‐binding protein 4 (circ‐LARP4) with tumor characteristics and prognosis, and its effect on chemosensitivity in breast cancer.
Methods
Circ‐LARP4 from tumor and adjacent tissues of 283 female breast cancer patients underwent resection was detected by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Tumor features, disease‐free survival (DFS), and overall survival (OS) were recorded. In vitro, circ‐LARP4 in human normal mammary epithelial cells (HMEC) and breast cancer cell lines was detected by RT‐qPCR. MCF‐7 and MDA‐MB‐231 cells were transfected with circ‐LARP4 overexpression plasmid (as OE‐Circ group) and control overexpression plasmid (as OE‐Control group). Relative cell viability under different concentrations of doxorubicin was measured.
Results
Circ‐LARP4 was decreased in tumor tissues than adjacent tissues (P < .001). Tumor circ‐LARP4 negatively correlated with tumor size (P = .001), T stage (P = .009), N stage (P = .006), and TNM stage (P < .001), whereas positively correlated with DFS (P = .004) and OS (P < .001). In vitro, circ‐LARP4 was decreased MCF‐7, BT474, MDA‐MB‐231, and MDA‐MB‐468 cell lines than HMEC (all P < .001). Relatively cell viability of MCF‐7 cells (at 20 nmol/L [P < .05], 40 nmol/L [P < .01], 80 nmol/L [P < .05] of doxorubicin) and MDA‐MB‐231 cells (at 120 nmol/L [P < .05], 240 nmol/L [P < .05] of doxorubicin) was decreased in OE‐Circ group than OE‐Control group. IC50 value of doxorubicin was decreased in OE‐Circ group than OE‐Control group in MCF‐7 and MDA‐MB‐231 cell lines (both P < .01).
Conclusion
Circ‐LARP4 was a potential prognostic biomarker, which might improve the management of breast cancer.
Keywords: breast cancer, chemosensitivity, circular RNA LARP4, prognosis, tumor stage
1. INTRODUCTION
Breast cancer, one of the most frequent cancers worldwide, is a complex disease that exhibits largely heterogeneity.1, 2 Surgery is generally set as the primary option for the management of breast cancer in most cases.3 Meanwhile, the systematic therapy of breast cancer is also widely administrated along with the surgery (neoadjuvant/adjuvant therapy), which varies among endocrine therapy, chemotherapy, targeted therapy and immunotherapy depending on the molecular subtype of breast cancer and disease extent of patients.4, 5 Although the treatment of breast cancer has been improved greatly, breast cancer is still the leading cause of cancer‐related death in women, which causes over 500 000 cases of death annually.6, 7 Therefore, it is of great importance to search for novel biomarkers to assist the management of breast cancer patients, thus improving the prognosis of them.
Circular RNA La‐related RNA‐binding protein 4 (circ‐LARP4) is derived from the LARP4 gene, which displays anti‐tumor property in several cancers via sponging several microRNAs (miR) including miR‐424 and miR‐761 and activating several apoptosis‐related pathways such as p53/p21 signaling.8, 9 Moreover, circ‐LARP4 is reported to be a potential prognostic biomarker in ovarian cancer patients and osteosarcoma patients.10, 11 Therefore, we hypothesized that circ‐LARP4 might also act as a tumor suppressor in breast cancer, whereas relevant information is unclear. In this study, we investigated the correlations of circ‐LARP4 with cancer characteristics and prognosis in 283 female patients who underwent resection due to primary breast cancer, and explored the effect of circ‐LARP4 on chemosensitivity in breast cancer cell lines.
2. MATERIALS AND METHODS
2.1. Patients
This prospective study consisted of 283 female patients who underwent resection in The Central Hospital of Wuhan due to primary breast cancer between July 2014 and June 2018. All patients were newly diagnosed as primary breast cancer and prepared for resection. And the patients who were less than 18 years old or above 75 years old, presented with initial distant metastases, complicated with other malignancies, or received neoadjuvant therapy were excluded. Besides, pregnant or lactating women were also excluded from this study. The Ethics Committee of The Central Hospital of Wuhan approved this study, and all procedures were conducted following the Declaration of Helsinki. Each patient provided written informed consent before enrollment.
2.2. Collection of clinical features and samples
Tumor features including tumor differentiation, tumor size, T stage, N stage, TNM stage, hormone receptor (HR) status (including estrogen receptor (ER) status and progesterone receptor (PR) status) and human epithelial growth factor receptor‐2 (HER‐2) status of patients before resection were recorded. HR positive (HR+) was defined as the patients presented with ER positive (ER+) or/and PR positive (PR+), and HR negative (HR−) was defined as the patients presented with ER negative (ER−) and PR negative (PR−). Tumor tissue and adjacent tissue resected from surgery were stored in liquid nitrogen immediately, and the relative expression of circ‐LARP4 in tumor tissue and adjacent tissue was detected by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
2.3. Follow‐up
Regular follow‐up was carried out for all patients, and the last follow‐up date was June 30, 2019. The follow‐up duration was ranging from 2.0 months to 60.0 months, and the median follow‐up duration was 39.0 months. During follow‐up, the relapse events or death events were documented. Disease‐free survival (DFS) was defined as the duration from resection to relapse or death. Overall survival (OS) was defined as the duration from resection to death. For the patients not known whether relapse or death at the last follow‐up date, they were censored on the date of the last visit or the date of last known to be alive.
2.4. Cell culture
Human breast cancer cell lines (MCF‐7, BT474, MDA‐MB‐468, and MDA‐MB‐231) and human normal mammary epithelial cells (HMEC) were purchased from the American Type Culture Collection. MCF‐7 and BT474 cells were cultured in 90% Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco) with 10% fetal bovine serum (FBS) (Gibco). The MDA‐MB‐468 and MDA‐MB‐231 cells were cultured in 90% Leibovitz's L‐15 medium (Gibco) supplemented with 10% FBS (Gibco). In addition, all cells were maintained at 37°C with 95% air and 5% CO2. After culture, the relative expression of circ‐LARP4 in the cells was detected by RT‐qPCR.
2.5. Transfection and chemosensitivity
The circ‐LARP4 overexpression plasmid was constructed using pCD5‐ciR vector (Geneseed), and then, the overexpression plasmid was transfected into MCF‐7 and MDA‐MB‐231 cells using HilyMax (Dojindo). Meanwhile, a control overexpression plasmid was also constructed and transfected into MCF‐7 and MDA‐MB‐231 cells as the method described above. The cells transfected with circ‐LARP4 overexpression plasmid were defined as OE‐Circ group, and the cells transfected with control overexpression plasmid were defined as OE‐Control group. After transfection for 48 hours, both OE‐Circ cells and OE‐Control cells were treated with different concentrations of doxorubicin for additional 48 hours. Then, Cell Counting Kit‐8 (CCK‐8, Dojindo Molecular Technologies) was used to determine the viability of cells incubated with different concentrations of doxorubicin. The procedures were performed according to the manufacturer's manual. The relative cell viability was calculated referring the viability of cells treated by 0 nmol/L doxorubicin in each group. Finally, the doxorubicin concentration required to inhibit growth by 50% (IC50) value was estimated by Probit regression analysis using SPSS 24.0 software (IBM).
2.6. Detection of circ‐LARP4
The relative expression of circ‐LARP4 in tissue samples and cells was detected by RT‐qPCR. Total RNA was extracted using TRIzol™ Reagent (Invitrogen™). RNase R (Epicentre) was used to digest linear RNAs for circ‐LARP4 detection. Reverse transcription was conducted by iScript™ cDNA Synthesis Kit (Bio‐Rad). qPCR was performed using QTaq™ DNA Polymerase Mix (Clontech). All reagents or kits were used according to a previous research12 with modifications or under the manufacturers' instructions. GAPDH was set as the internal reference according to previous studies.11, 13, 14 For GAPHD detection, total RNA was treated as the above‐mentioned, but the digestion of linear RNAs was skipped. The relative expression of circ‐LARP4 was calculated by the 2−ΔΔ C t formula. The original sequence of human has‐circ‐LARP4 in breast tumor tissue as well as adjacent tissue was obtained from Tissue‐Specific CircRNA Database (http://gb.whu.edu.cn/TSCD/), and the primers were designed with Primer Premier 6 (PREMIER Biosoft). Primers (5′‐3′): circ‐LARP4 forward primer, GAGACCAAGTCATAAGCGTTGTATT; reverse primer, AAACCAGTTCCTTTAGATGCTACCT. GAPDH forward primer, TGACCACAGTCCATGCCATCAC; reverse primer, GCCTGCTTCACCACCTTCTTGA.
2.7. Statistical analysis
Statistical analysis was performed using SPSS 24.0 software (IBM), and figure was plotted using GraphPad Prism 7.00 software (GraphPad Software). Comparison of circ‐LARP4 between paired groups (tumor tissue and adjacent tissue) was determined by Wilcoxon signed‐rank test. Comparison of baseline characteristics between circ‐LARP4 high group and circ‐LARP4 low group was determined by Student's t test, chi‐square test, or Wilcoxon rank‐sum test. Multiple comparisons of circ‐LARP4 relative expression among different cell lines were determined by Dunnett's t test (HMEC as control). Comparison of relative cell viability between OE‐Control group and OE‐Circ group under the same concentration of doxorubicin was determined by Student's t test. Comparison of IC50 value between OE‐Control group and OE‐Circ group was determined by Student's t test. Kaplan‐Meier curve was plotted to illustrate DFS and OS, and the differences of DFS and OS between circ‐LARP4 high group and circ‐LARP4 low group were determined by log‐rank test. P value <.05 was considered as significant.
3. RESULTS
3.1. Circ‐LARP4 in tumor tissue and adjacent tissue
The median value of circ‐LARP4 was 0.244 (0.121‐0.409) in tumor tissue and 1.000 (0.672‐1.441) in adjacent tissue, which was decreased in tumor tissue compared to adjacent tissue in breast cancer patients (P < .001) (Figure 1). Furthermore, we compared circ‐LARP4 between tumor and adjacent tissue in subgroup patients: in HR+/HER2−, HR+/HER2+, HR−/HER2+, and HR−/HER2− subgroup patients, circ‐LARP4 was all decreased in tumor tissue compared to adjacent tissue (all P < .001) (Figure 2A‐D).
Figure 1.
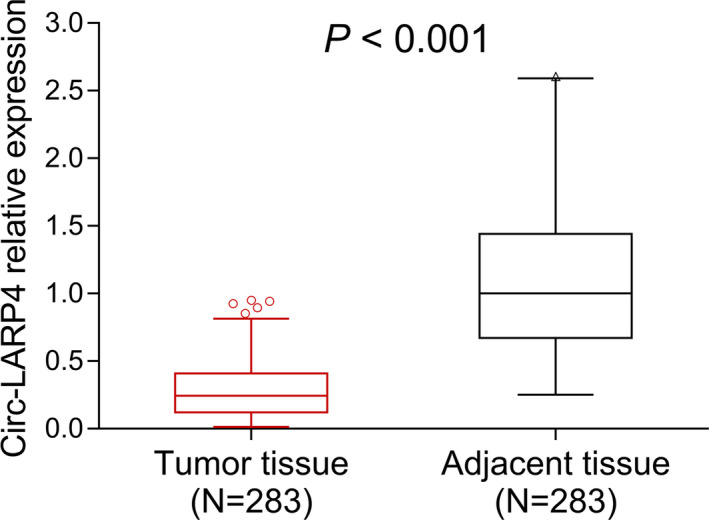
Circ‐LARP4 expression. Circ, circular RNA; LARP4, La‐related RNA‐binding protein 4
Figure 2.
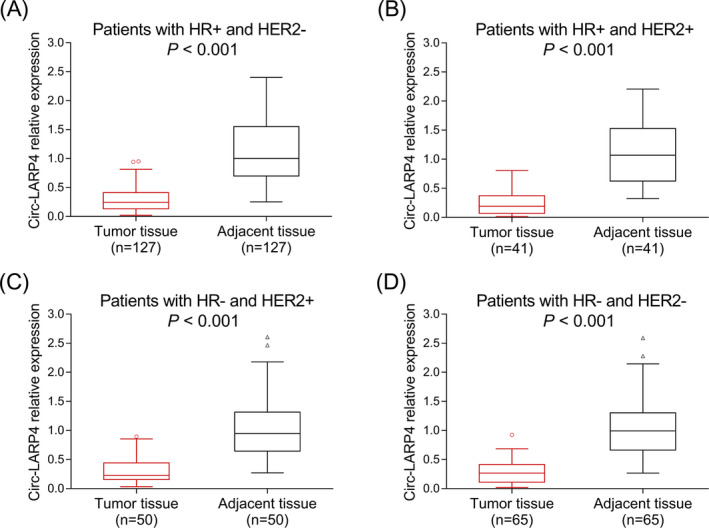
Circ‐LARP4 in patients with different subtypes of breast cancer. A, Circ‐LARP4 in patients with HR+ and HER2−. B, Circ‐LARP4 in patients with HR+ and HER2+. C, Circ‐LARP4 in patients with HR− and HER2+. D, Circ‐LARP4 in patients with HR− and HER2−. Circ, circular RNA; HER2, human epithelial growth factor receptor‐2; HR, hormone receptor; LARP4, La‐related RNA‐binding protein 4
3.2. Correlation of circ‐LARP4 with clinical characteristics
According to the median value of tumor circ‐LARP4 in breast cancer patients, they were divided into circ‐LARP4 high group and circ‐LARP4 low group; then, the clinical characteristics were compared between these two groups. Data revealed that tumor circ‐LARP4 was negatively correlated with tumor size (P = .001), T stage (P = .009), N stage (P = .006), and TNM stage (P < .001). However, no correlation was found in tumor circ‐LARP4 with age (P = .215), tumor differentiation (P = .107), ER status (P = .849), PR status (P = .590), or HER2 status (P = .235) in breast cancer patients (Table 1).
Table 1.
Baseline characteristics
| Items | Total patients (N = 283) | Circ‐LARP4 | P value | |
|---|---|---|---|---|
| High (n = 142) | Low (n = 141) | |||
| Age (years), mean ± SD | 54.3 ± 11.7 | 55.2 ± 11.8 | 53.5 ± 11.6 | .215 |
| Differentiation, No. (%) | ||||
| Well | 62 (21.9) | 31 (21.8) | 31 (22.0) | .107 |
| Moderate | 195 (68.9) | 103 (72.5) | 92 (65.2) | |
| Poor | 26 (9.2) | 8 (5.6) | 18 (12.8) | |
| Tumor size (cm), mean ± SD | 3.0 ± 2.0 | 2.6 ± 1.0 | 3.4 ± 2.6 | .001 |
| T stage, No. (%) | ||||
| T1 | 122 (43.1) | 70 (49.3) | 52 (36.9) | .009 |
| T2 | 137 (48.4) | 66 (46.5) | 71 (50.3) | |
| T3 | 24 (8.5) | 6 (4.2) | 18 (12.8) | |
| N stage, No. (%) | ||||
| N0 | 157 (55.5) | 89 (62.7) | 68 (48.2) | .006 |
| N1 | 73 (25.8) | 35 (24.6) | 38 (27.0) | |
| N2 | 48 (17.0) | 16 (11.3) | 32 (22.7) | |
| N3 | 5 (1.8) | 2 (1.4) | 3 (2.1) | |
| TNM stage, No. (%) | ||||
| I | 67 (23.7) | 46 (32.4) | 21 (14.9) | <.001 |
| II | 155 (54.8) | 76 (53.5) | 79 (56.0) | |
| III | 61 (21.6) | 20 (14.1) | 41 (29.1) | |
| ER status, No. (%) | ||||
| Negative | 118 (41.7) | 60 (42.3) | 58 (41.1) | .849 |
| Positive | 165 (58.3) | 82 (57.7) | 83 (58.9) | |
| PR status, No. (%) | ||||
| Negative | 133 (47.0) | 69 (48.6) | 64 (45.4) | .590 |
| Positive | 150 (53.0) | 73 (51.4) | 77 (54.6) | |
| HER2 status, No. (%) | ||||
| Negative | 192 (67.8) | 101 (71.1) | 91 (64.5) | .235 |
| Positive | 91 (32.2) | 41 (28.9) | 50 (35.5) | |
Abbreviations: ER, estrogen receptor; HER2, human epithelial growth factor receptor 2; PR, progesterone receptor; SD, standard deviation.
3.3. Correlation of circ‐LARP4 with DFS and OS
Disease‐free survival was increased in circ‐LARP4 high group compared with circ‐LARP4 low group (P = .004) (Figure 3A). Similarly, OS was also increased in circ‐LARP4 high group compared to circ‐LARP4 low group (P = .001) in breast cancer patients (Figure 3B).
Figure 3.
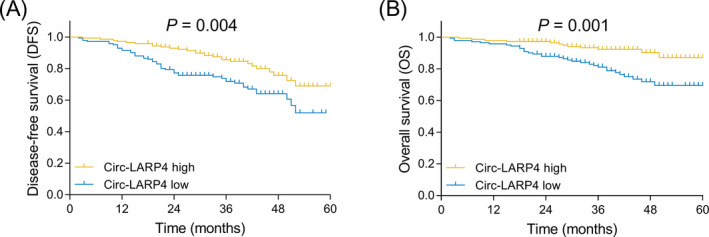
Correlation of circ‐LARP4 with prognosis in breast cancer patients. A, Correlation of tumor circ‐LARP4 with DFS. B, Correlation of tumor circ‐LARP4 with OS. Circ, circular RNA; DFS, disease‐free survival; LARP4, La‐related RNA‐binding protein 4; OS: overall survival
3.4. Circ‐LARP4 in mammary epithelial cells and breast cancer cell lines
In order to further clarify circ‐LARP4 expression in breast cancer cell lines and the role of circ‐LARP4 in the chemosensitivity of breast cancer cell lines, we conducted several in vitro assays. According to the RT‐qPCR, circ‐LARP4 was decreased in breast cancer cell lines including MCF‐7, BT474, MDA‐MB‐468, and MDA‐MB‐231 compared to mammary epithelial cells HMEC (all P < .001) (Figure 4). And we chose MCF‐7 and MDA‐MB‐231 for the following experiments.
Figure 4.
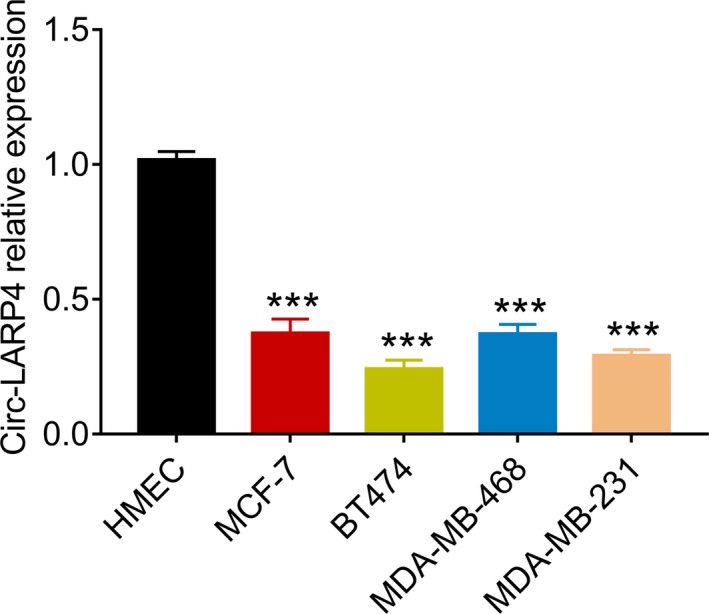
Difference of circ‐LARP4 expression in mammary epithelial cells and breast cancer cell lines. Circ, circular RNA; LARP4, La‐related RNA‐binding protein 4
3.5. Effect of circ‐LARP4 overexpression on breast cancer cell lines’ sensitivity to doxorubicin
In MCF‐7 cells, relative cell viability was decreased in OE‐Circ group compared to OE‐Control group when treated with 20 nmol/L (P < .05), 40 nmol/L (P < .01), and 80 nmol/L (P < .05) of doxorubicin (Figure 5A). And the IC50 value of doxorubicin was decreased in OE‐Circ group compared to OE‐Control group (P < .01) (Figure 5B). In MDA‐MB‐231 cells, relative cell viability was also decreased in OE‐Circ group compared to OE‐Control group when treated with 120 nmol/L (P < .05) and 240 nmol/L (P < .05) of doxorubicin (Figure 5C). And the IC50 value of doxorubicin was decreased in OE‐Circ group compared to OE‐Control group as well (P < .01) (Figure 5D). These data revealed that circ‐LARP4 overexpression increased the sensitivity of breast cancer cell lines to doxorubicin.
Figure 5.
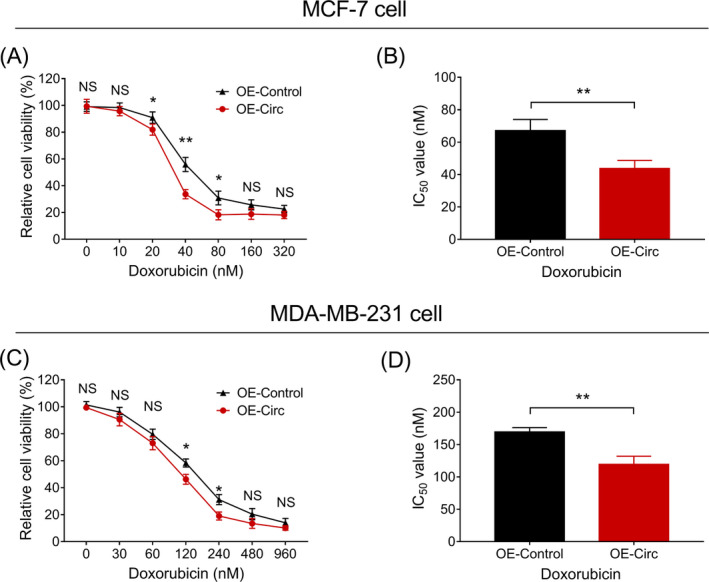
Effect of circ‐LARP4 on chemosensitivity of breast cancer cell lines. A, Relative cell viability of MCF‐7 cells under doxorubicin treatment with different concentrations. B, IC50 value of doxorubicin in treating MCF‐7 cells. C, Relative cell viability of MDA‐MB‐231 cells under doxorubicin treatment with different concentrations. D, IC50 value of doxorubicin in treating MDA‐MB‐231 cells. Circ, circular RNA; LARP4, La‐related RNA‐binding protein 4; NS, non‐significant
4. DISCUSSION
The role of circ‐LARP4 as a tumor suppressor in several cancers has been identified in previous researches.10, 13, 15 For example, it is proposed that circ‐LARP4 inhibits DNA synthesis, cell survival, and cell motility in gastric cancer cell lines by sponging miR‐424.13 Similar results are also observed in osteosarcoma cell line MG63, and it is reported that the overexpression of circ‐LARP4 promotes the chemosensitivity of MG63 cells.10 In hepatocellular carcinoma cell line, it is revealed that circ‐LARP4 suppresses cell proliferation and induces cell‐cycle arrest by increasing p53 and p21 expression. Further mechanism study indicates that circ‐LARP4 inhibits the progression of hepatocellular carcinoma cell lines by sponging miR‐761, thereby up‐regulates runt‐related transcription factor 3 and activates the p53/p21 signaling.15 Together, these studies suggest the anti‐tumor property of circ‐LARP4 in several cancers.
Clinically, the down‐regulation of circ‐LARP4 in tumor tissues (including hepatocellular carcinoma tissues, osteosarcoma tissues, and ovarian cancer tissues) is reported by previous researches.10, 11, 15 However, whether circ‐LARP4 is also dysregulated in breast cancer tissues is unclear. According to the above‐mentioned studies that circ‐LARP4 displays anti‐tumor property in several cancers, we hypothesized that circ‐LARP4 was decreased in breast cancer tissues and was a potential tumor suppressor in breast cancer. Therefore, we conducted this study and found that circ‐LARP4 was decreased in breast cancer tissues compared to adjacent tissues. Moreover, circ‐LARP 4 was negatively associated with tumor size, T stage, N stage, and TNM stage in breast cancer patients. Possible explanations might be that: (a) Circ‐LARP4 might also serve as a tumor inhibitor in breast cancer and prohibited cell proliferation by inhibiting DNA synthesis and activating p53/p21 signaling (as in gastric cancer, osteosarcoma, and hepatocellular carcinoma), and down‐regulation of circ‐LARP4 might promote the progression of breast cancer by enhancing proliferation of breast cancer cells. Therefore, circ‐LARP4 was decreased in breast tumor tissues. (b) In this study, circ‐LARP4 might also inhibit cell proliferation, cell invasion, and migration probably by sponging miR‐424 and miR‐761 (both of which are reported to be oncogenes in breast cancer)16, 17 in breast cancer cells (as in gastric cancer cells and hepatocellular carcinoma cells) to inhibit tumor progression and the metastasis of the tumor to lymph nodes in breast cancer patients. Therefore, circ‐LARP4 was negatively correlated with tumor size and tumor stage in breast cancer patients.
As to the prognostic value of circ‐LARP4, it is reported that increased circ‐LARP4 is correlated with prolonged OS in gastric cancer patients and osteosarcoma patients.10, 13 However, the correlation of circ‐LARP4 with the prognosis of breast cancer patients was unclear. In the present study, positive correlations of circ‐LARP4 with both DFS and OS were observed in breast cancer patients. These data could be explained by that (a) increased circ‐LARP4 was correlated with reduced tumor size and decreased tumor stage (mentioned above), which resulted in better DFS and OS in breast cancer patients. (b) Circ‐LARP4 might sponge miR‐424 (which is reported to be closely related to chemoresistance in breast cancer) 18, 19 to reduce chemoresistance (as in gastric cancer cells), thus promoted the effect of chemotherapy in breast cancer patients and prolonged their DFS and OS. Therefore, circ‐LARP4 was positively correlated with DFS and OS in breast cancer patients. However, several limitations existed in this study. (a) This study only included newly diagnosed breast cancer patients. However, patients with recurrent breast cancer were not included and further study should be conducted to investigate the role of circ‐LARP4 in them. (b) Patients with distant metastasis were not included; therefore, the correlation of circ‐LARP4 with distant metastasis in breast cancer patients was not clear, which needed further inquiry. (c) Since circular RNAs attracted the attention of researchers not long ago, the time from the initiation of this study to the end of follow‐up was relatively short; therefore, we could only evaluate the predictive value of circ‐LARP4 for the short‐term prognosis of breast cancer patients.
Since we found that circ‐LARP4 was a prognostic biomarker in breast cancer patients, we speculated that it might be involved in the sensitivity of patients to chemotherapy. Therefore, we further evaluated the effect of circ‐LARP4 on the sensitivity of breast cancer cell lines to chemotherapeutics. Data revealed that circ‐LARP4 was decreased in breast cancer cell lines, and overexpression of circ‐LARP4 increased the sensitivity of cells to doxorubicin in MCF‐7 and MDA‐MB‐231 cell lines. However, whether circ‐LARP4 enhanced chemosensitivity in breast cancer cell lines by sponging miR‐424 was unclear, which needed further investigation. Considering that doxorubicin is widely applied for the systematic treatment of breast cancer patients,20, 21 circ‐LARP4 as a novel therapeutic option might be promising in breast cancer patients, and studies should be conducted to substantiate our findings in the future.
To be conclusive, circ‐LARP4 was decreased in breast tumor tissues, and its down‐regulation was correlated with increased tumor size, enhanced clinical stage, and worse prognosis in breast cancer patients. Moreover, circ‐LARP4 overexpression promoted the chemosensitivity of breast cancer cell lines.
ACKNOWLEDGMENTS
This study was supported by Wuhan Municipal Health Commission (No. WX19Q49) and Health Commission of Hubei Province Scientific Research Project (No. WJ2019H396).
Zhang X, Su X, Guo Z, Jiang X, Li X. Circular RNA La‐related RNA‐binding protein 4 correlates with reduced tumor stage, as well as better prognosis, and promotes chemosensitivity to doxorubicin in breast cancer. J Clin Lab Anal. 2020;34:e23272 10.1002/jcla.23272
Xiaoyi Zhang and Xinyu Su contributed equally to this work.
REFERENCES
- 1. Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389(10074):1134‐1150. [DOI] [PubMed] [Google Scholar]
- 2. Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3(11):753‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de la Mare JA, Contu L, Hunter MC, et al. Breast cancer: current developments in molecular approaches to diagnosis and treatment. Recent Pat Anticancer Drug Discov. 2014;9(2):153‐175. [DOI] [PubMed] [Google Scholar]
- 4. McDonald ES, Clark AS, Tchou J, et al. Diagnosis and management of breast cancer. J Nucl Med. 2016;57(Supplement_1):9S–16S. [DOI] [PubMed] [Google Scholar]
- 5. Peart O. Breast intervention and breast cancer treatment options. Radiol Technol. 2015;86(5):535M‐558M; quiz 559–562. [PubMed] [Google Scholar]
- 6. Torre LA, Islami F, Siegel RL, et al. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26(4):444‐457. [DOI] [PubMed] [Google Scholar]
- 7. Tao Z, Shi A, Lu C, et al. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72(2):333‐338. [DOI] [PubMed] [Google Scholar]
- 8. Stavraka C, Blagden S. The La‐related proteins, a family with connections to cancer. Biomolecules. 2015;5(4):2701‐2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsiao KY, Sun HS, Tsai SJ. Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Y, Gu J, Shen H, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA‐424 in osteosarcoma. J Clin Lab Anal. 2019;34:e23045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zou T, Wang PL, Gao Y, et al. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178‐7182. [DOI] [PubMed] [Google Scholar]
- 12. Ganji SM, Saidijam M, Amini R, et al. Evaluation of microRNA‐99a and microRNA‐205 expression levels in bladder cancer. Int J Mol Cell Med. 2017;6(2):87‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR‐424‐5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan JY, Bao XL, Zhu K. Circular RNA_LARP4 inhibits cell proliferation and invasion of nasopharyngeal carcinoma by repressing ROCK1. Eur Rev Med Pharmacol Sci. 2019;23(22):9915‐9922. [DOI] [PubMed] [Google Scholar]
- 15. Chen Z, Zuo X, Pu L, et al. circLARP4 induces cellular senescence through regulating miR‐761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(2):568‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo GC, Wang JX, Han ML, et al. microRNA‐761 induces aggressive phenotypes in triple‐negative breast cancer cells by repressing TRIM29 expression. Cell Oncol (Dordr). 2017;40(2):157‐166. [DOI] [PubMed] [Google Scholar]
- 17. Zhang D, Shi Z, Li M, et al. Hypoxia‐induced miR‐424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5(6):e1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodriguez‐Barrueco R, Nekritz EA, Bertucci F, et al. miR‐424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu S, Tao Z, Hai B, et al. miR‐424(322) reverses chemoresistance via T‐cell immune response activation by blocking the PD‐L1 immune checkpoint. Nat Commun. 2016;7(1):11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia. 2017;59(5):368‐379. [DOI] [PubMed] [Google Scholar]
- 21. Ponde NF, Zardavas D, Piccart M. Progress in adjuvant systemic therapy for breast cancer. Nat Rev Clin Oncol. 2019;16(1):27‐44. [DOI] [PubMed] [Google Scholar]


