Abstract
We present automated continuous evolution (ACE), a platform for the hands-free directed evolution of biomolecules. ACE pairs OrthoRep, a genetic system for continuous targeted mutagenesis of user-selected genes in vivo, with eVOLVER, a scalable and automated continuous culture device for precise, multi-parameter regulation of growth conditions. By implementing real-time feedback-controlled tuning of selection stringency with eVOLVER, genes of interest encoded on OrthoRep autonomously traversed multi-mutation adaptive pathways to reach desired functions, including drug resistance and improved enzyme activity. The durability, scalability, and speed of biomolecular evolution with ACE should be broadly applicable to protein engineering as well as prospective studies on how selection parameters and schedules shape adaptation.
Keywords: continuous evolution, continuous culture, orthogonal replication, OrthoRep, eVOLVER
Continuous evolution has emerged as a powerful paradigm for the evolution of proteins and enzymes1–4 towards challenging functions.5,6 In contrast to classical directed evolution approaches that rely on stepwise rounds of ex vivo mutagenesis, transformation into cells, and selection,7 continuous evolution systems achieve rapid diversification and functional selection autonomously, often through in vivo targeted mutagenesis systems (Figure 1a).2,7–14 The result is a mode of directed evolution that requires only the basic culturing of cells, in theory, enabling extensive speed, scale, and depth in evolutionary search.3 In practice, however, developing a continuous evolution method that realizes all three properties has been challenging. Recently, our groups made two advances, OrthoRep and eVOLVER, that can pair to achieve continuous evolution at significant speed, scale, and depth.
Figure 1.
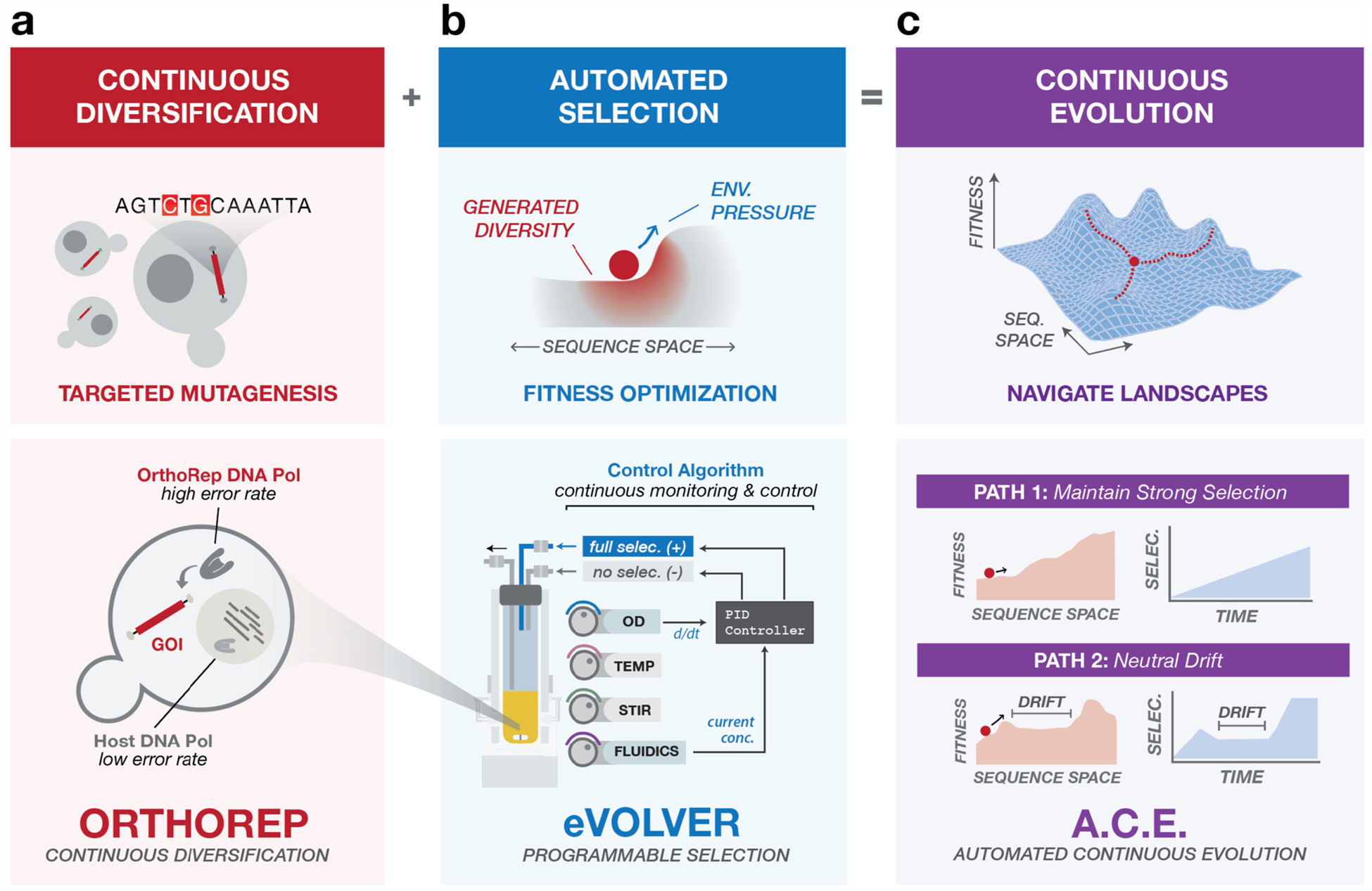
Automated Continuous Evolution (ACE). (a) OrthoRep enables continuous diversification of genes of interest (GOIs) via in vivo targeted mutagenesis in yeast. The basis of OrthoRep is an orthogonal DNA polymerase-plasmid pair that mutates GOIs ~100,000-fold faster than the genome. (b) eVOLVER is a continuous culturing platform for programmable, multiparameter control of selection conditions across many independent cultures. A PID control algorithm implemented with eVOLVER dynamically tunes selection pressure of populations as they adapt, precisely challenging them to achieve desired functions. PID control is achieved by tuning the ratio of full selection and no selection media inputs in response to growth rate. (c) By running OrthoRep in eVOLVER with PID control, ACE autonomously and rapidly navigates complex fitness landscapes. With a single framework, ACE can guide independent cultures through diverse trajectories.
OrthoRep is an engineered genetic system for continuous in vivo targeted mutagenesis of genes of interest (GOIs).2,14 OrthoRep uses a highly error-prone, orthogonal DNA polymerase-plasmid pair in yeast that replicates GOIs at a mutation rate of 10−5 substitutions per base (spb) without increasing the genomic mutation rate of 10−10 spb (Figure 1a). This ~100,000-fold increase in the mutation rate of GOIs drives their accelerated evolution (speed). Because the OrthoRep system functions entirely in vivo and culturing yeast is straightforward, independent GOI evolution experiments can be carried out in high-throughput (scale). In addition, long multi-mutation pathways can be traversed using OrthoRep, owing to the durability of mutagenesis over many generations (depth). However, to practically realize depth in evolutionary search, in vivo mutagenesis with OrthoRep must be coupled with a functional selection that can be tuned over the course of a continuous evolution experiment. This tuning is necessary to precisely and efficiently guide populations to the desired evolutionary search depth. For example, evolution of novel functions requiring long mutational trajectories may demand frequent modification of selection conditions in order to maintain strong selection,5,6,15 guide evolution through strategic intermediate functions,1,6 or impose periods of neutral drift or alternating selection to promote crossing of fitness valleys (Figure 1c).16,17 Yet, selection schedules cannot be determined a priori as the generation of beneficial mutations is a fundamentally stochastic process. Therefore, selection schedules should be adjusted dynamically based on how populations adapt, rendering manual implementation of continuous evolution experiments onerous. Further, each functional selection demands its own selection schedule, necessitating empirical probing of conditions that are appropriately stringent to generate selection pressures, yet sufficiently lenient to allow for mutational accumulation. Previous continuous evolution campaigns approached the challenge of optimizing selection schedules by either limiting the number of parallel evolution experiments being conducted so that selection can be manually tuned on the fly,1,5 or by setting a fixed but conservative selection schedule to buffer against variations in adaptation rate across a large number of replicate experiments.2 However, even with conservative selection schedules, a proportion of replicates in high-throughput evolution studies went extinct when the rate of selection stringency increase outpaced the rate of adaption.2 Indeed, streamlining selection schedules for experimental evolution remains an open challenge.18–20
To address this challenge, we turned to eVOLVER. eVOLVER is a versatile continuous culture platform that enables multiparameter control of growth and selection conditions across independent microbial cultures (Figure 1b).21 eVOLVER’s flexible hardware and software permit development of “algorithmic selection routines” that apply selective pressures based on real-time monitoring and feedback from culture growth characteristics. Additionally, eVOLVER’s robust framework ensures experimental durability over long timeframes, and its unique scalable design allows independent control over tens to hundreds of cultures. Combining OrthoRep and eVOLVER should therefore enable continuous evolution with speed, depth, and scale.
Here we describe this pairing of OrthoRep with eVOLVER to achieve Automated Continuous Evolution (ACE) (Figure 1c). By implementing a closed-loop feedback routine that dynamically adjusts the strength of selection for a desired function in response to growth rate changes of yeast populations diversifying a GOI on OrthoRep, we demonstrate completely automated continuous evolution over extended periods of time without manual intervention. To illustrate the performance and utility of ACE, we describe its application in two model protein evolution experiments, one yielding drug-resistant Plasmodium falciparum dihydrofolate reductases (PfDHFRs) and the other yielding variants of the thermophilic HisA enzyme from Thermotoga maritima (TmHisA) that operate well in mesophilic yeast hosts.
Results and Discussion
Establishment of ACE.
To establish ACE, we first reconfigured eVOLVER Smart Sleeves21 so that each culture vial receives two media inputs: (1) ‘no selection’ base media (e.g. media without drug or with the maximum concentration of nutrient in our cases) and (2) ‘full selection’ media (e.g. media with the maximum concentration of drug or without nutrient in our cases). Using eVOLVER software calculations, selection strength can be dynamically tuned by altering the ratios of the two media inputs as cultures are diluted over time (Figure 1b,c). We then implemented a closed-loop control system that seeks to achieve and maintain a target culture growth rate by dynamically adjusting selection strength. Briefly, culture growth rate is continuously measured based on real-time recordings of optical density (OD), and a proportional-integral-derivative (PID) control algorithm22 is used to determine the percentage of full selection media to add to the culture in order to minimize error between the actual growth rate and a target growth rate (or setpoint) (see Methods). Although simpler feedback algorithms18,19 have been previously used in microbial evolution experiments, these resulted in growth rate oscillations or excessive overshooting in our experiments, frequently driving cultures to extinction (Figure S1).
Evolution of PfDHFR resistance using ACE.
To validate ACE, we first repeated a continuous evolution experiment that we previously conducted using manual serial passaging. Specifically, we evolved Plasmodium falciparum dihydrofolate reductase (PfDHFR) to develop drug resistance to the antimalarial drug, pyrimethamine, by encoding PfDHFR on OrthoRep in a yeast strain that relies on PfDHFR activity for survival (Figure 2).2 We determined appropriate PID constants to tune the concentration of pyrimethamine (Figure 2b, S2) and keep the measured growth rate of cells at a target growth rate (Figure 2a, setpoint = dashed black line). This program forced cells to continuously experience a strong selection pressure imposed by pyrimethamine, which resulted in the rapid evolution of PfDHFR resistance (Figure 2c). We observed that after ~550 hours (~100 generations) of continuous hands-free operation of ACE, five out of six replicates adapted to 3 mM pyrimethamine, the highest concentration of pyrimethamine soluble in liquid media (Figure 2b, S3). ACE maintained cultures near the target growth rate over the entire course of the experiment (Figure 2a,b), demonstrating the effectiveness of the control loop. In contrast to the use of a fixed selection schedule2 or simpler control algorithms for selection (Figure S1) that resulted in occasional extinction caused by too-rapid increases in pyrimethamine concentration, all six ACE experiments reliably adapted to yield multi-mutation pyrimethamine-resistant PfDHFR variants. Validating our method, we found that populations converged on strong resistance mutations in PfDHFR – C50R, D54N, Y57H, C59R, C59Y, and S108N – as observed and characterized previously2 (Figure 2c). Additionally, the monotonically increasing pyrimethamine concentrations we observed for most replicates (Figure 2b) are consistent with step-wise fixation of beneficial mutations expected for the evolution of PfDHFR resistance under strong selection.2,21 Upon examination of one of the evolution replicates (V3), we noted a drop in pyrimethamine at ~200 hours, likely due to a mechanical error. Nevertheless, the selection self-adjusted, resulting in recovery in growth and demonstration of ACE’s control algorithm to robustly maintain selection. Finally, ACE demonstrated a substantial increase in speed over our previous evolution campaign performed by manual passaging; with ACE, culture growth rates in 5/6 vials stabilized at the maximum pyrimethamine concentration after ~550 hours, which is over 200 hours faster than for the manual evolution campaign done with serial passaging.2 Collectively, these results validate the ACE system and highlight its ability to enable reliable and rapid continuous evolution of proteins.
Figure 2.
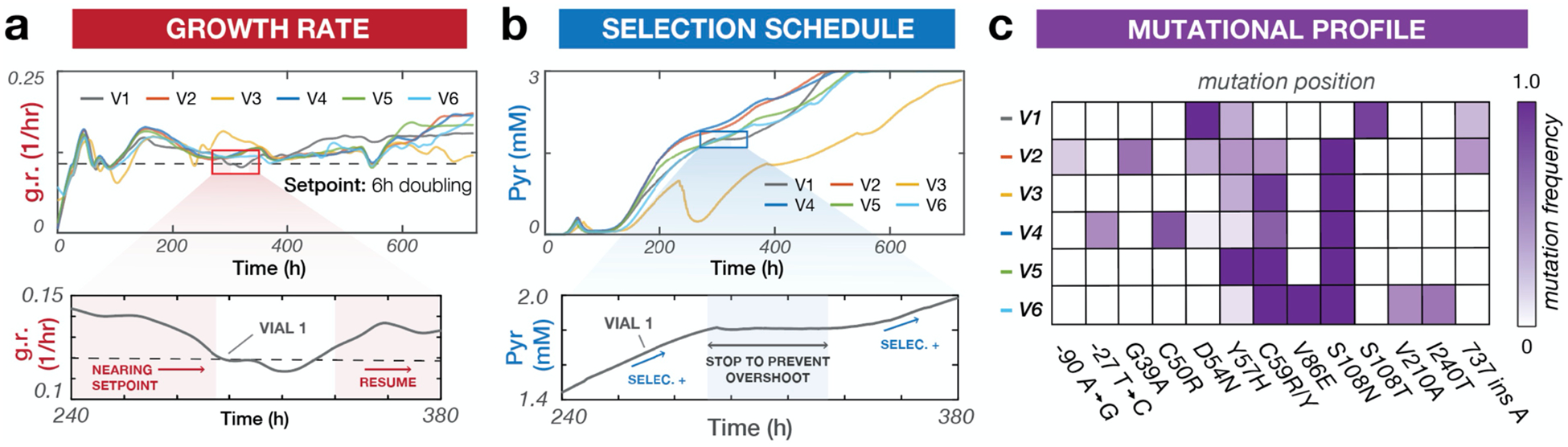
Automated continuous evolution of PfDHFR resistance to pyrimethamine. (a) Top: Growth rate traces for six independent OrthoRep cultures (V1–6) evolving PfDHFR resistance to pyrimethamine in eVOLVER using PID control. Bottom: A representative time window validating PID control. The growth rate (solid line) is controlled by automated tuning of pyrimethamine concentration (Figure 2b, bottom) to keep cultures constantly challenged at the setpoint growth rate (dashed line). (b) Top: Drug selection schedules for OrthoRep cultures evolving PfDHFR. Bottom: A representative time window demonstrating PID-based selection tuning. Pyrimethamine concentration autonomously adjusts in response to growth rate deviation from the setpoint (Figure 2a, bottom). (c) Promoter and PfDHFR mutations identified in six evolved populations. Mutation frequencies are estimated from SNP analysis of bulk Sanger sequencing traces.
Evolution of TmHisA activity using ACE.
We next applied ACE to evolve the thermophilic Thermotoga maritima HisA enzyme (TmHisA) to function in Saccharomyces cerevisiae at mesophilic temperatures. TmHisA, an ortholog of S. cerevisiae HIS6, catalyzes the isomerization of ProFAR to PRFAR in the biosynthesis of histidine. However, TmHisA does not effectively complement a his6 deletion in yeast when expressed from a medium-strength yeast promoter (Figure 3), likely due to the different temperature niches of S. cerevisiae and T. maritima (30°C and 80°C, respectively). We reasoned that ACE could readily drive the evolution of TmHisA to function in yeast Dhis6 strains by selecting for growth in media lacking histidine. This evolution serves as a valuable test of the capabilities of ACE for two reasons. First, adapting enzymes from non-model thermophiles to function in model mesophiles is useful for industrial biotechnology whose infrastructure is designed around model organisms like yeast and bacteria. Second, in contrast to drug resistance in PfDHFR, which is driven by a small number of large effect mutations,2 we reasoned that temperature and host adaptation of enzyme activity would involve a large number of small effect mutations, leading to a more complex fitness landscape. This would act as a more demanding test of ACE’s ability to achieve precise feedback-control during selection.
Figure 3.
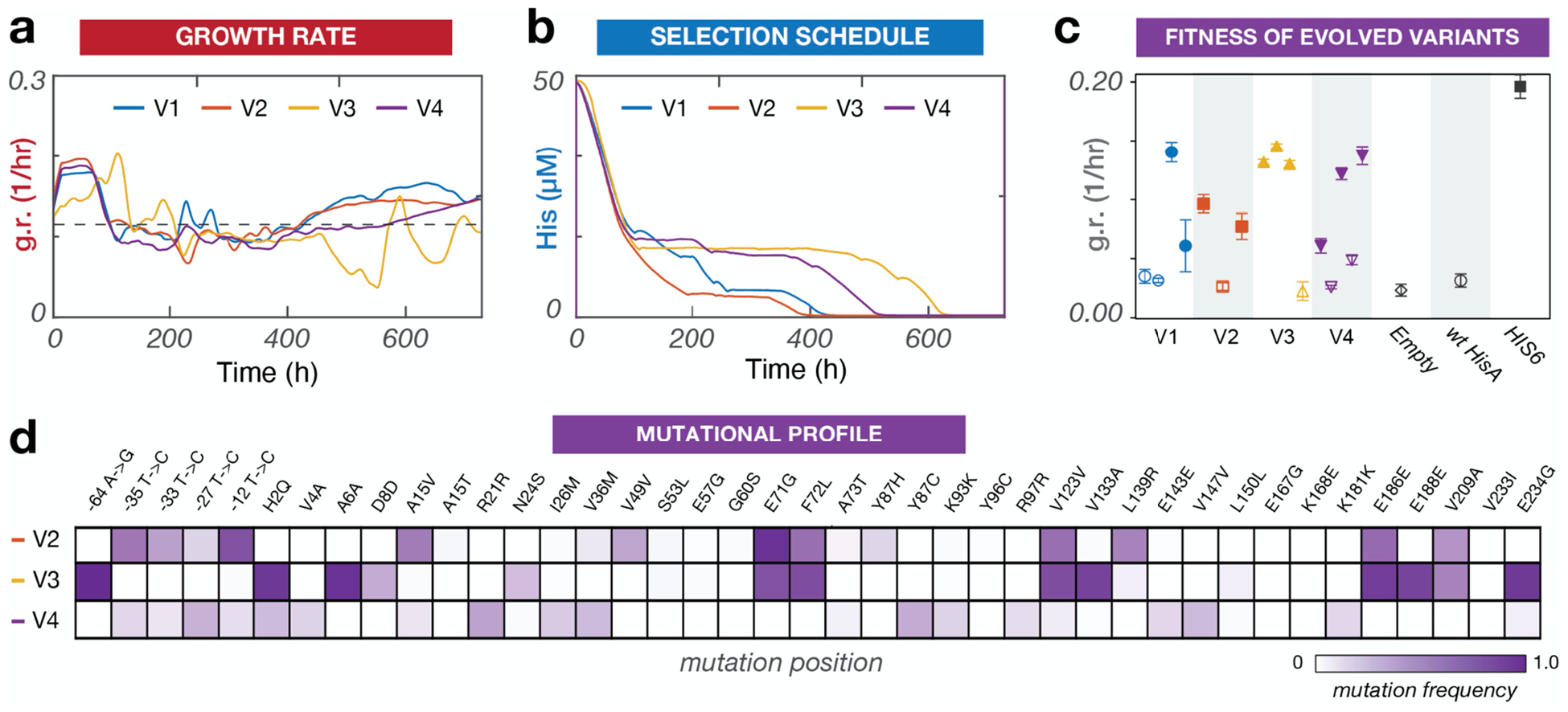
Automated continuous evolution of TmHisA to operate in mesophilic yeast. (a) Growth rate traces of four independent OrthoRep cultures (V1–4) evolving TmHisA to support growth of a Dhis6 yeast strain at 30 °C. (b) Nutrient concentration schedule of the four evolving cultures controlled via PID. (c) Growth rate analysis of individual TmHisA variants selected from adapted cultures. Three to five TmHisA variants were sampled from each replicate evolution experiment. The coding regions of the variants were cloned into a low-copy yeast plasmid under the control of a medium strength promoter. The ability of the variants to support growth of a Dhis6 strain was measured in triplicate in comparison to the ability of wt TmHisA and native yeast HIS6 to support growth. Shapes indicate means and error bars denote standard deviations. Darkened replicates indicate a p < 0.05 compared to wt TmHisA activity by one-way ANOVA. We note that a number of evolved variants sampled from our adapted replicates did not support growth, but this is likely because OrthoRep evolves TmHisA in the context of a multi-copy orthogonal plasmid, allowing inactive variants to hitchhike with active copies in the same cell. Such inactive variants could be sampled during the subcloning of TmHisA from orthogonal plasmids into the low-copy plasmids used for testing variants in fresh strains. We also note that variants corresponding to a growth rate below ~0.05 did not enter log phase growth during the 24-hour experiment. (d) Promoter and TmHisA mutations identified in evolved populations from V2–4. Mutation frequencies are estimated from SNP analysis of bulk Sanger sequencing traces. V1 Sanger sequencing traces are not included due to a technical mistake that rendered stocks of the evolved population inviable for revival and sequencing, although individual clones were fully analyzed (Figure 3c), as they were sampled before the stocking mistake.
We encoded TmHisA on OrthoRep in a Dhis6 strain and carried out ACE selection in four independent replicates for a total of 600 hours (~100 generations) (Figure 3, S4). At the beginning of the experiment, there was no detectable growth in the absence of histidine. At the end of the experiment, all four replicates successfully adapted to media lacking histidine. To confirm that TmHisA evolution was responsible for the observed adaptation, TmHisA variants were isolated from OrthoRep and characterized for their ability to complement a his6 deletion in fresh yeast strains. Indeed, the evolved TmHisA variants we sampled were able to support growth in media lacking histidine in contrast to wild-type TmHisA (Figure 3c). Consistent with a model of a more complex fitness landscape, growth rate traces for the four replicate cultures were noisier (Figure 3a) than those of PfDHFR (Figure 2a), full adaptation occurred only after a long period of neutral drift (hours ~100–500 in Figure 3b), and the sequences of independently evolved TmHisAs were diverse (Figure 3d, Table S1). Nevertheless, ACE was able to autonomously adapt TmHisA in all four replicates within 120 fewer hours than manual passaging experiments (unpublished results). Sequencing of sampled clones revealed TmHisA variants harboring between 6 and 15 mutations (Table S1), again demonstrating the durability of ACE in carrying out long evolutionary searches to discover high-activity multi-mutation enzyme variants.
Conclusion
In summary, we have developed a fully automated, in vivo continuous evolution setup termed ACE that couples OrthoRep-driven continuous mutagenesis and eVOLVER-enabled programmable selection. We demonstrated the evolution of drug resistance in PfDHFR and mesophilic operation of TmHisA, showcasing the ability of ACE to individually control selection schedules in multi-replicate GOI evolution experiments based on real-time measures of adaptation. We further validate the value and generalizability of a PID controlled selection scheme that successfully drives two mechanistically different selections. The result is a system that offers unprecedented speed, depth, and scalability for conducting evolutionary campaigns to achieve ambitious protein functions.
ACE paves the way for an array of complex evolution experiments that can advance both basic and applied protein and enzyme evolution. For example, eVOLVER can be used to program multidimensional selection gradients across OrthoRep experiments, test the effects of selection strength or different population sizes (and beneficial mutation supply) on the outcomes of adaptation, or explore the relationship between timescales of drift and adaptation. Real-time feedback on growth metrics to adjust selection stringency can ensure that every evolving population is being constantly challenged appropriately or allowed to drift, which is especially relevant when evolving biomolecules with rugged fitness landscapes where predefined selection strategies are prone to driving populations to extinction or local fitness maxima. In the future, many other algorithmic selection routines may be implemented with ACE to more efficiently and intelligently navigate fitness landscapes. For example, machine learning algorithms can take the outcomes of replicate evolution experiments carried out under different selection schedules to train ACE selection programs themselves. Finally, the automated, open-source nature of ACE is well-suited for integration with other open-source hardware and wetware tools to create larger automation pipelines. Overall, we foresee ACE as an enabling platform for rapid, deep, and scalable continuous GOI evolution for applied protein engineering and studying the fundamentals of protein evolution.
Methods
Cloning.
All plasmids used in this study are listed in Table S2. Plasmids were cloned using either restriction enzymes if compatible sites were available or using Gibson cloning23 with 20–40 bps of overlap. Primers and gBlocks were ordered from IDT Technologies. Enzymes for PCR and cloning were purchased from NEB. Plasmids were cloned into either Top10 E. coli cells from Thermo Fisher or SS320 E. coli from Lucigen.
Yeast transformation and DNA extraction.
All yeast strains used in this study are listed in Table S3. Yeast transformations were done with roughly 100 ng – 1 μg of plasmid or donor DNA via the Gietz high-efficiency transformation method.24 For integration of genes onto the orthogonal plasmid (pGKL1), cassettes were linearized with ScaI and subsequently transformed as described previously.2,14 Standard preparations of YPD and drop-out synthetic media were obtained from US Biological. When necessary, the following were supplemented at their respective concentrations: 5-FOA at 1 mg/mL, G418 at 400 μg/mL, and Nourseothricin at 200 μg/mL. Yeast DNA extraction of orthogonal plasmids were done as previously reported.2,14
eVOLVER feedback control configuration.
ACE experiments were performed using the previously described eVOLVER continuous culture system,21 modified to enable an additional media input into each culture. Specifically, each vessel consists of three connected pumps (two input, one efflux) and are actuated programmatically to implement a so-called “morbidostat” algorithm where the selection stringency is adjusted to maintain a particular rate of cell growth. The custom script of eVOLVER (custom_script.py) was extensively modified to change the behavior of eVOLVER from the default turbidostat to a morbidostat. Briefly, in the new morbidostat mode, eVOLVER dilutes the growing cultures after a defined time, which we set to an hour. At the time of dilution, the growth rate since the last dilution is calculated by fitting the OD measurements to an exponential equation y = A · eBx where B is the growth rate. Using the current and historical growth rate, a dilution parameter, r(t) was calculated as described below to dilute the morbidostat. The morbidostat algorithm and eVOLVER experimental code are written in Python and included in the supplemental files.
The efflux pump for each vessel is actuated whenever either of the influx pumps are triggered and stay ON for an additional 5 seconds. Therefore, the volume of the culture vessel is determined by the length of the efflux straw and estimated to be at 30 mL. The flow rate of each media input was individually calibrated for accurate metering of drug or nutrient into the culture.
Before each experiment, 40 mL borosilicate glass vessels (Chemglass), stir bars (Fisher), and fluidic straws were assembled and autoclaved. Fluidic lines were sterilized by flushing with 10% bleach and 70% ethanol before use. Culture vessel assemblies were connected to fluidic lines after sterilization and slotted into an eVOLVER Smart Sleeve for monitoring of OD and control of temperature and stir rate.
PID algorithm development and tuning.
To control the rate of dilution, we used the following equation to determine the percentage of selection media to add:
where KP, KI, KD, and KO, are empirically determined constant multipliers of proportional integral, derivative, and offset terms, and e(t) is the difference between the actual growth rate and the target growth rate. To estimate KP, KI, KD, and KO, we used the the Ziegler-Nichols method25 for initially tuning the parameters with the pre-evolution strain, ZZ-Y323. KI and KD were first set to zero and KP was increased until regular oscillations in growth rate were observed (Figure S2). This resulted in a KP = 4.
Using the parameters obtained during the oscillation and the Ziegler-Nichols estimation:
These initial values were empirically tuned to achieve the final values of KP = 0.07, KI = 0.05, KD = 0.2 and KO = 0.
These constants were then used to calculate r(t) at any given point during evolution. r(t) would then be used to determine the ratios of media to add during each dilution step by controlling the pump runtime. For example, if an r(t) = 0.25 was determined with a pump runtime of 5 seconds, the pump for the base media would run for [1 − r(t)] * 5 seconds = 3.75 seconds while the pump for the full selection media would run for r(t) * 5 seconds 1.25 seconds.
The integral error was reset at every instance the proportional error (e(t)) become negative, and the offset (KO) was updated to equal r(t) at that time. This was done to allow the PID controller to be more sensitive to the integral error and to avoid the bias that would result from the initial conditions having minimal selection pressure.
PfDHFR evolution.
eVOLVER was set to morbidostat mode with the PID settings described above, a target doubling time of 8 hours, and one dilution step per hour. A culture of ZZ-Y435 was grown to saturation in SC-HW and then inoculated 1:50 in eVOLVER vials. SC-HW served as the base media, while SC-HW + 3 mM pyrimethamine served as the full selection media. (3mM was previously determined as the maximum soluble concentration of pyrimethamine in media.2) After inoculation, the eVOLVER PID script was initiated and evolution commenced. During evolution, the only user intervention was media exchange and periodic sampling of cultures. After 725 hrs, all cultures achieved growth rates near wild-type levels in the full selection condition (Figure S3), so the experiment was stopped and cultures were frozen in glycerol stocks.
TmHisA evolution.
eVOLVER was set to morbidostat mode with the PID settings described above, a target doubling time of 8 hours, and one dilution step per hour. A culture of ZZ-Y323 was grown to saturation in SC-UL and then inoculated 1:50 in eVOLVER vials. SC-ULH + 7.76 mg/L (50 μM) histidine served as the base media, while SC-ULH served as the full selection media. After inoculation, the eVOLVER PID script was initiated and evolution commenced. During evolution, the only user intervention was media exchange and periodic sampling of cultures. After 715 hrs, all cultures achieved growth rates near wild-type levels in the full selection condition (Figure S4), so the experiment was stopped and cultures were frozen in glycerol stocks.
Bulk DNA sequencing and characterization.
Final evolution timepoints of PfDHFR and TmHisA were regrown in SC-HW and SC-ULH media, respectively, from glycerol stocks. The orthogonal plasmids encoding evolved PfDHFR or TmHisA were extracted from the bulk cultures as described above, PCR amplified, and sequenced via Sanger sequencing. Mutation frequencies were calculated from Sanger sequencing files with QSVanalyzer as previously described.2 However, V1 from TmHisA evolution could not be revived from the glycerol stock due to a stocking mistake and was not included for bulk DNA sequencing.
TmHisA isolated mutant cloning.
Final evolution time-points of TmHisA were streaked onto SC-ULH solid media. Individual colonies were regrown in SC-ULH media and the orthogonal plasmid DNA was extracted from the cultures as described above. The evolved TmHisA sequences were sequenced and cloned into a nuclear CEN6/ARS4 expression vector under control of the pRPL18B promoter and with the LEU2 selection marker. Since each colony can have different TmHisA mutants due to the multicopy nature of the orthogonal plasmid in OrthoRep, the cloned plasmids were sequenced again to determine the exact mutant of TmHisA being characterized. The resulting plasmids were transformed into ZZ-Y354, which lacks his6, for growth rate measurements.
TmHisA growth rate measurements.
Yeast strains containing each TmHisA mutant, WT TmHisA, S. cerevisiae HIS6, or none of the above expressed from a nuclear plasmid were grown to saturation in SC-L and diluted 1:100 in SC-LH. Three 100 uL replicates of each strain were placed into a 96 well clear-bottom tray, sealed, and grown at 30 °C. Cultures were continuously shaken and OD600 was measured every 30 minutes automatically for 24 hours (Tecan Infinite M200 Pro) according to a previously described protocol.26 A custom MATLAB script (growthassayV3.m), included in supplemental files, was used to calculate growth rates from raw OD600 data. The script carries out a logarithmic transformation of the OD600 data. The linear region of the transformed data as a function of time corresponds to log phase growth. A sliding window approach is used to find and fit this linear region in order to calculate the doubling time during log phase growth. This doubling time (T) is converted to the continuous growth rate plotted in Figure 3c by the formula ln(2)/T.
Statistical analysis.
Statistical analysis was done using GraphPad Prism and one-way ANOVA with multiple comparisons versus wild-type TmHisA and corrected for multiple comparisons. Results are reported at p < 0.05.
Supplementary Material
Figure S1. Sample evolution experiments in eVOLVER using the control algorithm derived from Toprak et al., 2013.
Figure S2. Oscillation of growth rate of ZZ-Y323 during the empirical determination of PID settings.
Figure S3. Adaptation history for all six replicates during PfDHFR evolution.
Figure S4. Adaptation history for all four replicates during TmHisA evolution.
Table S1. TmHisA mutants characterized.
Table S2. List of plasmids used in this study.
Table S3. List of yeast strains used in this study.
Acknowledgements
We thank members of the Liu and Khalil groups for helpful discussions. This work was funded by NIH (1DP2GM119163-01), NSF (MCB1545158), and DARPA (HR0011-15-2-0031) to CCL, and NIH (1DP2AI131083-01, 1R01EB027793-01), NSF (CCF-1522074), and DARPA (HR0011-15-C-0091) to ASK. We especially thank Dr. Justin Gallivan and the DARPA Biological Robustness in Complex Settings program, which sponsored the construction of an eVOLVER system in the Liu lab.
Abbreviations
- ACE
automated continuous evolution
- GOI
gene of interest
- spb
substitutions per base
- PfDHFR
Plasmodium falciparum dihydrofolate reductase
- TmHisA
Thermotoga maritima HisA enzyme
- ProFAR
N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide
- PRFAR
N′-((5′-phosphoribulosyl) formimino)-5-aminoimidazole-4-carboxamideribonucleotide
Footnotes
Supplementary code
growthassayV3.m – MATLAB code used to calculate growth rates from OD600 data. main_eVOLVER.py – python script to start eVOLVER eVOLVER_module.py – python script to containing basic eVOLVER functions custom_script.py – python script to implement PID function for eVOLVER
Conflict of Interest
The authors hold patents on OrthoRep and eVOLVER technologies. BGW, AR, ASK, and CCL are forming a company based on automated OrthoRep-driven evolution. BGW and ASK are founders of Fynch Biosciences, a manufacturer of eVOLVER hardware.
References
- (1).Esvelt KM, Carlson JC, Liu DR (2011) A System for the Continuous Directed Evolution of Biomolecules. Nature, 472, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ravikumar A, Arzumanyan GA, Obadi MKA, Javanpour AA, Liu CC (2018) Scalable , Continuous Evolution of Genes at Mutation Rates above Genomic Error Thresholds Resource Scalable , Continuous Evolution of Genes at Mutation Rates above Genomic Error Thresholds. Cell, 175, 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wellner A, Ravikumar A, Liu CC (accepted) Continuous Evolution of Proteins in vivo In Protein Engineering (Zhao H Ed.), Wiley-VCH. [Google Scholar]
- (4).Fact C, Tan ZL, Zheng X, Wu Y, Jian X, Xing X, Zhang C (2019) In Vivo Continuous Evolution of Metabolic Pathways for Chemical Production. Microb. Cell Fact, 18, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Badran AH, Guzov VM, Huai Q, Kemp MM, Vishwanath P, Kain W, Nance AM, Evdokimov A, Moshiri F, Turner KH, et al. (2016) Continuous Evolution of Bacillus Thuringiensis Toxins Overcomes Insect Resistance. Nature, 533, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Packer MS, Rees HA, Liu DR (2017) Phage-Assisted Continuous Evolution of Proteases with Altered Substrate Specificity. Nat. Commun, 8, 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Packer MS, Liu DR (2015) Methods for the Directed Evolution of Proteins. Nat. Rev. Genet, 16, 379–394. [DOI] [PubMed] [Google Scholar]
- (8).Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV, Dueber JE (2018) CRISPR-Guided DNA Polymerases Enable Diversification of All Nucleotides in a Tunable Window. Nature, 560, 248–252. [DOI] [PubMed] [Google Scholar]
- (9).Finney-Manchester SP, Maheshri N (2013) Harnessing Mutagenic Homologous Recombination for Targeted Mutagenesis in Vivo by TaGTEAM. Nucleic Acids Res, 41, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Crook N, Abatemarco J, Sun J, Wagner JM, Schmitz A, Alper HS (2016) In Vivo Continuous Evolution of Genes and Pathways in Yeast. Nat. Commun, 7, 13051–13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hess GT, Frésard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC (2016) Directed Evolution Using dCas9-Targeted Somatic Hypermutation in Mammalian Cells. Nat. Methods, 13, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X (2016) Targeted AID-Mediated Mutagenesis (TAM) Enables Efficient Genomic Diversification in Mammalian Cells. Nat. Methods, 13, 1029–1035. [DOI] [PubMed] [Google Scholar]
- (13).Moore CL, Papa LJ, Shoulders MD (2018) A Processive Protein Chimera Introduces Mutations across Defined DNA Regions in Vivo. J. Am. Chem. Soc, 140, 11560–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ravikumar A, Arrieta A, Liu CC (2014) An Orthogonal DNA Replication System in Yeast. Nat. Chem. Biol, 10, 175–177. [DOI] [PubMed] [Google Scholar]
- (15).Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH (2008) Evolutionary History of a Specialized P450 Propane Monooxygenase. J. Mol. Biol, 383, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Steinberg B, Ostermeier M (2016) Environmental Changes Bridge Evolutionary Valleys. Sci. Adv, 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bershtein S, Goldin K, Tawfik DS (2008) Intense Neutral Drifts Yield Robust and Evolvable Consensus Proteins. J. Mol. Biol, 379, 1029–1044. [DOI] [PubMed] [Google Scholar]
- (18).Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R (2012) Evolutionary Paths to Antibiotic Resistance under Dynamically Sustained Drug Selection. Nat. Genet, 44, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Toprak E, Veres A, Yildiz S, Pedraza JM, Chait R, Paulsson J, Kishony R (2013) Building a Morbidostat: An Automated Continuous-Culture Device for Studying Bacterial Drug Resistance under Dynamically Sustained Drug Inhibition. Nat. Protoc, 8, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).DeBenedictis EA, Chory EJ, Gretton D, Wang B, Esvelt K (2020) A High-Throughput Platform for Feedback-Controlled Directed Evolution. bioRxiv Epub Apr 2, 2020 DOI: 10.1101/2020.04.01.021022. [DOI] [Google Scholar]
- (21).Wong BG, Mancuso CP, Kiriakov S, Bashor CJ, Khalil AS (2018) Precise, Automated Control of Conditions for High-Throughput Growth of Yeast and Bacteria with eVOLVER. Nat. Biotechnol, 36, 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ang KH, Chong G, Li Y (2005) PID Control System Analysis, Design, and Technology. IEEE Trans. Control Syst. Technol, 13, 559–576. [Google Scholar]
- (23).Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA, Smith HO (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nature Meth, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- (24).Gietz RD, Schiestl RH (2018) High-Efficiency Yeast Transformation Using the LiAc / SS Carrier DNA / PEG Method. Nature Prot., 2, 31–35. [DOI] [PubMed] [Google Scholar]
- (25).Mccormack AS, Godfrey KR (1998) Rule-Based Autotuning Based on Frequency Domain Identification. IEEE Transactions on Control Systems Technology, 6, 43–61. [Google Scholar]
- (26).Jung PP, Christian N, Kay DP, Skupin A, Linster CL (2015) Protocols and Programs for High-Throughput Growth and Aging Phenotyping in Yeast. PLoS ONE Epub Mar 30, 2015 DOI: 10.1371/journal.pone.0019807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sample evolution experiments in eVOLVER using the control algorithm derived from Toprak et al., 2013.
Figure S2. Oscillation of growth rate of ZZ-Y323 during the empirical determination of PID settings.
Figure S3. Adaptation history for all six replicates during PfDHFR evolution.
Figure S4. Adaptation history for all four replicates during TmHisA evolution.
Table S1. TmHisA mutants characterized.
Table S2. List of plasmids used in this study.
Table S3. List of yeast strains used in this study.


