Abstract
The survival and recurrence of dormant tumour cells following therapy is a leading cause of death in cancer patients. The metabolic properties of these cells are likely distinct from those of rapidly growing tumours. Here we show that Her2 down-regulation in breast cancer cells promotes changes in cellular metabolism, culminating in oxidative stress and compensatory upregulation of the antioxidant transcription factor, NRF2. NRF2 is activated during dormancy and in recurrent tumours in animal models and breast cancer patients with poor prognosis. Constitutive activation of NRF2 accelerates recurrence, while suppression of NRF2 impairs it. In recurrent tumours, NRF2 signalling induces a transcriptional metabolic reprogramming to re-establish redox homeostasis and upregulate de novo nucleotide synthesis. The NRF2-driven metabolic state renders recurrent tumour cells sensitive to glutaminase inhibition, which prevents reactivation of dormant tumour cells in vitro, suggesting that NRF2-high dormant and recurrent tumours may be targeted. These data provide evidence that NRF2-driven metabolic reprogramming promotes the recurrence of dormant breast cancer.
Keywords: NRF2, ROS, Tumor metabolism, Residual disease, Breast cancer recurrence, Her2
Introduction
Tumor formation requires profound changes in cellular metabolism. These changes are mediated, in part, through the direct regulation of metabolic pathways by oncogenes and tumour suppressors1,2. For example, the PI3K-Akt pathway promotes glucose utilization via several mechanisms, including promoting the membrane localization of glucose transporters3. MYC induces transcription of glutaminase and glutamine transporters to induce glutamine utilization4. Her2/neu signalling, in addition to regulating glucose metabolism through the PI3K-Akt pathway, induces high rates of de novo fatty acid synthesis by regulating fatty acid synthase5 and lipid storage pathways6. These oncogenic pathways are necessary to sustain anabolic metabolism. Metabolic reprogramming is an early event in transformation and is required for tumorigenesis, in part to support the rapid proliferation characteristic of most tumour cells7,8.
Tumor dormancy describes a stage of tumorigenesis during which tumours do not exhibit an increase in size, and can result from either tumour-mass dormancy or cellular dormancy. Tumour-mass dormancy arise when there is a balance between tumour cell proliferation and cell death, which is often mediated by the immune system or insufficient vascularization9–11. In cellular dormancy, tumour cells persist in a slowly growing or non-proliferative state, often as a result of therapeutic intervention or dissemination9–11. In contrast to the extensive literature on the metabolism of rapidly proliferating tumour cells, much less is known about the metabolic properties of dormant or slowly proliferating tumour cells. Similarly, very little is known about the metabolism of recurrent tumours that form when dormant tumour cells re-enter the cell cycle or re-initiate rapid proliferation. Considering the intimate relationship between cancer cell proliferation and metabolism, we reasoned that the metabolism of cancer cells may change dramatically during these transitions between proliferation and dormancy. However, it is not known whether sustained activity of particular metabolic pathways is functionally important in regulating tumour dormancy and recurrence.
Several recent studies have begun to address these issues. For instance, it has been shown that inhibition of diverse oncogenes – including Her29,11, Ras10,11, Myc11, and Wnt11 – causes an immediate drop in glucose uptake both in vitro9,10 and in vivo11. In response to decreased glucose metabolism, cells that survive the loss of oncogenic Her212 and KRAS13 signalling upregulate fatty acid oxidation (FAO) pathways12,13. Additionally, both decreased glucose metabolism and increased FAO following oncogene inhibition results in increased levels of reactive oxygen species (ROS)12–14, which can impair tumour cell viability15,16. These results suggest that tumour cells that survive therapy and eventually develop resistance must adapt to overcome therapy-induced metabolic stress. However, the pathways that promote metabolic adaptations during tumour dormancy and recurrence remain unknown.
To gain insight into how tumour cell metabolism changes following oncogene inhibition, and how these changes contribute to the survival and recurrence of residual cells following therapy, we used a transgenic mouse model of Her2-driven breast cancer17–19. In this model, doxycycline (dox) administration to bitransgenic MMTV-rtTA;TetO-Her2 (MTB/TAN) mice leads to Her2 expression and the formation of invasive mammary adenocarcinomas. Removal of doxycycline causes Her2 downregulation and induces tumour regression. However, a small population of cells persists in a dormant state19–21 before eventually re-initiating proliferation to form a recurrent tumour. Using this model, we interrogated the metabolic changes that accompany Her2 downregulation, dormant residual disease, and tumour recurrence. We identified the NRF2 pathway as a critical mediator of metabolic reprogramming following oncogene inhibition. NRF2 promotes the recurrence of dormant tumours by promoting redox homeostasis and nucleotide metabolism. As a consequence of this metabolic reprogramming, recurrent tumours with high NRF2 are sensitive to glutaminase inhibition, suggesting a therapeutic strategy for treating recurrent tumours.
Results
Her2 downregulation induces metabolic changes and loss of redox homeostasis
To examine the metabolic consequences of Her2 downregulation, we used an in vitro model in which tumour cells maintain their dependency upon Her2. Primary Her2-driven tumours were digested and cultured as mammospheres in non-adherent, serum-free conditions in the presence of dox. Removal of dox from these cells led to a rapid decrease in Her2 levels and in downstream components of the MAPK and Akt-mTOR pathways (Extended Data Fig. 1a,b). This was associated with a loss of proliferation that was evident as early as 2 days (Fig. 1a), and an induction of apoptosis starting at 7 days following dox withdrawal (Fig. 1b,c). This apoptosis was transient, and by 13 days following dox withdrawal we identified surviving cells that were quiescent but non-apoptotic (Fig. 1d). These surviving cells were viable, because re-addition of dox to these mammospheres led to re-induction of proliferation (Fig. 1e). Taken together, these results indicate that these mammospheres faithfully model the behavior of tumours in vivo: tumour cells remain dependent upon Her2 for their growth and survival, but a population of cells can survive Her2 downregulation and persist in a viable, non-proliferative state.
Fig. 1 |. Her2 downregulation induces metabolic changes.
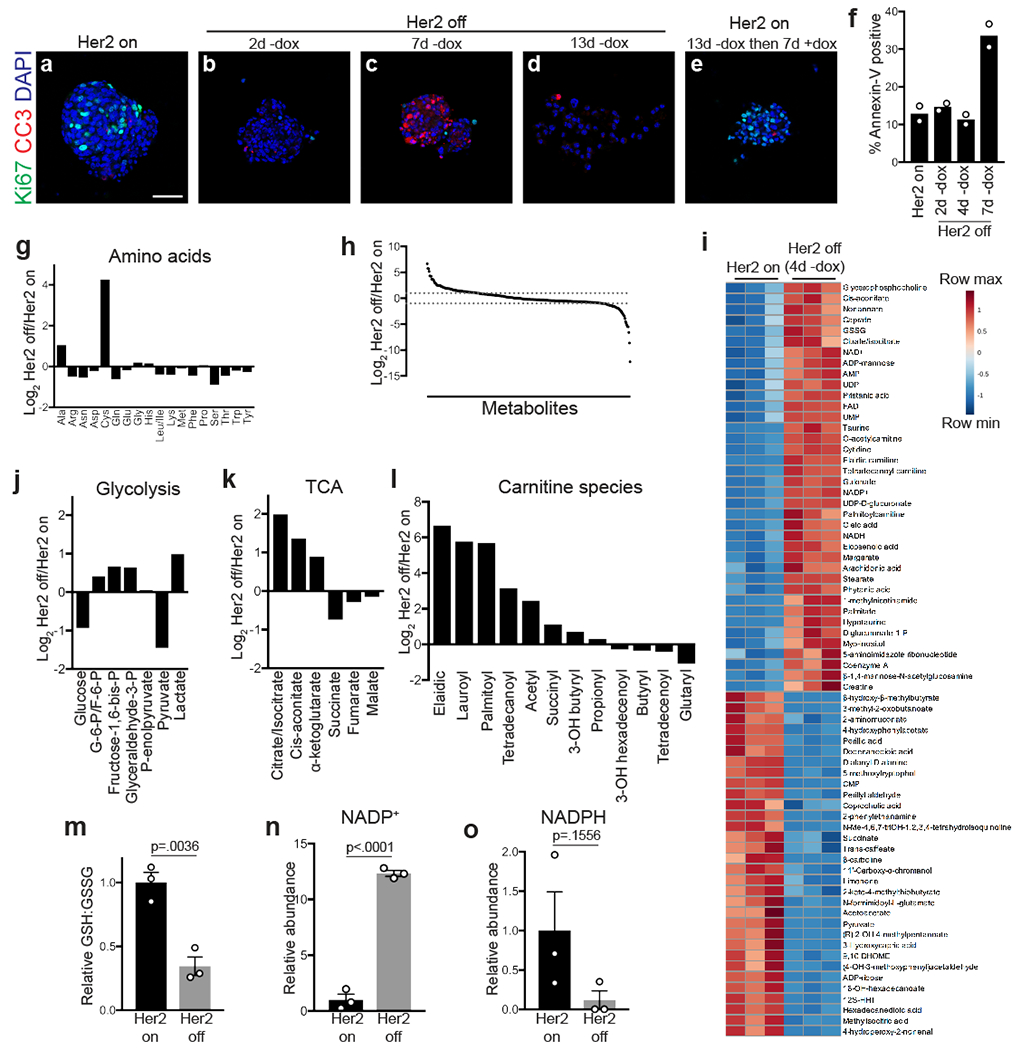
a-e, Immunofluorescence staining for Ki67 (green) and cleaved caspase-3 (red) in mammospheres cultured in the presence of dox (A; Her2 on), without dox (Her2 off) for 2 days (B), 7 days (C), and 13 days (D), and without dox for 13 days before re-addition of dox for 7 days (E). Scale bar, 50 μm. Data represent 3 independent experiments. f, Percentage of Annexin V-positive cells in mammospheres cultured in the presence of dox (Her2 on), or without dox (Her2 off) for 2, 4, and 7 days. Data are shown as the mean of n=2 biologically independent replicates and are representative of 3 independent experiments. g, Changes in amino acid levels between Her2 off and Her2 on conditions. h, Ratio of metabolite levels between Her2 off and Her2 on conditions. Dashed lines represent 2-fold changes. i, Heatmap showing top 70 altered metabolites following Her2 downregulation in mammospheres. j-l, Changes in glycolytic intermediates (j), citric acid cycle (TCA) intermediates (k), and carnitine species (l) between Her2 off and Her2 on conditions. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate. m, Relative ratio of reduced (GSH) to oxidized (GSSG) glutathione in cells with Her2 on or Her2 off. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test. n, Relative levels of oxidized NADP (NADP+) in cells with Her2 on or Her2 off. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test. o, Relative levels of reduced NADP (NADPH) in cells with Her2 on or Her2 off. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test.
We next used this model to characterize the metabolic changes that occur following Her2 downregulation. To avoid any confounding effects of apoptosis on cell metabolism, we chose a 4-day time-point, which is after the loss of Her2 signalling but prior to the onset of apoptosis (Fig. 1f). We reasoned that at this early time-point cells would be undergoing metabolic changes as they transitioned from a proliferative cell state. Thus, analyzing this time-point may allow for the identification of metabolic pathways that contribute to the fate (survival vs. apoptosis) of these cells after Her2 inhibition. We used liquid chromatography-mass spectrometry to examine changes in metabolite levels in mammospheres 4 days following Her2 downregulation. The majority of metabolites, including most amino acids (Fig. 1g), did not change following Her2 downregulation. Of the 317 measurable metabolites, 70 were significantly increased (fold-change >2, p<0.05) and 26 were significantly decreased (fold-change <0.5, p<0.05) (Fig. 1h,i). Glucose and pyruvate levels decreased following Her2 downregulation, while lactate levels increased (Fig. 1j). Consistent with this result, we observed an increase in the levels of phosphorylated pyruvate dehydrogenase (p-PDH) (Extended Data Fig. 1c), suggesting that Her2 downregulation diverts pyruvate away from citric acid cycle anaplerosis and toward conversion to lactate. We also observed increased steady-state levels of several citric acid cycle intermediates, including citrate/isocitrate, cis-aconitate, and α-ketoglutarate (Fig. 1k). Further, Her2 downregulation induced a considerable increase in the steady-state levels of many acylcarnitines (Fig. 1l). Because acylcarnitines are intermediates in fatty acid oxidation, this suggests that these cells have increased levels of fatty acid oxidation (FAO), which is in agreement with findings in similar systems12,13.
Interestingly, Her2 downregulation also induced profound changes in the steady-state levels of components of glutathione metabolism. There was a substantial decrease in the ratio between the reduced (GSH) and oxidized (GSSG) forms of glutathione (Fig. 1m), and this coincided with an increase in NADP+ (Fig. 1n) and a decrease in NADPH (Fig. 1o), resulting in an increase in the ratio of oxidized to reduced NADP (Extended Data Fig. 1d). Glutathione is a critical intracellular antioxidant, and it is maintained in its reduced form using reducing capacity of NADPH. Thus, the observed shifts in GSH:GSSG and NADP+:NADPH ratios indicate that Her2 downregulation is associated with a loss of cellular redox homeostasis. These changes are consistent with reduced levels of glycolytic intermediates and increased FAO intermediates following Her2 downregulation. Taken together, these results suggest that Her2 downregulation induces cells to use FAO for energy generation, which is associated with changes in redox homeostasis.
Her2 inhibition increases ROS levels and leads to ROS-dependent cell death
Changes in the redox states of glutathione and NADP are often indicative of oxidative stress caused by accumulation of reactive oxygen species (ROS). We therefore directly measured the levels of cellular ROS using 2’,7’–dichlorofluorescin diacetate (DCFDA) in mammospheres cultured with or without dox for four days. Her2 downregulation led to a substantial increase in ROS levels (Fig. 2a). To ensure that increased ROS was not a secondary consequence of apoptosis, we measured ROS levels two days following dox withdrawal, a time-point that precedes apoptosis in this model (see Fig. 1e,f), and found increased ROS at this early time-point as well (Extended Data Fig. 2a). To expand these results, we tested the effects of Her2 inhibition in two well-characterized human Her2-amplified breast cancer cell lines, BT474 and SKBR3 cells. Treatment with lapatinib, a small-molecule dual Her2/EGFR inhibitor, led to a dose-dependent increase in ROS levels in both Her2-amplified cell lines (Fig. 2b,c) but not in triple negative MDA-MB-231 cells that do not have Her2 amplification (Extended data Fig. 2b). To further test if this increase in ROS was caused by on-target effects of lapatinib, we treated cells with the structurally distinct pan-HER kinase inhibitor neratinib, which is an irreversible inhibitor of EGFR, Her2, and Her4. We again found that neratinib treatment induced ROS in Her2-amplified cell lines but not in MDA-MB-231 cells (Extended Data Fig. 2c). These results are in agreement with the results from genetic downregulation of Her2 in mammosphere cultures, and demonstrate that Her2 inhibition specifically induces the accumulation of ROS. We next assessed changes in mitochondrial ROS levels following Her2 inhibition using MitoSox, which specifically measures ROS levels in mitochondria. Mitochondrial ROS levels increased following Her2 downregulation in mammospheres (Fig. 2d) as well as in BT474 and SKBR3 cells following lapatinib treatment (Fig. 2e,f). Given the well-established role of ROS in inducing cell death, we next asked whether the increased oxidative stress we observed contributes to cell death following Her2 inhibition. Treatment with the antioxidant N-acetylcysteine (NAC) prevented ROS accumulation (Fig. 2g) and rescued cell viability following lapatinib treatment (Fig. 2h–j and Extended Data Fig. 3a). To further test that this effect was mediated by the antioxidant capacity of NAC, we also treated cells with exogenous GSH, which similarly rescued cell viability following lapatinib treatment (Fig. 2k).
Fig. 2 |. Her2 inhibition increases ROS levels and leads to ROS-dependent cell death.
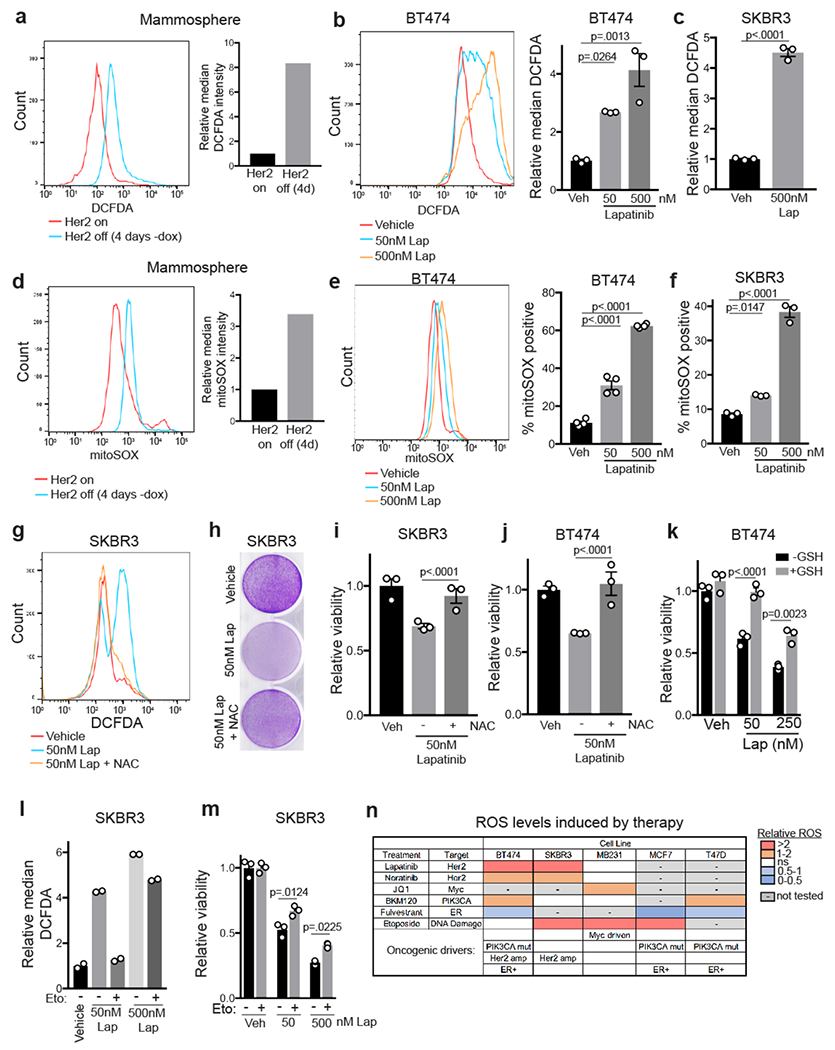
a, DCFDA staining showing ROS levels in mammospheres cultured with dox or without dox for 4 days. Data are representative of 3 independent experiments. b-c, DCFDA staining showing ROS levels in BT474 (b) and SKBR3 (c) cells treated with lapatinib for 48 hours. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by one-way ANOVA (Tukey’s multiple comparisons test) (b) and two-sided Student’s t Test (c). Data are representative of 3 independent experiments. d, MitoSOX staining showing mitochondrial superoxide levels in mammospheres cultured with dox or without dox for 4 days. Data are representative of 3 independent experiments. e-f, MitoSOX staining showing mitochondrial superoxide levels in BT474 (e) and SKRB3 (f) cells treated with lapatinib for 48 hours. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by one-way ANOVA (Tukey’s multiple comparisons test). Data are representative of 3 independent experiments. g, DCFDA staining showing ROS levels in SKBR3 cells treated with lapatinib and 5mM N-acetyl cysteine (NAC). Data are representative of 2 independent experiments. h, Crystal violet staining showing viability after treatment of SKBR3 cells with lapatinib and 5mM NAC for 6 days. i,j, Relative viability of SKBR3 cells (i) and BT474 cells (j) treated with lapatinib and 5mM NAC for 72 hours. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by one-way ANOVA (Tukey’s multiple comparisons test). k, Relative viability of BT474 cells treated with lapatinib and 10mM glutathione (GSH) for 72 hours. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). l, DCFDA staining showing ROS levels in SKBR3 cells treated with lapatinib and 100μM etomoxir (eto). Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 independent experiments. m, Relative viability of SKBR3 cells treated with lapatinib and 100μM etomoxir (eto) for 72 hours. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). n, Chart summarizing changes in ROS levels for a panel of breast cancer cell lines treated with indicated therapies. Color scale indicates the fold change in ROS levels. Known oncogenic drivers for each cell line are annotated in the bottom rows.
We next wanted to determine the metabolic basis for increased ROS following Her2 inhibition. Recent evidence has shown that inhibition of the glucose dependent oxidative pentose phosphate pathway (PPP), which generates NADPH, is sufficient to induce ROS and cell death in tumour cells22. Additionally, the induction of FAO in cells that survive oncogene downregulation has been shown to cause ROS accumulation12,13. Interestingly, we found that treatment with two independent inhibitors of the PPP, dehydroepiandrosterone (DHEA) and 6-aminonicotinamide (6-An), was not sufficient to increase ROS levels in breast cancer cell lines (Extended Data Fig. 3b,c) or mammospheres (Extended Data Fig. 3d). Further, PPP inhibition did not exacerbate induce ROS in combination with lapatinib (Extended Data Fig. 3c). On the other hand, we found that treatment of cells with the FAO inhibitor, etomoxir, prevented ROS accumulation following Her2 inhibition (Fig. 2l) and partially rescued cell viability (Fig. 2m and Extended Data Fig. 3e). This suggests that Her2 inhibition leads to the induction of FAO, consistent with our observation that acylcarnitines and mitochondrial ROS are enriched following Her2 downregulation (Fig. 1l and Fig. 2d–f). To explore this further, we tested whether Her2 inhibition induced expression of genes required for FAO. In two independent mammosphere cultures, Her2 downregulation resulted in significant increases in the mitochondrial acylcarnitine transporters, Cpt1a and Cpt1b, and the fatty acid transporter, CD36 (Extended Data Fig. 3f). Additionally, staining of mammospheres with the neutral lipid marker, boron-dipyrromethene (BODIPY), showed that mammospheres contained abundant lipid droplets (Extended Data Fig. 3g). Taken together, these results suggest that the induction of FAO following Her2 inhibition is responsible for ROS accumulation.
We next tested if other therapies induced oxidative stress similarly to Her2 inhibition. We first tested the effects of the PI3K inhibitor, BKM120, on ROS levels, and we found that BKM120 treatment increased ROS levels in PIK3CA-mutant cell lines (BT474 and T47D) but not in PIK3CA-wild type cell lines (SKBR3 and MDA-MB-231) (Extended Data Fig. 3h). We also tested the effects of the MYC inhibitor JQ1 in MYC-driven triple negative MDA-MB-231 cells, and found that JQ1 caused a modest increase in ROS levels (Extended Data Fig. 3i). In contrast, fulvestrant, a selective estrogen receptor degrader, did not increase ROS levels in 3 ER+ breast cancer cell lines (MCF7, T47D and BT474; Extended Data Fig. 4a). Finally, we tested whether a cytotoxic chemotherapeutic drug would also induce ROS. We found that the DNA damaging agent etoposide induced robust ROS levels in many, but not all, cell lines tested (Extended Data Fig. 4b). Taken together, these results demonstrate that many common therapies induce oxidative stress in breast cancer cells, and also that therapy-induced ROS is not specific to a particular breast cancer subtype (Fig. 2n).
The NRF2 antioxidant transcriptional program is activated in dormant and recurrent tumours
The finding that Her2 inhibition leads to ROS-dependent cell death suggests that dormant tumour cells that are able to survive Her2 downregulation may have activated intrinsic cellular antioxidant pathways. The transcription factor NRF2 (nuclear factor (erythroid-derived 2)-like 2, or Nfe2l2) is a critical mediator of the cellular adaptive antioxidant response. NRF2 becomes stabilized in response to oxidative stress and activates genes involved in restoring redox homeostasis23. NRF2 has been established as a driver of tumour progression24, resistance to therapy14,25,26, and metastasis27. We first assessed whether NRF2 is stabilized following Her2 downregulation in vivo. Western blotting of tumour samples from MTB/TAN mice with primary tumours or shortly after dox withdrawal showed that NRF2 protein levels increase following Her2 downregulation (Fig. 3a). Additionally, qRT-PCR analysis showed that the NRF2 target genes, Gclm, Nqo1, and Hmox1, were increased in these tumours (Extended Data Fig. 5a). Similar results were obtained using immunofluorescence staining for NRF2. NRF2 was expressed at low or undetectable levels in primary tumours, but was stabilized and located in the nucleus of tumour cells following Her2 downregulation (Fig. 3b). These results indicate that Her2 downregulation leads to NRF2 stabilization in vivo, likely as a result of increased ROS.
Fig. 3 |. The NRF2 antioxidant transcriptional program is activated in dormant and recurrent tumours.

a, Western blot for NRF2 in primary tumours and tumours 48 and 96 hours after dox withdrawal (Her2 off). Blot is representative of 2 independent experiments. b, Immunofluorescence staining for NRF2 in primary tumours and tumours 48 hours after dox withdrawal (Her2 off). Scale bars represent 50μm (left) and 25μm (right). White box on left panels indicates magnified section shown in right panels. Images are representative of 3 biologically independent experiments. c, Immunofluorescence staining for NRF2 in primary tumours and 56 days after dox withdrawal (residual tumour). Scale bars represent 50μm (left) and 25μm (right). White box on left panels indicates magnified section shown in right panels. d, Quantification of nuclear NRF2 staining from (c) comparing primary tumours and tumours 56 days after dox withdrawal (residual tumours). p-value was determined by two-sided Student’s t test of the average values for each group. e, Western blots for Her2 and NRF2 in primary (n=4) and recurrent (n=9) tumours. Data are representative of 3 biologically independent experiments. f, Western blots for NRF2 and NQO1 in two independent primary tumour cell lines and two independent recurrent tumour cell lines with or without treatment with the electrophile TBHQ (25μM). Data are representative of 3 biologically independent experiments.
A small population of tumour cells survives Her2 downregulation and persists in a dormant state prior to seeding recurrent tumours21. We next wanted to determine if NRF2 expression remains elevated in these residual tumour cells. Residual tumours were readily identifiable in previously tumour-bearing mammary glands (Extended Data Fig. 5b), and tumour cells within these residual tumours were predominantly Ki67-negative (Extended Data Fig. 5c,d). Strikingly, immunofluorescence staining for NRF2 in dormant tumours revealed that residual tumours exhibited high nuclear NRF2 staining, suggesting that this antioxidant response persisted in dormant tumours (Fig. 3c,d).
Fig. 5 |. NRF2 suppression impairs recurrent tumour growth in vivo.

a, qRT-PCR analysis of NRF2 (Nfe2l2), Nqo1, and Hmox1 expression in recurrent tumour cells expressing scrambled or NRF2-targeting shRNA. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). Data are representative of 2 independent experiments. b, Western blot for NRF2 and NQO1 in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. Blot is representative of two biologically independent experiments. c, Population doublings in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. n=3 biological replicates for each time point. Significance was determined by Analysis of Covariance (ANCOVA) between the slope (population doublings/time) for each condition. d,e, Tumor growth curves (d) and area under the curve analysis (e) for tumours generated from control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. Significance was determined by one-way ANOVA using the mean tumour volumes at experiment endpoint (n=16 per condition, Tukey’s multiple comparisons test). Data are representative of 2 independent experiments.
Residual tumour cells persist for several months before spontaneously re-initiating proliferation to give rise to recurrent tumours. We asked whether these recurrent tumours maintain elevated NRF2. Western blotting showed that the majority of recurrent tumours had high NRF2 levels (Fig. 3e). We next compared NRF2 levels in cells derived from independent primary and recurrent tumours. NRF2 levels were elevated in recurrent tumour cells as compared to primary cells (Fig. 3f), mimicking the behavior of primary and recurrent tumours in vivo. Similarly, NRF2 target genes were elevated in recurrent tumour cells cultured in vitro (Extended Data Fig. 5e). Taken together, these results demonstrate that tumours upregulate the NRF2 antioxidant pathway in response to metabolic changes caused by Her2 downregulation, and NRF2 activation persists during dormancy and in recurrent tumours.
Constitutive NRF2 activity promotes tumour recurrence
In light of our observation that NRF2 is frequently activated in cancer cells in response to the metabolic stress induced by Her2 inhibition, and remains elevated in the majority of residual and recurrent tumours, we next sought to determine if NRF2 activity promotes tumour recurrence. To this end, we took a gain of function approach to induce constitutive NRF2 activity in all residual tumour cells. We ectopically expressed a constitutively active NRF2 (caNRF2) – which lacks the N-terminal domain that binds its negative regulator, KEAP128,29 (Fig. 4a) – in primary tumour cells. Western blotting and qRT-PCR analysis showed that caNRF2 was expressed in primary tumour cells (Fig. 4b) and was transcriptionally active, evidenced by a marked increase in expression of its target genes Nqo1, Slc7a11, Gclm, and Hmox1 (Extended Data Fig. 6a). Additionally, basal ROS levels were modestly decreased in cells expressing caNRF2 (Extended Data Fig. 6b). Cohorts of recipient mice on dox were injected with primary tumour cells expressing either empty vector or caNRF2 (Fig. 4c). Expression of caNRF2 had no effect on the rate of primary tumour formation (Extended Data Fig. 6c). Following primary tumour formation, mice were then removed from dox to induce Her2 downregulation and tumour regression. Both control and caNRF2 tumours fully regressed (greater than 95% reduction in tumour volume) following dox withdrawal. Mice with residual tumours were palpated weekly to monitor the emergence of recurrent tumours (Fig. 4c). Tumors expressing caNRF2 recurred nearly 3 weeks sooner than control tumours (Fig. 4d). Importantly, caNRF2 and its target, NQO1, remained highly expressed throughout the time course of recurrence (Extended Data Fig. 6d). Interestingly, recurrent tumours from the control cohort activated endogenous NRF2, evidenced by increased NRF2 and NQO1 protein levels (Extended Data Fig. 6e) and increased NRF2 target gene expression (Fig. 4e), underscoring the importance of NRF2 for tumour recurrence. Taken together, these results indicate that NRF2 activity promotes the formation of recurrent tumours.
Fig. 4 |. Constitutive NRF2 activity promotes tumour recurrence.
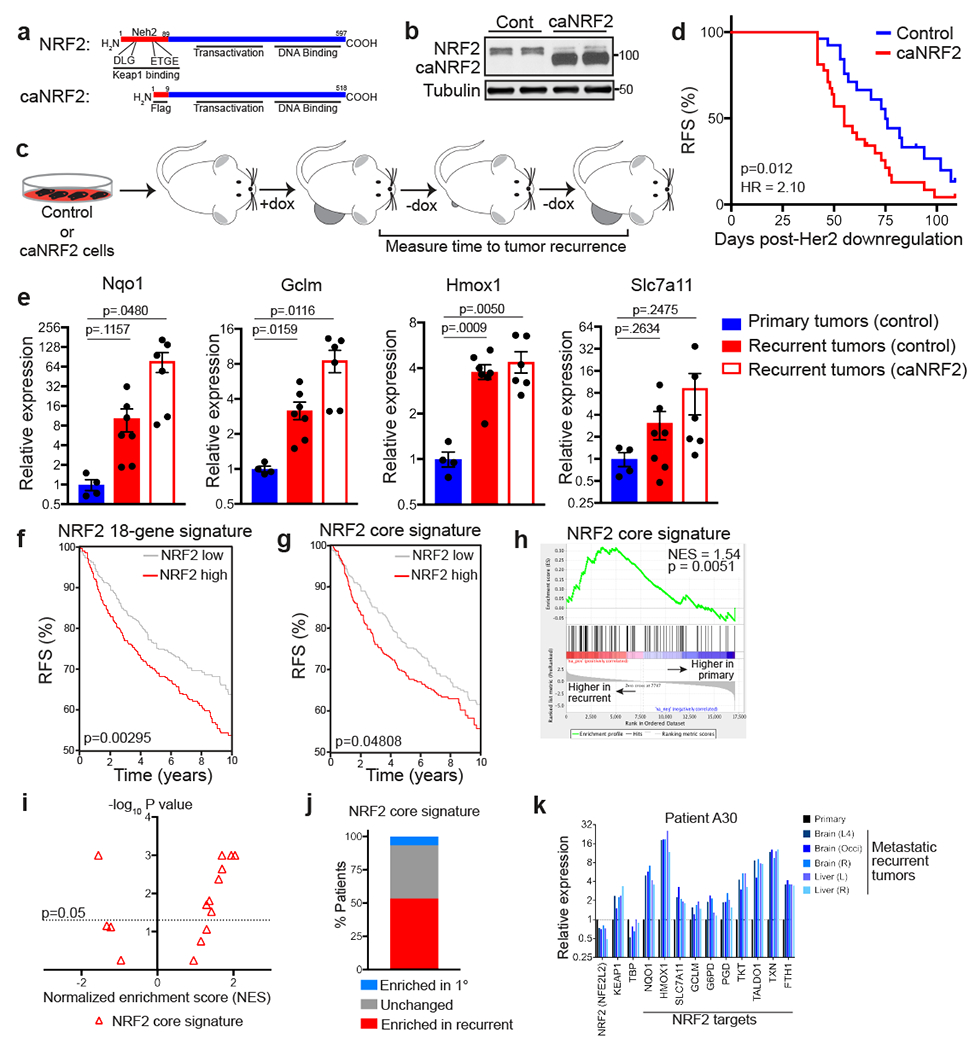
a, Schematic showing wild-type (top) and constitutively active NRF2 (caNRF2, bottom) proteins. caNRF2 lacks the first 86 amino acids containing the KEAP1 binding domains, DLG and ETGE. b, Western blot for NRF2 in primary tumour cells expressing empty vector (Cont) or caNRF2. Blot is representative of 3 biologically independent experiments. c, Schematic showing in vivo assay for tumour recurrence. d, Kaplan-Meier plot showing recurrence-free survival (RFS) for mice bearing control tumours (n=27 independent tumours) or caNRF2 tumours (n=32 independent tumours). Hazard ratio and p-value were determined by two-sided log-rank (Mantel-Cox) test. e, qRT-PCR analysis of Nqo1, Gclm, Hmox1, and Slc7a11 expression in control primary tumours (n=4 independent tumours) and control recurrent tumours (n=6 independent tumours) and caNRF2 recurrent tumours (n=6 independent tumours) from (d). Significance was determined by two-sided Student’s t test. f, Kaplan-Meier plot showing recurrence-free survival (RFS) for breast cancer patients whose tumours have low (gray, n=487 independent tumours) or high (red, n=427 independent tumours) expression of the NRF2 18-gene signature. Hazard ratio and p-value were determined by two-sided log-rank (Mantel-Cox) test. g, Kaplan-Meier plot showing recurrence-free survival (RFS) for breast cancer patients whose tumours have low (gray, n=523 independent tumours) or high (red, n=391 independent tumours) expression of the NRF2 core signature. Hazard ratio and p-value were determined by two-sided log-rank (Mantel-Cox) test. h, Gene set enrichment analysis of matched primary and recurrent breast tumours (n=15 patients) showing enrichment of the NRF2 core signature in recurrent tumours. Enrichment scores were calculated using the Kolmogorov-Smirnov statistic and p-values were calculated using permutation testing with 1000 permutations. i, Volcano plot showing enrichment scores for the NRF2 core signature in recurrent tumours for n=15 individual patients. j, The percentage of patients whose recurrent tumours had significantly enriched (red), unchanged (grey), or significantly decreased (blue) expression of the NRF2 core signature. k, Expression of NRF2, KEAP1, TBP, and 10 canonical NRF2 target genes in the primary tumour and matched recurrent tumours from a representative patient.
We next wanted to determine if NRF2 expression was correlated with poor clinical prognosis in breast cancer patients. We used a recently curated NRF2 gene signature30 (NRF2 core signature), consisting of 108 genes, and a smaller gene set consisting of 18 canonical target genes (NRF2 18-gene signature). High expression of both gene sets was correlated with an increased risk of recurrence in a cohort of more than 1,800 breast cancer patients31 (Fig. 4f,g). These results suggest that high NRF2 activity in primary tumours may promote recurrence in breast cancer. We next wanted to determine if there was direct evidence for NRF2 activation in recurrent breast cancers in humans. For this, we used RNA sequencing data from a rapid autopsy study that collected matched primary and metastatic recurrent tumours32. We first ranked genes according to their fold-change between primary and recurrent tumours in each patient, averaged the fold-change in expression across all patients, and then performed gene set enrichment analysis (GSEA) to identify gene expression changes in recurrent tumours. GSEA showed that the NRF2 core gene signature was significantly enriched in recurrent tumours (Fig. 4h). Notably, this analysis also showed that the “Reactive Oxygen Species Pathway” and “Fatty Acid Metabolism” hallmark gene signatures were enriched in recurrent tumours (Extended Data Fig. 7a,b). We then repeated GSEA in individual patients to determine the frequency of NRF2 enrichment in recurrent tumours. GSEA showed that the NRF2 core signature was significantly enriched in recurrent tumours in 8 patients (53.3%), not significantly changed in 6 patients (40%), and significantly decreased in only 1 patient (6.7%) (Fig. 4i,j). A similar analysis with two unrelated transcriptional signatures (“Hallmark Notch Signaling” and the “Hallmark WNT Beta Catenin Signaling”) did not show consistent enrichment of these signatures in recurrent tumours (Extended Data Fig. 7c–h), suggesting that the NRF2 transcriptional response is specifically activated in recurrent tumours. To further illustrate this, we focused on the expression of canonical NRF2 target genes in 4 individual patients whose recurrent tumours had enrichment for a NRF2 signature. The expression of NRF2 itself (NFE2l2), KEAP1, and a housekeeping gene, TBP, were not significantly altered in patients with high NRF2 enrichment scores. However, NRF2 target genes were consistently upregulated in recurrent tumours from these patients as compared to matched primary tumours (Fig. 4k and Extended Data Fig. 7i). Together, these results show that NRF2 activity is frequently increased in recurrent breast cancer in humans.
Fig. 7 |. Identification of essential NRF2-regulated pathways in recurrent tumours.
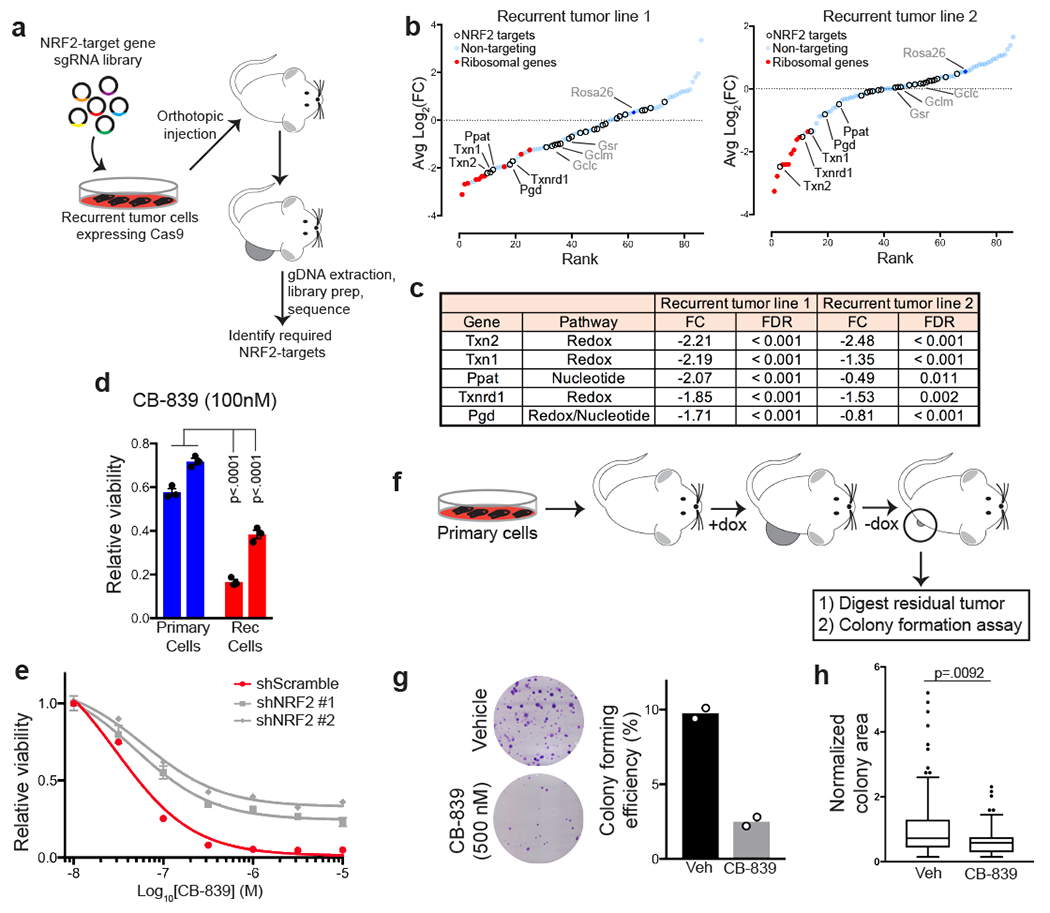
a, Schematic showing the design of an in vivo CRISPR screen to identify NRF2-regulated pathways essential for recurrent tumour growth. b, Fold-change in gRNA abundance between injected cells and tumours for two independent recurrent tumour cell lines. Genes in NRF2-regulated pathways are represented as hollow circles, essential ribosomal genes are represented as red dots, and non-targeting gRNAs are represented as blue dots. Genes are ranked from most to least depleted in tumours. Genes labeled in black were significantly depleted in tumours from both cell lines. Genes labeled in gray were not significantly altered in tumours from either cell lines. Significance was determined using Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK). c, Table showing the fold-change (FC, injected cells/tumours) and NRF2-regulated pathway for 5 genes that were essential (FDR < 0.05) in tumours from both cell lines (n=5 tumours for recurrent tumour line 1; n=12 for recurrent tumour line 2). Significance was determined using MAGeCK. d, Relative viability of primary and recurrent cell lines treated with the glutaminase inhibitor, CB-839, normalized to vehicle-treated condition. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). p < 0.001 for all pairwise comparisons between primary and recurrent cells. e, Dose-response curves showing relative viability in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells treated with indicated doses of CB-839. f, Schematic showing in vivo generation of residual tumours, which were subsequently digested and used for a colony formation assay. g, Crystal violet staining of representative colony formation assays from residual tumour cells treated with or without 500nM CB-839 for 2 weeks (left), and quantification of colony forming efficiency (right). Data are shown as the mean of n=2 biologically independent replicates and are representative of a single experiment. h, Normalized colony area of colonies in (g). Data are shown as the mean of n=2 biologically independent replicates and are representative of a single experiment.
NRF2 suppression impairs recurrent tumour growth in vivo
Having established that expression of constitutively active NRF2 promotes tumour recurrence, we next asked whether NRF2 suppression slows the growth of recurrent tumours. To do this, we knocked down NRF2 expression in recurrent tumour cells with high NRF2 (see Fig. 3f). Expression of independent shRNAs targeting NRF2 suppressed NRF2 mRNA levels in recurrent cells by 75-90% (Fig. 5a). Consistent with this, there was a significant reduction in the mRNA expression of NRF2 target genes, Nqo1 and Hmox1 (Fig. 5a) and a reduction in the protein levels of NRF2 and NQO1 (Fig. 5b). We next assessed the effect of NRF2 knockdown on tumour cell proliferation in vitro and tumour growth in vivo. NRF2 knockdown had no effect on the proliferation rate of recurrent tumour cells in vitro, as measured by the rate of population doublings (Fig. 5c). However, when implanted into the mammary fat pad of mice, the growth of NRF2-knockdown tumours was significantly impaired (Fig. 5d,e). Interestingly, tumours in NRF2-knockdown cohorts restored expression of NRF2 and its target genes at endpoint, further underscoring the importance of the NRF2 pathway for the growth of these tumours (Extended Data Fig. 7j). These results show that NRF2 activation sustains the growth of recurrent tumours in vivo.
NRF2 is required for redox homeostasis in recurrent tumour cells
We next examined the metabolic basis of the dependency for NRF2 activity in recurrent tumours. We reasoned that NRF2 expression in recurrent tumours may control a similar set of metabolic pathways as Her2 does in primary tumour cells. To gain insight into these pathways, we performed metabolomics on recurrent tumour cells with and without NRF2 knockdown, as well as primary tumour cells grown in the presence or absence of dox (Her2 on or off). We first sought to identify global changes in metabolic pathways between control and NRF2-knockdown cells using pathway enrichment analysis. We found that “Glutathione Metabolism” was one of the top altered pathways following NRF2 knockdown in recurrent tumour cells (Fig. 6a). We focused on the GSH:GSSG ratio as a readout of this pathway, and we found that GSH:GSSG levels decreased following Her2 downregulation in primary tumour cells (Fig. 6b). Recurrent tumour cells had re-established a high GSH:GSSG ratio, and in fact it was more than 4-fold higher in recurrent tumour cells as compared to primary tumour cells (Fig. 6b). Importantly, NRF2 knockdown in recurrent tumour cells reverted the GSH:GSSG ratio to below that of primary cells (Fig. 6b), while caNRF2 expression in primary cells increased the GSH:GSSG ratio nearly 3-fold (Extended Data Fig. 8a). Taken together, these results suggest that Her2 signalling regulates redox homeostasis in primary cells, while NRF2 maintains redox homeostasis in recurrent tumour cells.
Fig. 6 |. NRF2 directs metabolic reprogramming in recurrent tumour cells.

a, Pathway analysis showing top altered metabolic pathways between control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells (n=3 biologically independent replicates per condition). Enrichment scores and p values were determined by MetaboAnalyst (see methods) using GlobalTest and GlobalAncova algorithms. b, Relative GSH:GSSG ratio in primary tumour cells cultured with dox (Her2 on) and without dox for 7 days (Her2 off), and in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test. c,d, qRT-PCR analysis of Gclm and Slc7a11 (c) and Txnrd1 (d) expression in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cell lines. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). e, DCFDA staining showing ROS levels in control (shScr) and NRF2-knockdown (shNRF2 #1 and 2) recurrent tumour cells. Data are shown as the mean of n=2 biologically independent replicates and are representative of 3 independent experiments. f, Schematic showing NRF2 regulation of redox homeostasis through regulation of Txnrd1, Slc7a11, and Gclm. g, Waterfall plot showing the fold-change in nucleotide levels between primary (1°) and recurrent tumour cells. h, Waterfall plot showing the fold-change in nucleotide levels between control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. i, qRT-PCR analysis of Pgd expression in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cell lines. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). j, Relative levels of the pentose phosphate pathway metabolites, 6-Phosphogluconate (6PG), Erythrose 4-phosphate (E4P), Sedoheptulose 7-phosphate (S7P), and Ribose 5-phosphate (R5P), in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test. k, Relative levels of nucleotide precursors 5’-Phosphoribosyl-N-formylglycineamide (FGAR) and Inosine monophosphate (IMP) in control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells. Data are shown as mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test. l, Schematic showing NRF2 regulation of the oxidative pentose phosphate pathway gene, Pgd, and precursors of pyrimidine and purine biosynthesis (6PG, E4P, S7P, R5P, FGAR, and IMP). m, Log2 fold-change in metabolite levels between Keap1 mutant (mut) and wild type (WT) cell lines from the Cancer Cell Line Encyclopedia. Metabolites above the axis are enriched in Keap1 mutant cell lines.
We next examined the mechanistic basis of these changes, focusing on NRF2’s function as a transcription factor. NRF2 knockdown in recurrent tumour cells led to a large reduction in the expression of enzymes involved in de novo glutathione biosynthesis, including a subunit of glutamate-cysteine ligase, Gclm, and the cystine transporter Slc7a11 (Fig. 6c). NRF2 knockdown in primary cells, however, had no effect on Gclm expression and only a modest effect on Slc7a11 expression (Extended Data Fig. 8b), demonstrating that NRF2 specifically regulates glutathione synthesis in recurrent tumour cells. Further, NRF2 knockdown in recurrent cells, but not primary cells, led to a substantial decrease in expression of the antioxidant enzyme Txnrd1, which is involved in maintaining the redox state of protein sulfhydryls (Fig. 6d and Extended Data Fig. 8c). Given the decrease in glutathione levels, we speculated that NRF2 knockdown may lead to oxidative stress. Indeed, DCFDA staining showed a 2-fold increase in ROS levels in NRF2 knockdown cells (Fig. 6e). These results indicate that NRF2 stabilization is essential for maintenance of redox homeostasis in recurrent tumour cells (Fig. 6f).
NRF2 directs metabolic reprogramming in recurrent tumour cells
We next examined additional metabolic pathways regulated by NRF2 in recurrent tumour cells. Nearly half of the differentially regulated pathways between control and NRF2-knockdown cells related to nucleotide metabolism (“Pyrimidine Metabolism,” “Purine Metabolism,” “Pentose Phosphate Pathway,” and “Nucleotide Sugars Metabolism”) (Fig. 6a). Levels of both purine and pyrimidine nucleotides were higher in recurrent tumour cells, which have high NRF2 expression, as compared to primary cells (Fig. 6g). Consistent with this, NRF2 knockdown in recurrent tumour cells reduced the levels of these nucleotides (Fig. 6h), indicating that NRF2 drives high nucleotide levels in recurrent tumour cells.
The pentose phosphate pathway shunts glucose carbon from glycolysis to form ribose sugars, which are critical for de novo nucleotide biosynthesis33, and NRF2 has been reported to regulate enzymes of this pathway34. We next explored whether NRF2 regulation of genes in this pathway is responsible for its role in maintaining nucleotide levels in recurrent cells. qRT-PCR analysis showed that expression of the oxidative PPP enzyme Pgd was decreased following NRF2 knockdown (Fig. 6i). Many metabolite intermediates of the PPP were decreased following NRF2 knockdown (Fig. 6j), suggesting that NRF2 directs glucose carbon from glycolysis to form ribose-5-phosphate, which is essential for both purine and pyrimidine synthesis. Finally, we examined intermediates of purine biosynthesis, and found that the metabolites FGAR (5’-Phosphoribosyl-N-formylglycineamide) and IMP (Inosine monophosphate) were also decreased in NRF2 knockdown cells (Fig. 6k). Together, these results demonstrate that NRF2-dependent expression of PPP enzymes in recurrent tumour cells results in increased levels of nucleotide precursors and nucleotides.
The results above suggest that NRF2 regulates the de novo nucleotide synthesis pathway in recurrent tumour cells. We next addressed how this pathway is regulated in primary tumour cells. NRF2 knockdown in primary cells had no effect on Pgd expression (Extended Data Fig. 8d). Similarly, caNRF2 expression in primary cells had no effect on 6PG and R5P levels (Extended Data Fig. 8e) and had only a modest effect on nucleotide levels (Extended Data Fig. 8f). In contrast, Her2 downregulation in primary cells decreased the PPP intermediates 6PG and R5P (Extended Data Fig. 8g). Together, this suggests that Her2 regulates the nucleotide synthesis pathway in primary tumours and NRF2 regulates this pathway in recurrent tumours. Together, these findings suggest that NRF2 activity is required in recurrent tumours both to maintain redox homeostasis and to drive anabolic pathways that enable tumour growth (Fig. 6l).
To extend these findings to human cancer cells, we analyzed data from a recent study that performed metabolomics on a large panel of cells in the Cancer Cell Line Encyclopedia35. We used KEAP1 mutation as a proxy for NRF2 activation, and compared metabolite levels between KEAP1-wild-type and KEAP1-mutant cancer cells. We found that NADP, GSH, and GSSG were the most enriched metabolites in KEAP1-mutant cells, and all five nucleotides (AMP, GMP, CMP, UMP, and dCMP) in the dataset were also substantially enriched in KEAP1-mutant cancer cells (Fig. 6m).
NRF2-regulated redox and nucleotide metabolism are essential for recurrent tumour growth
Our findings above suggest that NRF2 regulates redox and nucleotide metabolism in recurrent tumours. NRF2 can also regulate genes involved in detoxification. To directly test which NRF2-regulated pathways are functionally important for recurrent tumour growth, we performed a CRISPR-Cas9 screen with a focused library of gRNAs targeting genes in the redox, detoxification, and nucleotide synthesis pathways (Extended Data Fig. 9a). This library also contained non-targeting gRNAs as negative controls and gRNAs targeting essential ribosomal genes as positive controls. We used this library to transduce two independent recurrent tumour cell lines, and after selection and expansion in culture, these cells were implanted orthotopically into recipient mice. After tumour formation, tumours were harvested, and next-generation sequencing was used to determine the change in gRNA abundance between the implanted cells and the tumours (Fig. 7a). This analysis identified 5 genes – Txn1, Txn2, Txnrd1, Pgd, and Ppat – that were required for tumour formation in both recurrent tumour cell lines. Txn1, Txn2, and Txnrd1 are thioredoxin enzymes that regulate the redox state of the proteome. gRNAs targeting these genes were consistently depleted in tumours arising from both recurrent tumour cell lines, indicating that these genes are essential for recurrent tumour formation (Fig. 7b,c). The other two genes we identified as essential for tumour growth in both recurrent cell lines were Pgd and Ppat (Fig. 7b,c). Pgd is in the oxidative PPP upstream of ribose-5-phosphate production, while Ppat is in the purine biosynthesis pathway downstream of ribose-5-phosphate. These hits suggest that NRF2-regulated nucleotide synthesis is functionally important for recurrent tumour growth. Importantly, gRNAs targeting Txn1, Txn2, Txnrd1, Pgd and Ppat were highly abundant in the injected cell population, in contrast to gRNAs targeting essential ribosomal genes, which were already partially depleted in vitro (Extended Data Fig. 9b). This suggests that these genes are required for tumour growth in vivo, but may not be required for tumour cell viability in vitro. Together, these results demonstrate that both NRF2-regulated redox homeostasis through thioredoxin reductase and nucleotide biosynthesis through the PPP are essential for recurrent tumour growth.
Glutaminase inhibition prevents reactivation of dormant tumour cells and recurrent tumour cell growth
We next asked if the dependency of recurrent tumour cells for NRF2 activity induces metabolic vulnerabilities that might be targeted to prevent the growth of recurrent tumours. It has recently been shown in non-small cell lung cancer models that constitutive NRF2 activity induced by Keap1 loss confers sensitivity to glutaminase inhibition30,36. We therefore asked whether the elevated NRF2 expression found in recurrent tumour cells renders these cells sensitive to glutaminase inhibition. In agreement with this, the glutaminase inhibitors CB-839 and BPTES inhibited the growth of recurrent tumour cells but not primary tumour cells (Fig. 7d and Extended Data Fig. 9c). To test if this sensitivity is mediated by high NRF2 levels, we measured the response of recurrent tumour cells with NRF2 knockdown to glutaminase inhibition. NRF2 knockdown partially rescued cell viability in response to CB-839 and BPTES (Fig. 7e and Extended Data Fig. 9d). Interestingly, caNRF2 expression in primary tumour cells was not sufficient to induce sensitivity to glutaminase inhibition (Extended Data Fig. 9e). Together, these results indicate that the NRF2-driven metabolic state renders recurrent breast cancer cells sensitive to glutaminase inhibition.
Glutaminase catalyzes the conversion of glutamine to glutamate, which can be used as an anaplerotic citric acid cycle substrate, for glutathione synthesis, or for secretion by the Xct antiporter in exchange for cystine uptake, which is also used for glutathione synthesis. We wanted to determine if glutaminase activity was required for maintaining redox homeostasis through GSH synthesis in recurrent tumour cells. Interestingly, BPTES treatment resulted in a decrease in ROS levels (Extended Data Fig. 9f), indicating that glutaminase inhibition does not induce oxidative stress. We next asked if replenishing cells with an anaplerotic substrate downstream of glutaminase would rescue viability after glutaminase inhibition. Treatment of cells with dimethyl-α-ketoglutarate (dm-αKG), a cell-permeable form of the TCA intermediate α-ketoglutarate, partially rescued recurrent tumour cell viability following CB-839 treatment (Extended Data Fig. 9g). This is consistent with a recently published model36, whereby cells with high NRF2 direct glutamate for GSH synthesis and Xct export, thereby limiting the glutamate available for anaplerosis and rendering cells hypersensitive to decreases in glutaminase activity.
We next tested if glutaminase inhibition could prevent the reactivation of dormant tumour cells. We implanted primary tumour cells into recipient mice on dox and allowed tumours to form before removing dox to induce tumour regression. After 4 weeks, residual dormant tumours were enzymatically digested and used for a colony formation assay in the presence of serum and growth factors, with or without the addition of CB-839 (Fig. 7f). Under these culture conditions, approximately 10% of dormant cells could proliferate to form colonies under control conditions (Fig. 7g), and CB-839 caused nearly a 4-fold decrease in colony forming efficiency (Fig. 7g). Additionally, colonies that grew in the presence of CB-839 were smaller than control colonies (Fig. 7h), suggesting that CB-839 also decreased the growth rate of these colonies. Together, these results show that CB-839 can prevent the reactivation of dormant tumour cells in the presence of mitogenic signals in vitro.
NRF2 is activated in recurrent tumours through a noncanonical mechanism
We next examined the mechanism of constitutive NRF2 protein expression in recurrent tumour cells. NRF2 is canonically regulated by KEAP1, which sequesters NRF2 in the cytoplasm and targets it for ubiquitylation by the CUL3 E3 ubiquitin ligase, leading to its rapid degradation by the proteasome under basal conditions37. Upon oxidative stress, however, cysteine residues on KEAP1 becomes oxidized, and this induces a conformational change that limits NRF2 ubiquitylation. Therefore, we first wanted to determine if high NRF2 levels in recurrent tumours cells are a consequence of high ROS levels in these cells. We found that ROS levels were not consistently higher in recurrent tumour cells (Fig. 8a), and in fact the ratio of GSH:GSSG was higher in recurrent tumour cells than primary cells (see Fig. 6b). Together, this indicates that persistent ROS is not the cause of elevated NRF2 in recurrent tumours. We additionally demonstrated that elevated NRF2 levels were not due to changes in NRF2 mRNA expression or KEAP1 expression (Extended Data Fig. 10a–c), and we did not detect mutations in either Keap1 or NRF2 in recurrent tumours. Furthermore, inhibition of the proteasome equalized NRF2 levels between primary and recurrent tumours, suggesting that high NRF2 expression in recurrent tumours is due to impaired protein degradation and not altered rates of protein production (Fig. 8b).
Fig. 8 |. NRF2 is activated in recurrent tumour cells through a KEAP1-independent mechanism.

a, DCFDA staining showing ROS levels in two independent primary cell lines and two independent recurrent (Rec) cell lines (n=3 independent biological replicates per cell line). b, Western blots for NRF2 and NQO1 in primary or recurrent cell lines treated with MG-132 for the indicated times. c,d,e, Western blots for KEAP1 (c), NRF2 (d), and NQO1 (e) in primary or recurrent cell lines expressing a control gRNA (Rosa) or one of two gRNAs targeting KEAP1. f, Western blots for NRF2 or V5 in recurrent tumour cells expressing a control vector or constitutively active V5-tagged GSK-3β (S9A). b,c,d,e,f, Blots are representative of 3 biologically independent experiments.
To address the role of KEAP1 in NRF2 regulation, we used CRISPR-Cas9 with two independent gRNAs to genetically knock out KEAP1 in primary and recurrent cells (Fig. 8c). KEAP1 knockout caused an increase in NRF2 and NQO1 in both cell lines, but both NRF2 and NQO1 levels remained more elevated in KEAP1-knockout recurrent cells as compared to KEAP1-knockout primary cells (Fig. 8c–e). Consistent with this, the oxidizing agent TBHQ, which inhibits KEAP1, increased NRF2 levels in both primary and recurrent tumours, but absolute NRF2 levels remained higher in recurrent tumours (Extended Data Fig. 10d,e). These results indicate that increased NRF2 levels and transcriptional activity in recurrent cells are independent of KEAP1.
NRF2 stability can also be regulated through Akt38 and GSK-3β-dependent phosphorylation39, and so we addressed the contribution of these pathways to NRF2 stabilization in recurrent tumours. Inhibition of Akt with the allosteric inhibitor MK2206 had no effect on NRF2 levels or expression of its target NQO1 in recurrent tumour cells (Extended Data Fig. 10f,g). In contrast, expression of a constitutively active GSK-3β S9A mutant, which prevents GSK-3β inactivation by phosphorylation of serine 9, caused a partial reduction in NRF2 levels in recurrent tumour cells (Fig. 8f). Consistent with this, p-GSK-3β was higher in recurrent tumour cells lines (Extended Data Fig. 10h). These results suggest that decreased GSK-3β activity is partially responsible for NRF2 stabilization in recurrent tumour cells.
Discussion
In this study we examined how tumour cell metabolism changes in response to oncogene inhibition, and elucidated the mechanism of metabolic reprogramming in recurrent tumours. We found that inhibition of Her2 signalling induced substantial and specific changes in cellular metabolism, including changes in metabolites suggestive of a switch from glycolysis to fatty acid oxidation. These metabolic changes resulted in a loss of redox homeostasis and increased ROS, findings consistent with recent reports in similar models12,13. A PI3K inhibitor and a DNA damaging agent both increased ROS, suggesting that oxidative stress is induced by many common breast cancer therapies. We found that treatment with antioxidants could rescue cell death following Her2 inhibition, suggesting that tumour cells that are able to adapt and overcome redox perturbations might be more likely to survive therapy and form recurrent tumours. Consistent with this, we found that tumours responded to Her2 downregulation by activating the antioxidant transcription factor NRF2, which promoted tumour recurrence. We additionally found that a NRF2 transcriptional program is frequently activated in human metastatic recurrent breast cancer, and high expression of NRF2 gene signatures was correlated with poor prognosis, demonstrating the clinical relevance of these metabolic changes. Interestingly, tumour cell dissemination is accompanied by increased ROS13, and antioxidant pathways are critical for metastasis40. This suggests that the NRF2 transcriptional program may promote both local recurrence, as we show here, as well as recurrence at distant sites. This study highlights an important role for tumour cell adaptation to therapy-induced metabolic stress, and identifies a novel role for NRF2 in promoting breast cancer recurrence by reprogramming cellular metabolism.
Antioxidant pathways are necessary for tumorigenesis24,41 and tumour progression42,43. Recent studies have shown that residual tumour cells experience oxidative stress12,13. Consistent with these results, we found that Her2 inhibition led to dysregulation of GSH metabolism and resulted in ROS accumulation and impaired viability. This suggests that re-establishing redox homeostasis after oncogene inhibition represents a metabolic barrier that tumour cells need to overcome in order to form recurrent tumours. Our results demonstrate that NRF2 activation is a frequent mechanism by which tumour cells accomplish this. As a result, recurrent tumours acquired a dependency on NRF2, which reinforces high levels of GSH metabolism and bolsters their antioxidant defense through high Txnrd1 expression. This finding is in agreement with several recent studies that have demonstrated the importance of antioxidant pathways for acquired resistance to targeted therapies in breast cancer cell lines42,44. Similarly, genetic activation of NRF2 has been recently shown to promote resistance to targeted therapies in lung cancer cells14. These studies support our finding that restoring redox homeostasis through NRF2 activation is imperative for tumour recurrence.
NRF2 can have pleiotropic effects on cell metabolism beyond regulating redox metabolism and combating ROS. In particular, NRF2 can activate anabolic pathways, including the nucleotide synthesis34 and serine synthesis pathways45. We found that NRF2 transcriptionally activates genes in the oxidative PPP. This pathway oxidizes glucose-6-phosphate to generate both NADPH, which can be utilized to maintain thioredoxin activity and GSH:GSSG levels, and ribose sugars required for de novo nucleotide synthesis. We showed that NRF2 maintains high levels of nucleotide pools in recurrent tumour cells, suggesting that NRF2 orchestrates anabolic metabolism in addition to regulating redox metabolism in these tumours. We speculate that residual tumour cells with high NRF2 activity may more readily access anabolic pathways, such as nucleotide metabolism, and NRF2 activation during residual disease might potentiate tumour recurrence by priming cells for anabolic metabolism required to resume proliferation. According to this model, NRF2 is not sufficient to induce the proliferation of dormant cells, but instead cooperates with other mitogenic pathways by creating a metabolic state permissive for proliferation.
Our results also demonstrate that the function of NRF2 in mammary tumours is context specific. Primary tumours had low basal levels of NRF2, and ectopic expression of a constitutively active NRF2 did not accelerate tumour growth. Additionally, NRF2 expression in primary tumour cells was not sufficient to induce sensitivity to glutaminase inhibition. In contrast, NRF2 expression accelerated tumour recurrence, and NRF2 knockdown significantly impaired recurrent tumour growth. These results suggest that, in primary tumours, oncogenic Her2 signalling is sufficient to orchestrate antioxidant and anabolic metabolism required for rapid proliferation. This notion is consistent with previous findings that Her2 expression rescues metabolic defects and oxidative stress in mammary acini16. It is possible that NRF2 and Her2 regulate a convergent set of metabolic pathways, such that NRF2 does not provide an additional benefit to primary tumours and does not impart a dependency on glutaminase. After Her2 downregulation, however, tumour cells become reliant on NRF2 activity to maintain redox homeostasis and de novo nucleotide synthesis, and these pathways are indispensable for sustained proliferation.
While KEAP1 and NRF2 mutations are frequently found in lung cancer46–48, they are not common in breast cancers, though recurrent breast cancers have not been extensively sequenced. Interestingly, we found that NRF2 is constitutively activated in recurrent mouse tumours even in the absence of activating genetic mutations or persistent ROS. We showed that NRF2 activation in recurrent tumour cells was mediated through a KEAP1-independent mechanism. Furthermore, expression of constitutively active GSK-3β reduced NRF2 levels in recurrent tumour cells, suggesting that low GSK-3β activity is partially responsible for NRF2 activation in recurrent tumour cells. Interestingly, Akt inhibition in recurrent tumour cells did not affect GSK-3β phosphorylation or NRF2 levels, suggesting that GSK-3β may be phosphorylated by an alternative kinase in these cells. A number of other kinases have been proposed to phosphorylate GSK-3β at serine 9, including S6K1, Aurora A, p90RSK, and PKA49. Our findings thus identify two modes of NRF2 activation during tumour recurrence: acute activation of NRF2 following oncogene inhibition or targeted therapies is likely mediated through increased ROS, while the persistent NRF2 activation observed in recurrent tumours is independent of ROS and KEAP1. Together, these findings highlight the importance of KEAP1-independent regulation of NRF2 in breast cancer.
Breast cancer recurrence poses a substantial clinical problem, and therapeutic methods to treat recurrent tumours are currently unavailable. We found that NRF2 promotes both the formation and growth of recurrent tumours, but direct inhibition of NRF2 remains challenging. To identify potentially targetable pathways downstream of NRF2, we conducted a focused in vivo CRISPR screen to identify and prioritize the NRF2-regulated pathways that are most critical for the growth of recurrent tumours. We identified thioredoxins (Txn1, Txn2, and Txnrd1), but not glutathione synthesis enzymes, as redox mediators that are essential for tumour growth in vivo. We additionally identified Pgd, of the oxidative PPP, and Ppat, which is required for the purine synthesis pathway, as potential targets downstream of NRF2 that impair recurrent tumour growth. We found that NRF2-driven recurrent tumour cells also acquired a metabolic dependency on glutaminase, which is in agreement with recent reports describing that KEAP1-mutant lung cancers are sensitive to glutaminase inhibition30,36. We found that glutaminase inhibition prevented the reactivation of dormant tumour cells ex vivo and impaired the growth of recurrent tumour cells in vitro, though it is important to note that glutamine metabolism and glutaminase dependency can differ between in vitro and in vivo contexts50. Together, these results suggest novel therapeutic approaches for the treatment of recurrent breast tumours that form following anti-Her2 therapies.
Methods
Tissue culture and reagents
Mammosphere cultures were generated by digesting primary tumours as previously decribed1,2 and plating single cell suspensions on Poly(2-hydroxyethyl methacrylate) (Poly-hema, Sigma) coated plates. Briefly, tumour chunks were digested with EBSS (without phenol red) supplemented with collagenase (300U/mL), hyaluronidase (100U/mL), 2% FBS, gentamycin (100μg/mL), 100U/mL Pen/Strep, and doxycycline (2μg/mL) at 37°C for 4 hours. Cells were then resuspended in Dispase II (5mg/mL) and DNase I (100μg/mL) and filtered before plating. Mammosphere cultures were grown in RPMI-1640 media supplemented with 2μg/mL doxycycline, B27 (Invitrogen 17504-044), 10ng/mL murine EGF (Sigma E4127), 20ng/mL bFGF (Invitrogen 13256-029), Pen/Strep (Gibco 15140-122) and 2mM L-Glutamine (Gibco 25030-081). For microscopy experiments in Fig. 1D and S2A, EGF and bFGF were excluded from the culture media. Mammospheres were passaged using enzymatic dissociation to re-plate as single cell suspensions. Doxycycline was excluded from the media where specified.
Primary and recurrent tumour cell lines were generated as previously described1,2. Briefly, tumours were enzymatically digested and plated on standard tissue-culture treated plates with the media described below. Primary tumour cells were cultured in DMEM with 10% super calf serum, 1% Penicillin/Streptomycin, and 1% L-Glutamine supplemented with 10 ng/ml EGF, 5μg/ml insulin, 1μg/ml hydrocortisone, 5μg/ml prolactin, 1μM progesterone and 2μg/ml doxycycline to maintain HER2/neu expression. Recurrent tumour cells were cultured in DMEM with 10% SCS, 1% Penicillin/Streptomycin, and 1% L-Glutamine supplemented with 10 ng/ml EGF and 5 μg/ml insulin. Human breast cancer cells were obtained from the Duke University Cell Culture Facility, and sub-cultured per ATCC protocols.
For proteasome inhibitor experiments, 1×106 primary and recurrent cell lines were plated for each time-point in duplicate. The next day, media before media were replaced with either vehicle or 5μM MG-132. After the indicated amount of time, samples were collected by scraping cells in ice cold PBS, centrifuged, and harvested for western blotting. For constitutively active GSK-3β experiments, recurrent tumour cells were transfected (nucleofection, Amaxa) to transiently express a V5-tagged GSK-3β S9A mutant (see plasmids section). After two days, cells were harvested and prepared for western blotting.
Plasmids and viral transduction
To generate a lentiviral construct for expression of caNRF2, NRF2 (Nfe2l2) was PCR amplified from recurrent tumour cell cDNA to generate a truncated gene fragment lacking the first 88 codons, and including a Kozak sequence, start codon, and flag epitope tag sequence (see Reagents table for primer sequences). This was first cloned into the gateway entry vector pEntr4 (Thermo, A10465) using restriction digest (SalI and NotI), and then cloned into the gateway destination vector, pLenti PGK Neo DEST (w531-1), (a gift from Eric Campeau & Paul Kaufman; Addgene plasmid # 19067)3. The empty vector control was created by SalI restriction digest of the pLenti PGK Neo DEST (w531-1) destination vector to remove the ccdb cassette. The backbone was gel extracted and re-ligated. NRF2-targeting shRNAs in pLKO were obtained from Sigma (shNRF2#1: TRCN0000054659 and shNRF2#2: TRCN0000054658). Scramble shRNA was a gift from David Sabatini (Addgene plasmid # 1864)4. Lenti-Cas9-Blast was a gift from Feng Zhang (Addgene plasmid #52962)5 and was used to transduce cell with Cas9. Guide RNAs targeting Keap1 were cloned into lentiGuide-puro by Gibson Assembly (Addgene #52963) as described6 (sgKeap1 #1 – GGTTCGGTTACCGTCCTGCG ; #2 – CCGAAGCCCGTTGGTGAACA). pBabe-puro GSK-3β (S9A) was a gift from Brendan Manning (Addgene plasmid # 14128) and was electroporated into recurrent tumour cells using the Amaxa Nucleofector.
To generate lentivirus, HEK293T cells were transfected with psPAX2 and pMDG.2 packaging plasmids (gifts from Didier Trono, EPFL, Lausanne, Switzerland; Addgene plasmids 12559 and 12660) and the lentiviral expression construct. Viral supernatant was collected after 48 and 72 hours and filtered. This virus was used to transduce cells with 6μg/mL polybrene (MilliporeSigma).
Animals
Animal care and animal experiments were performed with the approval of, and in accordance with, guidelines of the Duke University IACUC. Mice were housed under barrier conditions with 12-hour light/12-hour dark cycles. Bitransgenic MMTV-rtTA;TetO-Her/neu (MTB;TAN) mice on an FVB/N background were obtained from Lewis Chodosh (University of Pennsylvania). Mice were bred and tumours were induced as previously described2. Briefly, 2 mg/mL doxycycline was provided in the drinking water, and tumour growth was monitored by palpation. For recurrent tumour formation, doxycycline was removed from the drinking water, and regressed tumours were monitored until recurrent tumours formed. For orthotopic tumour studies, outbred athymic mice (J:nu) were obtained from the Duke breeding core.
Orthotopic recurrence assays
Orthotopic tumour recurrence assays were performed as described2. Cohorts of 6-week old immunocompromised mice (nu/nu) under dox administration were injected orthotopically in the 4th inguinal mammary fat pad with 1×106 primary tumour cells (expressing either caNRF2 or an empty vector). Once tumours reached 5 mm (2-3 weeks), dox was be removed to initiate oncogene down-regulation and tumour regression. Cohorts were blinded , and mice were palpated biweekly to monitor tumour recurrence, which was defined as reaching 5×5mm. Differences in recurrence-free survival between control (n=27) and caNRF2 (n=32) cohorts were compared using Kaplan-Meier survival curves, and evaluated by the hazard ratio and p-value calculated from a log-rank (Mantel-Cox) test2.
Orthotopic tumour growth assays
Cohorts of 14 6-week old immunocompromised mice (nu/nu) were injected orthotopically in the 4th inguinal mammary fat pad with 1×105 recurrent tumour cells (expressing either a scrambled or NRF2 targeting shRNA). Cohorts were blinded, and tumour growth was monitored by palpation. Tumor AUC was calculated using the formula [(vol1 + vol2)/2]*(day2-day1).
Residual tumour samples for immunofluorescent and immunohistochemical staining
Primary tumours were generated in MTB;TAN mice provided 0.1mg/mL dox water. After primary tumour formation, dox was removed to induce tumour regression. Mammary glands previously bearing primary tumours were harvested 56 days after dox withdrawal and either frozen in OCT (for immunofluorescent staining) or fixed in 10% normal formalin for 16 hr, then washed twice with PBS and transferred to 70% ethanol before embedding in paraffin.
Immunohistochemical staining
Hematoxylin and Eosin (H&E) staining was performed using a Leica H&E slide staining instrument. Immunohistochemical staining for Ki67 was developed using a Dako immunohistochemistry autostainer. Specific antibodies used are in the Reagents table. Ki67 was quantified using imageJ.
Immunofluorescence
Mammospheres were collected by centrifugation onto coverslips. Frozen tissues were cut in 8μm sections (Fig. 1) or 12μm sections (Fig. 3) using a cryostat. Tissue sections or mammospheres were fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton-X 100, and blocked in 3% BSA and 10% normal goat serum. Slides were incubated with primary antibodies for 1 hour at room temperature, washed, and incubated in with secondary AlexaFluor conjugated antibodies for 1 hour at room temperature. For lipid droplet imaging, cells were incubation with BODIPY for 30 min at room temperature before mounting coverslips with Prolong Gold. For experiments using Hoechst, Hoechst was included in the secondary antibody incubation. For experiments using DAPI, cells were mounted using Prolong Gold with DAPI. Slides were imaged using a Lecia SP5 inverted microscope. NRF2 nuclear intensity was quantified using CellProfiler (Broad Institute). For each primary tumours (n=3), 3 fields of view were imaged, and the nuclear NRF2 intensity was quantified for a range of 3734-4320 cells per tumour. For each residual tumours (n=3), 5 fields of view were imaged, and the nuclear NRF2 intensity was quantified for a range of 2787-6184 cells per tumour. Reagent concentrations and supplier information are included in the Reagent table.
Metabolite extraction
For mammosphere metabolomics experiment, 3×105 cells for +dox condition and 6×105 cells for -dox condition were plated in poly-hema coated 6-well plates. Mammospheres were collected by brief centrifugation (2 min, 300g), media were aspirated, and pellets were immediately resuspended in 1mL −80°C extraction solvent (80%MeOH/water). Tubes were moved to −80°C for 15 minutes before centrifugation (20,000g for 10 min at 4°C). Supernatant was transferred to a new tube, and samples were dried using a SpeedVac. Samples were stored at −80°C prior to LC-MS analysis. For primary and recurrent tumour cell adherent cultures, 3.5×105 cells were plated in 6-well culture dishes. 24 hours later, media were quickly aspirated, and plates were placed on dry ice. 1mL −80°C extraction solvent (80% MeOH/water) was added to each well, and plates were moved to −80°C for 15 min. Plates were then scraped and collected in Eppendorf tubes for centrifugation (20,000g for 10 min at 4°C). Supernatant was transferred to a new tube, and samples were dried using a SpeedVac. Samples were stored at −80°C prior to LC-MS analysis.
Liquid chromatography and mass spectrometry
Metabolite profiling was performed as previously described7,8. Ultimate 3000 UHPLC (Dionex) is coupled to Q Exactive Plus-Mass spectrometer (QE-MS, Thermo Scientific) for metabolite profiling. A hydrophilic interaction chromatography method (HILIC) employing an Xbridge amide column (100 × 2.1mm i.d., 3.5μm; Waters) is used for polar metabolite separation. Detailed LC method was described previously7 except that mobile phase A was replaced with water containing 5 mM ammonium acetate (pH 6.8). The QE-MS is equipped with a HESI probe with related parameters set as below: heater temperature, 120 °C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 3.0 kV for the positive mode and 2.5 kV for the negative mode; capillary temperature, 320 °C; S-lens, 55; A scan range (m/z) of 70 to 900 was used in positive mode from 1.31 to 12.5 minutes. For negative mode, a scan range of 70 to 900 was used from 1.31 to 6.6 minutes and then 100 to 1,000 from 6.61 to 12.5 minutes; resolution: 70000; automated gain control (AGC), 3×106 ions. Customized mass calibration was performed before data acquisition.
Metabolomic data analysis
LC-MS peak extraction and integration were performed using commercially available software Sieve 2.2 (Thermo Scientific). The peak area was used to represent the relative abundance of each metabolite in different samples. The missing values were handled as described previously7. The metabolite heatmap and pathway analysis were performed using MetaboAnalyst 4.0. For the heatmap, top 70 metabolites are displayed. For pathways analysis, all metabolites that had a 2-fold increase or decrease between recurrent cells expressing control or NRF2 targeting hairpin were analyzed.
Keap1 mutant cancer cell line metabolomic analysis
Metabolomic data comparing cancer cell lines with or without KEAP1 genetic mutations were published by Li et al9. Log2 fold change (KEAP-mut/KEAP1-WT) values were ranked, and redox metabolites (NADP, GSH, and GSSG) and all reported nucleotides (AMP, CMP, GMP, UMP, dCMP) were identified and labeled.
Cell viability assays
Unless otherwise indicated, cell viability assays were performed using CellTiter-Glo (Promega) according to manufacturer instructions. For adherent cell cultures, 1,000-5,000 cells were plated on an opaque 96-well plate. The next day, media were replaced with indicated drug treatments (see supplemental table for drug supplier and catalog numbers). For mammosphere cell viability experiment, cells were plated 2 days prior to drug treatment, and cells were collected by centrifugation and re-plated in media with or without dox and with or without 100μM etomoxir. The +dox samples were collected on day 3, and the -dox samples were collected on day 7. Media were refreshed once for the 7-day time point. Cells were collected by centrifugation and resuspended in CellTiter-Glo. They were then incubated at room temperature with shaking for 20 minutes before transferring to an opaque 96-well plate for luminescence measurement. For the SKBR3 Crystal Violet experiment, 1.25×105 cells were plated on a 6-well plates. The next day, media were replaced with indicated media. After 6 days, media were removed, cells were rinsed once with cold PBS before incubating in crystal violet (0.5% crystal violet in 25% MeOH) for 5 min. Plates were rinsed in water 6 times before allowing to dry. For growth curve of recurrent tumour cells expressing scrambled or NRF2 targeting shRNA, 5×104 cells were plated in 10cm dishes. Cells were counted the next day, and this was used as day 0. Cells were then counted on days 2, 4, and 6. Cells were counted using a hemocytometer. For the dormant tumour cell colony formation assay, a 6-week old immunocompromised mouse (nu/nu) under dox administration was injected orthotopically in the 4th inguinal mammary fat pad with 1×106 primary tumour cells. Once tumours reached 5 mm (2-3 weeks), dox was be removed to initiate oncogene down-regulation and tumour regression. 4 weeks after dox was removed, dormant tumours were enzymatically digested as described in the Tissue Culture and Reagents section above1,2. 1,000 single cells were plated, and media were replaced the next day with media containing either vehicle or 500nM CB-839. Media were refreshed every 3 days for 2 weeks before staining with crystal violet. Colonies were counted using imageJ, and colony forming efficiency was calculated using the formula: Colony forming efficiency = 100*(colonies formed)/(cells plated). Colony areas were quantified using imageJ.
Immunoblotting and qRT-PCR
Western blotting was performed as previously described2. Primary and secondary antibodies used in this study are shown in Supplemental Table 1. For fluorescent antibodies membranes were imaged using Odyssey infrared imaging system (Li-Cor), and for HRP conjugated antibodies, membranes were treated with ECL reagents (Bio-Rad or Millipore Sigma) developed using autoradiography film (Genesee Scientific). Band intensities were quantified using ImageJ. RNA was extracted using the RNeasy kit (Qiagen). cDNA was synthesized using the ImProm-II Reverse Transcription System (Promega). Taqman probes used for qPCR are listed in Supplemental Table 1, and qPCR was performed using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Actb and Gapdh were used for normalization controls. Reagent concentrations and supplier information are included in the Reagent Table.
DCFDA and mitoSOX assay
For DCFDA staining, cells were trypsinized and incubated in phenol red-free media containing 10% serum and 10μM DCFDA (Abcam) on a rocker at 37°C in the dark for 45 minutes. Cells were then collected by centrifugation, rinsed once with cold PBS, and resuspended in PBS supplemented with 0.1% serum. For MitoSOX staining, cells were incubated in phenol red-free media containing 10% serum and 2.5μM mitoSOX Red (Thermo) at 37°C for 10 minutes. Cells were then trypsinized, rinsed once with cold PBS, and resuspended in PBS supplemented with 0.1% serum. Both DCFDA and mitoSOX samples were immediately analyzed using a BD FACSCanto II flow cytometry. For gating, cells were first gated by size using FSC and SSC, and then gated by FSC-height by FSC-area to exclude doublets before measuring fluorescence. When indicated, samples were also imaged using Zeiss Axio Imager Widefield Fluorescence Microscope.
Human survival analysis
A publicly available online tool for correlating gene sets with survival in breast cancer was used. Gene expression-based Outcome for Breast cancer Online (GOBO10) uses 4 datasets (GSE1456, GSE3494, GSE6532, and GSE7390), and it is available at http://co.bmc.lu.se/gobo/gsa.pl. Both the “NRF2 core gene signature” (previously compiled11) and the “NRF2 18-gene signature” were used for input. These gene sets are included in the Gene Signature table.
Gene set enrichment analysis (GSEA)
A publicly available RNA sequencing dataset of matched primary and metastatic tumours (GSE11059012) was downloaded. Expression values for metastases were averaged for each individual patient, and the difference (delta) between the primary tumour value and average metastasis value was calculated for each gene. Any genes with no value for the primary tumour were not included for that patient. A rank ordered list of the deltas for each individual patient was created, and the “NRF2 core gene signature” was used for GSEA. This gene set is included in the Gene Signature table. The differences for all individual patients were averaged (average delta), and a rank ordered list was generated. The “NRF2 core gene signature” and “Hallmarks” gene signatures13 were used for GSEA. Enrichment scores were calculated using the Kolmogorov-Smirnov statistic and p-values were calculated using permutation testing with 1000 permutations.
CRISPR screening
A custom CRISPR library targeting 35 mouse genes and the Rosa26 locus with five sgRNAs each and 50 non-targeting sgRNAs (see Supplemental Table 1) was designed using GUIDES14. Pooled oligos (Twist Biosciences) were PCR amplified and cloned into lentiGuide-puro by Gibson Assembly (Addgene #52963) as described6. Assembled sgRNA vectors were then transformed into Endura competent cells (Lucigen) with resulting library representation of >280-fold. Library DNA was isolated using ZymoPURE midiprep kits. A total of 15 clones were also isolated after transformation for Sanger sequencing to verify correct assembly. Library lentivirus was prepared by co-transfecting HEK293T cells with lentivector, psPAX2 and pMD2.g and harvesting supernatant 48 hours post-transfection.
Two independent Cas9 expressing recurrent tumour cell lines were transduced at a multiplicity of infection of 0.3, and after two days, cells were treated with puromycin to select for transduced cells. After selection and expansion (day 6 after infection), cells were collected. At this time-point, cell pellets were frozen to analyze library coverage and determine the baseline guide RNA distribution (n=2 for each cell line), and 5×105 cells (representing >2000X library coverage) were injected orthotopically in the 4th inguinal mammary fat pad of 6-week old immunocompromised mice (nu/nu) (n=5 for Recurrent tumour line 1; n=12 for Recurrent tumour line 2). After reaching 8×8mm, tumours were harvested and gDNA was extracted using a previously reported salt-precipitation protocol15, and a library was prepared for sequencing as previously published16. Briefly, two PCR reactions were performed (see Reagents table for primer sequences). For the first PCR (18 cycles), lentiGuide-puro DNA was amplified using primers that flank the guide RNA sequence. 8 PCR reactions using 1μg of genomic DNA each were performed for each sample to maintain >5000X coverage. These PCR products were pooled for each sample. The products from PCR1 were used for PCR2 (18 cycles), which attached Illumina adaptors, sample barcodes, and a variable length sequence to increase library complexity. PCR2 products were gel purified, quantified, mixed, and sequenced using a NovaSeq6000 (50bp, paired end, S-Prime flow cell). Barcoded reads were matched to samples and a guide RNA level read counts table was generated using MAGeCK17. To minimize the effects of random clonal dominance in tumours, the top ten most abundant guides were excluded for each sample. This clonal correction was used for determining log2 fold changes only, and nothing was excluded for determining statistical significance (determined by MAGeCK, see Statistical Analysis). Read counts were normalized to the median count number for all samples, so each normalized sample had the same number of total reads to allow for comparison across samples. The read count for each guide was averaged for day 0 samples and for tumour samples, and the log2 fold change (tumour/day 0) was calculated. These were then collapsed to the gene level by averaging the log2 fold change values for all five guide RNAs for each gene. These were ranked from lowest to highest and plotted.
Statistics and reproducibility
Plots and statistical analyses were generated using Prism 7 software. Specific statistical tests are identified in figure legends for each experiment. For all western blots and qRT-PCR experiments, a single experiment is shown that is representative of results from at least 2 independent experiments, unless otherwise noted in the figure legend. Metabolomics experiments are shown as single experiments with n=3 biologically independent replicates. All other experiments were repeated multiple times and results are shown as the mean of at least 3 independent replicates, unless noted otherwise in the figure legend. For CRISPR screening experiments, FDR values were generated using MAGeCK17 to determine statistical significance for depleted genes. Additional information about reagents used for this study can be found in Supplementary Table 1. NRF2 gene signatures used in this study can be found in Supplementary Table 2. Additionally, information about reagents, animals, and software used in this study can be found in the Nature Reporting Summary.
Data availability
RNA sequencing data comparing primary and recurrent MTB;TAN-derived tumour cell lines is available online using National Center for Biotechnology Information’s Short Read Archive (SRA) under project accession number 626PRJNA50583918. Prognostic analysis for NRF2 gene signatures was performed using Gene expression-based Outcome for Breast cancer Online (GOBO10), which uses 4 datasets (GSE1456, GSE3494, GSE6532, and GSE7390), available at http://co.bmc.lu.se/gobo/gsa.pl. RNA Sequencing data used for human recurrent breast cancer GSEA was accessed using NCBI’s GEO database (GSE11059012). Metabolomic data for KEAP1 mutant cancer cell lines was accessed using the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) and supplementary tables from Li, H et al. (2019)9. Metabolomics datasets generated in this study are available in Supplementary Table 3s.
Extended Data
Extended Data Fig. 1. Signaling changes induced by Her2 downregulation.
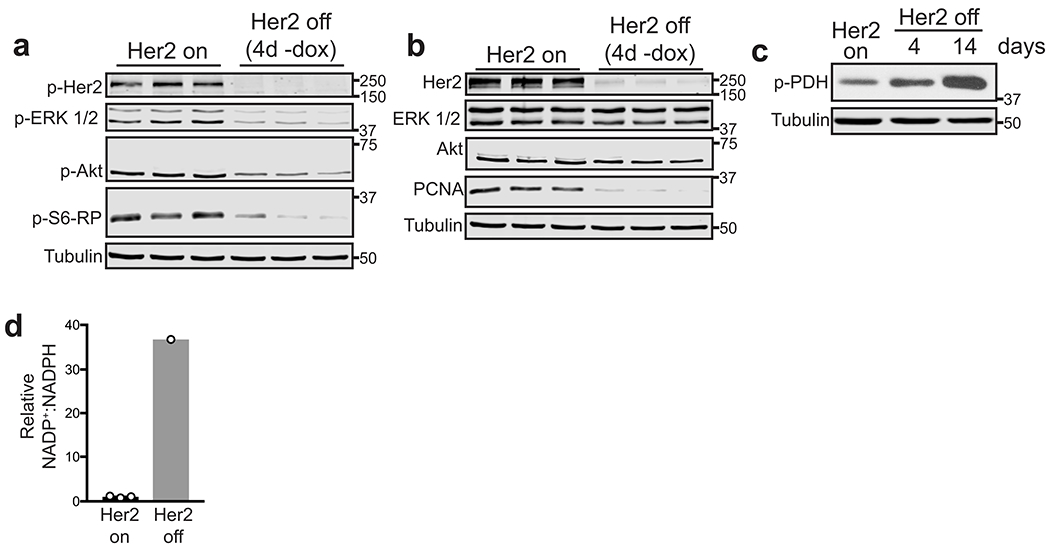
a, Western blots for p-Her2 (Tyr1221/1222), p-ERK1/2 (Thr202/Tyr204), p-Akt (Ser473), and p-S6-RP (Ser240/244) in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off). Data represent 3 independent experiments. b, Western blots for Her2, ERK1/2, Akt, and PCNA in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off). Data represent 3 independent experiments. c, Western blot for p-PDH (Ser293) in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off) for 4 and 14 days. Data represent 3 independent experiments. d, Relative ratio of oxidized (NADP+) to reduced NADP (NADPH) between Her2 off and Her2 on conditions. For –dox cells, the NADP+:NADPH ratio is shown for a single sample, since the NADPH level for the other samples was below the limit of detection, and therefore the ratio is undefined.
Extended Data Fig. 2. Her2-targeted therapies induce ROS in breast cancer cells.

a, DCFDA staining showing ROS levels in mammospheres cultured with dox or without dox for 2 days. Significance was determined by two-sided Student’s t test, and data are represented as mean ± SEM (n=6 fields of view). Scale bars represent 100μm. b, DCFDA staining showing ROS levels in MDA-MB-231 cells treated with 500nM lapatinib for 48 hours. Data are representative of n=3 biologically independent replicates for a single experiment. c, DCFDA staining showing ROS levels in SKBR3, BT474, and MDA-MB-231 cells treated with 500nM neratinib for 48 hours. Data are shown as the mean ± SEM for n=3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 3. PI3K inhibition induces ROS in breast cancer cells.

Relative viability of HCC1954 cells treated with lapatinib and 5mM NAC for 72 hours. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). b, DCFDA staining showing ROS levels in BT474 and SKBR3 cells treated with lapatinib or 100μM DHEA for 48 hours. BT474 cells were treated with 100μM tert-butyl hydrogen peroxide (TBHP) for 12 hours as a positive control. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 biologically independent experiments. c, DCFDA staining showing ROS levels in BT474 cells treated with lapatinib and 50μM 6-An for 48 hours. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 biologically independent experiments. d, DCFDA staining showing ROS levels in mammospheres treated with 50μM 6-An or 100μM TBHP for 24 hours. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 biologically independent experiments. e, Relative viability of mammospheres cultured with or without dox in the presence or absence of 100μM etomoxir (eto) for 7 days. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons test. f, qRT-PCR analysis of Cpt1a, Cpt1b, and CD36 expression in 2 independent mammosphere cultures grown in the presence of dox or without dox for 4 days. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons test. g, BODIPY staining showing lipid droplets (green) in mammospheres cultured in the presence of dox. Scale bar, 25 μm. h, DCFDA staining showing ROS levels in T47D, BT474, MDA-MB-231, and SKBR3 cells treated with 500nM BKM120 for 48 hours. Data are representative of n=3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test. i, DCFDA staining showing ROS levels in MDA-MB-231 cells treated with 500nM JQ1 for 48 hours. Data are representative of n=3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 4. DNA damage therapy induces ROS in breast cancer cells.

a, DCFDA staining showing ROS levels in T47D, BT474, and MCF7 cells treated with 1μM fulvestrant for 48 hours. Data are representative of n=3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test. b, DCFDA staining showing ROS levels in MCF7, SKBR3, MDA-MB-231, BT474, and T47D cells treated with indicated doses of etoposide for 48 hours. Data are representative of n=3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 5. Dormant and recurrent tumor cells upregulate NRF2.
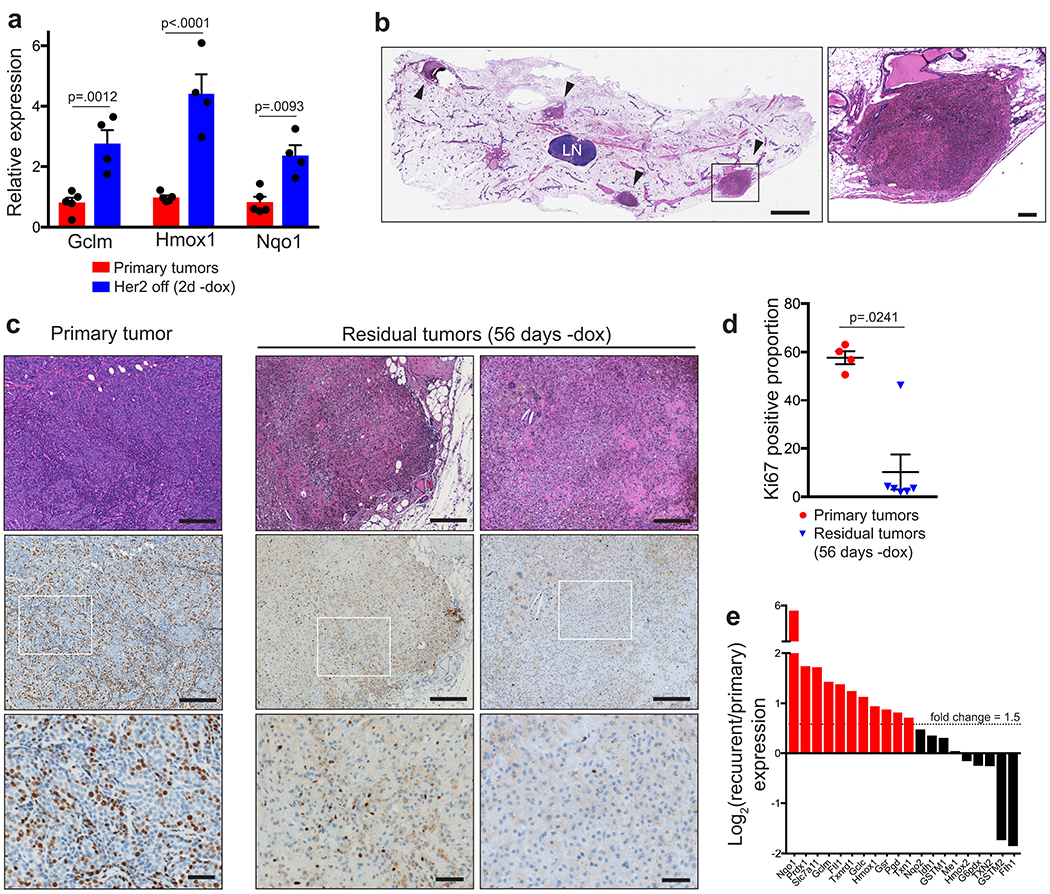
a, qRT-PCR analysis of Gclm, Nqo1, and Hmox1 expression in primary tumours (n=5) and tumours 48 hours after dox withdrawal (Her2 off; n=4). Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). b, Hematoxylin and eosin staining of a mammary gland with residual tumours, indicated by black arrowheads. LN indicates the lymph node. Black box on left panel indicates magnified residual tumour shown in right panel. Scale bars represent 2mm (left) and 200μm (right). Data are representative of 3 biologically independent experiments. c, Hematoxylin and eosin staining (top panels) and Ki67 immunohistochemical staining (brown, middle and bottom panels) of serial tissue sections from a representative primary tumour and representative residual tumours. White boxes indicate magnified section shown in bottom panels. Scale bars represent 200μm (top and middle panels) and 50μm (bottom panels). d, Quantification of the proportion of Ki67-positive cells identified by immunohistochemical staining of primary (n=4) and residual (n=6) tumours. Significance was determined by two-sided Student’s t test. e, Waterfall plot of RNA sequencing data showing the log2 fold-change in expression of NRF2 target genes between primary tumour cell lines (n=2) and recurrent tumour cell lines (n=2). Red bars indicate genes whose expression is greater than 1.5-fold higher in recurrent cells.
Extended Data Fig. 6. NRF2 expression and activity during tumor recurrence.

a, qRT-PCR analysis of Nqo1, Slc7a11, Gclm, and Hmox1 expression in primary tumour cells expressing and empty vector (Control) or caNRF2. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). Data are representative of 2 independent experiments. b, DCFDA staining showing ROS levels in primary tumour cells expressing an empty vector (Cont) or caNRF2. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 independent experiments. c, Kaplan-Meier plot showing primary tumour-free survival for mice bearing control tumours (n=27 independent tumours) or caNRF2 tumours (n=32 independent tumours). Hazard ratio and p-value were determined by two-sided log-rank (Mantel-Cox) test. d, Western blot for NRF2 and NQO1 in caNRF2-expressing primary and recurrent tumours. Blot is representative of a single experiment. e, Western blots for NRF2 and NQO1 in control primary and recurrent tumours. Blot is representative of a single experiment.
Extended Data Fig. 7. NRF2 is upregulated in human recurrent breast tumors.
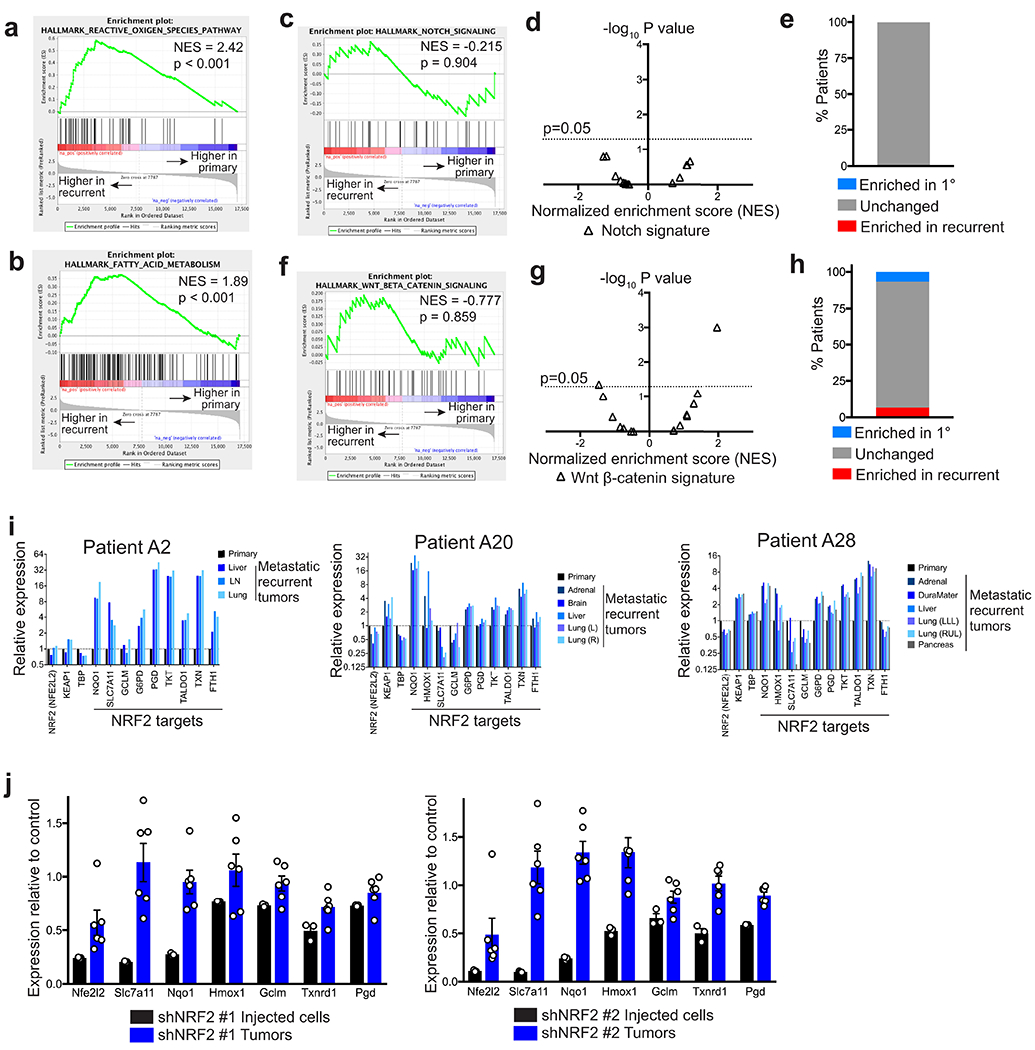
a, Gene set enrichment analysis of matched primary and recurrent breast tumours (n=15 patients) showing enrichment of “Reactive Oxygen Species Pathway” signature in recurrent tumours. b, Gene set enrichment analysis of matched primary and recurrent breast tumours (n=15 patients) showing enrichment of “Fatty Acid Oxidation” signature in recurrent tumours. c, Gene set enrichment analysis of matched primary and recurrent breast tumours (n=15 tumours) showing non-significant of the “Notch Signaling” signature in recurrent tumours. d, Volcano plot showing enrichment scores for the “Notch Signaling” signature in recurrent tumours in n=15 individual patients. e, The percentage of patients whose recurrent tumours had a significantly enriched (red), unchanged (grey), or significantly decreased (blue) “Notch Signaling” signature. f, Gene set enrichment analysis of matched primary and recurrent breast tumours (n=15 tumours) showing non-significant of the “Wnt Beta Catenin Signaling” signature in recurrent tumours. g, Volcano plot showing enrichment scores for the “Wnt Beta Catenin Signaling” signature in recurrent tumours in n=15 individual patients. h, The percentage of patients whose recurrent tumours had a significantly enriched (red), unchanged (grey), or significantly decreased (blue) “Wnt Beta Catenin Signaling” signature. i, Expression of NRF2, KEAP1, TBP, and 10 canonical NRF2 target genes in matched primary and recurrent tumours from representative patients. j, qRT-PCR analysis showing expression of NRF2 (Nfe2l2) and its target genes, Slc7a11, Nqo1, Hmox1, Gclm, Txnrd1, and Pgd, in shNRF2 #1 and shNRF2 #2 injected cells and tumour samples from Fig. 5d,e relative to shScramble control. a,b,c,f, Enrichment scores were calculated using the Kolmogorov-Smirnov statistic and p-values were calculated using permutation testing with 1000 permutations.
Extended Data Fig. 8. NRF2 has limited effects on metabolism in primary tumor cells.

a, Relative GSH:GSSG ratio in primary tumour cells expressing empty vector (control) or caNRF2. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two sided Student’s t test. b,c,d, qRT-PCR analysis of Gclm and Slc7a11 (b), Txnrd1 (c), and Pgd (d) expression in control (shScr) and NRF2-knockdown (shNRF2) primary tumour cell lines. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). e, Relative levels of the pentose phosphate pathway metabolites 6-Phosphogluconate (6PG) and Ribose 5-phosphate (R5P) in primary tumour cells expressing an empty vector (control) or caNRF2. Data are shown as the mean ± SEM for n=3 biologically independent replicates. p values were determined by two-sided Student’s t test. f, Waterfall plot showing the fold-change in nucleotide levels between primary tumour cells expressing an empty vector (control) or caNRF2. g, Relative levels of the pentose phosphate pathway metabolites 6-Phosphogluconate (6PG) and Ribose 5-phosphate (R5P) in primary tumour cells cultured with dox (Her2 on) and without dox for 7 days (Her2 off). Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 9. Identification of essential NRF2-regulated pathways in recurrent tumors.
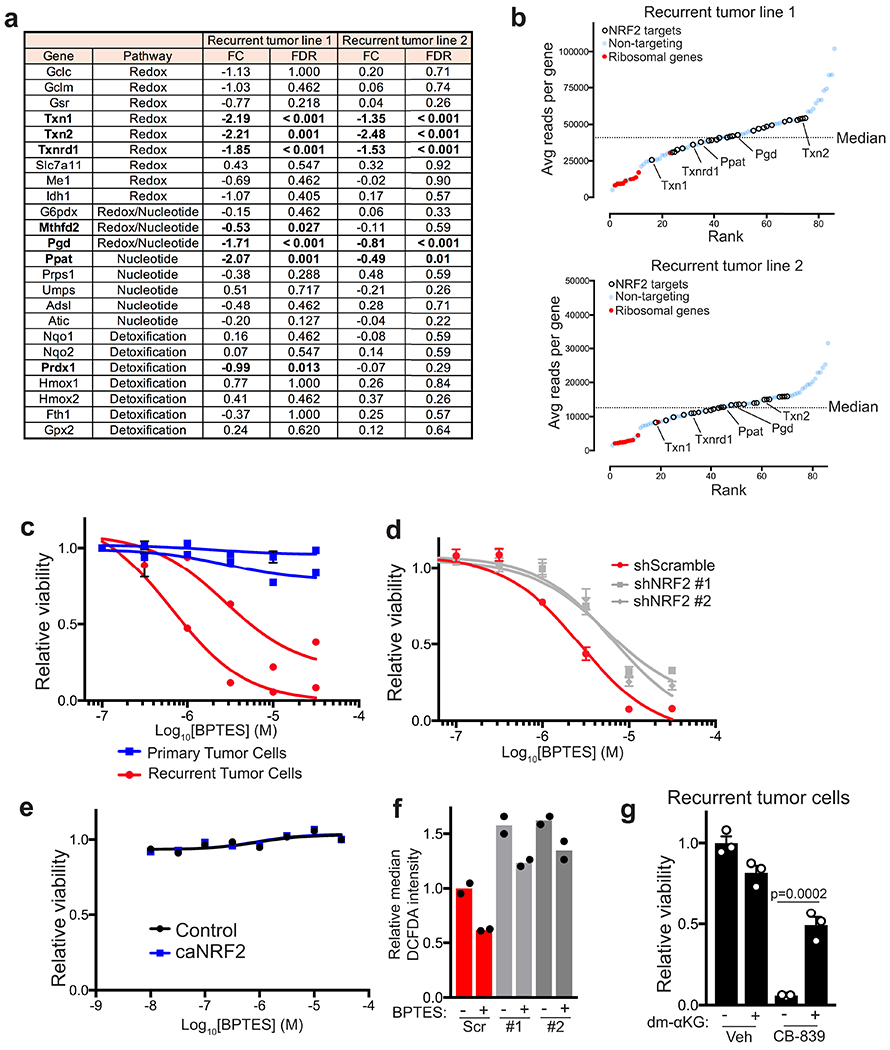
a, Table showing the fold change (FC, injected cells/tumours) and NRF2-regulated pathway for all genes tested for both cell lines (n=5 tumours for recurrent tumour line 1; n=12 for recurrent tumour line 2). Significance was determined using Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK). b, Average reads per gene for injected cell samples for each recurrent cell line. Genes in NRF2 regulated pathways are represented as hollow circles. Essential ribosomal genes are represented as red dots. Non-targeting sgRNA are represented as blue dots. Genes are ranked from fewest to most reads. c, Dose-response curves showing relative viability of two primary and two recurrent tumour cell lines treated with indicated doses of BPTES for 48 hours. Data points represent mean ± SEM for n=3 biologically independent replicates. d, Dose-response curves showing relative viability of control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells treated with indicated doses of BPTES for 72 hours. Data points represent mean ± SEM for n=3 biologically independent replicates. e, Dose response curves showing relative viability of primary tumour cells expressing an empty vector (control) or constitutively active NRF2 (caNRF2) treated with indicated doses of BPTES for 72 hours. Data points represent mean ± SEM for n=3 biologically independent replicates. f, DCFDA staining showing ROS levels in control (shScr) and NRF2-knockdown (shNRF2) recurrent cells treated with 3 μM BPTES for 48 hours. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 independent experiments. g, Relative viability of recurrent cell lines treated with CB-839 and dimethyl-α-ketoglutarate (dm-αKG), as indicated. Data are shown as the mean ± SEM for n=3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test).
Extended Data Fig. 10. Mechanism of NRF2 activation in recurrent tumors.

a, qRT-PCR analysis of NRF2 (Nfe2l2;) expression in two independent primary cell lines and two independent recurrent (Rec) cell lines. Data are shown as the mean ± SEM for n=3 biologically independent replicates. b, qRT-PCR showing mRNA expression of Keap1 in two independent primary and recurrent (Rec) cell lines. Data are shown as the mean of n=2 biologically independent replicates and are representative of 2 independent experiments. c, Western blot for KEAP1 in two independent primary cell lines and two independent recurrent (Rec) cell lines. Blot is representative of 3 independent experiments. d, Western blots for NRF2 and NQO1 in a primary cell line and a recurrent cell line treated with the indicated dose of TBHQ for 16 hours. Blot is representative of 2 independent experiments. e, Quantification of NRF2 expression (normalized to Tubulin) from (d). f, Western blot showing p-Akt (Ser473) levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 30 minutes. Blot is representative of 2 independent experiments. g, Western blot showing p-Akt (Ser473), p-GSK-3β (Ser9), and p-PRAS40 levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 3 days. Blot is representative of 2 independent experiments. h, Western blot showing NRF2 and NQO1 protein levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 3 days. Blot is representative of 2 independent experiments. i, Western blot showing p-GSK-3β (S9) levels in two independent primary cell lines and two independent recurrent cell lines. Blot is representative of 2 independent experiments.
Supplementary Material
Supplementary Table 1 - Reagents list
Supplementary Table 2 – NRF2 gene signatures
Unprocessed metabolomics data
Acknowledgements
We thank So Young Kim from the Duke Functional Genomics Core, Nicolas Devos from the Duke Center for Genomic and Computational Biology, and Yasheng Gao from the Duke Light Microscopy Core. We would also like to thank Nathaniel Mabe for his advice on bioinformatic analysis. This work was funded by National Cancer Institute grants R01CA208042 (JVA), R01CA193256 (JWL), F31CA228321 (DBF), the V-Foundation, Golfers Against Cancer, the Integrative metabolomics shared resource, and by startup funds from the Duke Cancer Institute, the Duke University School of Medicine and the Whitehead Foundation (to JVA).
Footnotes
Declaration of Interests
The authors declare no competing interests
References
- 1.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boroughs LK & DeBerardinis RJ Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17, 351–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elstrom RL et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64, 3892–9 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Wise DR et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A 105, 18782–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menendez JA et al. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc Natl Acad Sci U S A 101, 10715–20 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Sun Y, Wong J & Conklin DS PPARgamma maintains ERBB2-positive breast cancer stem cells. Oncogene 32, 5512–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–74 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Tennant DA, Duran RV, Boulahbel H & Gottlieb E Metabolic transformation in cancer. Carcinogenesis 30, 1269–80 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Klein CA Framework models of tumor dormancy from patient-derived observations. Current Opinion in Genetics & Development 21, 42–49 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Sosa MS, Bragado P & Aguirre-Ghiso JA Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 14, 611–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh AC & Ramaswamy S Mechanisms of Cancer Cell Dormancy--Another Hallmark of Cancer? Cancer Res 75, 5014–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havas KM et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J Clin Invest 127, 2091–2105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viale A et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krall EB et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chio IIC & Tuveson DA ROS in Cancer: The Burning Question. Trends Mol Med 23, 411–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schafer ZT et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moody SE et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, 451–61 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Moody SE et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Alvarez JV et al. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell 24, 30–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walens A et al. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. Elife 8(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y et al. SPSB1 promotes breast cancer recurrence by potentiating c-MET signaling. Cancer Discov 4, 790–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen T, Nioi P & Pickett CB The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284, 13291–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeNicola GM et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshimori N, Oristian D & Fuchs E TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang T et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res 70, 5486–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med 8, 334ra51 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kohler UA et al. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology 60, 670–8 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi M et al. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7, 807–20 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Romero R et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23, 1362–1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringner M, Fredlund E, Hakkinen J, Borg A & Staaf J GOBO: gene expression-based outcome for breast cancer online. PLoS One 6, e17911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel MB et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 128, 1371–1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane AN & Fan TW Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res 43, 2466–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuishi Y et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Li H et al. The landscape of cancer cell line metabolism. Nat Med 25, 850–860 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayin VI et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furukawa M & Xiong Y BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol 25, 162–71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lien EC et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol 18, 572–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada P et al. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31, 1121–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piskounova E et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris IS et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 27, 211–22 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Deblois G et al. ERRalpha mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat Commun 7, 12156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi N et al. Cancer Cells Co-opt the Neuronal Redox-Sensing Channel TRPA1 to Promote Oxidative-Stress Tolerance. Cancer Cell 33, 985–1003 e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S et al. ERRalpha-Regulated Lactate Metabolism Contributes to Resistance to Targeted Therapies in Breast Cancer. Cell Rep 15, 323–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeNicola GM et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47, 1475–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padmanabhan B et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21, 689–700 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Hast BE et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res 74, 808–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagini S, Sophia J & Mishra R Glycogen synthase kinases: Moonlighting proteins with theranostic potential in cancer. Semin Cancer Biol 56, 25–36 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Muir A, Danai LV & Vander Heiden MG Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis Model Mech 11(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Moody SE et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Alvarez JV et al. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell 24, 30–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campeau E et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Guertin DA, Ali SM & Sabatini DM Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–101 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joung J et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Ser Z & Locasale JW Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem 86, 2175–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liberti MV et al. A Predictive Model for Selective Targeting of the Warburg Effect through GAPDH Inhibition with a Natural Product. Cell Metab 26, 648–659 e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H et al. The landscape of cancer cell line metabolism. Nat Med 25, 850–860 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ringner M, Fredlund E, Hakkinen J, Borg A & Staaf J GOBO: gene expression-based outcome for breast cancer online. PLoS One 6, e17911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero R et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23, 1362–1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel MB et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 128, 1371–1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier JA, Zhang F & Sanjana NE GUIDES: sgRNA design for loss-of-function screens. Nat Methods 14, 831–832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabe NW et al. G9a Promotes Breast Cancer Recurrence Through Repression of a Pro-inflammatory Program. bioRxiv, 2020.01.09.900183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 - Reagents list
Supplementary Table 2 – NRF2 gene signatures
Unprocessed metabolomics data
Data Availability Statement
RNA sequencing data comparing primary and recurrent MTB;TAN-derived tumour cell lines is available online using National Center for Biotechnology Information’s Short Read Archive (SRA) under project accession number 626PRJNA50583918. Prognostic analysis for NRF2 gene signatures was performed using Gene expression-based Outcome for Breast cancer Online (GOBO10), which uses 4 datasets (GSE1456, GSE3494, GSE6532, and GSE7390), available at http://co.bmc.lu.se/gobo/gsa.pl. RNA Sequencing data used for human recurrent breast cancer GSEA was accessed using NCBI’s GEO database (GSE11059012). Metabolomic data for KEAP1 mutant cancer cell lines was accessed using the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) and supplementary tables from Li, H et al. (2019)9. Metabolomics datasets generated in this study are available in Supplementary Table 3s.


