Abstract
Mutational signatures can reveal the history of mutagenic processes that cells were exposed to before and during tumorigenesis. We expect that as-yet-undiscovered mutational processes will shed further light on mutagenesis leading to carcinogenesis. With this in mind, we analyzed the mutational spectra of 36 Asian oral squamous cell carcinomas. The mutational spectra of two samples from patients who presented with oral bacterial infections showed novel mutational signatures. One of these novel signatures, SBS_AnT, is characterized by a preponderance of thymine mutations, strong transcriptional strand bias, and enrichment for adenines in the 4 bp 5′ of mutation sites. The mutational signature described in this manuscript was shown to be caused by colibactin, a bacterial mutagen produced by E. coli carrying the pks-island. Examination of publicly available sequencing data revealed SBS_AnT in 25 tumors from several mucosal tissue types, expanding the list of tissues in which this mutational signature is observed.
Mutagenesis is one of the major causes of cancer. A thorough mapping of mutational signatures promises to illuminate the mechanisms of carcinogenesis and help identify carcinogenic mutagenic compounds and processes. In recent years, the field of mutational-signature analysis has made huge strides in identifying distinct mutational processes. Currently, 65 distinct single-base substitution (SBS) signatures have been described (Alexandrov et al. 2020). Most of these stem from defects in DNA repair and replication, endogenous mutagenic processes, or exposure to mutagenic compounds such as benzo[a]pyrene or aristolochic acid. However, the etiology of 20 mutational signatures remains unknown (Alexandrov et al. 2020).
Although the mutational signatures of most common mutational processes are known, we expect that there are additional mutational processes that contribute to small numbers of tumors. An example of such a rare signature is SBS42, owing to occupational exposure to haloalkanes (Mimaki et al. 2016; Alexandrov et al. 2020). This signature was not discovered in the original COSMIC signatures (Forbes et al. 2017) but was only discovered in cholangiocarcinomas from patients who worked at a printing company. SBS42 was extremely rare in other cancer types (Alexandrov et al. 2020). This example suggests that there are more rare mutational processes that are caused by rare occupational exposures, dietary exposures, or genetic variants affecting DNA repair or replication mechanisms. Rare mutational processes will be challenging to find, but they will point to cancers that could be prevented if the responsible mutagens can be identified and exposure to them avoided. We might expect populations that have not been intensively studied to harbor such rare mutational signatures.
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, with more than 680,000 new cases every year (Ferlay et al. 2015). With 300,000 new cases per year, oral squamous cell carcinoma (OSCC) is the largest subtype (Ferlay et al. 2015). In OSCC, nine different mutational signatures have been detected, but >92% of mutations are owing to mutational signatures associated with aging (clock-like signatures SBS1 and SBS5), APOBEC cytidine deaminases (SBS2 and SBS13), and chewing tobacco (SBS29) (Alexandrov et al. 2020). With this in mind, we analyzed the whole-exome sequencing data of 36 Asian OSCCs to search for possible rare mutational processes.
Results
Bacterial infection–associated OSCCs show novel mutational signatures
We analyzed whole-exome sequencing data from 36 OSCCs treated in Singapore, including 18 previously published OSCCs (Vettore et al. 2015). Clinical information on these tumors is included in Supplemental Table S1. These tumors had significantly fewer somatic SBSs than did the OSCCs and HNSCCs analyzed by The Cancer Genome Atlas (TCGA) consortium (median 1.02 vs. 1.66 and 2.44 mutations per megabase; P = 4.11 × 10−5 and 4.85 × 10−10, respectively, Wilcoxon rank-sum tests) (Ellrott et al. 2018; Alexandrov et al. 2020). No difference in tumor mutation burden was observed between smokers and nonsmokers. The two tumors from patients that presented with strong bacterial infection (62074759 and TC1) showed a higher mutation burden, although not statistically significant (average mutation burden of 2.6 and 1.14 mutations per megabase, respectively; P=0.078, Wilcoxon rank-sum test). Experience has shown that mutational signature assignment to tumors with extremely low numbers of mutations is unreliable. Therefore, we excluded six tumors that had fewer than 10 SBSs from further analysis. The mutational spectra of the remaining 30 tumors are shown in Supplemental Figure S1.
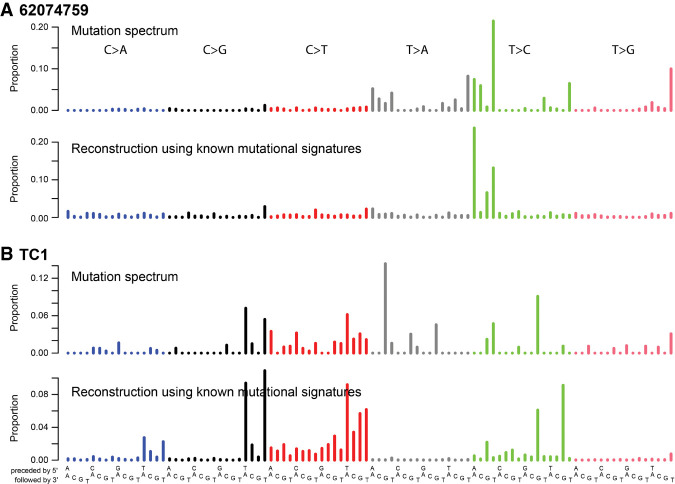
We computationally reconstructed the mutational spectra of the 30 tumors using the mutational signatures previously observed in HNSCCs and OSCCs (Supplemental Fig. S2A; Alexandrov et al. 2020). The spectra of 62074759 and TC1 were poorly reconstructed (Fig. 1A,B; Supplemental Fig. S2B). Examination of the pathology reports revealed that both 62074759 and TC1 had presented with strong oral bacterial infections, whereas none of the other 34 had mentions of bacterial infection (P = 0.0016, Fisher's exact test) (Supplemental Table S1). Both of these poorly reconstructed spectra showed unique distinctive mutation patterns. Clustering of the mutational spectra of the OSCC cohort together with the TCGA HNSCCs showed 62074759 and TC1 clustering apart, supporting these mutational spectra being distinct (Supplemental Fig. S3). This led us to hypothesize that each was caused predominantly by a single, novel, mutational process, which in the case of TC1 appeared to be combined with APOBEC-associated mutagenesis (Alexandrov et al. 2020). Both spectra showed T > A and T > C peaks with strong transcriptional strand bias but were clearly distinct.
Figure 1.
Two OSCC mutation spectra were poorly reconstructed using known mutational signatures. Mutational signature plots comparing the observed exome mutational spectra of 62074759 (A) and TC1 (B) to the corresponding reconstructed spectra.
The SBS mutational spectrum in 62074759
During routine visual inspection of the read alignments supporting the somatic variants in 62074759, we noticed that 51 out of the 84 T > C mutations were directly preceded by at least three adenines (three adenines directly 5′ of the T > C mutation). In addition, most of the TTT > TNT mutations were located within TTTT homopolymers. Because of the high risk of sequencing errors in and near homopolymers, we performed Sanger sequencing to validate 96 somatic SBSs detected in 62074759, all of which were confirmed.
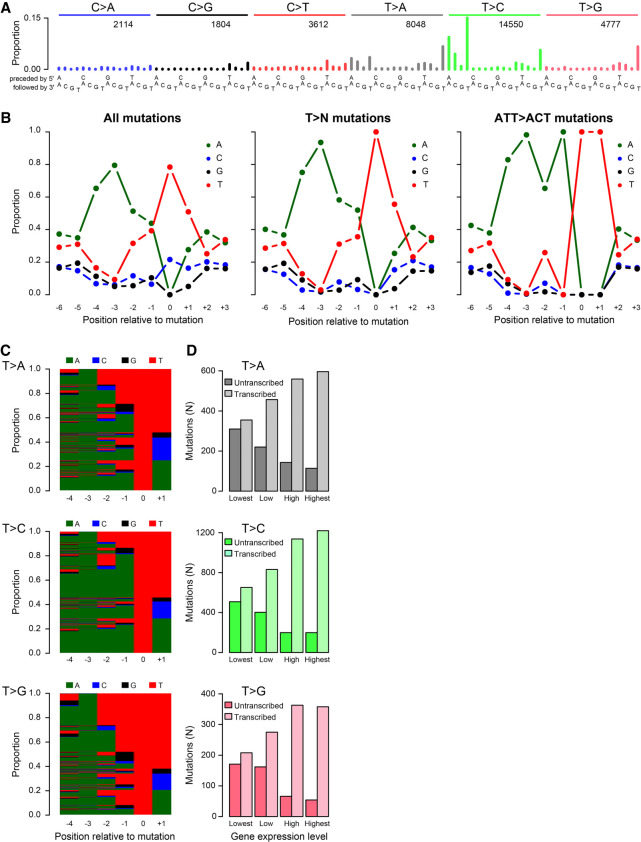
We next sequenced the whole genome of 62074759, identifying 34,905 somatic SBSs and 4037 small insertions and deletions (indels). The whole-genome SBS mutation spectrum confirmed the spectrum observed in the exome (Fig. 2A; Supplemental Fig. S1). The spectrum was dominated by AT> AA and AT> AC mutations, with a main peak at ATT > ACT, and by TTT > TAT, TTT > TCT, and TTT > TGT mutations. Similar to the exome data, the genome data showed an enrichment for adenines 5′ of T > C mutations. Among all SBSs, 79.5% had an adenine 3 bp 5′ of the mutation sites, and 65.3% had an adenine 4 bp 5′ of the mutation (Fig. 2B). Thymine mutations predominantly occurred in AAWWTW motifs, with 93.5% and 75.2% having adenines 3 bp and 4 bp 5′ of the mutation, respectively. ATT > ACT SBSs mainly occurred in AAWATT motifs, with 98.2% having an adenine 3 bp 5′ of the mutation. More broadly, we also observed strong enrichment for AAAA immediately 5′ of thymine SBSs (Fig. 2C). No enrichment of adenines 5′ of mutated cytosines was observed (Supplemental Fig. S4).
Figure 2.
In-depth characterization of SBS_AnT in the whole-genome data from tumor 62074759. (A) SBS spectrum. (B) SBS sequence context preferences, revealing strong preference for adenines 3 bp 3′ of mutated thymines. Thymine mutations predominantly occurred in AAWWTW motifs. (C) Per-mutation view of sequence context preferences of mutations from thymines. Each row represents one mutation, with bases indicated by color as in panel B. (D) Transcriptional strand bias as a function of gene expression level.
In 62074759, the mutational spectra of SBSs in trinucleotide context were essentially identical at a wide range of variant allele frequencies (VAFs) (Supplemental Fig. S5). The presence of this signature in mutations with high VAFs as well as lower VAFs suggests that the underlying mutational process continued for a considerable period of time, which included both tumor initiation and tumor expansion.
Mutational processes associated with large adducts are known to generate more mutations on the nontranscribed strands of genes than on the transcribed strands, owing to transcription-coupled nucleotide excision repair (TC-NER) of the adducts on transcribed strands (Mugal et al. 2009). Therefore, to investigate whether this novel signature might have been caused by large adducts, we examined its transcriptional strand bias. We observed very strong enrichment of mutations when thymine is on the transcribed strand (and adenine is on the nontranscribed strand), which is indicative of adduct formation on adenines. Consistent with the activity of TC-NER, the bias of T > A, T > C, and T > G mutations correlated strongly with transcriptional activity (P = 9.50 × 10−41, 6.33 × 10−91, and 5.69 × 10−33, respectively, chi-squared tests) (Fig. 2D). This, plus the absence of enrichment for adenines 5′ of cytosine mutations, suggests that the cytosine mutations in this sample were not caused by the same mutational process as the thymine mutations. In light of the preference for adenines 5′ of mutations from thymines in 62074759, we call this signature SBS_AnT.
Insertions, deletions, and dinucleotide substitutions associated with SBS_AnT
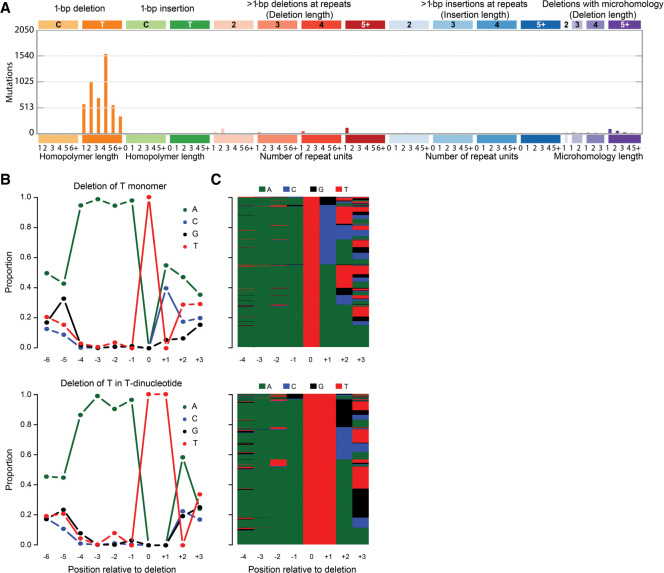
The vast majority of indels were deletions (98.6%), mainly of single thymines (Fig. 3A). The indel spectrum did not resemble any of the previously published indel signatures (Alexandrov et al. 2020). Like the SBSs, deletions of thymines in thymine mono- and dinucleotides showed strong enrichment for three preceding adenines (Fig. 3B,C). Thymine deletions in thymine tri- to octo-nucleotides had very strong enrichment for single adenines immediately 5′ of the thymine repeat, but enrichment for adenines further 5′ decreased rapidly for longer repeats (Supplemental Fig. S6). For thymine deletions outside of thymine repeats, we observed strand bias consistent with adenine adducts (P = 0.01, binomial test) (Supplemental Fig. S7). Contrastingly, thymine deletions in thymine tetranucleotides showed transcriptional strand bias in the opposite direction (P = 0.01, binomial test). Thymine deletions in thymine homopolymers of other lengths lacked transcriptional strand bias. We call this indel signature ID_AnT.
Figure 3.
In-depth characterization of the ID_AnT indel signature in the whole-genome data from tumor 62074759. (A) Spectrum of indels in the classification proposed by the PCAWG Consortium (Alexandrov et al. 2020). The indel spectrum is dominated by deletions of single thymines (orange). The numbers at the bottom indicate the lengths of the repeats in which the deletions occurred; “1” denotes deletions of thymines flanked by nonthymines, “2” denotes deletions of thymines in dinucleotides (TT > T), and so on. (B) Sequence contexts of deletions of single thymines not in a T-repeat (top) and in thymine dinucleotides (bottom). Supplemental Figure S6 provides analogous plots for thymine deletions in longer homopolymers. (C) Per-mutation view of sequence contexts of thymine deletions not in a T-repeat (top) and in a thymine dinucleotide (bottom); bases are indicated by colors as in panel B.
We detected 171 double-base substitutions (DBSs) in tumor 62074759, most of which were CC > NN and TC > NN substitutions (Supplemental Fig. S8). As the predominant mutational process in 62074759 caused almost exclusively thymine mutations, it is unlikely that these DBSs were caused by the same mutational process. The DBS spectrum best resembles the DBS spectrum observed in cisplatin exposed cell lines (cosine similarity of 0.859) (Boot et al. 2018). In concordance with cisplatin-associated mutagenesis inducing these DBSs, before the development of the tumor that we sequenced, which was a recurrence, the initial tumor had been treated with several chemotherapeutic drugs, including cisplatin.
SBS_AnT in publicly available sequencing data
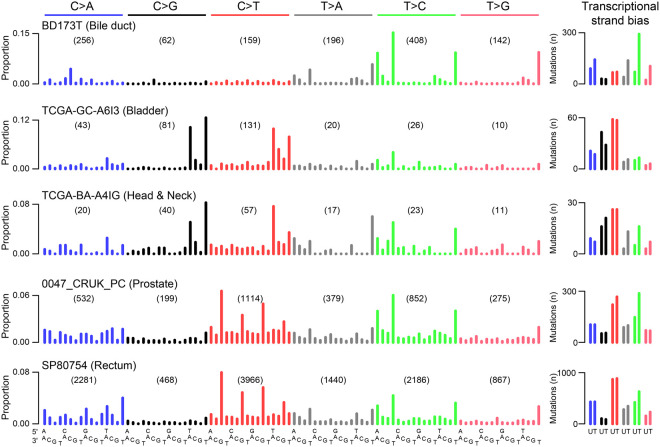
To investigate whether SBS_AnT was also present in other tumors, we investigated 4645 tumor genomes and 19,184 tumor exomes compiled for the PCAWG Mutational Signatures Working Group (Alexandrov et al. 2020). We searched for tumors that both show strong enrichment of adenines 3 and 4 bp 5′ of mutated thymines, as well as clear presence of the SBS_AnT mutational signature in the trinucleotide mutation spectrum. We found statistically significant enrichment for adenines 3 and 4 bp 5′ of thymine mutations in 39 whole-exome and 16 whole-genome samples (Supplemental Data S1). These included tumors of the bladder, colon, rectum, and prostate. Visual inspection of the mutation spectra showed POLE-associated mutagenesis (SBS10a) in 25 tumors, suggesting POLE-associated mutagenesis sometimes shows sequence context specificity similar to SBS_AnT (Supplemental Fig. S9).
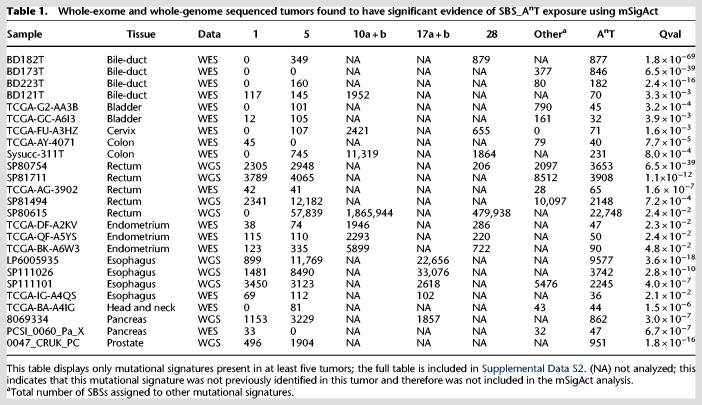
To further increase confidence in the selection of tumors showing SBS_AnT we used the mSigAct signature presence test to assess whether SBS_AnT (interpreted as a SBS signature in trinucleotide context) was needed to explain candidate observed spectra (Supplemental Data S2; Ng et al. 2017). By using the signature assignments previously reported for these tumors (Alexandrov et al. 2020), we compared reconstruction of the mutational spectra with and without SBS_AnT. We identified 25 tumors that were reconstructed significantly better when including SBS_AnT (Table 1). Example spectra of these tumors are shown in Figure 4. In these 25 tumors, we identified 53 somatic SBSs that were likely caused by SBS_AnT and that affected known oncogenes or tumor suppressor genes (Supplemental Table S2). Affected genes included TP53, PTEN, KMT2A, KMT2C, and EZH2. Among the 25 tumors with likely SBS_AnT mutations, indel information was only available for the six PCAWG whole genomes (The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium 2020). Five of these had thymine deletions with the expected sequence contexts (Supplemental Figs. S10, S11).
Table 1.
Whole-exome and whole-genome sequenced tumors found to have significant evidence of SBS_AnT exposure using mSigAct
Figure 4.
Discovery of SBS_AnT in publicly available mutation data. Example spectra of tumors positive for SBS_AnT. The right panel shows the transcriptional strand bias; for the whole-genome samples, only SBSs in transcribed regions were included. The bile duct, bladder, and HNSCC tumors are whole-exome data; the prostate and rectal tumors are whole-genome data. (U) Untranscribed strand; (T) transcribed strand.
Exploring the etiology of SBS_AnT
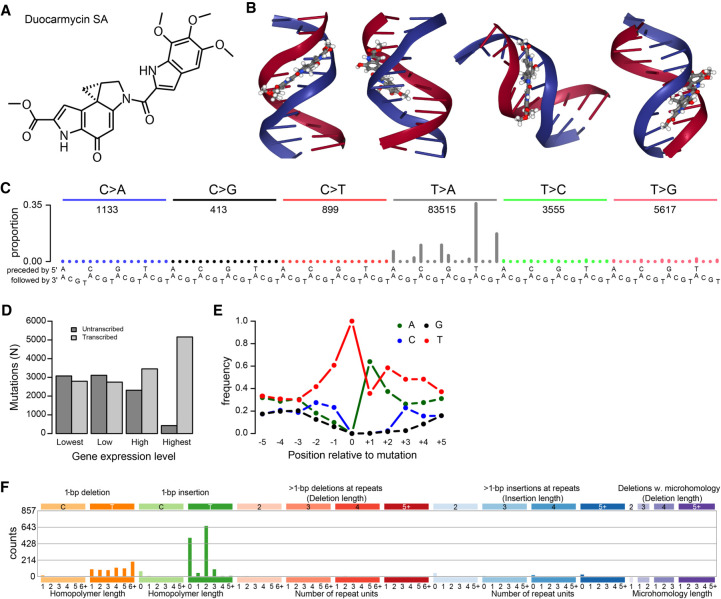
DNA repair defects can affect mutational signatures (Volkova et al. 2020). Therefore, we first checked for defects in DNA repair genes that could have transformed the appearance of a known mutational process to the mutational signature we observed. We observed MSH6 p.V878A and ATR p.L1483X substitutions (Supplemental Table S2). However, MSH6 p.V878A is predicted to be benign and ATR p.L1483X was only present at 7.4% VAF and therefore could not have accounted for the vast majority of SBS_AnT mutations that had higher VAFs. We therefore concluded that these variants did not play a role in shaping the SBS_AnT mutational signature. Moreover, none of the other 25 SBS_AnT-positive tumors showed mutations in these genes, nor did we observe any other recurrently affected DNA repair genes in these tumors (Supplemental Table S2). We next sought to identify the etiology of SBS_AnT. The enrichment of mutations of T > A on the transcribed strand is indicative of a large molecule that adducts on adenines. Additionally, it is also expected to be an exceptionally large adduct, large enough to reach to 4 bp 5′ of the mutated site. Through literature study, we identified a class of minor-groove binding compounds called duocarmycins, which are produced by several species of Streptomyces, a common class of bacteria that are known human symbionts (Hurley and Rokem 1983; Ichimura et al. 1991; Seipke et al. 2012). The molecular structure of duocarmycin SA (duoSA), a naturally occurring duocarmycin, is shown in Figure 5A. Figure 5B shows duoSA intercalated in the minor groove of the DNA helix (source: PDB ID: 1DSM) (Smith et al. 2000; Rose et al. 2018). Duocarmycins bind specifically to adenines in A/T-rich regions, which matches SBS_AnT's sequence context (Reynolds et al. 1985; Baraldi et al. 1999; Woynarowski 2002).
Figure 5.
Mutational signature of duocarmycin SA (duoSA). (A) duoSA, one of the naturally occurring duocarmycins. (B) Several views of the conformation of duoSA intercalated with DNA (source: PDB ID: 1DSM) (Smith et al. 2000; Rose et al. 2018). Duocarmycins slot into the minor groove of the DNA helix. (C) SBS mutation spectrum of one of the duoSA-treated HepG2 clones. (D) Transcriptional strand bias of T > A mutations induced by duoSA as a function of gene expression. (E) Extended sequence context specificity of T > A mutations induced by duoSA. (F) Indel spectrum of one of the duoSA-treated HepG2 clones.
To investigate whether duocarmycins could be causing SBS_AnT, we sequenced four duoSA-exposed HepG2 clones. The mutational spectra of duoSA-exposed HepG2 clones are shown in Figure 5C and Supplemental Figure S12. The mutational signature of duoSA is characterized by strong peaks of T > A mutations with always either an adenine directly 3′ or a thymine directly 5′ of the mutation site. DuoSA-associated mutagenesis showed strong transcriptional strand bias and showed extended sequence context preference of thymines 3′ of mutated thymines (Fig. 5D,E). Indels caused by duoSA treatment mainly comprised indels of single thymines (Fig. 5F; Supplemental Fig. S13). Insertions were mainly found either not next to thymines or in 2-bp thymine repeats (TT > TTT). Deletions occurred in any length of thymine repeats. In additional, we also observed TA > AT, CT > AA, and TT > AA DBSs in all clones (Supplemental Fig. S14). From these results, we concluded that SBS_AnT is not caused by duoSA. While this manuscript was under review, the mutational signature of colibactin was published (Pleguezuelos-Manzano et al. 2020). Colibactin is a mutagen produced by Escherichia coli carrying the pks-island. The SBS_AnT and ID_AnT signatures reported here are similar to the SBS and ID signatures caused by colibactin, including the extended sequence context.
Characterization of the mutational signature in TC1
We also sequenced the whole genome of TC1, identifying 5402 SBSs and 67 indels. Besides APOBEC-associated mutations, we observed prominent TG > AG peaks and a strong GTG > GCG peak, all with strong transcriptional strand bias (Supplemental Fig. S15). No extended sequence context preference was observed (Supplemental Fig. S15E). As only the signature of SBS mutations in trinucleotide context was distinctive, we screened for cosine similarity between the thymine (T > N) mutations and screened for T > A mutations specifically for all 23,829 tumors. We found no tumors in which presence of the TC1 mutational signature was visible in the mutation spectrum (Supplemental Fig. S16).
Identification of bacteria causing SBS_AnT and the TC1 mutational spectrum
To identify the bacterial species associated with SBS_AnT and the mutational spectrum observed in TC1, we extracted all reads from the WGS data that did not align to the human genome and mapped them to bacterial reference genomes. Fewer than 0.1% of reads from both normal samples as well as tumor 62074759 were nonhuman, opposed to 1.5% from tumor TC1. Of the nonhuman reads, only a small proportion aligned to any of the bacterial genomes (Supplemental Fig. S17). In the sequencing data of the adjacent normal tissue of 62074759, we observed a small proportion of reads aligning to the E. coli genome. By focusing on tumor TC1, we identified several genera of bacteria, including Lachnoanaerobaculum, Prevotella, Anaerococcus, and Streptococcus (Supplemental Fig. S17). All these bacterial genera are common oral symbionts (Downes et al. 2008; Labutti et al. 2009; Hedberg et al. 2012; Abranches et al. 2018). Because of the rareness of the mutational signatures discovered in this study, it is unlikely that such common oral bacteria would be causal. To explore whether other microorganisms (such as fungi) could be present, we also performed a nucleotide-BLAST on some of the nonhuman reads from all samples, but no high-confidence alignments were found.
Discussion
We analyzed the mutational signatures of 36 Asian OSCCs, hypothesizing that there were still rare mutational processes to be discovered. We identified two novel mutational signatures. These two OSCCs were also the only tumors from our cohort of OSCCs with pathology reports that mentioned high levels of bacterial infection. The rarity of these signatures was illustrated by the fact that we only found 25 additional tumors with SBS_AnT and no additional tumors with the TC1 signature after examining a total of 23,829 tumors. In tumors from tissue types in which we discovered SBS_AnT, only 0.4% showed SBS_AnT. All tumors in which SBS_AnT was detected were from mucosal tissues that harbor bacterial symbionts or that are in direct contact with tissues that harbor symbionts.
Since the initial publication of this manuscript on bioRxiv, SBS_AnT has also been reported in normal colonic crypts from healthy individuals (Lee-Six 2019). SBS_AnT was found to be predominantly active early in life, and different patterns of SBS_AnT activity distribution over the colon were observed. These results fit with the hypothesis that bacterial compounds could be causing this signature. “Patchy” exposure patterns are unlikely if there had been dietary or occupational exposure to chemicals, and occupational exposure is also improbable because SBS_AnT was found to be mostly active early in life. We postulate that early in life, while the microbiome is still being established, bacterial infections might have occurred in these patients. Later in life, microbiome homeostasis may have been established, preventing SBS_AnT mutagenesis later in life. For patient 62074759, we propose that the unusual initial treatment of the OSCC before surgery, which included three kinds of chemotherapy and radiotherapy, could have opened a window for bacterial infection after the oral microbiome had been disrupted by the treatments. The tumor sample we sequenced was a recurrence 9 mo after treatment. We can exclude the possibility of the treatments causing SBS_AnT, as the mutational signatures associated with 5-fluorouracil, cisplatin, and radiotherapy have already been published, and gemcitabine, a cytosine analog, would be unlikely to cause thymine mutations (Sherborne et al. 2015; Boot et al. 2018; Christensen et al. 2019).
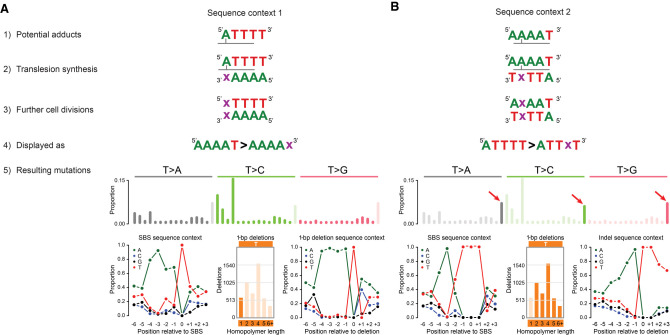
SBS_AnT shows strong transcriptional strand bias, which is commonly observed for mutational processes associated with bulky adducts (Huang et al. 2017; Ng et al. 2017; Boot et al. 2018). The depletion of adenine mutations on the transcribed strand (which corresponds to depletion of thymine mutations on the untranscribed strand) suggests that the mutational process causing SBS_AnT involves formation of a bulky adduct on adenine. Figure 6, A and B, shows a proposed model for adduct formation leading to SBS_AnT. The model assumes two independent adducts are formed, either directly adjacent to thymine homopolymers (Fig. 6A) or inside adenine homopolymers (Fig. 6B). We propose that adducts inside adenine homopolymers lead to T > A, T > C, and T > G mutations in a TTT context as well as deletions of single thymines in thymine homopolymers. Conversely, adducts on adenines directly adjacent to thymine homopolymers would lead to T > A and T > C mutations in the AAAAT context as well as deletions of single thymines not in homopolymers. The sequences for the adducts in the model are the reverse complement of each other, and we cannot exclude the possibility of an inter-strand crosslink. However, if this were the case, we would expect to also observe multiple pairs of SBSs separated by two unaffected bases, which we did not.
Figure 6.
Proposed model for adduct formation leading to the mutation patterns of SBS_AnT and ID_AnT. (A) Potential adduct 1. (1) Adducts are formed on adenines directly 5′ of thymine homopolymers. (2) During translesion synthesis, an incorrect nucleotide (x) is incorporated opposite the adducted adenine. (3) During further cell divisions, the mutation is maintained. (4) Following the conventions of the mutational signature field, we display mutations as occurring from the pyrimidine of the Watson–Crick base pair. (5) Potential adduct 1 would lead to SBSs directly adjacent to TTT trinucleotides and deletions of single thymines not in thymine homopolymers. Both SBSs and deletions resulting from potential adduct 1 would be enriched for adenines up to 4 bp 5′ of the mutation site. (B) Potential adduct 2. (1) Adducts are formed in adenine homopolymers with a thymine directly 3′. (2) During translesion synthesis, an incorrect nucleotide (x) is incorporated opposite the adducted adenine. (3) During further cell divisions, the mutation is maintained. (4) Following the conventions of the mutational signature field, we display mutations as occurring from the pyrimidine of the Watson–Crick base pair. (5) Potential adduct 2 would lead to SBSs inside TTT trinucleotides and deletions of single thymines inside thymine homopolymers. SBSs resulting from potential adduct 2 would be strongly enriched for adenines 3 bp 5′ of the mutation site. Deletions resulting from potential adduct 2 would be strongly enriched for adenines up to 4 bp 5′ of the mutation site. The latter is owing to the possible different locations of the adduct inside the homopolymers. We believe that for longer homopolymers (more than three thymines), the adduct will nearly always be situated opposite the third thymine, making the −3 position (relative to the adduct) the −1 position relative to the thymine homopolymers. This explains the ∼100% presence of adenines directly 5′ of the thymine homopolymers.
Based on literature research for compounds that could induce mutagenesis with the characteristics of SBS_AnT, we experimentally established the mutational signature of duoSA. DuoSA is a naturally occurring minor-groove binding DNA alkylating agent produced by a subset of Streptomyces species. As reported, duoSA was highly mutagenic, causing T > A transversions in A/T-rich regions (Woynarowski 2002). However, the mutational spectrum was clearly distinct from that of SBS_AnT. In addition, in contrast to SBS_AnT, duoSA-associated mutagenesis showed sequence context enrichment 3′ of the mutated site. Indels induced by duoSA were dominated by insertions of thymines, whereas SBS_AnT showed exclusively thymine deletions. We concluded that SBS_AnT was not caused by duoSA. After the initial submission of our manuscript, a study of a large set of metastatic solid tumors was published that detected the mutational signature of duoSA in two patients (Priestley et al. 2019). These patients had been treated with SYD985, a duocarmycin-based antibody-drug conjugate.
To identify the bacteria associated with SBS_AnT and the mutational spectrum observed in TC1, we examined the whole-genome sequencing data for reads that map to bacterial genomes. The TC1 tumor data had a very high number of nonhuman sequencing reads. However, alignment to a set of 209 bacterial genomes failed to identify the bacteria associated with the mutational spectrum observed in TC1. Possibly a different genus of bacteria is present in this patient, for which the reference genome sequence is yet to be elucidated. In the 62074759 tumor data, we also observed a low number of reads aligning to the same genera of bacteria also observed in tumor TC1. The absence of large numbers of nonhuman reads in tumor 62074759 is likely because of sampling; if the DNA we sequenced was from the center of the tumor mass opposed to the edge, less contamination would be expected. Ideally, we would have tested the saliva of patients 62074759 and TC1 to identify the bacteria that cause SBS_AnT and the mutational spectrum observed in TC1, but no saliva samples were stored for these patients. After learning that SBS_AnT is caused by colibactin, we checked specifically for reads from 62074759 that might align to the E. coli genome. A very small proportion of reads in the normal tissue indeed aligned to the E. coli genome, which we had not originally noted, but no reads aligned to the pks-island. As we do not have saliva samples from this patient, we cannot determine whether E. coli in this patient's saliva carried the pks-island. We also have no samples that would let us determine if pks + E. coli might have been present for some period of time before surgery.
While this manuscript was under review, experimental data were published confirming that SBS_AnT is caused by colibactin (Pleguezuelos-Manzano et al. 2020). The similarities between the colibactin mutational signature and SBS_AnT leave little doubt that these reflect the same mutational process. Beyond the clear similarities of the 96-channel mutational signature together with strong transcriptional strand bias, the colibactin mutational signature also reports enrichment of adenines 5′ of mutated thymines. Additionally, the indels reported with both signatures are also highly similar. In particular, we point to the similarity between the Supplementary Figure 4 by Pleguezuelos-Manzano et al. (2020) and our Supplemental Figures S6 and S11.
Bacteria have long been known to be associated with cancer. However, for most associations, such as the association between Salmonella and gallbladder and colon cancer and the association between Chlamydia and cervical carcinoma, only epidemiological evidence exists (van Elsland and Neefjes 2018). The only bacterium for which experimental evidence exists that it causes cancer is Heliobacter pylori, which has been shown to cause gastric cancer in gerbils (Watanabe et al. 1998). H. pylori, as well as most other cancer-associated bacteria, is thought to stimulate carcinogenesis through the inflammation associated with the infection (van Elsland and Neefjes 2018). However, some bacteria have been reported to produce toxins able to induce double-strand DNA breaks (van Elsland and Neefjes 2018). For OSCC, the association with bacterial infection is well known, but no mutagenic compounds have been reported to be produced by these bacteria (Karpinski 2019).
Because mutations from ATN > ACN are prominent in both SBS_AnT and Signature 16, which is associated with ethanol exposure in several cancer types, we considered whether ethanol might also contribute to SBS_AnT, possibly via bacteria that metabolize ethanol to acetaldehyde (Yokoi et al. 2015; Letouzé et al. 2017; Li et al. 2018; Tagaino et al. 2019). However, the resemblance between SBS_AnT and Signature 16 is superficial: Signature 16 lacks the extended sequence context and the associated deletion signature of SBS_AnT. Furthermore, we are not aware of any evidence that acetaldehyde elevates T:A > C:G mutations. Thus, it seems highly unlikely that ethanol, either directly or via bacterially metabolized acetaldehyde, contributes to SBS_AnT. This is especially the case given the strong evidence linking SBS_AnT to colibactin.
In summary, we identified two novel mutational signatures in Asian OSCCs that had presented with strong oral bacterial infections. In the other 34 Asian OSCCs, of which none had presented with strong bacterial infections, no novel mutational signatures were discovered. While our manuscript was in revision, a preprint was released that described the sequence context specificity of double-strand breaks induced by the bacterial toxin colibactin (Dziubańska-Kusibab et al. 2020). Colibactin adduct–induced double-strand breaks were strongly enriched in AT-rich regions, with the AAWWTT motif to be the most enriched at colibactin-induced double-strand breaks, which fits exactly with the sequence context specificity we observed for the thymine mutations in 62074759 as shown in Figure 2B, panel 2, positions −4 to +1 relative to the mutation site. Subsequently, experimental evidence was published confirming that SBS_AnT is caused by colibactin (Pleguezuelos-Manzano et al. 2020). Our pan-cancer analysis identified several additional tumor types in which SBS_AnT is observed that were not previously detected. Additionally, owing to the high load of SBS_AnT mutations in 62074759, we were able to perform more precise quantification of the sequence context specificity of this mutagenic process, especially with respect of the sequence context specificity of deletions of single thymines.
Methods
Samples
Deidentified fresh-frozen tissue samples and matching whole blood were collected from OSCC patients operated on between 2012 and 2016 at the National Cancer Centre Singapore. In accordance with the Helsinki Declaration of 1975, written consent for research use of clinical material and clinicopathologic data was obtained at the time of surgery. This study was approved by the SingHealth Centralized Institutional Review Board (CIRB 2007/438/B).
Whole-exome and whole-genome sequencing
Whole-exome sequencing was performed at Novogene on a HiSeq X Ten instrument with 150-bp paired-end reads. Whole-genome sequencing was performed at BGI (Hong Kong) on the BGIseq500 platform, generating 100-bp paired-end reads.
Alignment and variant calling
Sequencing reads were trimmed by Trimmomatic (Bolger et al. 2014). Alignment and variant calling and filtering were performed as described previously (Boot et al. 2018). Reads were aligned to GRCh37. We are confident that mapping reads to GRCh38 would not alter the conclusions of this manuscript, as the trinucleotide frequencies are essentially the same in GRCh37 and GRCh38, and the analysis presented does not depend on any particular regions of the human genome or genome annotations that were updated between GRCh37 and GRCh38. Annotation of somatic variants was performed using ANNOVAR (Wang et al. 2010). Sequencing reads that did not align to the human genome were subsequently aligned to 209 bacterial reference genomes from Ensembl (ftp://ftp.ensemblgenomes.org/pub/bacteria/release-35/fasta/bacteria_183_collection/). To detect the presence of reads aligning to the pks-island (AM229678.1), sequencing data were aligned to the human reference genome with the pks-island sequence (AM229678.1) added as a separate contig. For driver gene analysis, only variants inside Tier 1 genes of the cancer gene census were considered (Sondka et al. 2018).
Validation of SBSs by Sanger sequencing
We performed Sanger sequencing to validate 96 variants detected in the whole-exome sequencing of sample 62074759. We selected variants with >15% allele frequency to avoid variants below the detection limit of Sanger sequencing and excluded variants immediately adjacent to a homopolymer of ≥9 bp. PCR product purification and Sanger sequencing were performed at GENEWIZ.
Signature assignment
We assigned mutational signatures to the mutational spectra of the 30 OSCCs with 10 or more mutations using SigProfiler and the SigProfiler reference mutational signatures (Alexandrov et al. 2020). As OSCC is a subset of HNSCC, all mutational signatures that were identified in HNSCCs and OSCCs in the International Cancer Genome Consortium's Pan Cancer Analysis Working Group (PCAWG) analysis were included for reconstruction (Alexandrov et al. 2020). As the PCAWG mutational signatures are based on the trinucleotide abundance of the human genome, when analyzing whole-exome sequencing data, we adjusted to the mutational signatures for exome trinucleotide frequency.
Gene expression data
Single-cell gene expression data for OSCC were downloaded from NCBI GSE103322 (Puram et al. 2017). We took the median gene expression for all tumor cells as the representative expression level of OSCCs.
Identification of additional tumors with the signature in 62074759
Previously compiled whole-exome (N = 19,184) and whole-genome (N = 4645) sequencing data were screened for presence of the signature in 62074759 (Alexandrov et al. 2020). This included 2780 whole genomes from the Pan Cancer of Whole Genomes Consortium (The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium 2020) and 9493 whole exomes from the TCGA Consortium (Ellrott et al. 2018). We examined tumors with 50 or more (exomes) or with 500 or more (genomes) thymine mutations to identify enrichment for mutations with the 5′ sequence context characteristic of the signature in 62074759 (Supplemental Data S1).
We then used the mSigAct signature presence test to test for the signature in 62074759 among the candidate tumors identified in the previous step (Supplemental Data S2; Ng et al. 2017; Boot et al. 2018). This test provides a P-value for the null hypothesis that a signature is not needed to explain an observed spectrum compared with the alternative hypothesis that the signature is needed.
In vitro duoSA exposure
Exposure of HepG2 cells to duoSA was performed as described previously (Boot et al. 2018). In short, HepG2 cells were exposed to 100 pM and 250 pM duoSA for 2 mo followed by single-cell cloning. For each concentration, two clones were whole-genome sequenced. duoSA (CAS 130288-24-3) was obtained from BOC Sciences.
Data access
Sanger sequencing results validating 96 variants observed in tumor 62074759 are included in Supplemental Data S3. The sequencing FASTQ files generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/) under accession number ERP116345 (duoSA-treated HepG2 clones) and the European Genome-phenome Archive (EGA; https://ega-archive.org/) under accession number EGAS00001003131 (patient data).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The results here are partly based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and data assembled by the International Cancer Genome Consortium Pan Cancer Analysis Working Groups. This study was funded by the National Medical Research Council, NMRC/CIRG/1422/2015 to S.G.R.
Author contributions: A.B. and S.G.R. designed the study, drafted the manuscript, and prepared figures. A.B., A.W.T.N., and W.Y. performed bioinformatics analyses. S.-C.H. performed cell line experiments. F.T.C., D.S.W.T., and N.G.I. contributed materials. All authors read and approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255620.119.
References
- Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral Streptococci. Microbiol Spectr 6 10.1128/microbiolspec.GPP3-0042-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578: 94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraldi PG, Cacciari B, Guiotto A, Romagnoli R, Zaid AN, Spalluto G. 1999. DNA minor-groove binders: results and design of new antitumor agents. Farmaco 54: 15–25. 10.1016/S0014-827X(98)00102-5 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot A, Huang MN, Ng AWT, Ho SC, Lim JQ, Kawakami Y, Chayama K, Teh BT, Nakagawa H, Rozen SG. 2018. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res 28: 654–665. 10.1101/gr.230219.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Van der Roest B, Besselink N, Janssen R, Boymans S, Martens JWM, Yaspo ML, Priestley P, Kuijk E, Cuppen E, et al. 2019. 5-Fluorouracil treatment induces characteristic T > G mutations in human cancer. Nat Commun 10: 4571 10.1038/s41467-019-12594-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes J, Hooper SJ, Wilson MJ, Wade WG. 2008. Prevotella histicola sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 58: 1788–1791. 10.1099/ijs.0.65656-0 [DOI] [PubMed] [Google Scholar]
- Dziubańska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, Katainen R, Cajuso T, Crosetto N, Orozco M, Aaltonen LA, et al. 2020. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med 10.1038/s41591-020-0908-2 [DOI] [PubMed] [Google Scholar]
- Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, et al. 2018. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 6: 271–281.e7. 10.1016/j.cels.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg ME, Moore ER, Svensson-Stadler L, Hörstedt P, Baranov V, Hernell O, Wai SN, Hammarström S, Hammarström ML. 2012. Lachnoanaerobaculum gen. nov., a new genus in the Lachnospiraceae: characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from the human small intestine, and Lachnoanaerobaculum orale sp. nov., isolated from saliva, and reclassification of Eubacterium saburreum (Prévot 1966) Holdeman and Moore 1970 as Lachnoanaerobaculum saburreum comb. nov. Int J Syst Evol Microbiol 62: 2685–2690. 10.1099/ijs.0.033613-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MN, Yu W, Teoh WW, Ardin M, Jusakul A, Ng AWT, Boot A, Abedi-Ardekani B, Villar S, Myint SS, et al. 2017. Genome-scale mutational signatures of aflatoxin in cells, mice, and human tumors. Genome Res 27: 1475–1486. 10.1101/gr.220038.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley LH, Rokem JS. 1983. Biosynthesis of the antitumor antibiotic CC-1065 by Streptomyces zelensis. J Antibiot (Tokyo) 36: 383–390. 10.7164/antibiotics.36.383 [DOI] [PubMed] [Google Scholar]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. 2020. Pan-cancer analysis of whole genomes. Nature 578: 82–93. 10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura M, Ogawa T, Katsumata S, Takahashi K, Takahashi I, Nakano H. 1991. Duocarmycins, new antitumor antibiotics produced by Streptomyces; producing organisms and improved production. J Antibiot (Tokyo) 44: 1045–1053. 10.7164/antibiotics.44.1045 [DOI] [PubMed] [Google Scholar]
- Karpiński TM. 2019. Role of oral microbiota in cancer development. Microorganisms 7: 20 10.3390/microorganisms7010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labutti K, Pukall R, Steenblock K, Glavina Del Rio T, Tice H, Copeland A, Cheng JF, Lucas S, Chen F, Nolan M, et al. 2009. Complete genome sequence of Anaerococcus prevotii type strain (PC1T). Stand Genomic Sci 1: 159–165. 10.4056/sigs.24194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Six H. 2019. “Somatic evolution in human blood and colon.” PhD thesis, University of Cambridge, Cambridge. [Google Scholar]
- Letouzé E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J, et al. 2017. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 8: 1315 10.1038/s41467-017-01358-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Wang MY, Yang M, Dai HJ, Zhang BF, Wang W, Chu XL, Wang X, Zheng H, Niu RF, et al. 2018. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann Oncol 29: 938–944. 10.1093/annonc/mdy011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimaki S, Totsuka Y, Suzuki Y, Nakai C, Goto M, Kojima M, Arakawa H, Takemura S, Tanaka S, Marubashi S, et al. 2016. Hypermutation and unique mutational signatures of occupational cholangiocarcinoma in printing workers exposed to haloalkanes. Carcinogenesis 37: 817–826. 10.1093/carcin/bgw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugal CF, von Grunberg HH, Peifer M. 2009. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol Biol Evol 26: 131–142. 10.1093/molbev/msn245 [DOI] [PubMed] [Google Scholar]
- Ng AWT, Poon SL, Huang MN, Lim JQ, Boot A, Yu W, Suzuki Y, Thangaraju S, Ng CCY, Tan P, et al. 2017. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med 9: eaan6446 10.1126/scitranslmed.aan6446 [DOI] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C, Puschhof J, Huber AR, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, et al. 2020. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580: 269–273. 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A, Onstenk W, et al. 2019. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575: 210–216. 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171: 1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds VL, Molineux IJ, Kaplan DJ, Swenson DH, Hurley LH. 1985. Reaction of the antitumor antibiotic CC-1065 with DNA: location of the site of thermally induced strand breakage and analysis of DNA sequence specificity. Biochemistry 24: 6228–6237. 10.1021/bi00343a029 [DOI] [PubMed] [Google Scholar]
- Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlić A, Rose PW. 2018. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 34: 3755–3758. 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke RF, Kaltenpoth M, Hutchings MI. 2012. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev 36: 862–876. 10.1111/j.1574-6976.2011.00313.x [DOI] [PubMed] [Google Scholar]
- Sherborne AL, Davidson PR, Yu K, Nakamura AO, Rashid M, Nakamura JL. 2015. Mutational analysis of ionizing radiation induced neoplasms. Cell Rep 12: 1915–1926. 10.1016/j.celrep.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Bifulco G, Case DA, Boger DL, Gomez-Paloma L, Chazin WJ. 2000. The structural basis for in situ activation of DNA alkylation by duocarmycin SA. J Mol Biol 300: 1195–1204. 10.1006/jmbi.2000.3887 [DOI] [PubMed] [Google Scholar]
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. 2018. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 18: 696–705. 10.1038/s41568-018-0060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaino R, Washio J, Abiko Y, Tanda N, Sasaki K, Takahashi N. 2019. Metabolic property of acetaldehyde production from ethanol and glucose by oral Streptococcus and Neisseria. Sci Rep 9: 10446 10.1038/s41598-019-46790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsland D, Neefjes J. 2018. Bacterial infections and cancer. EMBO Rep 19: e46632 10.15252/embr.201846632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore AL, Ramnarayanan K, Poore G, Lim K, Ong CK, Huang KK, Leong HS, Chong FT, Lim TK, Lim WK, et al. 2015. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med 7: 98 10.1186/s13073-015-0219-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova NV, Meier B, González-Huici V, Bertolini S, Gonzalez S, Vöhringer H, Abascal F, Martincorena I, Campbell PJ, Gartner A, et al. 2020. Mutational signatures are jointly shaped by DNA damage and repair. Nat Commun 11: 2169 10.1038/s41467-020-15912-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. 1998. Helicobacter pylori infection induces gastric cancer in Mongolian gerbils. Gastroenterology 115: 642–648. 10.1016/S0016-5085(98)70143-X [DOI] [PubMed] [Google Scholar]
- Woynarowski JM. 2002. Targeting critical regions in genomic DNA with AT-specific anticancer drugs. Biochim Biophys Acta 1587: 300–308. 10.1016/S0925-4439(02)00093-5 [DOI] [PubMed] [Google Scholar]
- Yokoi A, Maruyama T, Yamanaka R, Ekuni D, Tomofuji T, Kashiwazaki H, Yamazaki Y, Morita M. 2015. Relationship between acetaldehyde concentration in mouth air and tongue coating volume. J Appl Oral Sci 23: 64–70. 10.1590/1678-775720140223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.