Abstract
Objective
MicroRNA-126–3p (miR-126) is required for angiogenesis during development or the repair of injured arterial vasculature. The role of miR-126 in lung microvascular endothelial cells, which are essential for gas exchange and for lung injury repair and regeneration, remains poorly understood. Considering the significant heterogeneity between endothelial cells from different vascular beds, we aim to determine the role of miR-126 in regulating lung microvascular endothelial cell function and to elucidate its downstream pathways.
Approach and Results
Overexpression and knockdown of miR-126 in primary human lung microvascular endothelial cells (HLMVEC) were achieved via transfections of miR-126 mimics and antisense inhibitors. Increasing miR-126 levels in HLMVEC reduced cell proliferation, weakened tube formation, and increased cell apoptosis, whereas decreased miR-126 levels stimulated cell proliferation and tube formation. Whole genome RNA sequencing revealed that miR-126 was associated with an anti-angiogenic and pro-apoptotic transcriptomic profile. Using validation assays and knockdown approaches, we identified that the effects of miR-126 on HLMVEC angiogenesis was mediated by the L-type amino acid transporter 1 (LAT1), via regulation of mTOR signaling. Furthermore, downregulation of miR-126 in HLMVEC inhibited cell apoptosis and improved tube formation during exposure to environmental insults such as cigarette smoke.
Conclusion
miR-126 inhibits HLMVEC angiogenic function by targeting the LAT1-mTOR signaling axis, suggesting that miR-126 inhibition may be useful for conditions associated with microvascular loss, whereas miR-126 augmentation may help control unwanted angiogenesis.
Keywords: Pulmonary, endothelium, apoptosis, miRNA
Subject codes: Angiogenesis, Basic Science Research, Cell Biology/Structural Biology, Endothelium/Vascular Type/Nitric Oxide, Pulmonary Biology, Vascular Biology, Cell signaling/Signal Transduction, Gene Expression and Regulation
Introduction
Highly enriched in the endothelial cells, the microRNA-126–3p (herein referred to as miR-126) is essential for vascular development by maintaining vascular integrity and promoting angiogenesis1, 2, a highly regulated process involving cell proliferation, invasion, and migration3. A similar effect of miR-126 has been shown in cultured endothelial cells such as primary human umbilical vein endothelial cells (HUVEC)1, 2, human aortic endothelial cells (HAEC)1, and human coronary artery endothelial cells (HCAEC)4. However, the role of miR-126 in regulating the angiogenic function of microvascular endothelial cells has not been elucidated.
The mechanisms underlying the pro-angiogenic function of miR-126 involve direct inhibition of sprout-related, EVH1 domain-containing protein 1(SPRED1) and phosphoinositide-3-kinase regulatory subunit 2 (PIK3R2), with subsequent disinhibition of vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) signaling 1, 2, 4. However, endothelial cells are highly heterogenous, exhibiting distinct transcriptional and functional profiles that are influenced by the stage of organismal development, type of vascular bed, tissue or organ of origin, and pathogenic states 5–7. For example, anti-angiogenic and anti-proliferative effects of miR-126 have been reported in late endothelial progenitor cells (EPC) 8, in hematopoietic stem cell 9, and in pathological proliferative microvascular states such as choroidal neovascularization 10, diabetic retinopathy 11, or various cancers 12–14. These anti-angiogenic effects of miR-126 were reported to be mediated through distinct mRNA targets such as VEGF-A 10, 15, PIK3R2 9, 12, ADMA9 (disintegrin and metalloproteinase domain-containing protein 9) 16, and LAT1 (L-type amino acid transporter 1)17, 18, indicating that the function and targets of miR-126 are cell-type and disease-specific. We set out to determine the role of miR-126 in lung microvascular endothelial cells which serve as important structural and functional components of the alveoli-capillary membrane 19, 20. The pro-angiogenic function of lung microvascular cells may be required for the prevention and repair of lung injury in multiple diseases, including acute respiratory distress syndrome, pulmonary hypertension, and emphysema 19.
Using cultured primary human lung microvascular endothelial cells (HLMVEC), we report here that miR-126 inhibits microvascular endothelial angiogenic function through suppressing LAT1-mediated mTOR signaling.
Materials and methods
The authors declare that all supporting data are available within the article and the supplemental files provided.
Reagents
utilized are listed in Major Resource Table.
Cell Culture
Primary human lung microvascular endothelial cells (HLMVEC) were purchased from Lonza (CC-2527, LONZA) and maintained in microvascular endothelial growth medium EGM-2 MV supplemented with growth factors and 5% FBS (CC-3202, LONZA). All experiments were performed in HLMVEC between passages 3 to 7, from at least three distinct donors (biological replicates). Donor information is listed in the Major Resources Table.
MiRNA and SiRNA Transfections
We achieved miR-126 overexpression (126-OE) by transfecting HLMVEC at 80–90% confluency with miR-126–3p mimic (Dharmacon; 5nM for 16hrs), using lipofectamine RNAiMAX (Invitrogen). Similarly, we achieved miR-126 knockdown (126-KD) by transfection of antisense miR-126–3p inhibitors (Dharmacon; 5nM for 16hrs). We used as controls cells co-transfected with non-targeting miRNA mimics and antisense inhibitors (5nM each,16hrs) when both 126-OE and 126-KD were simultaneously studied. Alternatively, we used as controls cells transfected with either non-targeting miRNA mimics or non-targeting miRNA antisense inhibitor (5nM respectively, 16hrs) when studying only the effect of 126-OE or only the effect of 126-KD. We compared these three control conditions and noted no difference in their effect on miR-126 or LAT1 expression, or on S6 activity (Sup Fig I). To achieve LAT1 knockdown, HLMVEC were transfected with LAT1 siRNA (Dharmacon; 10nM, 16hrs), using a non-targeting siRNA as control (Dharmacon; 10nM, 16hrs). Transfection efficiencies were verified with RT-qPCR and corresponding protein expressions after knockdown were confirmed with Western Blot.
Cell Proliferation
Cell proliferation was determined by Ki67 immunofluorescence staining and by live cell counting. Following transfection with the indicated miRNA or siRNA, HLMVEC were seeded (0.3 × 105 cells per well) onto fibronectin-coated (1μg/cm2) Nunc Lab-Tek II chamber slides (Thermo Fisher) for immunostaining. After 24 hours, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 30 min at room temperature. Ki67 was stained with Alexa 488-conjugated mouse-anti human Ki67 antibody (sc-23900 AF488, Santa Cruz) and mounted with ProLong Gold with DAPI (Thermo Fisher). For cell counting, 10–15 images were taken per condition in a blinded fashion with Nikon Eclipse Ti-U inverted microscope with Nikon Elements BR v4.30.02 software. Ki67 positive cells and total number of cells (DAPI positive) were counted using Image J (1.52p) and the data were expressed as percentages of Ki67 positive cells. Representative images for figures were taken with Zeiss LSM700 Confocal with Zen Black software. For growth rate studies, transfected cells were seeded at 1.5 × 104/ well on gelatin-coated 24-well plates. After 24, 48 and 72 hours, cells were stained with Trypan blue stain (ThermoFisher, 0.4%) and counted using a hematocytometer. Each independent experiment was performed in technical duplicate.
In Vitro Tube Formation Assay
In vitro tube formation assay was modified from a published protocol 21. Briefly, following transfections of indicated miRNA or siRNA, HLMVEC were seeded between 0.65 to 0.75 × 104 cells per well in a 96-well plate, pre-coated with growth factor-reduced matrigel (Fisher Scientific). In order to visualize the structures formed, calcein AM (Fisher Scientific; 2μM, 15min) was added after 6 hours to each well and images were captured in a blinded fashion, using Nikon Eclipse Ti-U inverted microscope with Nikon Elements BR v4.30.02 software. Data were analyzed using Angiogenesis Analyzer in Image J. Each independent experiment was performed in technical triplicate.
Caspase-3/7 Activity
HLMVEC were cultured in an opaque 96-well plate with clear bottom and transfected as indicated. The caspase activity was measured using the Caspase-Glo-3/7 assay kit (Promega), following the manufacturer’s instructions.
Gene expression studies
Total RNA (including miRNA and mRNA) was isolated using the miRNeasy mini kit (Qiagen, Germantown, MD), following the manufacturer’s instructions. Real-time quantitative polymerase chain reaction (qPCR) was performed on the StepOnePlus System using Taqman Universal PCR Master Mix (ThermoFisher) and Taqman gene expression assays. For microRNA studies, RNU48 was used as reference gene. For mRNA studies, 18S was used as reference gene. All primers are listed in the Major Resource Table.
MiR-126 Standard Curve and Copy Number Calculation
Synthetic miR-126–3p or miR-126–5p mimics (Dharmacon) were diluted at 5 × 10−1, 5 × 10−2, 5× 10−3, 5 × 10−4, 5 × 10−5 ng and RT-qPCR was performed using Taqman miR-126–3p or −5p expression assay. The value of qPCR cycle threshold (Ct) of each dilution gradient was plotted with corresponding RNA concentration (log10 transformed) to generate a standard curve from which the amount (ng) of miR-126 can be calculated based on the Ct value of an unknown sample. To minimize the impact of loading variations, Ct of miR-126 was adjusted based on the Ct of reference gene U48. Using the amount of miR-126 derived from the standard curve, the copy number of miR-126 in each sample was calculated as follows: copy number = (ng of RNA x 6.022 × 1023)/(length of ssRNA x 109 × 340); The mature sequence of miR-126–3p is 22nt (UCGUACCGUGAGUAAUAAUGCG) and that of miR-126–5p is 21nt (CAUUAUUACUUUUGGUACGCG).
Ambient Air (AC) and Cigarette Smoke (CS) Extract Preparation and Exposure
CS extract was prepared by bubbling the smoke from two cigarettes (3R4F, University of Kentucky, Lexington, KY), burnt at 1 min/ cigarette, and AC extract was prepared by bubbling room air for 2 min, each into 20 mL of PBS (100% v/v). AC and CS extracts were then adjusted to pH=7.4 and filtered with 0.22-μm filters as previously described 22. HLMVEC were incubated with extracts (1% or 3% v/v; 6 hrs) in EGM-2 MV starvation media containing 2% FBS and growth factors (40% of concentrations used for full culture media).
Quantifications and Statistical Analysis
All data were presented as means ± standard error of the mean (SEM). Between 3–6 individual experiments were performed for each endpoint, which may affect the power of statistical tests due to small sample size. Normality and variance were not tested. Statistical analyses were performed with Prism v7.0 software (GraphPad Software, La Jolla, CA) using non-parametric test, one-way analysis of variance (ANOVA), or two-way ANOVA as indicated. P < 0.05 was considered statistically significant. To note, during one-way ANOVA analysis, Dunnett’s multiple comparisons test was used when comparing the mean of each experimental group to the control group, and Tukey’s post-hoc multiple comparisons test was used when comparing the means of all groups, regardless of control. The specific test used was noted in the respective figure legend.
Methods for Western blot23, RNA sequencing24–32, linear mixed-effect regression and Ingenuity Pathway Analysis (IPA) are listed in Supplemental Materials.
Results
MiR-126 inhibits cell proliferation and tube formation and triggers apoptosis in HLMVEC
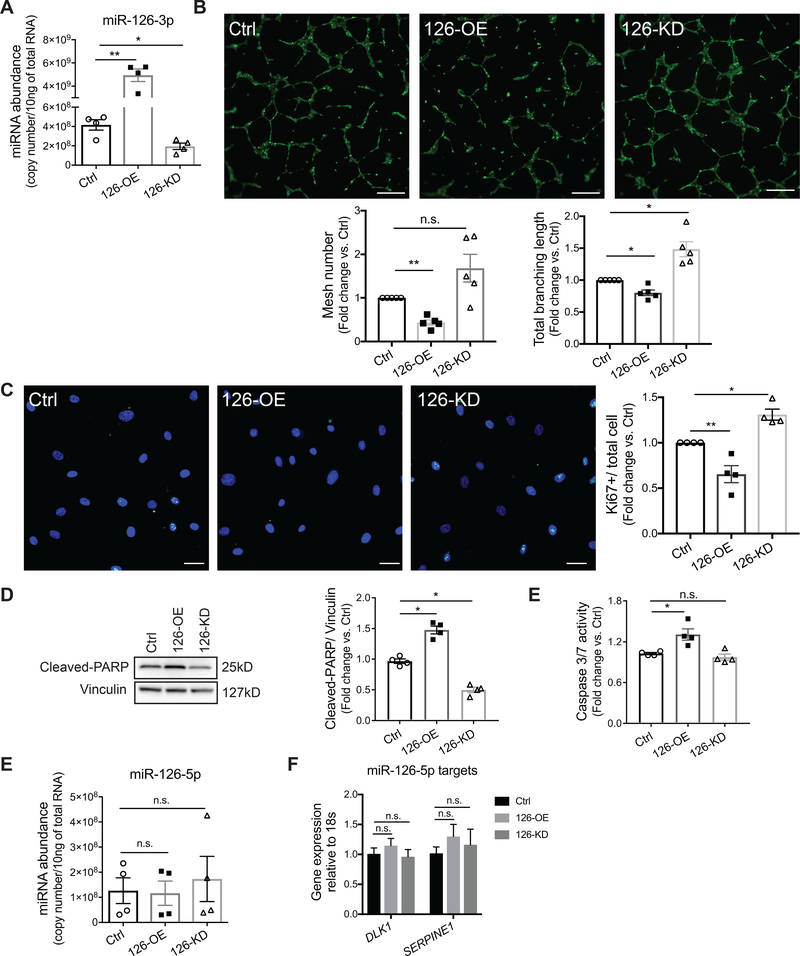
To study the function of miR-126 in HLMVEC, we transfected primary HLMVEC with miR-126–3p mimics for miR-126 overexpression (126-OE), antisense miR-126–3p inhibitors for miR-126 knockdown (126-KD) and co-transfected non-targeting mimics and inhibitors as negative control. After 16 hours, transfection efficiencies were determined by RT-qPCR (Fig 1A, Sup Fig I). To investigate HLMVEC angiogenic function, we performed an in vitro tube formation assay by seeding the transfected cell in matrigel and we observed branching and formation of mesh-like structures as early as 4 hours (data not shown), with a peak between 6 to 8 hours. At 6 hours, 126-OE markedly decreased the number of meshes by more than 50% and total branching length by 15% (Fig 1B). Although there was no significant difference between control and 126-KD conditions in the numbers of meshes formed, 126-KD increased the total branching length by more than 20% (Fig 1B). Because cell proliferation is a crucial step in angiogenesis, we next investigated whether miR-126 regulates HLMVEC proliferation. Using the same transfection strategy, the transfected cells were re-plated at equal number and Ki67 expression was measured 24 hours later using immunofluorescence. Compared with control cells, 126-OE significantly decreased whereas 126-KD increased the percentage of Ki67 positive cells (Fig 1C), suggesting that miR-126 inhibits HLMVEC proliferation. Since endothelial function can be affected by endothelial cell survival, we further evaluated the effect of miR-126 on HLMVEC apoptosis. Compared with control cells, 126-OE HLMVEC exhibited significantly higher levels of cleaved-poly (ADP-ribose) polymerase (cleaved-PARP) (Fig 1D) and caspase 3/7 activity (Fig 1E). 126-KD did not significantly alter the level of caspase 3/7 activity compared to that of the control cells (Fig 1E), but it reduced the cleaved-PARP levels by almost 50% (Fig 1D). In addition, there were no significant changes in autophagy markers such as Atg5, Atg7, and SQSTM1 (p62) among control, 126-OE, and 126-KD conditions (Sup Fig II A). These results indicated that miR-126 has anti-angiogenic and pro-apoptotic effects in HLMVEC.
Figure 1. Effect of miR-126–3p (miR-126) on human lung microvascular endothelial cell (HLMVEC) proliferation, tube formation, and apoptosis.
A, Relative miR-126–3p copy number in HLMVEC 24hrs after transfection with a mixture of miRNA non-targeting mimics and inhibitor controls (Ctrl), miR-126–3p mimics (126-OE) or antisense inhibitors (126-KD). B, Top: Images of mesh-like structures formed by HLMVEC in matrigel 6hrs after seeding; cells were stained with calcein AM (Green). Scale bar: 200μm. Bottom: Quantifications of total mesh number (left) and total branching length (right) analyzed by the Angiogenesis Analyzer in Fuji. Each individual experiment was performed in technical triplicate. C, Left: Immunofluorescence (IF) images of HLMVEC stained for Ki67 (green) and DAPI (blue) 24hrs after seeding. Scale bar: 50μm. Right: Quantification of Ki67 expression by first normalizing the number of Ki67 positive cells to the total number of cells (DAPI) and then to the Ctrl condition. At least 10 images were analyzed per individual experiment. D-E, Cell apoptosis measured by cleaved-PARP level using Western Blot (D) and by caspase 3/7 activity measured using a Promega luminescence assay (E). F-G, Relative miR-126–5p copy number and expression levels of miR-126–5p targets, DLK1 and SERPINE1, determined by RT-qPCR 24hrs after transfection of miR-126–3p mimics (126-OE) or inhibitors (126-KD). All results presented as mean ± SEM; *p<0.05; **p<0.01; No statistical significance was detected in F and G; One-way ANOVA followed by Dunnett’s multiple comparisons test.
The passenger strand of miR-126, miR-126–5p, is also known to regulate endothelial angiogenic function10, 33 and can be altered during miR-126–3p manipulation due to feedback regulation. We therefore determined that the transfection of miR-126 (3p) mimics or inhibitors did not significantly change the expression levels of miR-126–5p, or that of its main targets (Fig 1F–G), suggesting a lack of miR-126–5p contribution to our functional studies.
Global transcriptome profiling reveals unique angiogenic targets of miR-126 in HLMVEC
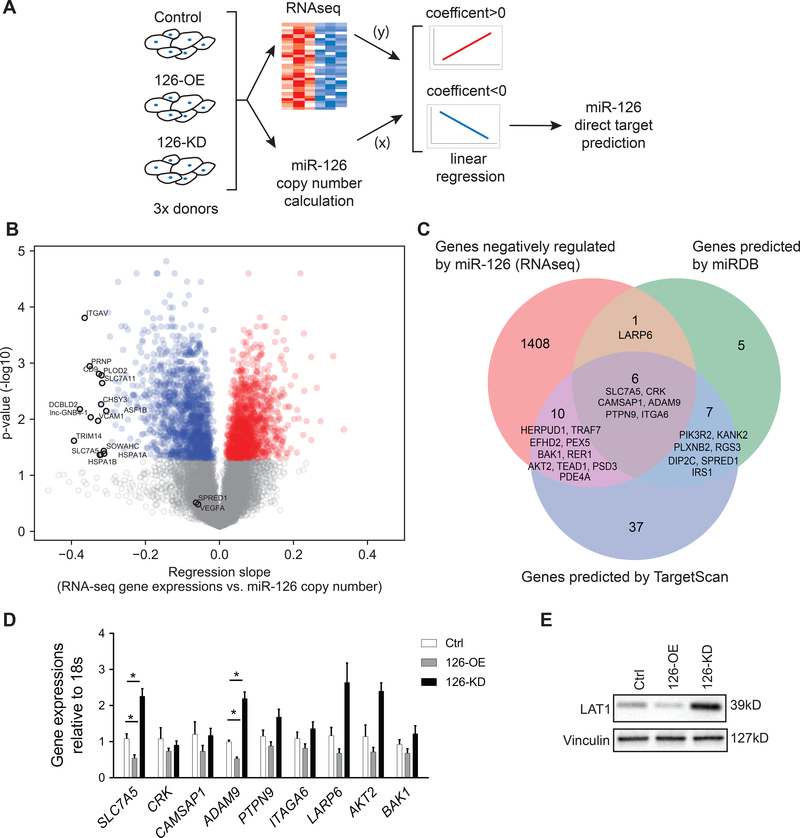
We hypothesized that the distinct miR-126 effect on HLMVEC, as compared to published data in endothelial cells from other vascular beds (HUVEC or HAEC)1, is due to the inhibition of a unique set of target mRNAs. Therefore, following the transfections of HLMVEC from three individual donors with non-targeting negative controls (Ctrl), miR-126 mimics (126-OE), or miR-126 antisense inhibitors (126-KD), we calculated the copy number concentration of miR-126 in each sample using a PCR standard curve (Sup Table I) and performed genome-wide RNA sequencing on these samples (Fig 2A). Plotting the gene expression data for each transfection condition (Ctrl, 126-OE and 126-KD) against their respective miR-126 concentrations, we performed a linear regression analysis to discover genes that were significantly affected by changes in miR-126 levels. We identified that 1258 genes were positively correlated, and 1415 genes were inversely correlated with miR-126 levels (Fig 2B). Of note, we did not observe significant correlations between miR-126 and SPRED1 or PIK3R2 (Fig 2B, labelled in grey), the miR-126 targets that mediate its pro-angiogenic effect in HUVECs and HAECs 1, 2. In addition, miR-126 did not significantly correlate with VEGFA (Fig 2B, labelled in grey), a miR-126 target reported in various type of tumors 12, 15. Consistent with the results from our in vitro functional assays (Fig 1), Ingenuity Pathway Analysis showed that high level of miR-126 stimulated an anti-angiogenic and pro-apoptotic profile (Sup Table II), featuring negative association with pathways involved in growth factor signaling (FGF, VEGF, hepatocyte growth factor (HGF), angiopoietin), cell growth (eIF4 and p70S6K), and cell motility (RhoA, actin cytoskeleton, and ephrin receptor signaling). In contrast, miR-126 was positively associated with activation of pathways related to cell cycle arrest (p53 and PTEN signaling) and apoptosis (TNFR1 and ceramide signaling). These results suggest that miR-126 promotes a quiescent state of HLMVEC which lacks signals that promote cell growth and angiogenesis and may even induce apoptosis.
Figure 2. Global profiling of genes regulated by miR-126 followed by bioinformatics prediction of miR-126 direct targets.
A, Schematic overview of the study; HLMVEC from three individual donors were used as biological replicates. B, Volcano plot of genes positively (red) or negatively (blue) associated with miR-126 in HLMVEC with adjusted p< 0.05. Genes with adjusted p value > 0.05 are labelled in grey. The top 15 genes with the highest regression coefficient are annotated, as well as SPRED1 and VEGFA, known direct targets of miR-126 in HUVEC and cancer cells, respectively. C, Venn diagram of genes negatively associated with miR-126 identified in our RNA-seq analysis (left, orange), genes predicted as direct targets of miR-126 by miRDB (right, green) and by TargetScan (middle, blue). The overlapped genes are annotated and listed based on Aggregate PCT (highest to lowest) calculated by the computational algorithms of the two databases. D, RT-qPCR quantification of the six genes identified by all three approaches (center of the Venn graph in C) as well as additional three candidate genes (LARP6, AKT2 and BAK1) with angiogenic functions (n=5). E, Western blot quantification of SLC7A5 (LAT1) protein abundance. Results presented as mean ± SEM; *p<0.05; Statistical significance was determined by Friedman non-parametric test followed by Dunn’s multiple comparisons test in D.
Bioinformatics analysis identifies SLC7A5 and ADAM9 as miR-126 targets in HLMVEC
The functions of miRNAs are carried out through post-transcriptional inhibition, which is mediated by the interaction between the “seed sequence” of a miRNA and the 3’-UTRs of its mRNA targets 34, 35. A single miRNA can target multiple mRNAs at the same time and these interactions can be predicted using computational algorithms available through databases such as miRDB 36, 37 and TargetScan 38. To identify direct targets of miR-126 in HLMVEC, we focused on genes identified by our regression analysis to have significant inverse correlation with miR-126 levels (correlation coefficient <0, adjusted p<0.05; Fig 2A). Of these 1436 genes, SLC7A5, CRK, CAMSAP1, ADAM9, PTPN9, and ITGA6 were identified to be direct targets of miR-126 by both miRDB and TargetScan databases (Fig 2C), with SLC7A5 ranked as the top predicted target using Aggregate PCT 39. In addition, genes reported to regulate endothelial function and cell death such as LARP6, AKT2 and BAK1 40–42, were identified as miR-126 target by either miRDB or by TargetScan (Fig 2C). We assessed the expression of these 9 genes in Ctrl, 126-OE, and 126-KD HLMVEC using RT-qPCR and noted that all genes tended to decrease with 126-OE and increase with 126-KD (Fig 2D). However, only the expression levels of SLC7A5 (solute carrier family 7 member 5, also known as LAT1) and ADAM9 were significantly downregulated in 126-OE HLMVEC, by nearly 50% and upregulated in 126-KD HLMVEC, by more than 2-fold (Fig 2D). We also determined that LAT1 protein levels were regulated by miR-126 in a similar fashion (Fig 2E). Since it has been previously validated that miR-126 directly interacts with the 3’ UTRs of LAT1 and ADAM911, 16, 43, our results indicate that LAT1 and ADAM9 are direct targets of miR-126 in HLMVEC.
MiR-126 signals via the LAT1- mTOR axis in HLMVEC
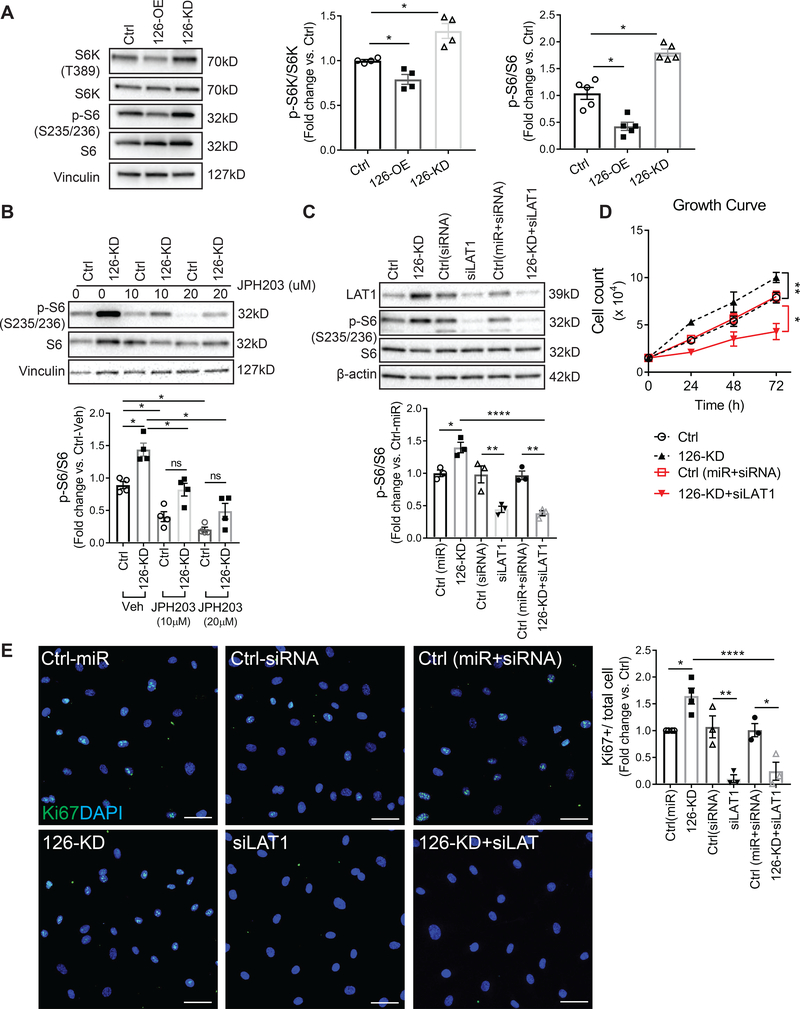
We next focused our studies on LAT1, since it was the highest ranked miR-126 target by TargetScan (Fig 2C), and the signaling pathway by which LAT1 may mediate miR-126 effects remains unclear. LAT1 is an L-glutamine/essential amino acid (EAA) antiporter, which mediates extracellular EAA uptake and subsequently activates mTOR signaling 44. To investigate if mir-126 regulates mTOR, we measured the phosphorylation of mTOR effectors p70 ribosomal protein S6 kinase (S6K) and ribosomal protein S6 (S6) in 126-OE and 126-KD HLMVEC. We found that cells with 126-OE showed marked decrease in the phosphorylation levels of both S6K and S6 whereas cells with 126-KD showed increased S6K and S6 phosphorylation (Fig 3A). To establish that the increased S6K and S6 phosphorylation in these cells were mediated by mTOR, following transfection with miR-126 antisense inhibitors, we suppressed mTOR activity with rapamycin (0.5nM or 1nM, 6hrs). We noted that in the presence of rapamycin, 126-KD cells failed to increase S6 phosphorylation compared to cells treated with vehicle (Sup Fig III A). To determine if LAT1 was required for the mTOR activation in 126-KD cells, we inhibited LAT1 pharmacologically with JPH203 or co-transfected 126-KD cells with LAT1 siRNA. JPH203 (10μM or 20μM, 4hrs) dose-dependently reduced S6 activity in 126-KD cells (Fig 3B). Co-transfection of antisense miR-126 (5nM, 16hrs) and LAT1 siRNA (10nM, 16hrs) reduced, as expected, miR-126 and LAT1 levels compared with non-targeting controls (Sup Fig IV A–B), and markedly inhibited S6 phosphorylation compared with cells transfected with 126-KD alone (Fig 3C). These findings suggest that inhibition of miR-126 activates mTOR signaling in a LAT-1-dependent fashion.
Figure 3. Signaling mechanism of miR-126 knockdown on HLMVEC proliferation.
A, Activity of mTOR signaling pathway measured by the phosphorylation levels of S6K and S6. Left: representative Western blot images. Middle, right: phosphorylation levels of S6K and S6 quantified by normalizing the density of phosphorylated S6K (p-S6K) and phosphorylated S6 (p-S6) to total S6K and S6, respectively. B-C, Activity of S6 in cells with 126-KD after treatment with LAT1 inhibitor JPH203 (10μM or 20μM, 4hrs, B) or after co-transfection with LAT1 siRNA (10nM, 16hrs, C). Top: representative Western blot images. Bottom: Densitometry quantification of S6 phosphorylation. D, Growth rates obtained by live cell counts at 24, 48, and 72 hrs. E, Left: representative IF images of HLMVEC stained for Ki67 (green) and DAPI (blue). Scale bar: 50μm. Right: quantification of Ki67 expression by first normalizing Ki67 positive cells to the total number of cells (DAPI) and then to the Ctrl condition. 10 images were analyzed per transfection condition per individual experiment. Results presented as mean ± SEM; *p<0.05; **p<0.01; ****p<0.0001; One-way ANOVA followed by Tukey’s post-hoc multiple comparisons.
MiR-126 knockdown promotes HLMVEC proliferation via LAT1
Since mTOR signaling plays an essential role in the regulation of cell proliferation, growth and survival 45, 46, we asked whether the pro-angiogenic and anti-apoptotic effects of 126-KD were due to LAT1-mediated mTOR activation. We found that rapamycin significantly reduced cell proliferation even in cells with 126-KD (Sup Fig III B–C), suggesting that the proliferative effect of 126-KD is mTOR dependent. HLMVEC co-transfected with miR-126 antisense inhibitors and LAT1 siRNA (5nM and 10nM respectively, 16hrs) were seeded at the same number as controls and, cell proliferation was determined by growth curves and Ki67 staining. Compared to cells transfected with 126-KD alone, which exhibited a high proliferative rate, cells that had both 126-KD and LAT1 knockdown exhibited a much lower proliferative rate (Fig 3D–E). Interestingly, we did not observe any difference in the cleaved-PARP levels between 126-KD alone and 126-KD with LAT1 siRNA (Sup Fig IV C). These results indicate that although LAT1 mediates the proliferative effect of 126-KD, it may not be involved in the anti-apoptotic effect of miR-126 knockdown.
MiR-126 knockdown ameliorates the apoptotic and anti-angiogenic effects of cigarette smoke (CS) exposure
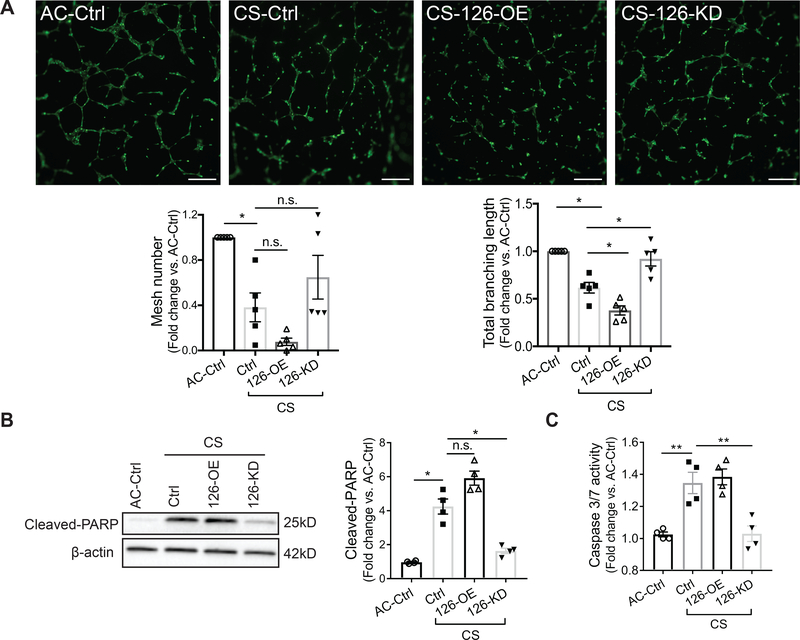
We and others have reported that CS exposure induces apoptosis and impairs angiogenesis of the lung microvasculature 47, 48. To determine whether the newly discovered function of miR-126 in HLMVEC can be employed to modulate their response to injury induced by CS exposure, HLMVEC transfected with negative controls, miR-126 mimics and miR-126 antisense (5nM respectively, 16hrs) were exposed to aqueous CS or ambient air control (AC) extracts. Tube formation assay was performed to assess angiogenesis during AC or CS exposure (1% v/v, 6hrs). We found that relative to AC, the number of mesh and the total branching length were significantly reduced by CS (Fig 4A). Compared with CS-exposed control cells, the total branching length was further reduced by 126-OE, and was improved by 126-KD, the latter also tended to increase mesh number (Fig 4A). In addition, CS exposure (3% v/v, 6 hrs) increased the levels of cleaved-PARP and caspase 3/7 activity in HLMVEC, as expected (Fig 4B–C). In CS-exposed cells, 126-OE did not further increase cell apoptosis, but 126-KD significantly reduced the levels of cleaved-PARP and caspase 3/7 activity, by more than 50% (Fig 4B–C). Our results suggest that miR-126 knockdown improves HLMVEC angiogenic functions and reduces cell death during CS exposure.
Figure 4. Effect of decreased miR-126 on cigarette smoke (CS)-exposed HLMVEC.
A, Top: tube formation by HLMVEC seeded in matrigel with 1% (v/v) ambient air (AC) or CS extract for 6hrs and stained with calcein AM (2μM, 15 min). Bottom: mesh number (left) and total branching length (right) analyzed by Angiogenesis Analyzer in Fuji. Each individual experiment was performed in technical triplicate. Scale bar: 200μm. B-C, Cell apoptosis measured by cleaved-PARP levels (left and middle, B) and caspase 3/7 activity (right, C) after treating the cells with AC or CS extract (3% v/v, 6hrs). Results presented as mean ± SEM; *<0.05; **p<0.01; One-way ANOVA followed by Tukey’s post-hoc multiple comparisons.
Discussion
In this study, we demonstrate that miR-126 acts as a potent inhibitor of primary HLMVEC proliferation, angiogenesis and when over-expressed, promotes apoptosis. In turn, downregulation of miR-126 induces a pro-angiogenic, pro-survival phenotype which is in part dependent on the disinhibition of LAT1 and subsequent mTOR activation. Moreover, we were able to leverage the pro-angiogenic effects of decreased miR-126 to protect HLMVEC from environmental insults such as CS exposure.
To our knowledge, we are the first to report an anti-angiogenic function of miR-126 in the lung microvascular endothelial cells and to identify the underlying mechanism of this effect. When analyzed in light of previous studies reporting a pro-angiogenic role of miR-126 1, 2, 49, we interpret our results as an indication of the intrinsic diversity of the endothelial cells from the macro- and micro-vasculature beds, and the distinct developmental stage of the vasculature 5–7. Indeed, similar to our findings in HLMVEC, an anti-angiogenic function of miR-126 was reported in human late EPC 8, also known as the late outgrowth endothelial cells that are highly proliferative and angiogenic 50. HLMVEC share these attributes, relative to other endothelial cell types such as the pulmonary artery endothelial cells 51. In addition, high miR-126 level drives the hematopoietic stem cell quiescence 9. The anti-angiogenic effects of miR-126 have also been reported in pathological conditions that result from hyperproliferative microvasculature beds including choroidal neovascularization 10, diabetic retinopathy 11, and various forms of cancer 12, 13, 15. All these conditions feature excessive angiogenesis with high endothelial proliferative rate and excessive vascular branching which can be inhibited by the overexpression of miR-126. These results, together with our current study, suggest that the function of miR-126 is endothelial cell type-specific. Interestingly, although not selectively deleted in the microvascular beds, miR-126 knockouts in mouse and zebrafish 1, 2 show embryonic lethality and major vascular development defects. Results from these studies, coupled with our results obtained at miR-126 levels ~25–40% of normal, indicate that the functional effects of miR-126 may be dependent on a specific range abundance. Although the passenger strand of miR-126, miR-126–5p, may also regulate endothelial cell proliferation33, we are confident that the functional effects described here were specific to miR-126–3p since no significant change was detected in miR-126–5p or its principal targets DLK1 and SERPINE1. However, these results do not rule out important type-specific function of miR-126–5p in this cell type, which could be investigated in future studies.
Our findings provide a mechanistic connection between the anti-proliferative effect of miR-126 and inhibition of LAT1, which controls mTOR activity by regulating EAA uptake 44. Since mTOR is a key regulator of fundamental cellular processes including protein synthesis, cell growth and survival 45, 46, miR-126 is critically responsible for the microvascular endothelial cell fate. Additionally, consistent with previous study that inhibition of mTOR dampens cellular response to angiogenic growth factors in endothelial cells 44, 52, our pathway analysis showed simultaneous inhibition of mTOR and FGF, HGF, VEGF signaling pathways in the cells with high miR-126 expression. In addition to LAT1, we identified and validated ADAM9 as another miR-126 target which, may also contribute to the inhibition of growth factor signaling by miR-126 given its critical role in the growth factor and receptor interaction53, 54.
However, our result that miR-126 knockdown reduced cell apoptosis independent of LAT-1 indicates that LAT1 may not mediate all miR-126-induced effects in HLMVEC. Also, despite the key role of mTOR in regulating autophagy 44, 55, 126-OE or 126-KD did not affect autophagy markers that would indicate autophagosome initiation and elongation, such as Atg5 and Atg7, or formation and degradation autophagosomes by lysosomes, such as SQSTM1 (p62)56. These results suggest alternative mechanisms of miR-126-mediated cell apoptosis. The top candidates, based on our pathway analysis, include the ceramide, PTEN and p53, and the TNFR1 signaling pathways, which were all markedly activated by high miR-126 level, and previously shown to be involved in the induction of programmed cell death 57–59.
Besides elucidating the role of miR-126 during HLMVEC homeostasis, our findings may inform the design of interventions promoting the survival and fitness of these cells during harmful environmental exposures such as CS. Chronic CS exposure causes extensive lung endothelial cell death which contributes to the pathogenesis of emphysema, a phenotype of COPD characterized by net loss of alveolar gas exchange area 60. It remains unclear if miRNA-126 plays a role in the development of CS-induced emphysema. We have previously reported that acute exposure to CS increases the release of miR-126 within exosomes and decreases the intracellular miR-126 levels in HLMVEC61. In light of our data, the decrease in intracellular miR-126 is likely an adaptive survival response, which is consistent with previous reports that short-term CS exposure activates pro-survival signaling such as AKT and mTOR 62, 63. In our study, HLMVEC with reduced miR-126 level were protected from CS-induced cell death and dysfunction (measured as weaker tube formation). It remains to be determined if this effect will extrapolate to animal models and whether miR-126 depletion following CS exposure has a similar beneficial effect.
Our findings demonstrate a cell type-specific role of miR-126 in primary human lung microvascular cells involving negative regulations of targets and pathways essential for angiogenesis and cell survival, which may in turn have potential implication on the pathogenesis of microvasculature-based lung diseases and provide basis for site-specific therapeutic treatment.
Supplementary Material
Highlights.
High levels of miR-126 in HLMVEC drive an anti-angiogenic and pro-apoptotic transcriptomic profile that causes inhibition on cell proliferation, angiogenesis and induces apoptosis.
Reducing miR-126 in HLMVEC enhances cell proliferation and angiogenesis by targeting LAT1, with subsequent activation of mTOR signaling.
Reducing miR-126 in HLMVEC inhibits cell apoptosis.
Induction of a pro-angiogenic, anti-apoptotic phenotype by miR-126 knockdown reduces cell death and improves angiogenesis in HLMVEC exposed to cigarette smoke.
Acknowledgements
DC conceived research, performed experiments, analyzed data, interpreted results, and wrote the manuscript. AJR performed experiments and analyzed data. AMM and ELB performed experiments. KCA performed bioinformatics analysis. KK helped design experiments and interpreted results. AMM and DC generated schematics. IP conceived the research, provided reagents /materials /analysis tools, interpreted results, and wrote the manuscript.
Sources of Funding: This study was supported by NIH/NHLBI (RO1HL077328) to IP; AHA predoctoral fellowship (16PRE31310015) and the University of Colorado, Anschutz Medical Campus, RNA Bioscience initiative predoctoral fellowship to DC.
Abbreviations
- AC
air control
- ADAM9
disintegrin and metalloproteinase domain-containing protein 9
- CS
cigarette smoke
- HLMVEC
human lung microvascular endothelial cells
- KD
knockdown
- LAT1
L-type amino acid transporter 1
- miR
microRNA
- mTOR
mammalian target of rapamycin
- OE
Overexpression
- PARP
poly (ADP-ribose) polymerase
- S6
ribosomal protein S6
- S6K
p70 ribosomal protein S6 kinase
- VEGF
vascular endothelial growth factor
Footnotes
Disclosure: All authors declare no conflict of interest.
Reference
- 1.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microrna mir-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. Mir-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol. 2011;27:563–584 [DOI] [PubMed] [Google Scholar]
- 4.Jansen F, Yang X, Hoelscher M, Cattelan A, Schmitz T, Proebsting S, Wenzel D, Vosen S, Franklin BS, Fleischmann BK, Nickenig G, Werner N. Endothelial microparticle-mediated transfer of microrna-126 promotes vascular endothelial cell repair via spred1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128:2026–2038 [DOI] [PubMed] [Google Scholar]
- 5.Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, van de Rijn M, Botstein D, Brown PO. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003;100:10623–10628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolan DJ, Ginsberg M, Israely E, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goerke SM, Kiefer LS, Stark GB, Simunovic F, Finkenzeller G. Mir-126 modulates angiogenic growth parameters of peripheral blood endothelial progenitor cells. Biol Chem. 2015;396:245–252 [DOI] [PubMed] [Google Scholar]
- 9.Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, Bader GD, Dick JE, Naldini L. Attenuation of mir-126 activity expands hsc in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q, Anderson C, Hanus J, Zhao F, Ma J, Yoshimura A, Wang S. Strand and cell type-specific function of microrna-126 in angiogenesis. Mol Ther. 2016;24:1823–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai Y, Bai X, Wang Z, Zhang X, Ruan C, Miao J. Microrna-126 inhibits ischemia-induced retinal neovascularization via regulating angiogenic growth factors. Exp Mol Pathol. 2011;91:471–477 [DOI] [PubMed] [Google Scholar]
- 12.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y. Endothelial-specific intron-derived mir-126 is down-regulated in human breast cancer and targets both vegfa and pik3r2. Mol Cell Biochem. 2011;351:157–164 [DOI] [PubMed] [Google Scholar]
- 13.Ebrahimi F, Gopalan V, Smith RA, Lam AK. Mir-126 in human cancers: Clinical roles and current perspectives. Exp Mol Pathol. 2014;96:98–107 [DOI] [PubMed] [Google Scholar]
- 14.Lechman ER, Gentner B, Ng SWK, et al. Mir-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, Kirita T, Kuniyasu H. Downregulation of mir-126 induces angiogenesis and lymphangiogenesis by activation of vegf-a in oral cancer. Br J Cancer. 2012;107:700–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, Huang E, Xiong S, Xu F, Liu G, Ge D, Chu Y. Dnmt1-microrna126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via adam9-egfr-akt signaling. Clin Cancer Res. 2015;21:854–863 [DOI] [PubMed] [Google Scholar]
- 17.Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, Bacso Z, Scholtz B. Mir-126 inhibits proliferation of small cell lung cancer cells by targeting slc7a5. FEBS Lett. 2011;585:1191–1196 [DOI] [PubMed] [Google Scholar]
- 18.Li H, Chen S, Liu J, Guo X, Xiang X, Dong T, Ran P, Li Q, Zhu B, Zhang X, Wang D, Xiao C, Zheng S. Long non-coding rna pvt1–5 promotes cell proliferation by regulating mir-126/slc7a5 axis in lung cancer. Biochem Biophys Res Commun. 2018;495:2350–2355 [DOI] [PubMed] [Google Scholar]
- 19.Lammers S, Scott D, Hunter K, Tan W, Shandas R, Stenmark KR. Mechanics and function of the pulmonary vasculature: Implications for pulmonary vascular disease and right ventricular function. Compr Physiol. 2012;2:295–319 [DOI] [PubMed] [Google Scholar]
- 20.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnaoutova I, Kleinman HK. In vitro angiogenesis: Endothelial cell tube formation on gelled basement membrane extract. Nat Protoc. 2010;5:628–635 [DOI] [PubMed] [Google Scholar]
- 22.Schweitzer KS, Johnstone BH, Garrison J, et al. Adipose stem cell treatment in mice attenuates lung and systemic injury induced by cigarette smoking. Am J Respir Crit Care Med. 2011;183:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike K, Berdyshev EV, Mikosz AM, Bronova IA, Bronoff AS, Jung JP, Beatman EL, Ni K, Cao D, Scruggs AK, Serban KA, Petrache I. Role of glucosylceramide in lung endothelial cell fate and emphysema. Am J Respir Crit Care Med. 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Lei R, Ding SW, Zhu S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews S. Fastqc A quality control tool for high throughput sequence data. 2010 [Google Scholar]
- 26.Bates DM Martin; Bolker Ben; Walker Steve. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. 2015;67 [Google Scholar]
- 27.Benjamini YH Yosef.,Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Royal Statist. Soc., Series B. 1995;57 [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. Star: Ultrafast universal rna-seq aligner. Bioinformatics. 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuznetsova AB, Per B; Christensen Rune H. B. Lmertest package: Tests in linear mixed effects models. Journal of Statistical Software. 2017;82 [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930 [DOI] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul Flicek MRA, Daniel Barrell, Kathryn Beal, et al. Ensembl 2014. Nucleic Acids Research. 2014;42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. Microrna-126–5p promotes endothelial proliferation and limits atherosclerosis by suppressing dlk1. Nat Med. 2014;20:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297 [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong N, Wang X. Mirdb: An online resource for microrna target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Wang X. Prediction of functional microrna targets by integrative modeling of microrna binding and target expression data. Genome Biol. 2019;20:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell. 2005;120:15–20 [DOI] [PubMed] [Google Scholar]
- 39.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams JM, Cory S. The bcl-2 protein family: Arbiters of cell survival. Science. 1998;281:1322–1326 [DOI] [PubMed] [Google Scholar]
- 41.Shiojima I, Walsh K. Role of akt signaling in vascular homeostasis and angiogenesis. Circ Res. 2002;90:1243–1250 [DOI] [PubMed] [Google Scholar]
- 42.Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One. 2012;7:e42697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Chen H, Wong N, Haynes W, Baker CM, Wang X. Pseudohypoxia induced by mir-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett. 2017;394:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mtor and autophagy. Cell. 2009;136:521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laplante M, Sabatini DM. Mtor signaling at a glance. J Cell Sci. 2009;122:3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxton RA, Sabatini DM. Mtor signaling in growth, metabolism, and disease. Cell. 2017;169:361–371 [DOI] [PubMed] [Google Scholar]
- 47.Demedts IK, Demoor T, Bracke KR, Joos GF, Brusselle GG. Role of apoptosis in the pathogenesis of copd and pulmonary emphysema. Respir Res. 2006;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voelkel NF, Douglas IS, Nicolls M. Angiogenesis in chronic lung disease. Chest. 2007;131:874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microrna-126 by apoptotic bodies induces cxcl12-dependent vascular protection. Sci Signal. 2009;2:ra81. [DOI] [PubMed] [Google Scholar]
- 50.Hur J, Yoon CH, Kim HS, Choi JH, Kang HJ, Hwang KK, Oh BH, Lee MM, Park YB. Characterization of two types of endothelial progenitor cells and their different contributions to neovasculogenesis. Arterioscler Thromb Vasc Biol. 2004;24:288–293 [DOI] [PubMed] [Google Scholar]
- 51.Basile DP, Yoder MC. Circulating and tissue resident endothelial progenitor cells. J Cell Physiol. 2014;229:10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dormond O, Madsen JC, Briscoe DM. The effects of mtor-akt interactions on anti-apoptotic signaling in vascular endothelial cells. J Biol Chem. 2007;282:23679–23686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guaiquil V, Swendeman S, Yoshida T, Chavala S, Campochiaro PA, Blobel CP. Adam9 is involved in pathological retinal neovascularization. Mol Cell Biol. 2009;29:2694–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weskamp G, Kratzschmar J, Reid MS, Blobel CP. Mdc9, a widely expressed cellular disintegrin containing cytoplasmic sh3 ligand domains. J Cell Biol. 1996;132:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crespo JL, Hall MN. Elucidating tor signaling and rapamycin action: Lessons from saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2002;66:579–591, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of pten transcription by p53. Mol Cell. 2001;8:317–325 [DOI] [PubMed] [Google Scholar]
- 58.Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the tnf superfamily. Biochem Pharmacol. 2003;66:1403–1408 [DOI] [PubMed] [Google Scholar]
- 59.Petrache I, Natarajan V, Zhen L, Medler TR, Richter AT, Cho C, Hubbard WC, Berdyshev EV, Tuder RM. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat Med. 2005;11:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuder RM, Yoshida T, Arap W, Pasqualini R, Petrache I. State of the art. Cellular and molecular mechanisms of alveolar destruction in emphysema: An evolutionary perspective. Proc Am Thorac Soc. 2006;3:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serban KA, Rezania S, Petrusca DN, et al. Structural and functional characterization of endothelial microparticles released by cigarette smoke. Sci Rep. 2016;6:31596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshida T, Mett I, Bhunia AK, et al. Rtp801, a suppressor of mtor signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat Med. 2010;16:767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrusca DN, Van Demark M, Gu Y, Justice MJ, Rogozea A, Hubbard WC, Petrache I. Smoking exposure induces human lung endothelial cell adaptation to apoptotic stress. Am J Respir Cell Mol Biol. 2014;50:513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.