Abstract
With renewed interest in free energy methods in contemporary structure-based drug design there is a pressing need to validate against multiple targets and force fields to assess the overall ability of these methods to accurately predict relative binding free energies. We computed relative binding free energies using GPU accelerated Thermodynamic Integration (GPU-TI) on a dataset originally assembled by Schrödinger, Inc.. Using their GPU free energy code (FEP+) and the OPLS2.1 force field combined with the REST2 enhanced sampling approach, these authors obtained an overall MUE of 0.9 kcal/mol and an overall RMSD of 1.14 kcal/mol. In our study using GPU-TI from AMBER with the AMBER14SB/GAFF1.8 force field but without enhanced sampling, we obtained an overall MUE of 1.17 kcal/mol and an overall RMSD of 1.50 kcal/mol for the 330 perturbations contained in this data set. A more detailed analyses of our results suggested that the observed differences between the two studies arise from differences in sampling protocols along with differences in the force fields employed. Future work should address the problem of establishing benchmark quality results with robust statistical error bars obtained through multiple independent runs and enhanced sampling, which is possible with the GPU-accelerated features in AMBER.
Graphical Abstract
1. Introduction
Protein-ligand binding affinity calculation has drawn a lot of attention for many years because of its ability to significantly accelerate drug discovery by focusing experimental effort on high quality leads.1–4 Using molecular dynamics (MD) or Monte Carlo algorithms to perform the necessary sampling, many methods have been developed to address this task5–19: These include the linear interaction energy (LIE) approach5–6, the Molecular Mechanics-Poisson−Boltzmann/Surface Area (MM-PBSA) and Molecular Mechanics-Generalized Born/Surface Area (MM-GBSA)7–10 approaches, and physical pathway methods such as the Umbrella Sampling (US) method11–12 combined with Weighted Histogram Analysis (WHAM)20–21 or the variational free energy profile (vFEP) methods22–23.
Another class of methods is the alchemical methods, where the thermodynamic path between two end states is defined and the free energy change is calculated based on statistical mechanical analysis of the simulations. Based on the theoretical foundation of Kirkwood, Zwanzig and Bennett among others,15–19, 24–25 the three most widely used alchemical methods are Thermodynamic Integration (TI),16 Free Energy Perturbation (FEP),24 and (Multistate) Bennett Acceptance Ratio (MBAR).15, 19 FEP is based on the Zwanzig equation by considering the process of transforming A → B. The free energy change can be written as:
| (1) |
where UB and UA are the potential energy of A and B, kB is the Boltzmann constant, and T is the temperature. From a practical perspective, this method is limited to small perturbations due to convergence and end state catastrophe issues, which to some extent is addressed via introducing a coupling parameter λ that varies from 0 to 1. The intermediate state λ has a potential energy U(λ), which is equal to U(A) when λ is 0, and to U(B) when λ is 1. TI computes the free energy change of this transformation by integrating the Boltzmann averaged ∂U(λ)/∂λ:
| (2) |
Different functional forms of U(λ) have been constructed and tested.26–27 MBAR calculates the free energy difference between neighboring intermediate states using:
| (3) |
where w is a function of F(λ) and F(λ + 1). The equation is solved iteratively to give the free energy change of neighboring states ΔF(λ → λ + 1), which via combination yield the overall free energy change.
Another notable method is λ-dynamics,17 pioneered by Brooks and co-workers. It combines the idea of alchemical methods and US method by treating the λ variable dynamically, and generates a potential of mean force in λ-space. The initial application studies of alchemical methods were performed a few decades ago by a number of groups.2, 28–29 Since this time, a number of applications in both academia and industry have been reported.30–43 However, to more reliably drive decisions in lead optimization, there are two major issues that alchemical methods should deal with, namely, adequate sampling of relevant configurations and force field accuracy.3 To address the sampling issues methods like Hamiltonian exchange and replica exchange have been developed,44–46 while improvements in force fields, like CGenFF,47 AMOEBA,48 OPLS49 and AMBER/GAFF,50–51 have been an ongoing effort and will continue to be for the foreseeable future.
Recently, a FEP protocol was described that was able to predict the relative binding affinity over a broad range of protein-ligand complexes.1 This approach used replica exchange with solute tempering (REST2) in combination with the OPLS2.1 force field that was carefully parameterized using an extensive training set of relevant torsional and covalent interactions.1 The AMBER ff14SB/GAFF force fields50, 52 are widely used, but have not been extensively validated against such a large data set (8 systems and 330 perturbations). To fill this gap we employed the recently implemented53–54 GPU-accelerated TI module of Amber18 to repeat the calculations for these systems in order to assess the capabilities of another force field model on this same data set. Overall, our computed results have larger average errors than the FEP+ result. The overall mean unsigned error (MUE) and root mean square deviation (RMSD) for FEP+ versus AMBER are 0.90 versus 1.17 kcal/mol, and 1.14 versus 1.50 kcal/mol, respectively. This indicates that the GPU-TI module of AMBER using the AMBER ff14SB/GAFF 1.8 force field is an alternative choice for high-throughput relative binding free energy calculations, albeit with slightly larger overall errors.
2. Methods
2.1. System preparation
All of the protein and ligand PDBs were obtained from the SI of the Wang et al. publication.1 The atom names of the ligands were modified manually so that the common atoms of the ligands in each protein system have the same name and the unique atoms have different names. The protonation states of all the charged residues as well as Histidine residues were maintained as reported in Wang et al.. The AMBER FF14SB force field55 was employed to describe the proteins and GAFF (version 1.8) was50 used for the ligands. Restrained electrostatic potential (RESP) charges for the ligands were calculated at the HF/6–31G(d) level of theory using the Gaussian 09 program56 and AMBERTools16. The parmchk utility from AMBERTools16 was used to generate the missing parameters for the ligands. The systems were solvated using the SPC/E57 water model using cubic simulation cells. 5 Å and 10 Å were used as the minimum distance between the edge of the cell and the solute atoms of the protein and ligand systems, respectively. The resulting solvated protein/ligand systems were then charge neutralized by adding Na+ or Cl− ions58. The particle mesh Ewald (PME) method59–60 was used to treat the long-range electrostatic interactions. All bonds involving hydrogen atoms were constrained using SHAKE61. The AMBER16 package62 was used to run the MD simulations. MD simulations for each protein-ligand system were performed to equilibrate the systems. Five steps of minimization were performed to remove close contacts. The first step minimizes the water molecules and counter ions, with the protein restrained. The second, third and fourth step restrains the heavy atoms, backbone heavy atoms, backbone carbon and oxygen atoms of the protein, while the last step minimizes the entire system. Each minimization step consisted of 20000 cycles of minimization using the steepest descent method. Afterwards the system was heated from 0 K to 300 K using the Langevin thermostat with a collision frequency of 2.5 ps−1. The solute was restrained using a 5 kcal/(mol*Å2) restraining potential. Finally, the system was equilibrated at 300 K for 5 ns employing the NPT ensemble using a Langevin thermostat with a collision frequency of 1 ps−1. The Berendsen barostat was used for the pressure control with a pressure relaxation time of 10 ps. The time step was 2 fs and the nonbonded cutoff was 12 Å. The last snapshot was used to generate a pdb file. Using the generated pdb file, all the protein atoms were duplicated along with the common atoms of the second ligand. The unique atoms of the second ligand were added according to the mol2 file of the second ligand, the coordinates of which were obtained from the input files from Wang, et al. The “timerge” function of the parmed.py utility of the AMBER 14 package was used to generate the dual ligand topology.
2.2. TI simulations
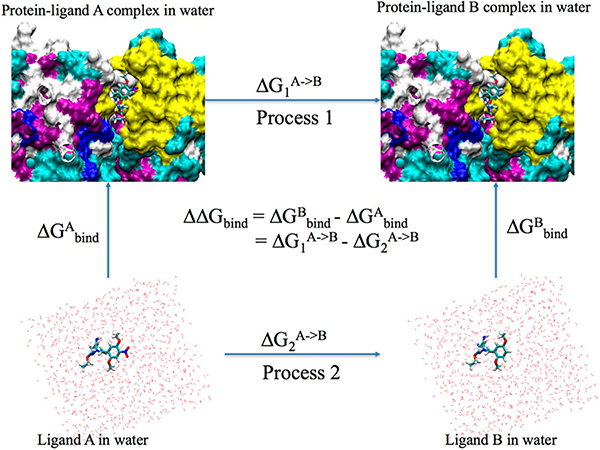
As shown in Figure 1, the relative binding free energy (ΔΔG) can be calculated as the difference between the free energies (ΔGs) of changing one ligand to the other in the protein matrix and in solution. Therefore, TI simulations for both process 1 and 2 were performed. For each process, the one-step protocol was adopted, i.e. disappearing one ligand and appearing the other ligand simultaneously. The common atoms of the two ligands were linearly transformed and the unique atoms were in the softcore region. Both the charge and vdW interactions between the disappearing (or appearing) unique atoms with the surrounding atoms were described by softcore potentials. Alternatively one can use the 3-step protocol, which consists of three steps: disappearing the charge interaction of one ligand, changing the vdW and bonded terms, and then appearing the charge of the second ligand. The one-step protocol not only takes less steps but also has the same charge for the initial and final state of the TI simulation. However, for the 3-step protocol, the charge of the system may change during the decharging/charging steps, which may affect the long-range electrostatic interactions via the use of a neutralizing background plasma in AMBER.
Figure 1.
Thermodynamic cycle used for the calculation of the relative binding free energy between protein-ligand system A and protein-ligand system B.
2.2.1. The one-step protocol
The detailed TI simulation protocol is as follows: First, using the dual ligand topology parameter file, 50000 steps of steepest descent minimization was performed. Then the system was heated from 0 to 300 K at the ps timescale, followed by 1 ns NVT equilibration at 300 K. Afterwards 1 ns pf NPT equilibration at 300 K and 1 bar was performed to equilibrate the density. These simulations were performed at λ=0.5 to equilibrate the system63–64. No restraint was applied for these simulations and all structures were visually checked. For some perturbations, multiple runs had to be performed in order to obtain a stable starting structure. Afterwards the equilibrated structure was used as the starting structure for 12 λ windows (0.00922, 0.04794, 0.11505, 0.20634, 0.31608, 0.43738, 0.56262, 0.68392, 0.79366, 0.88495, 0.95206, 0.99078). For each λ, 1 ns of NVT equilibration was performed with the initial velocities randomly generated to give a temperature of 300 K. Afterwards 5 ns of NVT simulation was performed to collect ∂U/∂λ data. A 12-point Gaussian quadrature was used for the numerical integration of ∂U/∂λ to obtain all necessary ΔG values. The non-bonded interaction cutoff was 9.0 Å and a softcore potential26–27 was used. The parameter α and β of softcore potential was 0.5 and 12 Å2, respectively. The time step was 1 fs for all simulations and SHAKE was not used. All TI simulations used the Berendsen thermostat with a coupling constant of 2 ps, except for the NPT equilibration step, which used the Langevin thermostat with a collision frequency of 2 ps−1. We note that the Langevin thermostat is generally preferred over the Berendsen thermostat. The Berendsen barostat was used for NPT equilibration with a pressure relaxation time of 2 ps. NPT equilibration was performed using the CPU version of pmemd from the Amber14 package. The obtained results for all eight systems can be found in the Supplemental Information (SI). The input files are available at GitHub: https://github.com/linfranksong/Input_TI
3. Results and Discussion
3.1. Overall results
The ΔΔG values directly obtained from the TI calculations can be found in the SI. The mean unsigned error (MUE) and root mean square deviation (RMSD) of these values compared to experiment are summarized in SI Table 1. After obtaining the ΔΔG values, we employed the cycle closure convergence strategy described previously65 and obtained our final ΔΔG values. Thus, the following analysis is based on the cycle-closure ΔΔG values. Table 1 summarizes the MUE and RMSD compared to experiment. The overall MUE obtained with GPU-TI of AMBER using the AMBER FF14SB/GAFF1.8 force field (AMBER for short) is 1.17 kcal/mol (0.27 kcal/mol larger than FEP+. Similarly, the RMSD is a bit higher for AMBER: 1.50 kcal/mol versus 1.14 kcal/mol for FEP+. Moreover, in our current work, we did not apply replica exchange, which could help enhance the overall sampling and improve the quality of the computed free energies. Future work will explore the role sampling (both in λ-space and phase space) plays on these systems versus the effect of force field errors.
Table 1.
Summary of the MUE and RMSD, R2 and Kendall's tau coefficient (τ) of the eight systems based on cycle closure ΔΔG values.
| System | # of ligands | # of perturbations | FEP+/OPLS 2.1 (kcal/mol) | AMBER GPU-TI/AMBER FF14SB + GAFF (1.8) (kcal/mol) | Difference* (kcal/mol) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MUE | RMSD | R2 | τ | MUE | RMSD | R2 | τ | MUE | RMSD | |||
| Thrombin | 11 | 16 | 0.76 | 0.93 | 0.17 | 0.21 | 0.46 | 0.62 | 0.50 | 0.54 | −0.30 | −0.31 |
| Tyk2 | 16 | 24 | 0.75 | 0.93 | 0.48 | 0.54 | 1.07 | 1.27 | 0.24 | 0.26 | 0.32 | 0.34 |
| Jnk1 | 21 | 31 | 0.78 | 1.00 | 0.35 | 0.44 | 1.07 | 1.45 | 0.05 | 0.23 | 0.29 | 0.45 |
| CDK2 | 16 | 25 | 0.91 | 1.11 | 0.15 | 0.30 | 0.97 | 1.13 | 0.35 | 0.46 | 0.06 | 0.02 |
| PTP1B | 23 | 49 | 0.89 | 1.22 | 0.43 | 0.55 | 1.06 | 1.40 | 0.35 | 0.51 | 0.17 | 0.18 |
| BACE | 36 | 58 | 0.84 | 1.03 | 0.37 | 0.36 | 1.20 | 1.47 | 0.27 | 0.31 | 0.36 | 0.44 |
| MCL1 | 42 | 71 | 1.16 | 1.41 | 0.26 | 0.35 | 1.52 | 1.83 | 0.16 | 0.28 | 0.36 | 0.42 |
| P38a | 34 | 56 | 0.80 | 1.03 | 0.62 | 0.60 | 1.20 | 1.56 | 0.31 | 0.39 | 0.40 | 0.53 |
| Overall | 199 | 330 | 0.90 | 1.14 | 0.36 | 0.44 | 1.17 | 1.50 | 0.23 | 0.34 | 0.27 | 0.36 |
The difference is calculated as AMBER MUE or RMSD minus Schrodinger MUE or RMSD.
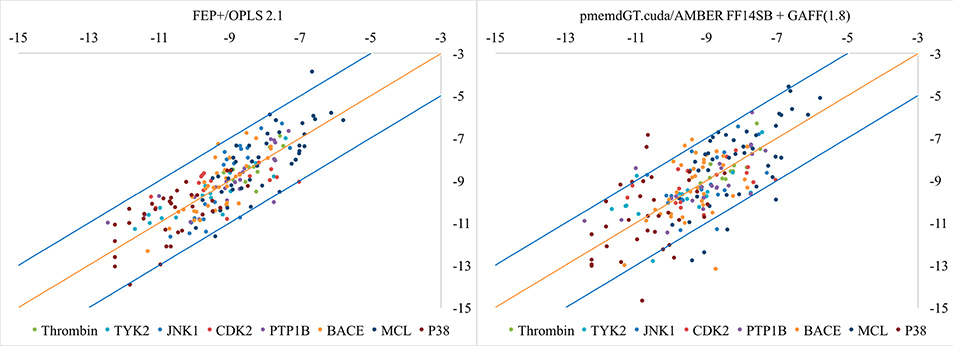
With the cycle-closure ΔΔG values, we obtained the ΔG values following the procedure of Wang, et al.. In short, in this procedure all of the ligands’ experimental values were used as a reference, and the sum of the predicted ΔG values was set to be equal to the sum of the experimental ΔG values. Though this way of calculating the offset can artificially improve the overall results, we adopted this procedure in order to better compare with Wang, et al. The predicted ΔG values were plotted against experimental ΔG values in Figure 2. We can see AMBER performs worse than FEP+. Out of the 199 ligands, 5 ligands (2.5%) for Schrödinger and 18 ligands (9.0%) for AMBER are more than 2kcal/mol off from experiment. The R2 and Kendall’s tau coefficient are listed in Table 2. Figure 3 shows the individual plots of predicted versus experimental ΔG values for each of the 8 systems for both FEP+ and AMBER TI.
Figure 2.
Correlation between predicted binding free energies and experimental data for the eight systems.
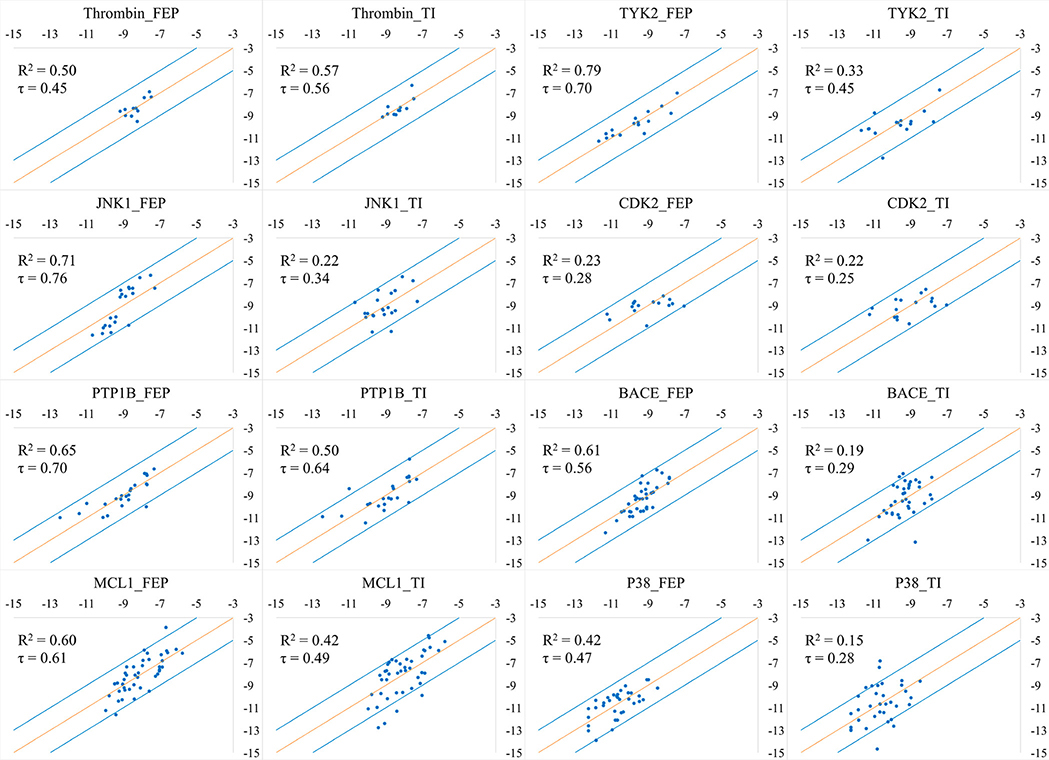
Table 2.
R2 and Kendall's tau coefficient for the correlation between predicted binding free energies and experimental data for the eight systems; τ represents the Kendall's tau coefficient.
| System | # of ligands | # of perturbations | FEP+/OPLS 2.1 (kcal/mol) | AMBER GPU-TI/AMBER FF14SB + GAFF (1.8) (kcal/mol) | ||
|---|---|---|---|---|---|---|
| R2 | τ | R2 | τ | |||
| Thrombin | 11 | 16 | 0.50 | 0.45 | 0.57 | 0.56 |
| Tyk2 | 16 | 24 | 0.79 | 0.70 | 0.33 | 0.45 |
| Jnk1 | 21 | 31 | 0.71 | 0.76 | 0.22 | 0.34 |
| CDK2 | 16 | 25 | 0.23 | 0.28 | 0.22 | 0.25 |
| PTP1B | 23 | 49 | 0.65 | 0.70 | 0.50 | 0.64 |
| BACE | 36 | 58 | 0.61 | 0.56 | 0.19 | 0.29 |
| MCL1 | 42 | 71 | 0.60 | 0.61 | 0.42 | 0.49 |
| P38a | 34 | 56 | 0.42 | 0.47 | 0.15 | 0.28 |
| Overall | 199 | 330 | 0.66 | 0.62 | 0.44 | 0.48 |
Figure 3:
Correlation between the predicted binding free energies and experimental data for the eight systems studied herein. X axis: Experimental ΔG (kcal/mol); Y axis: Predicted ΔG (kcal/mol). τ is the Kendall’s tau coefficient.
3.2. Uncertainty estimate
To estimate the uncertainty in the calculations, we randomly selected 2 perturbations from each of the 8 systems and repeated the calculations described in section 2.2.1 twice. From Table 3, we can see most of the perturbations have standard deviations of less than 0.5 kcal/mol, except 4 of the perturbations. The overall standard deviation is 0.33 kcal/mol.
Table 3.
Estimate of the uncertainty of the calculations.
| System | Ligand 1 | Ligand 2 | Run_1 (kcal/mol) | Run_2 (kcal/mol) | Run_3 (kcal/mol) | Average (kcal/mol) | Standard Deviation (kcal/mol) |
|---|---|---|---|---|---|---|---|
| Thrombin | 1d | 1c | −0.20 | −0.15 | −0.15 | −0.17 | 0.03 |
| 6e | 6b | 0.60 | 0.75 | 0.75 | 0.70 | 0.09 | |
| TYK2 | ejm 31 | ejm 46 | −0.75 | −0.85 | −0.65 | −0.75 | 0.10 |
| jmc 28 | jmc 30 | −2.00 | −2.00 | −1.90 | −1.97 | 0.06 | |
| JNK1 | 18626–1 | 18624–1 | 1.50 | 0.95 | 1.05 | 1.17 | 0.29 |
| 18659–1 | 18634–1 | −0.95 | −1.10 | −0.35 | −0.80 | 0.40 | |
| CDK2 | 22 | lhlr | −0.55 | −0.90 | −0.70 | −0.72 | 0.18 |
| 1oiy | 1h1q | 1.65 | 1.70 | 2.85 | 2.07 | 0.68 | |
| PTP1B | 23466 | 23475 | −1.50 | −1.60 | −2.65 | −1.92 | 0.64 |
| 20670(2qbs) | 23330(2qbq) | −1.65 | −1.40 | −1.85 | −1.63 | 0.23 | |
| BACE | CAT-13a | CAT-17g | 1.95 | 1.10 | 1.65 | 1.57 | 0.43 |
| CAT-4p | CAT-13k | −1.45 | −1.20 | −0.85 | −1.17 | 0.30 | |
| MCL1 | 26 | 57 | −0.85 | −1.05 | −1.00 | −0.97 | 0.10 |
| 68 | 45 | −0.75 | −0.70 | −0.85 | −0.77 | 0.08 | |
| P38 | p38a_2aa | p38a_2bb | −1.35 | −0.20 | 0.65 | −0.30 | 1.00 |
| p38a_3fly | p38a 3fmh | 0.00 | −0.35 | 0.85 | 0.17 | 0.62 | |
| Overall | 0.33 | ||||||
3.3. The “problematic cases”
As alluded to in section 2.2.1, for some perturbations, multiple runs at λ=0.5 had to be run in order to obtain a stable starting structure; for example, the ligand significantly moved in the binding pocket or the conformation of the protein changed. In order to understand the origin of this problem better, we visually checked the initial structures and the structures after minimization, and found that there were a few cases that had close contacts in the initial structure, but after minimization, the structures had improved. No clashes between the ligand and the binding site of the protein were observed after minimization. We next hypothesized that our heating protocol was too fast, which caused the observed structural issues. Hence, we repeated the “problematic cases” with a more rigorous minimization, heating and equilibration procedure. Five steps of minimization were performed to remove close contacts. The first step minimized the water molecules and the counter ions, with the protein restrained. The second, third and fourth step restrained the heavy atoms, backbone heavy atoms, backbone carbon and oxygen atoms of the protein, while the last step minimized the entire system. Each minimization step consisted of 20000 cycles of minimization using the steepest descent method. Afterwards the system was heated from 0 K to 300 K gradually over 1 ns with a coupling restraint of 5 kcal/(mol*Å2) on the solute, followed by equilibration at 300 K using the NPT ensemble for 200 ps with the same restraint. Then another 200 ps of NPT equilibration with a weaker restraint (2 kcal/(mol*Å2)) was performed. Finally the restraint was released and the system was equilibrated using NPT conditions for 600 ps. With these settings, the simulations successfully finished and the structures appeared fine after visual inspection. With the equilibrated structure, 12 λ windows were used for data collection with similar settings except: 1) the initial velocity was taken from the equilibrated structure as well as the coordinates; 2) the two end windows (0.00922 and 0.99078) used the velocity and coordinates from the equilibrated structure of the neighboring window (0. 04794 and 0. 95206). A few other differences between these new simulations and the former simulations include: 1) parmchk2 was used to generate the missing bond/angle/dihedral parameters for the ligands; 2) 22 and 12 Å was used as the minimum distance between the edge of the solvated cell and the ligand and protein/ligand systems respectively; 3) the protein/ligand system was thermalized more gradually and more steps of equilibration were used; 4) the Langevin thermostat was used with a collision frequency of 2 ps−1 for all the TI simulations; 5) the CPU version of the AMBER 18 package was used instead of the AMBER 14 package for the TI simulations under NPT conditions. The overall MUE and RMSD for these perturbations are about the same: 1.61 kcal/mol and 2.09 kcal/mol for the new protocol versus 1.52 kcal/mol and 1.93 kcal/mol for the former protocol. Even so, some of the individual changes were significant, but given the differing box sizes, thermalization protocols, thermostats, etc. this wasn’t particularly surprising. Nonetheless, the average performance is relatively insensitive to the protocol employed. These data are summarized in the spreadsheet provided in the SI.
3.4. The 3-step protocol
A recent publication highlighted differences between the one-step protocol and a 3-step protocol when employing AMBER TI calculations.66 In order to explore the impact of using one protocol over the other we performed 3-step calculations for one of the systems, i.e. the JNK1 system. As discussed above, the 3-step protocol consists of disappearing the charge, a vdW change and a charge reappearance step. For each step, the same minimization, heating and equilibration was performed at λ=0.5 as described in section 3.3. The equilibrated structure and velocities were used for the 12λ window TI calculation. The remaining settings for the TI calculations were the same as in section 2.2.1. We found that the MUE and RMSD is nearly the same as the one step protocol: 1.11 versus 1.07 kcal/mol, 1.43 versus 1.45 kcal/mol, respectively. This suggests that although there are differences between the two protocols that are worthy of in-depth exploration, the overall performance using either protocol is about the same, using the current code base and force fields. These data are summarized in SI spread sheet “FEP_vs_GTI-dG-SI”.
3.5. Discussion
In the SI (see Trend.xlsx) we summarize all of the 330 perturbations. Overall, we find that AMBER performs reasonably well for perturbations between halogens and H, CH3 or CH2CH3: 44 of 49 perturbations have errors less than 2 kcal/mol, 34 of which have an error less than 1 kcal/mol. Perturbations involving large van der Waals radii changes, like Br to H or I to H, tend to have larger errors. We further analyzed the perturbations based on the “size” of the perturbation; whether there is ring appearance/disappearance or whether there is a ring type change (for example, pyridine to benzene). We classified perturbations that involved 3 heavy atoms changing or more as “big change” perturbations, and the others as “small change” perturbations. AMBER performs well for “big change” as well as “small change” perturbations: 151 of the 194 “big change” perturbations (~80%) have errors less than 2 kcal/mol, 99 of which have errors less than 1 kcal/mol; 107 of the 136 “small change” perturbations have errors less than 2 kcal/mol, 99 of which have errors less than 1 kcal/mol. Compared to “big change” perturbations, a larger percentage of “small change” perturbations have errors less than 1 kcal/mol: 69% for “small change” vs 51% for “big change” perturbations. Moreover, ring disappearance/appearance and ring type changes are also often seen in perturbation studies and they’re present in this data set as well. From our analysis, we find that AMBER performs well for both: 54 of 68 ring disappearance/appearance perturbations have errors less than 2 kcal/mol, 35 of which have an error less than 1 kcal/mol; 52 of 78 ring type change perturbations have errors less than 2 kcal/mol, 29 of which have an error less than 1 kcal/mol. While it would have been helpful to find systematic issues within certain classes of perturbations when using the AMBER class of force fields, in order to help guide force field improvement efforts, we found this wasn’t the case in the present data set.
4. Conclusions
We repeated the relative binding free energy calculations on the data set described in previous work.1 Comparing to the Schrödinger FEP/OPLS 2.1 force field, GPU TI with AMBER FF14SB and the GAFF (1.8) force field performs reasonably well on this data set, with errors above those seen using the FEP/OPLS 2.1 force field. For the 330 perturbations, AMBER has MUE and RMSDs of 1.17 kcal/mol and 1.50 kcal/mol, which is a few tenths of kcal/mol larger than the reported values (0.90 kcal/mol and 1.14 kcal/mol).1 For the 199 ligands, most of the binding free energy values are within 2 kcal/mol, except for 18 ligands (versus 5 reported previously1). Interestingly, a null model, which assumes all the ΔΔG values are 0 kcal/mol, gives similar results (see SI Figure 1): 8 ligands are not within 2 kcal/mol. This is due to the small range of the experimental ΔG values: the widest range of ΔG values is 5.13 kcal/mol. To better demonstrate and test free energy approaches, data sets with larger experimental ΔG ranges should be explored. Future work will also explore the use of replica exchange and other features within AMBER to enhance the sampling (both in λ-space and orthogonal degrees of freedom). Along with technical advances we will also explore the capabilities of the next generation GAFF2 and protein force fields. Finally, test procedures for creating benchmark quality results with meaningful error estimates that can be used as a baseline for other comparisons will be explored.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pengfei Li (University of Yale, ORCID iD: https://orcid.org/0000-0002-2572-5935) for helpful discussions. This work was supported by NIH grants GM107485 (DMY). Computational resources were provided by the MSU HPC.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available:
FEP_vs_GTI-ddG-SI: ΔΔG values, cycle closure errors, uncertainty estimates, problematic perturbations.
FEP_vs_GTI-dG-SI: ΔG values.
SI_GPU-TI: MUE and RMSD based on ΔΔG values directly obtained from FEP or TI calculations; plot of predicted ΔG versus experimental ΔG values for a null model setting ΔΔG values to 0.
Perturbation graph-SI: Perturbation graph plotted based on Wang, et al.
Reference
- 1.Wang L; Wu YJ; Deng YQ; Kim B; Pierce L; Krilov G; Lupyan D; Robinson S; Dahlgren MK; Greenwood J; Romero DL; Masse C; Knight JL; Steinbrecher T; Beuming T; Damm W; Harder E; Sherman W; Brewer M; Wester R; Murcko M; Frye L; Farid R; Lin T; Mobley DL; Jorgensen WL; Berne BJ; Friesner RA; Abel R, Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. Journal of the American Chemical Society 2015, 137 (7), 2695–2703. [DOI] [PubMed] [Google Scholar]
- 2.Merz KM; Kollman PA, Free-Energy Perturbation Simulations of the Inhibition of Thermolysin - Prediction of the Free-Energy of Binding of a New Inhibitor. Journal of the American Chemical Society 1989, 111 (15), 5649–5658. [Google Scholar]
- 3.Mobley DL; Gilson MK, Predicting Binding Free Energies: Frontiers and Benchmarks. Annu Rev Biophys 2017, 46, 531–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mobley DL; Klimovich PV, Perspective: Alchemical free energy calculations for drug discovery. Journal of Chemical Physics 2012, 137 (23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aqvist J; Luzhkov VB; Brandsdal BO, Ligand binding affinities from MD simulations. Accounts Chem Res 2002, 35 (6), 358–365. [DOI] [PubMed] [Google Scholar]
- 6.Guitierrez-de-Teran H; Aqvist J, Linear Interaction Energy: Method and Applications in Drug Design. Methods Mol Biol 2012, 819, 305–323. [DOI] [PubMed] [Google Scholar]
- 7.Kollman PA; Massova I; Reyes C; Kuhn B; Huo SH; Chong L; Lee M; Lee T; Duan Y; Wang W; Donini O; Cieplak P; Srinivasan J; Case DA; Cheatham TE, Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts Chem Res 2000, 33 (12), 889–897. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn B; Kollman PA, Binding of a diverse set of ligands to avidin and streptavidin: An accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J Med Chem 2000, 43 (20), 3786–3791. [DOI] [PubMed] [Google Scholar]
- 9.Li Y; Liu ZH; Wang RX, Test MM-PB/SA on True Conformational Ensembles of Protein-Ligand Complexes. J Chem Inf Model 2010, 50 (9), 1682–1692. [DOI] [PubMed] [Google Scholar]
- 10.Rastelli G; Del Rio A; Degliesposti G; Sgobba M, Fast and Accurate Predictions of Binding Free Energies Using MM-PBSA and MM-GBSA. J Comput Chem 2010, 31 (4), 797–810. [DOI] [PubMed] [Google Scholar]
- 11.Souaille M; Roux B, Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput Phys Commun 2001, 135 (1), 40–57. [Google Scholar]
- 12.Torrie GM; Valleau JP, Non-Physical Sampling Distributions in Monte-Carlo Free-Energy Estimation - Umbrella Sampling. J Comput Phys 1977, 23 (2), 187–199. [Google Scholar]
- 13.Velez-Vega C; Gilson MK, Overcoming Dissipation in the Calculation of Standard Binding Free Energies by Ligand Extraction. J Comput Chem 2013, 34 (27), 2360–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ytreberg FM, Absolute FKBP binding affinities obtained via nonequilibrium unbinding simulations. Journal of Chemical Physics 2009, 130 (16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett CH, Efficient Estimation of Free-Energy Differences from Monte-Carlo Data. J Comput Phys 1976, 22 (2), 245–268. [Google Scholar]
- 16.Kirkwood JG, Statistical mechanics of fluid mixtures. Journal of Chemical Physics 1935, 3 (5), 300–313. [Google Scholar]
- 17.Kong XJ; Brooks CL, lambda-Dynamics: A new approach to free energy calculations. Journal of Chemical Physics 1996, 105 (6), 2414–2423. [Google Scholar]
- 18.Lee FS; Chu ZT; Bolger MB; Warshel A, Calculations of Antibody Antigen Interactions - Microscopic and Semimicroscopic Evaluation of the Free-Energies of Binding of Phosphorylcholine Analogs to Mcpc603. Protein Eng 1992, 5 (3), 215–228.1409541 [Google Scholar]
- 19.Shirts MR; Chodera JD, Statistically optimal analysis of samples from multiple equilibrium states. Journal of Chemical Physics 2008, 129 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S; Bouzida D; Swendsen RH; Kollman PA; Rosenberg JM, The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. J Comput Chem 1992, 13 (8), 1011–1021. [Google Scholar]
- 21.Kumar S; Rosenberg JM; Bouzida D; Swendsen RH; Kollman PA, Multidimensional Free-Energy Calculations Using the Weighted Histogram Analysis Method. J Comput Chem 1995, 16 (11), 1339–1350. [Google Scholar]
- 22.Lee TS; Radak BK; Pabis A; York DM, A New Maximum Likelihood Approach for Free Energy Profile Construction from Molecular Simulations. J Chem Theory Comput 2013, 9 (1), 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TS; Radak BK; Huang M; Wong KY; York DM, Roadmaps through Free Energy Landscapes Calculated Using the Multidimensional vFEP Approach. J Chem Theory Comput 2014, 10 (1), 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwanzig RW, High-Temperature Equation of State by a Perturbation Method .1. Nonpolar Gases. Journal of Chemical Physics 1954, 22 (8), 1420–1426. [Google Scholar]
- 25.Zwanzig RW; Kirkwood JG; Oppenheim I; Alder BJ, Statistical Mechanical Theory of Transport Processes .7. The Coefficient of Thermal Conductivity of Monatomic Liquids. Journal of Chemical Physics 1954, 22 (5), 783–790. [Google Scholar]
- 26.Steinbrecher T; Mobley DL; Case DA, Nonlinear scaling schemes for Lennard-Jones interactions in free energy calculations. Journal of Chemical Physics 2007, 127 (21). [DOI] [PubMed] [Google Scholar]
- 27.Steinbrecher T; Joung I; Case DA, Soft-Core Potentials in Thermodynamic Integration: Comparing One- and Two-Step Transformations. J Comput Chem 2011, 32 (15), 3253–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollman P, Free-Energy Calculations - Applications to Chemical and Biochemical Phenomena. Chem Rev 1993, 93 (7), 2395–2417. [Google Scholar]
- 29.Jorgensen WL, Free-Energy Calculations - a Breakthrough for Modeling Organic-Chemistry in Solution. Accounts Chem Res 1989, 22 (5), 184–189. [Google Scholar]
- 30.Luccarelli J; Michel J; Tirado-Rives J; Jorgensen WL, Effects of Water Placement on Predictions of Binding Affinities for p38 alpha MAP Kinase Inhibitors. J Chem Theory Comput 2010, 6 (12), 3850–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinbrecher T; Case DA; Labahn A, A multistep approach to structure-based drug design: Studying ligand binding at the human neutrophil elastase. J Med Chem 2006, 49 (6), 1837–1844. [DOI] [PubMed] [Google Scholar]
- 32.Stembrecher T; Hrenn A; Dormann KL; Merfort I; Labahn A, Bornyl (3,4,5-trihydroxy)-cinnamate - An optimized human neutrophil elastase inhibitor designed by free energy calculations. Bioorgan Med Chem 2008, 16 (5), 2385–2390. [DOI] [PubMed] [Google Scholar]
- 33.Lawrenz M; Wereszczynski J; Amaro R; Walker R; Roitberg A; McCammon JA, Impact of calcium on N1 influenza neuraminidase dynamics and binding free energy. Proteins-Structure Function and Bioinformatics 2010, 78 (11), 2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy MR; Erion MD, Calculation of relative binding free energy differences for fructose 1,6-bisphosphatase inhibitors using the thermodynamic cycle perturbation approach. Journal of the American Chemical Society 2001, 123 (26), 6246–6252. [DOI] [PubMed] [Google Scholar]
- 35.Palma PN; Bonifacio MJ; Loureiro AI; Soares-da-Silva P, Computation of the Binding Affinities of Catechol-O-methyltransferase Inhibitors: Multisubstate Relative Free Energy Calculations. J Comput Chem 2012, 33 (9), 970–986. [DOI] [PubMed] [Google Scholar]
- 36.Erion MD; Dang Q; Reddy MR; Kasibhatla SR; Huang J; Lipscomb WN; van Poelje PD, Structure-guided design of AMP mimics that inhibit fructose-1,6-bisphosphatase with high affinity and specificity. Journal of the American Chemical Society 2007, 129 (50), 15480–15490. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn B; Tichy M; Wang L; Robinson S; Martin RE; Kuglstatter A; Benz J; Giroud M; Schirmeister T; Abel R; Diederich F; Hert J, Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors. J Med Chem 2017, 60 (6), 2485–2497. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Benito L; Keranen H; van Vlijmen H; Tresadern G, Predicting Binding Free Energies of PDE2 Inhibitors. The Difficulties of Protein Conformation. Sci Rep 2018, 8 (1), 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciordia M; Perez-Benito L; Delgado F; Trabanco AA; Tresadern G, Application of Free Energy Perturbation for the Design of BACE1 Inhibitors. J Chem Inf Model 2016, 56 (9), 1856–71. [DOI] [PubMed] [Google Scholar]
- 40.Steinbrecher TB; Dahlgren M; Cappel D; Lin T; Wang L; Krilov G; Abel R; Friesner R; Sherman W, Accurate Binding Free Energy Predictions in Fragment Optimization. J Chem Inf Model 2015, 55 (11), 2411–20. [DOI] [PubMed] [Google Scholar]
- 41.Raman EP; Vanommeslaeghe K; Mackerell AD Jr., Site-Specific Fragment Identification Guided by Single-Step Free Energy Perturbation Calculations. J Chem Theory Comput 2012, 8 (10), 3513–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cappel D; Hall ML; Lenselink EB; Beuming T; Qi J; Bradner J; Sherman W, Relative Binding Free Energy Calculations Applied to Protein Homology Models. J Chem Inf Model 2016, 56 (12), 2388–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park H; Lee S, Homology modeling, force field design, and free energy simulation studies to optimize the activities of histone deacetylase inhibitors. J Comput Aided Mol Des 2004, 18 (6), 375–88. [DOI] [PubMed] [Google Scholar]
- 44.Jiang W; Roux B, Free Energy Perturbation Hamiltonian Replica-Exchange Molecular Dynamics (FEP/H-REMD) for Absolute Ligand Binding Free Energy Calculations. J Chem Theory Comput 2010, 6 (9), 2559–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khavrutskii IV; Wallqvist A, Improved Binding Free Energy Predictions from Single-Reference Thermodynamic Integration Augmented with Hamiltonian Replica Exchange. J Chem Theory Comput 2011, 7 (9), 3001–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L; Berne BJ; Friesner RA, On achieving high accuracy and reliability in the calculation of relative protein-ligand binding affinities. P Natl Acad Sci USA 2012, 109 (6), 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I; MacKerell AD, CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J Comput Chem 2010, 31 (4), 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ponder JW; Wu CJ; Ren PY; Pande VS; Chodera JD; Schnieders MJ; Haque I; Mobley DL; Lambrecht DS; DiStasio RA; Head-Gordon M; Clark GNI; Johnson ME; Head-Gordon T, Current Status of the AMOEBA Polarizable Force Field. J Phys Chem B 2010, 114 (8), 2549–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harder E; Damm W; Maple J; Wu CJ; Reboul M; Xiang JY; Wang LL; Lupyan D; Dahlgren MK; Knight JL; Kaus JW; Cerutti DS; Krilov G; Jorgensen WL; Abel R; Friesner RA, OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J Chem Theory Comput 2016, 12 (1), 281–296. [DOI] [PubMed] [Google Scholar]
- 50.Wang JM; Wolf RM; Caldwell JW; Kollman PA; Case DA, Development and testing of a general amber force field. J Comput Chem 2004, 25 (9), 1157–1174. [DOI] [PubMed] [Google Scholar]
- 51.Case DA, I. Y. B-S, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro WD V., Darden TA,; Duke RE, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Huang Y,; Izadi S, A. K., Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, M. M. K, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, R. R. D, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, SalomonFerrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DMand Kollman PA, AMBER 2018, University of California, San Francisco: 2018. [Google Scholar]
- 52.Case DA, R. MB, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H,; Goetz AW, N. H., Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, T. L., Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, A. O., Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J,; Walker RC, J. W., Wolf RM, Wu X, Xiao L and Kollman PA, AMBER 2016, University of California, San Francisco: 2016. [Google Scholar]
- 53.Lee TS; Hu Y; Sherborne B; Guo Z; York DM, Toward Fast and Accurate Binding Affinity Prediction with pmemdGTl: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration. J Chem Theory Comput 2017, 13 (7), 3077–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee TS; Cerutti DS; Mermelstein D; Lin C; LeGrand S; Giese TJ; Roitberg A; Case DA; Walker RC; York DM, GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J Chem Inf Model 2018, 58 (10), 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Case DA, V. B., Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE III, Darden TA, R.E.; Duke HG, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A,; Lee TS, S. L., Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C,; Salomon-Ferrer R, G. S., Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, X.; Kollman W. a. P. A., AMBER 14, University of California, San Francisco: 2014. [Google Scholar]
- 56.Frisch MJ, G. W. T., Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, and Fox DJ, Gaussian 09 (Gaussian, Inc., Wallingford CT, 2009). [Google Scholar]
- 57.Berendsen HJC; Grigera JR; Straatsma TP, The Missing Term in Effective Pair Potentials. J Phys Chem-Us 1987, 91 (24), 6269–6271. [Google Scholar]
- 58.Joung IS; Cheatham TE, Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B 2008, 112 (30), 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darden T; York D; Pedersen L, Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics 1993, 98 (12), 10089–10092. [Google Scholar]
- 60.Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG, A Smooth Particle Mesh Ewald Method. Journal of Chemical Physics 1995, 103 (19), 8577–8593. [Google Scholar]
- 61.Ryckaert JP; Ciccotti G; Berendsen HJC, Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J Comput Phys 1977, 23 (3), 327–341. [Google Scholar]
- 62.Case DA, R. M. B., Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H,; Goetz AW, N. H., Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, C.; Lin TL, Luo R, Madej B, Mermelstein D, K.M., Merz G, Monard H, Nguyen HT, Nguyen I; Omelyan AO, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J,; Walker RC, J. W., Wolf RM, Wu X, Xiao L and Kollman PA (2016),, AMBER 2016, University of California, San Francisco. [Google Scholar]
- 63.Ucisik MN; Hammes-Schiffer S, Relative Binding Free Energies of Adenine and Guanine to Damaged and Undamaged DNA in Human DNA Polymerase eta: Clues for Fidelity and Overall Efficiency. Journal of the American Chemical Society 2015, 137 (41), 13240–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaus JW; Pierce LT; Walker RC; McCammon JA, Improving the Efficiency of Free Energy Calculations in the Amber Molecular Dynamics Package. J Chem Theory Comput 2013, 9 (9), 4131–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang LL; Deng YQ; Knight JL; Wu YJ; Kim B; Sherman W; Shelley JC; Lin T; Abel R, Modeling Local Structural Rearrangements Using FEP/REST: Application to Relative Binding Affinity Predictions of CDK2 Inhibitors. J Chem Theory Comput 2013, 9 (2), 1282–1293. [DOI] [PubMed] [Google Scholar]
- 66.Loeffler HH; Bosisio S; Duarte Ramos Matos G; Suh D; Roux B; Mobley DL; Michel J, Reproducibility of Free Energy Calculations across Different Molecular Simulation Software Packages. J Chem Theory Comput 2018, 14 (11), 5567–5582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.