Abstract
Little is known about how human Y-Chromosome gene expression directly contributes to differences between XX (female) and XY (male) individuals in nonreproductive tissues. Here, we analyzed quantitative profiles of Y-Chromosome gene expression across 36 human tissues from hundreds of individuals. Although it is often said that Y-Chromosome genes are lowly expressed outside the testis, we report many instances of elevated Y-Chromosome gene expression in a nonreproductive tissue. A notable example is EIF1AY, which encodes eukaryotic translation initiation factor 1A Y-linked, together with its X-linked homolog EIF1AX. Evolutionary loss of a Y-linked microRNA target site enabled up-regulation of EIF1AY, but not of EIF1AX, in the heart. Consequently, this essential translation initiation factor is nearly twice as abundant in male as in female heart tissue at the protein level. Divergence between the X and Y Chromosomes in regulatory sequence can therefore lead to tissue-specific Y-Chromosome-driven sex biases in expression of critical, dosage-sensitive regulatory genes.
A wide range of diseases, collectively affecting all organ systems, manifest differentially in human males and females (Wizemann and Pardue 2001). The molecular mechanisms responsible for these differences remain poorly characterized. It was once assumed that all such differences were the products of circulating hormones (e.g., androgens, estrogens), but they are increasingly speculated to stem in part from the direct effects of sex-chromosome genes expressed in tissues throughout the body (Arnold 2012). With regard to the sex chromosomes, most attention has been paid to the X Chromosome, particularly those X-Chromosome genes that are expressed more highly in XX (female) than in XY (male) individuals because they escape X-Chromosome inactivation in XX cells (Deng et al. 2014; Tukiainen et al. 2017). Researchers often cite the Y Chromosome's paucity of genes and those genes’ presumed specialization for reproduction as reasons to look past the Y Chromosome, if it is considered at all. But recent studies indicate that the Y Chromosome retains conserved, dosage-sensitive regulatory genes expressed in tissues throughout the body (Bellott et al. 2014), which might underlie newly found associations between the Y Chromosome and disease (Tartaglia et al. 2012; Cannon-Albright et al. 2014; Eales et al. 2019).
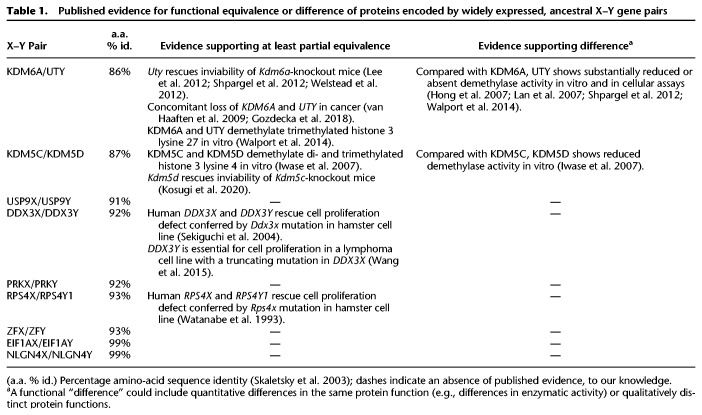
To better understand how Y-Chromosome genes might contribute to differences between XX and XY individuals, we sought to obtain a quantitative understanding of Y-Chromosome gene expression across the human body. We excluded Y-Chromosome genes in the two pseudoautosomal regions, where the X and Y Chromosomes are identical in sequence, and instead focused on genes in the Y Chromosome's male-specific region (MSY) (Fig. 1A; Supplemental Table S1; Skaletsky et al. 2003). For our purposes, it was useful to distinguish two groups of MSY genes—those that have similar but nonidentical homologs on the X Chromosome and those that do not. MSY genes without X homologs are the products of transposition or retrotransposition events that brought copies of autosomal genes to the MSY at various points during mammalian evolution (Saxena et al. 1996; Lahn and Page 1999b; Skaletsky et al. 2003). Because these MSY genes have no counterparts on the X, they could confer differences to XX and XY individuals in any tissue where they are robustly expressed. A different set of considerations pertains to the MSY genes with X homologs, most of which are remnants of the ancestral pair of autosomes from which the mammalian sex chromosomes evolved, and have survived millions of years of Y-Chromosome decay (Lahn and Page 1999a; Ross et al. 2005). Previous studies suggest that the X- and Y-linked members of these homologous X–Y gene pairs encode proteins that are at least partially equivalent in function (Table 1). Nevertheless, up- or down-regulated expression of the MSY gene in a particular tissue might lead to a quantitative difference between XX and XY individuals in the expression level of the X–Y gene pair overall. Because ancestral MSY genes with X homologs encode highly dosage-sensitive regulators of transcription, translation, and protein stability (Bellott et al. 2014; Naqvi et al. 2018), even small sex biases in expression could have cascading effects on genes across the genome.
Figure 1.
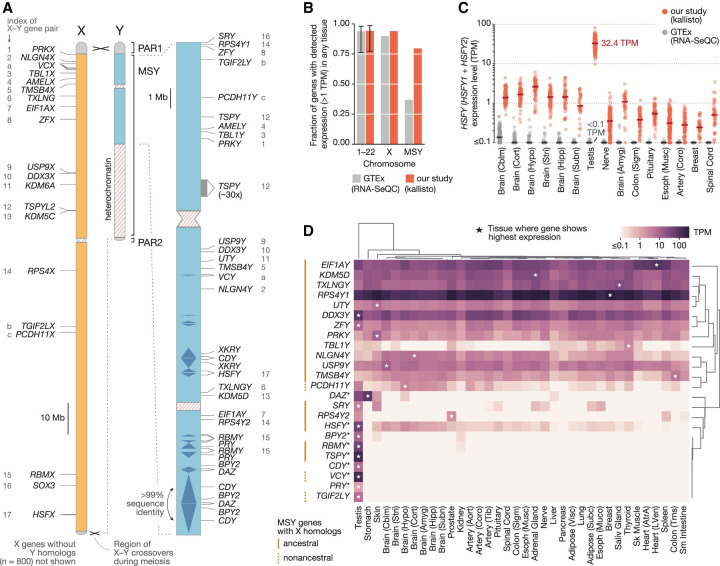
Estimates of MSY gene expression across 36 human tissues. (A) Outside of the two pseudoautosomal regions (PAR1, PAR2), the X and Y Chromosomes have diverged in sequence. The locations of protein-coding genes and multicopy gene families in the male-specific region of the human Y Chromosome (MSY; blue) are shown at right. The X-linked homologs of MSY genes are annotated in the nonpseudoautosomal region of the X (orange); numbers (ancestral X–Y pairs) and letters (acquired X–Y pairs) match MSY genes to their X-linked homologs. (B) Fraction genes on autosomal chromosomes (1–22), the X Chromosome, or the MSY expressed above 1 TPM in at least one tissue when multimapping RNA-seq reads are discarded (gray) or included (red). Error bars, minimum and maximum values among individual autosomes. (C) Each point shows estimated expression level of the HSFY gene family in a single sample when multimapping reads are included (red) and discarded (gray). Lines show median expression levels. The 15 tissues shown are those with the highest median expression level after discarding multimapping reads, in descending order. (D) Median expression levels of MSY genes and gene families (*) in each tissue, with row and column order determined by hierarchical clustering. Asterisks denote the tissue with the highest expression for a given gene.
Table 1.
Published evidence for functional equivalence or difference of proteins encoded by widely expressed, ancestral X–Y gene pairs
The current understanding of MSY gene expression is based on limited observations from humans and other mammals. Previous studies included only a few tissue types while using small sample sizes or suboptimal methodologies for analyzing MSY gene expression quantitatively. These studies established that some MSY genes show testis-specific expression whereas others are expressed widely across the body, but they could not detect more subtle quantitative differences in MSY gene expression between tissues (Lahn and Page 1997; Skaletsky et al. 2003; Bellott et al. 2014; Cortez et al. 2014). Other studies have found that MSY genes show lower expression levels than their corresponding X-linked homologs (Xu et al. 2002, 2008a,b; Johnston et al. 2008; Trabzuni et al. 2013; Cortez et al. 2014; Johansson et al. 2016). However, most such studies focused on small numbers of MSY genes or tissues or on nonhuman mammals. This has made it difficult to discern a consistent quantitative picture of MSY gene expression and its bearing on sex differences in humans. These efforts have been further complicated by complexities of the MSY's sequence. Homology with the X Chromosome and an abundance of complex segmental duplications pose various challenges for accurately measuring the expression of MSY genes at the transcript level. Even less is known about the expression of MSY genes at the protein level owing in large part to the difficulty of obtaining reagents that can distinguish X- and Y-encoded amino-acid sequences. We therefore set out to conduct a systematic and quantitative survey of MSY gene expression across a diversity of human tissues.
Results
Accurately estimating MSY gene expression levels
We obtained thousands of bulk-tissue RNA-sequencing (RNA-seq) samples released by the GTEx Consortium (The GTEx Consortium 2017), spanning 36 adult human tissues and hundreds of post-mortem donors. To generate a quantitative view of MSY gene expression, we sought a method that could accurately estimate the expression levels of Y-Chromosome genes using short RNA-seq reads, overcoming challenges inherent in the MSY's sequence. Some MSY genes show ∼99% identity with their corresponding X-linked homologs in nucleotide sequence (Skaletsky et al. 2003). Other MSY genes have been amplified into multicopy gene families, with genes in these families showing upwards of 99.9% nucleotide sequence identity. In an RNA-seq experiment, many short reads from these genes will map to multiple genomic locations. These multimapping reads are routinely discarded in RNA-seq analyses to avoid the uncertainty of their origins, but excluding them can lead to underestimates of gene expression (Robert and Watson 2015). We suspected that the expression of MSY genes had been disproportionately underestimated in the publicly available expression-level estimates released by the GTEx Consortium, for which multimapping reads were discarded. In these published estimates, a much smaller fraction of MSY genes appeared to be expressed (≥1 transcript per million [TPM]) than genes from other chromosomes (MSY: 38.8%; autosomes, Chr X: 78.2%–98.6%) (Fig. 1B; Supplemental Table S2), in line with the MSY's deficit of uniquely mappable sequence (Supplemental Fig. S1).
To obtain accurate expression-level estimates for all MSY genes, we re-estimated expression levels genome-wide from the GTEx raw data with kallisto (Bray et al. 2016), a program that jointly infers the most likely origins of uniquely and multimapping reads under a statistical model. In contrast to a procedure that discards multimapping reads, kallisto enabled us to accurately estimate the expression levels of MSY genes in simulated RNA-seq data sets (±7.3% for the average MSY gene, when simulated at 5 TPM; Methods), including the relative expression of Y- and X-linked homologs and the total expression of genes in multicopy families (Supplemental Fig. S2). The accuracy of kallisto in these tests implies that, for high levels of sequence identity (∼99%), enough uniquely mapping reads are present in GTEx RNA-seq libraries to inform the correct assignment of multimapping reads. We then applied kallisto to the raw RNA-seq data and found that 80% of MSY genes are expressed in at least one tissue, a number more typical of other chromosomes (Fig. 1B). In some cases, our re-estimates identified expression levels more than two orders of magnitude higher than previously reported (e.g., the HSFY gene family in testis, 32.4 TPM vs. <0.1 TPM) (Fig. 1C; Supplemental Table S2). These differences were most pronounced for the MSY's multicopy gene families. In contrast, ancestral single-copy MSY genes produced few if any multimapping reads; their expression levels were therefore not systematically underestimated (Supplemental Figs. S1, S2). Nevertheless, of the approaches tested, we found kallisto to yield the most accurate estimates overall (Supplemental Fig. S2).
After performing a series of quality control steps, including outlier-sample detection and expression-level adjustment for three indicators of sample quality (Methods), we retained 6358 RNA-seq samples spanning 36 adult tissues, collected from 337 XY donors and 178 XX donors, for our primary analysis. Overall, we detected expression of 24 of the 26 MSY genes and gene families in at least one tissue (Fig. 1D; Supplemental Fig. S3; Supplemental Table S3).
Most MSY genes without X homologs show testis-specific expression
MSY genes that lack X homologs belong to five multicopy gene families (BPY2, CDY, DAZ, PRY, XKRY) (Supplemental Table S1). We first asked if any of these gene families are robustly expressed in a nonreproductive tissue, that is, in a tissue found in both XX and XY donors. We identified one such instance. Genes of the DAZ gene family, which are generally viewed as testis-specific genes involved in spermatogenesis (Vogt et al. 2008), were expressed in testis samples but also showed robust (and even 2.5-fold higher) expression in the stomach (Fig. 1D; Supplemental Fig. S4A), replicating a similar observation from a recent, smaller study (Gremel et al. 2015). In contrast, the DAZ family's autosomal homolog and progenitor (Saxena et al. 1996), DAZL, was not expressed in stomach samples from XY or XX donors (Supplemental Fig. S4B). The DAZ genes’ expression in the stomach proved to be the exception among MSY genes without X-linked homologs. One of the four remaining gene families (XKRY) was not robustly expressed in any tissue, whereas the others (BPY2, CDY, PRY) showed exquisitely testis-specific expression (Fig. 1D; Supplemental Fig. S3). We conclude that, overall, MSY genes without X homologs are unlikely to contribute substantially to differences between XX and XY individuals outside of the reproductive system.
Quantitative differences between X- and Y-homolog expression in XY individuals
Next, we considered the expression of MSY genes with X homologs, focusing on those X–Y gene pairs in which the MSY gene is expressed predominantly in nonreproductive tissues. Because these MSY genes were typically expressed in the same tissues as their corresponding X homologs (Supplemental Figs. S5, S6; Supplemental Tables S4, S5), we specifically sought to characterize the quantitative differences in X- and Y-homolog expression.
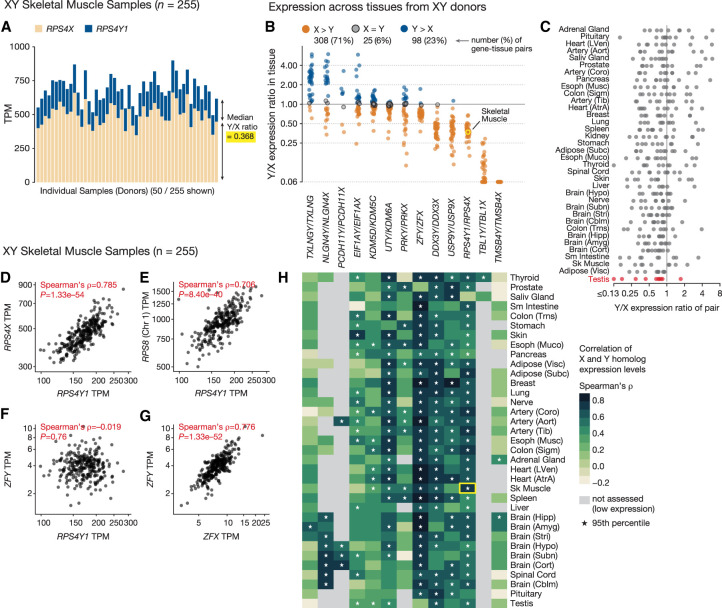
We first asked if the MSY genes are expressed at higher or lower levels than their X-linked homologs in tissues of XY individuals. We estimated the Y-homolog–to–X-homolog expression ratio (Y/X expression ratio) in each XY tissue sample and aggregated these into tissue-level estimates (Fig. 2A,B; Supplemental Table S6). We observed differences among the X–Y pairs in their average Y/X expression ratios. Two MSY genes (TMSB4Y, TBL1Y) showed substantially lower expression than their corresponding X-linked homologs in all tissues (Fig. 2B). However, for the remaining X–Y pairs, the expression levels of the Y- and X-linked homologs were more similar. Some MSY genes (e.g., DDX3Y, USP9Y, and RPS4Y1) were typically expressed at 30%–50% of the level of their X homolog, whereas others were often expressed at equal (e.g., KDM5D, EIF1AY) or higher (e.g., TXLNGY, NLGN4Y) levels. We replicated these Y/X-expression-ratio estimates using independently generated RNA-seq data spanning a subset of the GTEx tissues (Supplemental Fig. S7).
Figure 2.
Quantitative comparison of X- and Y-homolog expression in XY individuals. (A) Expression of RPS4Y1 (blue) and its X-linked homolog RPS4X (tan) in individual skeletal muscle samples (50 of 255 total samples are shown). (B,C) Each point shows Y/X expression ratio for one widely expressed X–Y gene pair in one tissue; points are grouped by gene pair (B) or tissue (C). In B, colors denote higher Y-homolog expression (blue), higher X-homolog expression (tan), or no significant difference (gray; Wilcoxon signed-rank test, FDR < 0.05). In C, Y/X expression ratios in testis are highlighted. Highlighted point (yellow) shows the summary of data in A. (D–G) Each point shows the coexpression of an MSY gene with another gene in a single skeletal muscle sample: RPS4Y1 versus RPS4X (D), RPS4Y1 versus RPS8 (E), RPS4Y1 versus ZFY (F), and ZFY versus ZFX (G). (H) Each cell shows the correlation coefficient (Spearman's ρ) of expression for X- and Y-linked members of an X–Y gene pair in one tissue. Asterisks indicate that a member of the X–Y pair shows more correlated expression with its homolog than with 95% of other genes in the genome. The highlighted cell (yellow) summarizes data in C and A.
Although some X–Y gene pairs had higher or lower Y/X expression ratios than others (Friedman test, P = 1 × 10–28), no one tissue had significantly higher or lower Y/X expression ratios overall (Friedman test, P = 0.42) (Fig. 2C; Supplemental Fig. S8). This implies that the expression of individual, widely expressed MSY genes largely reflects gene-specific regulation rather than an MSY-wide specialization for a biological process like reproduction. Indeed, despite the absence of substantial differences between tissues, when the tissues are ranked, testis was the tissue for which Y/X expression ratios are lowest on average (Fig. 2C; Supplemental Fig. S8).
For each X–Y gene pair, we next sought to determine if the expression of the X and Y homologs continues to be regulated by the same upstream factors. If so, variation in the activity of these factors from one sample to the next should yield correlated X- and Y-homolog expression. Indeed, we found that the X and Y homologs of most X–Y gene pairs showed highly correlated expression in many tissues (Fig. 2H; Supplemental Fig. S9; Supplemental Table S7). For example, the Y-linked ribosomal protein gene RPS4Y1 and its X-linked homolog RPS4X showed tightly correlated expression in most tissues across the body (Fig. 2D,H; Supplemental Fig. S9). RPS4Y1’s expression levels also correlated tightly with those of ribosomal protein genes on other chromosomes, such as RPS8 on Chromosome 1 (Fig. 2E), but not with those of Y-linked transcription factor ZFY (Fig. 2F), whose expression levels, instead, correlated with those of its X homolog ZFX (Fig. 2G). This suggests that RPS4Y1’s expression levels are determined in accordance with molecular function rather than chromosomal location. MSY genes that were typically expressed at only 30%–50% of the levels of their X homologs (e.g., RPS4Y1, DDX3Y, ZFY) still showed tightly correlated expression with their X homologs in many tissues (Fig. 2B,H; Supplemental Fig. S9; Supplemental Table S7). This highly correlated expression is not an artifact of read mismapping between the X and Y Chromosomes, as few reads mapped to both X and Y homologs of widely expressed X–Y gene pairs, and we could independently estimate their expression levels in simulated RNA-seq data sets (Supplemental Fig. S10). Thus, even though these Y homologs show diminished expression, the ancestral regulatory elements governing their expression likely remain intact and under considerable evolutionary constraint, despite millions of years of Y-Chromosome decay in the absence of regular recombination with the X Chromosome.
Evolutionary loss of a microRNA target site promoted elevated EIF1AY expression in the heart
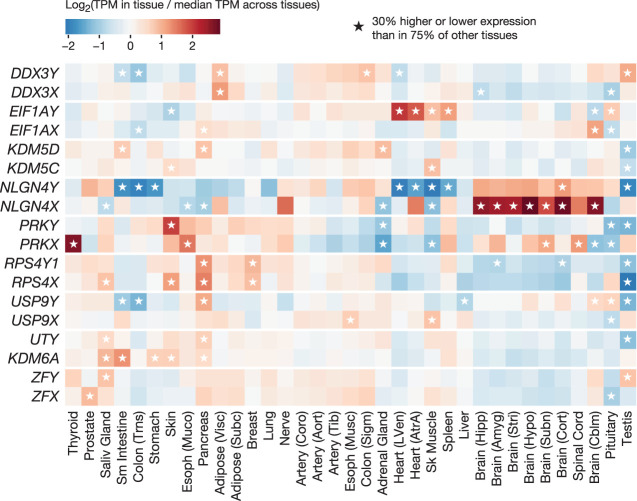
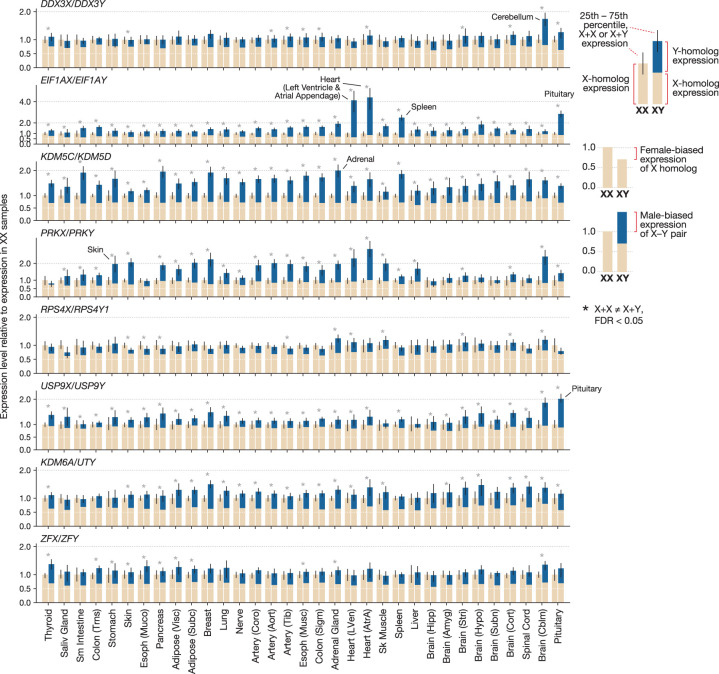
We also found evidence of tissue-specific divergence in the regulation of X- and Y-homolog expression. Individual X–Y pairs showed Y/X expression ratios in some tissues that differed substantially from their ratios in other tissues (e.g., USP9Y/USP9X = 1.1 in the pituitary compared with 0.2–0.6 in most tissues) (Fig. 2B), leading us to hypothesize that one member of the X–Y gene pair, but not the other, might be up- or down-regulated. To explore this possibility, for each X and Y homolog separately, we identified tissues where its expression level is 30% higher or lower than its expression level in most other tissues (Methods). All widely expressed MSY genes showed significantly higher or lower expression in at least one tissue (Fig. 3; Supplemental Table S8). We observed increased expression in a variety of tissues, including endocrine glands (e.g., pituitary, adrenal, pancreas), striated muscle (heart and skeletal), spleen, and skin.
Figure 3.
Tissue-specific up- and down-regulation of X and Y homologs. Each cell shows a gene's expression level in a tissue relative to its median expression level across all tissues. Asterisks denote tissues where a gene's expression level is significantly higher or lower (±30%; Welch's t-test, P < 0.001) than in 75% of other tissues.
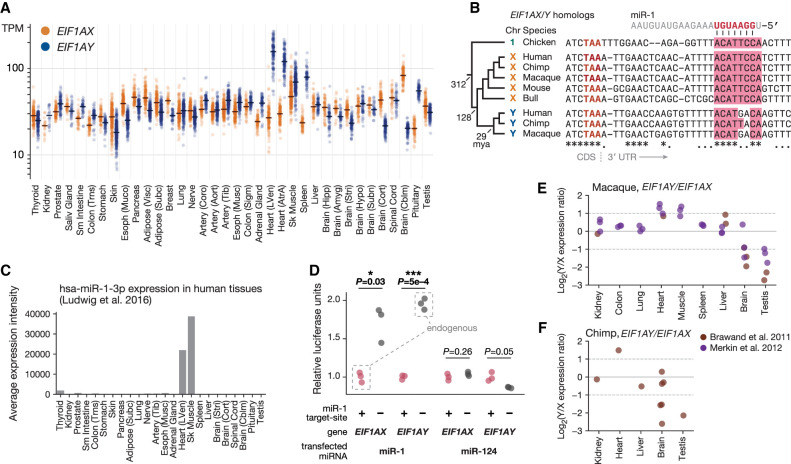
A prominent example of elevated expression of an MSY gene, without a corresponding increase in the expression of its X-linked homolog, is that of EIF1AY. EIF1AY encodes eukaryotic translation initiation factor 1A (EIF1A). EIF1A is one of 27 primary factors used to initiate protein synthesis in all eukaryotic linages (Hinnebusch 2014), and the only such factor encoded on both X and Y Chromosomes in primates (Bellott et al. 2014). The X and Y isoforms of EIF1A—encoded by EIF1AX and EIF1AY, respectively—are likely to be functionally equivalent: They differ by only a single amino acid, a conservative leucine-to-methionine substitution at a position outside of EIF1A's key functional domains, at which both leucine and methionine are observed in various vertebrate species (Supplemental Fig. S11). Although EIF1AY and its X-linked homolog EIF1AX are expressed at similar levels in most tissues, we found elevated expression of EIF1AY in the heart, skeletal muscle, spleen, and pituitary, causing EIF1AY expression levels to be as much as 5.8-fold higher than those of EIF1AX (Fig. 4A). We replicated this tissue-specific pattern of higher EIF1AY expression in human RNA-seq data from an independently generated data set (Supplemental Fig. S12).
Figure 4.
Y-specific loss of the miR-1 target site led to elevated EIF1AY expression in XY heart and tissue. (A) Each point shows expression level of EIF1AY (blue) or EIF1AX (gold) in a single tissue sample from an XY individual. Lines show median expression level. (B) Alignment of 3′ UTRs of EIF1AY, EIF1AX, and their orthologs; miR-1 target site in pink. Key branch points annotated with estimated divergence times in millions of years ago (mya). Fully conserved sites annotated with an asterisk; sites consistent with a single evolutionary substitution event annotated with a dot. (C) Quantile-normalized expression levels of miR-1 across human tissues. (D) Activity of luciferase reporter fused to 3′ UTR sequences of EIF1AX or EIF1AY with intact (+) or disrupted (−) miR-1 site in HEK293 cells, upon transfection with miR-1 or miR-124. Luciferase activity of each reporter with a disrupted miR-1 site is normalized to activity of corresponding reporter with intact site. P-values from two-sided Welch's t-test. (E,F) Each point shows Log2(Y/X expression ratio) for EIF1AY/EIF1AX orthologs in macaque (E) and chimpanzee (F).
We searched for factors that might explain EIF1AY’s elevated expression relative to EIF1AX in these tissues. Motivated by our previous studies (Naqvi et al. 2018), we wondered if these two genes might be differentially regulated by microRNAs (miRNAs), small regulatory RNAs that act as sequence-specific repressors of gene expression (Bartel 2018). A miRNA might specifically target EIF1AX, limiting its expression level in these tissues. When we searched the 3′ untranslated region (3′ UTR) of EIF1AX (Methods), the miRNA target site with the highest predicted efficacy was a match to miR-1 (Fig. 4B; Supplemental Table S9), a miRNA expressed abundantly and specifically in heart and skeletal muscle (Fig. 4C; Lim et al. 2005; Ludwig et al. 2016). At the homologous position in the 3′ UTR of EIF1AY, however, this miR-1 target site is disrupted by two nucleotide substitutions at positions critical for effective miRNA-mediated repression (Fig. 4B).
Two observations indicate that disruption of the miR-1 site in EIF1AY contributed to EIF1AY’s higher expression in the heart and skeletal muscle. First, using luciferase assays, we found that the 3′ UTR of EIF1AX, but not of EIF1AY, mediated approximately twofold repression of a reporter upon miR-1 transfection but not upon transfection with another miRNA (Fig. 4D). miR-1's repression of the EIF1AX-reporter construct required the target site to be intact, and repairing the two target-site substitutions within the EIF1AY-reporter construct was sufficient to confer miR-1–mediated repression. Second, the status of the miR-1 site predicts the expression pattern of EIF1AX and EIF1AY orthologs across species (Supplemental Table S10). In other primates, which both retain an intact EIF1AY gene and possess the disrupted miR-1 site, EIF1AY showed approximately twofold higher expression than EIF1AX specifically in heart and skeletal muscle (Fig. 4E,F). This expression pattern was likely acquired by primate EIF1AY, as EIF1AX orthologs in and outside of mammals do not show elevated heart expression (Supplemental Fig. S13). Together, these observations suggest that two nucleotide substitutions within an EIF1AY regulatory element contributed to tissue-specific up-regulation of EIF1AY.
Male-biased expression of X–Y gene pairs at the transcriptional level
We next asked if the divergent expression we observed within XY individuals leads to differences in expression between XX and XY individuals. We found that the X-linked members of the eight most widely expressed X–Y gene pairs typically showed XX-biased expression, that is, higher expression in tissue samples from XX individuals than in the same tissue type from XY individuals (Fig. 5). This XX-biased expression is expected because the X homologs of widely expressed X–Y gene pairs are not subject to X-Chromosome inactivation in XX cells and thus are expressed biallelically (Carrel and Willard 2005; Tukiainen et al. 2017). In all cases, the magnitude of XX bias was 2.0-fold and typically less than 1.5-fold (Fig. 5). This is consistent with past observations that the X-linked allele on the otherwise inactivated X Chromosome shows lower expression than the X-linked allele on the fully active X (Cotton et al. 2013; Berletch et al. 2015; Tukiainen et al. 2017). Next, for each X–Y gene pair, we compared the summed expression level of the X and Y homologs in XY samples to the expression level of the X homolog in XX samples. When accounting for Y-homolog expression, the X–Y gene pairs typically showed slightly XY-biased expression, with differences in expression less than 2.0-fold. However, in tissues where the X and Y homologs of a given pair showed uncorrelated expression (Supplemental Fig. S14) and in tissues with elevated Y-homolog expression, the XY-biased expression was more prominent. For example, KDM5D showed elevated expression in the adrenal gland (Fig. 3), leading to 2.1-fold XY-biased expression of KDM5C/KDM5D (Fig. 5; Supplemental Table S11). In the pituitary gland, elevated expression of USP9Y, together with depleted expression of USP9X, yields 2.0-fold XY-biased expression of USP9X/USP9Y (Figs. 3, 5). Up-regulated EIF1AY expression in the heart leads to 5.2-fold higher expression of EIF1AX/EIF1AY in XY heart (left ventricle) tissue. Thus, at the transcriptional level, the Y-linked members of human X–Y gene pairs typically show higher expression in XY cells than the second copy of their X-linked homologs in XX cells, causing the X–Y gene pairs to show at least subtly, and sometimes substantially, male-biased expression.
Figure 5.
Tissue-specific, male-biased expression of X–Y gene pairs at the transcriptional level. Each pair of bars shows the median expression level of an X–Y gene pair in XX (left) and XY (right) samples from one tissue. Expression is normalized to the level in XX samples. In XY samples, the sum of X-homolog (tan) and Y-homolog (blue) expression is shown. Error bars, 25th and 75th percentiles. An asterisk indicates X-homolog expression in XX samples is significantly different from the summed X- and Y-homolog expression in XY samples: FDR < 0.05.
Male-biased expression of EIF1A in the heart at the protein level
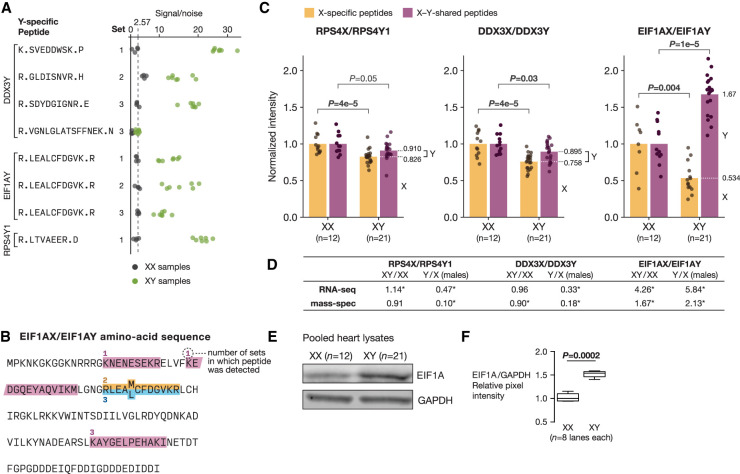
We sought to assess whether the male-biased expression of X–Y gene pairs at the transcript level further manifests as male-biased expression at the protein level. We generated proteome-wide measurements of protein abundance in 21 XY and 12 XX heart (left ventricle) tissue samples by multiplexed, tandem mass tag (TMT)–based mass spectrometry (Methods) (Supplemental Fig. S15). These samples, which we obtained from the GTEx tissue biobank, were selected through rigorous histological review to ensure that XX and XY samples showed minimal pathology and similar cell-type composition (Methods). At a 0.22% false-discovery rate (FDR), we detected peptides that specifically match seven X or Y protein isoforms encoded by widely expressed X–Y gene pairs (RPS4X, RPS4Y1, EIF1AX, EIF1AY, DDX3X, DDX3Y, USP9X) (Fig. 6A,B; Supplemental Fig. S16; Supplemental Table S12). Each of these proteins (except RPS4Y1) was supported by multiple, independent observations of isoform-specific peptides. Moreover, Y-specific peptides from all Y isoforms showed only background levels of signal in XX samples (Fig. 6A). Together, these observations provide strong evidence that these seven proteins are present in heart tissue. The absence of peptides from the remaining 11 proteins was consistent with their lower expression levels at the transcript level and the overall rate at which we recovered peptides from expressed genes across the genome (7/18 X–Y pair genes vs. 4788/11,936 expressed genes; one-tailed Fisher's exact test, P ≈ 1.0) (Supplemental Fig. S17). Thus, whether these 11 remaining proteins are present in human heart tissue remains an open question.
Figure 6.
Male-biased expression of EIF1A protein in the heart. (A) Signal/noise values for Y-specific peptides in XX (gray) and XY (green) samples. Set refers to the 11-plex experiment (out of three total) in which the peptide was detected. The dotted line shows average signal/noise value in XX samples. (B) Amino-acid sequence of EIF1AX/Y: X- and Y-specific amino acids are superscripted and subscripted, respectively. X-specific (gold), Y-specific (blue), and X–Y shared (purple) peptides detected by mass spectrometry are shown, along with the number of 11-plex experiments in which each peptide was detected. (C) Relative abundance of X and Y protein isoforms in XX (n = 12) and XY (n = 21) heart tissue samples by mass spectrometry. For each X-Y pair, points show the levels of the X isoform (gold) or the total level of the X and Y isoform (purple) in XX samples compared with XY samples, from which the relative proportion of X and Y isoform expression in XY samples can be inferred (dotted white line). P-values by estimated by permutation. (D) Comparison of estimated Y/X expression ratios and sex-biased expression from RNA-seq and mass spectrometry. Asterisks indicate statistical significance in the corresponding analysis. (E) Abundance of EIF1A and GAPDH by western blot in pooled XX and XY protein lysates. (F) Quantification of EIF1A levels in pooled XX and XY samples by western blot; P-value by Welch's t-test.
For the three X–Y gene pairs from which both X- and Y-specific peptides were detected (DDX3X/DDX3Y, EIF1AX/EIF1AY, RPS4X/RPS4Y1), we asked if their expression is sex biased at the protein level. For each X–Y pair, we first used signal from X-specific peptides to estimate the sex bias of the X isoform; we next used signal from peptides that match both X and Y isoforms (X–Y-shared peptides) to estimate the sex bias of the X–Y pair overall, accounting for the contribution of the Y isoform (Supplemental Fig. S15; Supplemental Table S13). These two expression ratios then allowed us to infer the relative abundances of X and Y isoforms within XY tissue. This approach contrasts with the common practice in mass-spectrometric analysis of assigning nonunique peptides to the apparently most abundant protein (e.g., Cox and Mann 2008), which would conflate the expression of X and Y isoforms in these samples.
We found that the X isoforms of all three X–Y pairs showed XX-biased protein abundance (P < 5 × 10–3, by permutation; Methods), consistent with their escape from X-Chromosome inactivation (Fig. 6C). In contrast, proteins encoded by X-Chromosome genes that are subject to X-Chromosome inactivation showed no or only modest sex biases in protein abundance (Supplemental Fig. S18; Supplemental Table S14). For RPS4X/RPS4Y1 and DDX3X/DDX3Y, the combined expression levels of X and Y isoforms in XY tissues were slightly below the levels of the X isoforms in XX tissues on average (RPS4X/RPS4Y1: mean XY/XX ratio = 0.91, P = 0.05 by permutation; DDX3X/DDX3Y: mean XY/XX ratio = 0.90, P = 0.03 by permutation) (Fig. 6C), albeit at only nominally statistically significant levels, suggesting RPS4Y1 and DDX3Y mostly, if not entirely, compensate for the XX-biased expression of RPS4X and DDX3X. The combined expression of EIF1AX and EIF1AY, however, showed a 1.7-fold XY bias (P < 10–6, permutation), indicating that EIF1AY overcompensates for the XX-biased expression of EIF1AX. These estimates further imply that EIF1AY protein is 2.1-fold more abundant than EIF1AX in XY heart tissue. By using an EIF1A antibody that recognizes both EIF1AX and EIF1AY (Supplemental Fig. S19), we corroborated EIF1AX/EIF1AY's (i.e., EIF1A's) XY-biased expression in these same heart tissue samples by western blot (Fig. 6E,F). Although EIF1AY transcripts were 5.8-fold more abundant than EIF1AX transcripts in heart (left ventricle) tissue, EIF1AY protein was only 2.1-fold more abundant than EIF1AX (Fig. 6D). Nevertheless, EIF1AY’s up-regulated expression in the heart—a result of its noncoding divergence from EIF1AX—is sufficient to lead to a male-biased abundance of this essential translation initiation factor.
Discussion
How do human Y-Chromosome genes contribute to differences between XX and XY individuals beyond the reproductive system? It has been tempting to speculate that MSY genes encode proteins with male-specific effects (Arnold 2012), as the result of protein-coding sequence divergence between MSY genes and their corresponding X homologs. Such instances might yet be uncovered. However, given past evidence attesting to the functional interchangeability of X and Y protein isoforms (Table 1) and our observations of divergent X–Y expression herein, we propose that divergence of MSY genes from their X homologs in regulatory (i.e., noncoding) sequence is an important means by which the Y Chromosome could directly give rise to differences between XX and XY individuals. Because the X–Y gene pairs encode regulators of transcription, translation, and protein stability that are highly dosage sensitive (Bellott et al. 2014; Naqvi et al. 2018), small differences in their expression levels could contribute significantly to the widespread sex differences in gene expression observed across tissues (Naqvi et al. 2019) and ultimately to phenotypic differences between the sexes.
This focus on regulatory-sequence divergence, rather than protein-coding divergence, accords with prevailing views from complex trait genetics and evolutionary developmental biology. In these contexts, phenotypic variation within and across species is thought to flow in large part from noncoding substitutions that alter the expression of pleiotropic regulatory genes (Carroll 2008; Albert and Kruglyak 2015), genes very much like those encoded by ancestral X–Y pairs. In a similar manner, quantitative differences between males and females in disease susceptibility or morphometric traits might reflect regulatory-sequence divergence between the X and Y Chromosomes that yields sex-biased expression of the X–Y gene pairs. It is likely that many types of regulatory factors beyond miRNAs are involved in establishing these expression patterns. As miRNAs typically repress their targets by less than twofold, a factor other than miR-1, such as a heart-specific transcription factor, might additionally contribute to EIF1AY’s approximately fivefold higher expression over EIF1AX in the heart (Fig. 4D; Baek et al. 2008).
One speculation is that the XY-biased expression of EIF1A contributes to sex differences in diseases of the heart, many of which manifest with greater incidence or severity in one sex (Regitz-Zagrosek et al. 2010). As a core component of the 43S preinitiation complex in eukaryotes (Hinnebusch 2014), EIF1A impacts the translation of many if not all mRNA transcripts in the cell (Sehrawat et al. 2018). Changes in translational regulation are a prominent molecular feature of human heart tissue from individuals with dilated cardiomyopathy (van Heesch et al. 2019), a disease with a 1.5-fold higher incidence in males than in females (Towbin et al. 2006). Although it is currently unknown whether elevated levels of EIF1A are beneficial, harmful, or neutral in consequence, EIF1AY’s expression pattern and those of other MSY genes provide new motivation to examine the Y Chromosome's contribution to various quantitative traits.
Beyond these cases of divergent X- and Y-homolog regulation, our observations accord with the view that MSY genes encode proteins that function similarly to their X-encoded homologs and that these shared functions are dosage sensitive across a multitude of tissues. The tightly correlated expression of X and Y homologs we observe is typical of genes whose proteins must together be synthesized in precise quantities (Taggart and Li 2018). It is unlikely that the regulatory elements that enable the MSY genes to be expressed in this manner would survive by chance, after tens of millions of years of Y-Chromosome decay.
We have provided direct evidence that the proteins encoded by MSY genes are present in human heart tissue. Our detection of DDX3Y protein in the heart conflicts with earlier claims that DDX3Y is widely transcribed but only translated in the testis (Ditton et al. 2004). By using a DDX3Y-specific antibody, Ditton et al. (2004) detected DDX3Y protein in testis but not in brain or kidney. In our analysis, we find that DDX3Y shows lower transcript abundance in brain and kidney than in most other tissues (Supplemental Fig. S3; Supplemental Table S3), suggesting DDX3Y protein might have been present only at low levels (Gueler et al. 2012). DDX3Y transcripts might also be translated inefficiently (see below). Beyond our study, DDX3Y has been detected by western blot in a neuronal cell line (Vakilian et al. 2015), and DDX3Y was identified as an essential gene in a leukemia cell line through a genome-wide, unbiased screen (Wang et al. 2015). Although the only known phenotype for individuals with DDX3Y deletions is spermatogenic failure (Vogt et al. 2008), milder nonreproductive phenotypes have not been excluded. Recognizing that DDX3Y protein is present in nonreproductive tissues has important implications for studies of DDX3X—an intellectual disability gene (Snijders Blok et al. 2015) and therapeutic target (Bol et al. 2015; Valiente-Echeverría et al. 2015)—which have typically disregarded the impact of its Y homolog.
We found that the protein expression levels of DDX3Y, EIF1AY, and RPS4Y1 were 1.8-fold to 4.7-fold lower than their transcript expression levels, when measured against their X homologs. It is possible that these transcripts are translated less efficiently, or that their proteins are less stable, than those encoded by their X homologs. If true for other MSY genes, this could explain why X–Y gene pairs often show slightly male-biased expression at the transcript level (Fig. 5): Overexpression of the Y homolog at the transcript level might be needed to achieve the requisite level of protein abundance. However, we caution against extrapolating these results to other MSY genes and other tissues until many more protein-level measurements are made.
Ultimately, our analyses establish that mass spectrometry can be used, in an unbiased manner, to detect the expression of MSY proteins in a nonreproductive tissue and to quantify the levels of X–Y pair proteins across individuals, even when the X and Y isoforms differ by only a single amino acid. This will remain a challenge for the Y (and X) protein isoforms expressed at lower levels (Meyfour et al. 2017). However, as is the case with analyses of the Y Chromosome in DNA and RNA sequence, a distinct picture of the Y Chromosome emerges with appropriate analytical approaches. Deploying methods that can resolve subtle differences between genes as standard practice, whether for RNA-seq (Li and Dewey 2011; Bray et al. 2016; Patro et al. 2017) or mass spectrometry (Malioutov et al. 2019), promises a more complete understanding not only of sex-chromosome genes but also of all groups of homologous genes genome-wide.
Going forward, we anticipate that additional examples of up-regulated MSY gene expression will be revealed through expression profiling in other contexts. Particularly promising will be the application of single-cell approaches to observe MSY gene expression in rare cell types, whose contributions to the bulk-tissue estimates here are diluted. Indeed, a recent study found elevated expression of TBL1Y—a gene we found showed lower expression than its X homolog in all instances—in cells of the inner ear, with implications for syndromic hearing loss (Di Stazio et al. 2019). Given the differences in expression between MSY genes and their X homologs, it will be especially important to characterize how increases or decreases in the expression of proteins encoded in X–Y pairs lead to changes across the genome in specific cell types, tissues, and developmental stages.
Methods
Code used in analysis
Unless stated otherwise, all analyses were conducted in Python (v3.6.9), drawing upon software packages numpy (v1.17.2) (Oliphant 2006; van der Walt et al. 2011), scipy (v1.3.1) (Virtanen et al. 2020), pandas (v0.25.1) (McKinney 2010), scikit-learn (v0.21.3) (Pedregosa et al. 2011), statsmodels (v0.10.1) (Seabold and Perktold 2010, matplotlib (v3.1.1) (Hunter 2007), and seaborn (v0.9.0; seaborn.pydata.org). Code and Jupyter notebooks (https://jupyter.org) for recreating these analyses are available on GitHub (https://github.com/akg8/MSY-expression) and as Supplemental Code.
Abbreviated tissue names
Tissues with long names are abbreviated in figures as follows: adipose—subcutaneous (Subc) and visceral (Visc); artery—aorta (Aort), coronary (Coro), and tibial (Tib); brain—amygdala (Amyg), cerebellum (Cblm), cortex (Cort), hippocampus (Hipp), hypothalamus (Hypo), striatum (Stri), and substantia nigra (Subn); colon—sigmoid (Sigm) and transverse (Trns); esophagus—mucosa (Muco) and muscularis (Musc); heart—atrial appendage (AtrA) and left ventricle (LVen); skeletal muscle (Sk Muscle); and small intestine (Sm Intestine).
Human transcriptome annotation and MSY genes
All human analyses use transcript/gene models defined in a custom subset of the comprehensive GENCODE version 24 transcript annotation, based on our annotation of the male-specific region of the human Y Chromosome (Supplemental Table S1; Skaletsky et al. 2003). For further details, see Supplemental Methods.
Comparison of RNA-seq analysis methods
Simulated RNA-seq libraries were generated using RSEM (v1.2.22) (Li and Dewey 2011), using a GTEx testis sample as a template. The expression levels of MSY genes and their X-linked homologs were set to predetermined levels in each simulation. Three methods were then used to estimate the expression levels of Y-Chromosome genes and their X-linked homologs: (1) Reads were aligned to the genome using TopHat2 (Kim et al. 2013), and the number of uniquely mapping reads overlapping each gene was counted with featureCounts (Liao et al. 2014) (this “unique reads” approach is based on the GTEx Consortium's procedure); (2) reads were aligned with TopHat2 and expression levels were estimated with Cufflinks (Trapnell et al. 2010) in “multiread-correct” mode; (3) reads were input to kallisto (Bray et al. 2016), which estimated expression levels using the transcriptome annotation. For further details, see Supplemental Methods.
Estimating transcript expression levels from GTEx RNA-seq samples
GTEx (v7) raw data were obtained from dbGaP (dbGaP accession: phs000424.v7.p2). Transcript expression levels were then estimated in TPM units using kallisto with sequence-bias correction (‐‐bias); transcript expression levels were summed to obtain gene expression levels. The expression levels of genes in multicopy gene families (Supplemental Table S1) were summed to obtain family-level estimates, which were used in place of estimates at the gene level. Within each tissue, samples that appeared to be outliers based on their genome-wide expression profile were identified and removed (Supplemental Methods). Samples from male and female donors were verified to have likely XY and XX sex-chromosome constitutions through the expression of MSY genes and XIST. The final set of samples used for analysis is given in Supplemental File S1. Samples from some tissue subsites defined by the GTEx Consortium (e.g., brain–cerebellum and brain–cerebellar hemisphere) could not be easily distinguished by hierarchical clustering. In these cases, we merged the tissue labels, treating them as single tissue types (Supplemental Methods).
To reduce technical variation in expression levels and increase tissue-to-tissue comparability, linear regression was used to adjust expression levels for the effects of ischemic time, RNA integrity number (RIN), and the sample intronic read mapping rate (see Supplemental Methods). These adjusted expression levels were used in all analyses, except when comparing our estimated expression levels from kallisto to those released by the GTEx Consortium in Figure 1, B and C, and Supplemental Figure S1.
We estimated a gene's expression level in a tissue as its median expression level among samples from that tissue unless otherwise noted. For Figure 1C, the estimated expression levels of MSY genes were clustered hierarchically by average linkage using correlation distances (scipy.cluster.hierarchy.linkage, with method = “average,” metric = “correlation”).
Comparison with the GTEx Consortium's analysis based on uniquely mapped reads
The GTEx Consortium's gene expression level estimates (v7) were downloaded from the GTEx Portal (gtexportal.org: GTEx_ Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_tpm.gct). Genes in GENCODE version 19 were matched to genes in our version-24–based annotation by Ensembl gene ID. The fraction of uniquely mapping reads per gene was estimated by aligning all possible 76-nt reads from its longest transcript isoform to the transcriptome exhaustively (see Supplemental Methods).
Expression-level normalization across samples and tissues
For analyses in which the expression level of a gene was compared across samples, we applied a modified version of the between-sample, size-factor normalization used in DESeq (Anders and Huber 2010). For a set of n samples, the normalization factor, si, for sample i was calculated as
where ygi is the expression level (in TPM units) of gene g in sample i, and GC is a set of control genes. Rather than using all genes in the genome, we base our normalization factor on a set of 50 control genes that are expressed like housekeeping genes. These control genes were identified as the 50 genes, among all genes with mean expression levels between 10 and 100 TPM, with the most conserved expression-level ranks (i.e., whose expression-level ranks showed the lowest coefficient of variation across the samples). This approach helps to ensure that the genes driving the normalization have known properties even when comparing samples from two or more tissues in which the expression levels of many genes would be expected to differ.
Y/X expression ratios
For a given X–Y gene pair and tissue, we estimated the Y/X expression ratio in each sample as (Y-homolog TPM + 0.5)/(X-homolog TPM + 0.5), excluding samples in which both genes were expressed below 1 TPM; the median sample-level Y/X ratio was then used as the tissue-level estimate. A tissue-level Y/X ratio was not reported where both genes were expressed below 1 TPM. For a given X–Y pair and tissue, the difference in the X and Y homolog's expression levels was assessed with a two-sided Wilcoxon signed-rank test (Python function: scipy.stats.wilcoxon). After obtaining P-values for all X–Y pairs in all tissues, these P-values were adjusted for multiple hypotheses using the Benjamini–Hochberg procedure (Python function: statsmodels.stats.multitest.multipletests, method = “fdr_bh”). To test for differences between Y/X expression ratios among X–Y pairs and among tissues, the Friedman test was applied (Python function: scipy.stats.friedmanchisquare), using the Y/X expression ratios from the 28 tissues where a ratio was estimated for the 10 most widely expressed X–Y pairs (listed in Supplemental Fig. S8).
Replication of gene expression patterns
To assess expression patterns in an independent data set, we analyzed raw RNA-seq data from the Human Protein Atlas (HPA) Project (Uhlén et al. 2015) with kallisto using sequence-bias correction. For replication of Y/X expression ratios in Supplemental Figure S7, we used the HPA tissues matching a GTEx tissue where at least four HPA samples from male donors were present (colon, prostate, testis). For more detailed replication of EIF1AY’s expression pattern (Supplemental Fig. S12), we used samples from all HPA tissues matching a GTEx tissue. When an HPA tissue potentially matched multiple GTEx tissues (e.g., colon–transverse, colon–sigmoid), the best-matching tissue was selected by calculating correlation coefficients between samples from the two data sets using genome-wide gene expression levels.
Correlated expression of X and Y homologs
Analyses of pairwise gene coexpression were performed in each tissue with at least 30 samples from male donors. Each tissue was analyzed separately, considering only those genes with expression levels ≥5 TPM. The expression levels from each sample were first normalized by the housekeeping method described above and transformed to log2(TPM + 0.5) units. To control for unmodeled technical factors (e.g., batch effects) that might lead to spuriously correlated expression between the X and Y homologs of X–Y pairs, the principal components (PCs) of the N genes × M samples matrix were calculated (Python function: sklearn.decomposition.PCA): After mean-centering the expression levels of each gene, each sample's loading on the top PC was extracted. For each gene, variation in expression associated with this PC was removed by linear regression. The degree of coexpression between gene i and gene j was measured with Spearman's correlation coefficient, ρij, of their PC-adjusted expression levels. The procedure used to obtain the significance of X–Y coexpression is described in the Supplemental Methods.
Differential expression across tissues
The housekeeping normalization was first applied to all XY samples from all tissues. Then, for each gene of interest, its log2(TPM + 0.5) expression levels were compared in each pair of tissues (excluding tissues with fewer than 30 samples) by Welch's t-test (Python function: scipy.stats.ttest_ind with equal_var = False). A gene was considered to be significantly differentially expressed between two tissues if the P-value was <10–3 and its average expression levels in the two tissues differed by at least 30% (1.3-fold). A gene was considered to be up-regulated (or down-regulated) in a tissue if its expression in that tissue was significantly higher (or lower) than its expression in at least 75% of the other tissues (to allow for the possibility of up-/down-regulation in multiple tissues). This analysis was limited to the nine X–Y gene pairs where the Y homolog was robustly expressed in many tissues. TXLNG/TXLNGY was excluded because the regulation of TXLNGY expression appears to have diverged almost completely from the regulation of TXLNG.
miRNA analyses
Scripts from TargetScan 6.0 (Friedman et al. 2009) were used to identify and evaluate miRNA target sites in the 3′ UTRs of the X- and Y-linked homologs of each widely expressed X–Y gene pair. Sites identified in X homologs were validated using the latest TargetScan predictions (release 7.2) (Supplemental Table S9; Agarwal et al. 2015). miRNA expression patterns were evaluated using quantile-normalized expression values from Ludwig et al. (2016). Among target sites for tissue-specific, highly expressed miRNAs, the miR-1 target site in EIF1AX is the target site with the greatest predicted efficacy that is preserved in one homolog of an X–Y pair but not the other. For luciferase assays, EIF1AX’s miR-1 site was changed to shuffled sequence, and EIF1AY’s disrupted miR-1 site was changed to match that of EIF1AX, using the QuikChange II kit (Agilent). Further details on the computational identification of miRNA sites and experimental validation with luciferase assays are provided in the Supplemental Methods and Supplemental File S2.
Cross-species analyses of sequence and expression
Multiple sequence alignments of EIF1AX/Y 3′ UTR and amino-acid sequences were generated with PRANK (Löytynoja and Goldman 2005) using fixed species trees (with separate clades for mammalian X- and Y-linked genes). Expression levels of EIF1AX/Y homologs in male chimpanzee (Pan troglodytes), rhesus macaque (Macaca mulatta), mouse (Mus musculus), and chicken (Gallus gallus) tissues were estimated with kallisto, using RNA-seq data from Brawand et al. (2011) and Merkin et al. (2012). For further details, see Supplemental Methods.
Quantitative proteomic analysis of human heart tissue
Heart (left ventricle) samples from 21 male donors and 12 female donors were obtained from the GTEx tissue biobank for quantitative proteomic analysis after thoroughly screening all left ventricle samples by donor medical history and histopathological analysis (Supplemental Methods; Supplemental File S3). Quantitative proteomic analysis was performed in three 11-plex TMT experiments as previously described (Chick et al. 2016) and as detailed in the Supplemental Methods. The protein encoded by a Y-linked homolog of an X–Y gene pair (Y isoform) was determined to be present in heart tissue if at least one peptide with the following two properties was detected: (1) Its sequence specifically matched the Y isoform and no other protein; (2) it showed signal above background only in male samples. For proteins not encoded by X–Y gene pairs, protein abundance was estimated as previously described (Supplemental Methods; Supplemental Table S12; Chick et al. 2016). Protein abundances of the X and Y isoforms were estimated separately using X isoform–specific and X and Y isoform–shared peptides, as detailed in the Supplemental Methods.
Immunoblot experiments
Human heart-tissue lysates (from tissue obtained for the mass spectrometry analysis) were pooled by sex for immunoblotting. EIF1AX and EIF1AY protein was detected with an EIF1A primary antibody (Abcam Ab177939, anti-rabbit), with GAPDH (Ambion AM4300, anti-mouse) as a loading control. EIF1A levels were quantified using the Odyssey CLx imaging system (LI-COR). Four technical replicates were performed per sex. To verify that the EIF1A antibody recognizes both EIF1AX and EIF1AY, immunoblot experiments were performed with protein lysates from human lymphoblastoid cell lines with varying numbers of sex chromosomes (45,X; 46,XX; 46,XY; 47,XYY; 48,XXXY; 49,XXXXY; 49,XYYYY) and, correspondingly, varying levels of EIF1AX and EIF1AY. For further experimental details, see Supplemental Methods.
Data access
The proteomic data generated in this study have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al. 2019; https://www.ebi.ac.uk/pride/archive/) under accession number PXD017055. Processed data (re-estimated TPM matrices) have been submitted to Zenodo (https://zenodo.org) under DOI 10.5281/zenodo.3627233.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank A.K. San Roman for sharing sex-chromosome-aneuploidy cell lines, S.W. Eichhorn and S.E. McGeary for discussions about miRNA targeting, and current and former members of the Page laboratory for valuable input over the course of this project. We thank D.W. Bellott, J.F. Hughes, and A.K. San Roman for critical reading of the manuscript. This work was supported by Biogen, Inc., the American Heart Association, the National Institutes of Health (grants GM67945 and HL007627), the Whitehead Institute, the Howard Hughes Medical Institute, and generous gifts from Brit and Alexander d'Arbeloff and Arthur W. and Carol Tobin Brill.
Author contributions: A.K.G., H.S., and D.C.P. designed the study. A.K.G. performed computational analyses. S.N. performed computational and experimental analyses of miRNA target sites. R.N.M. performed histological evaluations on human heart tissue samples. J.M.C. and S.P.G. contributed proteomic analyses, with assistance from L.C. A.K.G. performed mass-spectrometric data analysis of X and Y isoform abundance. L.C. performed immunoblotting experiments. A.K.G. and D.C.P. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261248.120.
References
- Agarwal V, Bell GW, Nam J-W, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. 2012. The end of gonad-centric sex determination in mammals. Trends Genet 28: 55–61. 10.1016/j.tig.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2018. Metazoan microRNAs. Cell 173: 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, Koutseva N, Zaghlul S, Graves T, Rock S, et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499. 10.1038/nature13206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X. 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet 11: e1005079 10.1371/journal.pgen.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol GM, Xie M, Raman V. 2015. DDX3, a potential target for cancer treatment. Mol Cancer 14: 188 10.1186/s12943-015-0461-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. 10.1038/nature10532 [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Cannon-Albright LA, Farnham JM, Bailey M, Albright FS, Teerlink CC, Agarwal N, Stephenson RA, Thomas A. 2014. Identification of specific Y chromosomes associated with increased prostate cancer risk. Prostate 74: 991–998. 10.1002/pros.22821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404. 10.1038/nature03479 [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. 2016. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534: 500–505. 10.1038/nature18270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ. 2013. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol 14: R122 10.1186/gb-2013-14-11-r122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM. 2014. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet 15: 367–378. 10.1038/nrg3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stazio M, Collesi C, Vozzi D, Liu W, Myers M, Morgan A, D'Adamo PA, Girotto G, Rubinato E, Giacca M, et al. 2019. TBL1Y: a new gene involved in syndromic hearing loss. Eur J Hum Genet 27: 466–474. 10.1038/s41431-018-0282-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditton HJ, Zimmer J, Kamp C, Meyts ER-D, Vogt PH. 2004. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet 13: 2333–2341. 10.1093/hmg/ddh240 [DOI] [PubMed] [Google Scholar]
- Eales JM, Maan AA, Xu X, Michoel T, Hallast P, Batini C, Zadik D, Prestes PR, Molina E, Denniff M, et al. 2019. Human Y chromosome exerts pleiotropic effects on susceptibility to atherosclerosis. Arterioscler Thromb Vasc Biol 39: 2386–2401. 10.1161/ATVBAHA.119.312405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, Pardo M, Yu L, Choudhary JS, Metzakopian E, et al. 2018. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet 50: 883–894. 10.1038/s41588-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremel G, Wanders A, Cedernaes J, Fagerberg L, Hallström B, Edlund K, Sjöstedt E, Uhlén M, Pontén F. 2015. The human gastrointestinal tract-specific transcriptome and proteome as defined by RNA sequencing and antibody-based profiling. J Gastroenterol 50: 46–57. 10.1007/s00535-014-0958-7 [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueler B, Sonne SB, Zimmer J, Hilscher B, Hilscher W, Graem N, Rajpert-De Meyts E, Vogt PH. 2012. AZFa protein DDX3Y is differentially expressed in human male germ cells during development and in testicular tumours: new evidence for phenotypic plasticity of germ cells. Hum Reprod 27: 1547–1555. 10.1093/humrep/des047 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci 104: 18439–18444. 10.1073/pnas.0707292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088. 10.1016/j.cell.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Johansson MM, Lundin E, Qian X, Mirzazadeh M, Halvardson J, Darj E, Feuk L, Nilsson M, Jazin E. 2016. Spatial sexual dimorphism of X and Y homolog gene expression in the human central nervous system during early male development. Biol Sex Differ 7: 5 10.1186/s13293-015-0056-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. 2008. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 4: e9 10.1371/journal.pgen.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi M, Otani M, Kikkawa Y, Itakura Y, Sakai K, Ito T, Toyoda M, Sekita Y, Kimura T. 2020. Mutations of histone demethylase genes encoded by X and Y chromosomes, Kdm5c and Kdm5d, lead to noncompaction cardiomyopathy in mice. Biochem Biophys Res Commun 525: 100–106. 10.1016/j.bbrc.2020.02.043 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1997. Functional coherence of the human Y chromosome. Science 278: 675–680. 10.1126/science.278.5338.675 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1999a. Four evolutionary strata on the human X chromosome. Science 286: 964–967. 10.1126/science.286.5441.964 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1999b. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat Genet 21: 429–433. 10.1038/7771 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694. 10.1038/nature06192 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee S-K. 2012. UTX, a histone H3-Lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 22: 25–37. 10.1016/j.devcel.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci 102: 10557–10562. 10.1073/pnas.0409137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, et al. 2016. Distribution of miRNA expression across human tissues. Nucleic Acids Res 44: 3865–3877. 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malioutov D, Chen T, Jaffe J, Airoldi E, Budnik B, Slavov N. 2019. Quantifying homologous proteins and proteoforms. Mol Cell Proteomics 18: 162–168. 10.1074/mcp.TIR118.000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. 2010. Data structures for statistical computing in Python. In Proceedings of the Ninth Python in Science Conference (ed. van der Walt S, et al. ), pp. 56–61. SciPy 2010, Austin, TX. 10.25080/Majora-92bf1922-00a [DOI] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyfour A, Pooyan P, Pahlavan S, Rezaei-Tavirani M, Gourabi H, Baharvand H, Salekdeh GH. 2017. Chromosome-Centric Human Proteome Project allies with developmental biology: a case study of the role of Y chromosome genes in organ development. J Proteome Res 16: 4259–4272. 10.1021/acs.jproteome.7b00446 [DOI] [PubMed] [Google Scholar]
- Naqvi S, Bellott DW, Lin KS, Page DC. 2018. Conserved microRNA targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Genome Res 28: 474–483. 10.1101/gr.230433.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Godfrey AK, Hughes JF, Goodheart ML, Mitchell RN, Page DC. 2019. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 365: eaaw7317 10.1126/science.aaw7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2006. A guide to NumPy. Trelgol Publishing, USA. [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12: 2825–2830. [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regitz-Zagrosek V, Oertelt-Prigione S, Seeland U, Hetzer R. 2010. Sex and gender differences in myocardial hypertrophy and heart failure. Circ J 74: 1265–1273. 10.1253/circj.CJ-10-0196 [DOI] [PubMed] [Google Scholar]
- Robert C, Watson M. 2015. Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol 16: 177 10.1186/s13059-015-0734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337. 10.1038/nature03440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, Reijo R, Rozen S, Dinulos MB, Disteche CM, et al. 1996. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet 14: 292–299. 10.1038/ng1196-292 [DOI] [PubMed] [Google Scholar]
- Seabold S, Perktold J. 2010. Statsmodels: econometric and statistical modeling with Python. In Proceedings of the Ninth Python in Science Conference (ed. van der Walt S, et al. ), pp. 92–96. SciPy 2010, Austin, TX. 10.25080/Majora-92bf1922-011 [DOI] [Google Scholar]
- Sehrawat U, Koning F, Ashkenazi S, Stelzer G, Leshkowitz D, Dikstein R. 2018. Cancer-associated eukaryotic translation initiation factor 1A mutants impair Rps3 and Rps10 binding and enhance scanning of cell cycle genes. Mol Cell Biol 39: e00441-18 10.1128/MCB.00441-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Iida H, Fukumura J, Nishimoto T. 2004. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp Cell Res 300: 213–222. 10.1016/j.yexcr.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8: e1002964 10.1371/journal.pgen.1002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MRF, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. 2015. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am J Hum Genet 97: 343–352. 10.1016/j.ajhg.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart JC, Li G-W. 2018. Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst 7: 580–589.e4 10.1016/j.cels.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia NR, Ayari N, Hutaff-Lee C, Boada R. 2012. Attention-deficit hyperactivity disorder symptoms in children and adolescents with sex chromosome aneuploidy: XXY, XXX, XYY, and XXYY. J Dev Behav Pediatr 33: 309–318. 10.1097/DBP.0b013e31824501c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin JA, Lowe AM, Colan SD, Sleeper LA, Orav EJ, Clunie S, Messere J, Cox GF, Lurie PR, Hsu D, et al. 2006. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 296: 1867–1876. 10.1001/jama.296.15.1867 [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, Hardy J, Ryten M, Consortium NABE. 2013. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun 4: 2771 10.1038/ncomms3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, et al. 2017. Landscape of X chromosome inactivation across human tissues. Nature 550: 244–248. 10.1038/nature24265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Vakilian H, Mirzaei M, Sharifi Tabar M, Pooyan P, Habibi Rezaee L, Parker L, Haynes PA, Gourabi H, Baharvand H, Salekdeh GH. 2015. DDX3Y, a male-specific region of Y chromosome gene, may modulate neuronal differentiation. J Proteome Res 14: 3474–3483. 10.1021/acs.jproteome.5b00512 [DOI] [PubMed] [Google Scholar]
- Valiente-Echeverría F, Hermoso MA, Soto-Rifo R. 2015. RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev Med Virol 25: 286–299. 10.1002/rmv.1845 [DOI] [PubMed] [Google Scholar]
- van der Walt S, Colbert SC, Varoquaux G. 2011. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 13: 22–30. 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523. 10.1038/ng.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann C-L, et al. 2019. The translational landscape of the human heart. Cell 178: 242–260.e29. 10.1016/j.cell.2019.05.010 [DOI] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17: 261–272. 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PH, Falcao CL, Hanstein R, Zimmer J. 2008. The AZF proteins. Int J Androl 31: 383–394. 10.1111/j.1365-2605.2008.00890.x [DOI] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, Schofield CJ, Johansson C. 2014. Human UTY(KDM6C) is a male-specific Nɛ-methyl lysyl demethylase. J Biol Chem 289: 18302–18313. 10.1074/jbc.M114.555052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Zinn AR, Page DC, Nishimoto T. 1993. Functional equivalence of human X- and Y-encoded isoforms of ribosomal protein S4 consistent with a role in Turner syndrome. Nat Genet 4: 268–271. 10.1038/ng0793-268 [DOI] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci 109: 13004–13009. 10.1073/pnas.1210787109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizemann TM, Pardue M. 2001. Exploring the biological contributions to human health: Does sex matter? National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. 2002. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet 11: 1409–1419. 10.1093/hmg/11.12.1409 [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Disteche CM. 2008a. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3: e2553 10.1371/journal.pone.0002553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. 2008b. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci 28: 4521–4527. 10.1523/JNEUROSCI.5382-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.