Abstract
Salmonella Infantis is one of the five serovars most frequently causing human salmonellosis in Europe, mainly associated with poultry. A clone harbouring a conjugative plasmid of emerging S. Infantis (pESI)-like megaplasmid, carrying multidrug resistant (MDR) and extended-spectrum beta-lactamases (ESBL) genes, has spread in the Italian broiler chicken industry also causing human illness. This work is aimed at elucidating the molecular epidemiology of S. Infantis and pESI-like in Europe using whole-genome sequencing and bioinformatics analysis, and to investigate the genetic relatedness of S. Infantis clones and pESI-like from animals, meat, feed and humans provided by institutions of nine European countries. Two genotyping approaches were used: chromosome or plasmid SNP-based analysis and the minimum spanning tree (MST) algorithm based on core-genome multilocus sequence typing (cgMLST). The European S. Infantis population appeared heterogeneous, with different genetic clusters defined at core-genome level. However, pESI-like variants present in 64.1 % of the isolates were more genetically homogeneous and capable of infecting different clonal lineages in most of the countries. Two different pESI-like with ESBL genes (n=82) were observed: bla CTX-M-1-positive in European isolates and bla CTX-M-65-positive in American isolates (study outgroup). Both variants had toxin-antitoxin systems, resistance genes towards tetracyclines, trimethoprim, sulphonamides and aminoglycosides, heavy metals (merA) and disinfectants (qacEΔ). Worryingly, 66 % of the total isolates studied presented different gyrA chromosomal point mutations associated with (fluoro)quinolone resistance (MIC range 0.125–0.5 mg/L), while 18 % displayed transferable macrolide resistance mediated by mph, mef and erm(B) genes. Proper intervention strategies are needed to prevent further dissemination/transmission of MDR S. Infantis and pESI-like along the food chain in Europe.
Keywords: Salmonella Infantis, pESI-like, megaplasmids, ESBL (Extended Spectrum Beta-Lactamases), whole genome sequencing, multidrug resistance
Data Summary
Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession numbers PRJEB23728 and PRJEB34620. Individual genome accession numbers are reported in Table S1 (available with the online version of this article).
Impact Statement.
Insights into plasmids such as the pESI-like megaplasmid and other mobile genetic elements across and beyond Europe, and into predictors of their success in animal productions, are considered very helpful in the surveillance of zoonotic disease agents. The success of this ‘parasitic’ megaplasmid in different S. Infantis clones is likely to have been favoured by the fact that while providing determinants enhancing colonization, resistance, virulence and fitness, it displays post-segregational toxin-antitoxin based killing mechanisms that prevent its cure. These genetic elements often provide to zoonotic bacterial hosts additional traits of virulence, fitness and resistance to various antimicrobial drugs, including those classified as highest priority critically important to humans. The presence of pESI-like may be associated with the success of S. Infantis in the European poultry system.
Introduction
Salmonella enterica serovar Infantis (S. Infantis) was reported as the most frequent serovar in broilers flocks (45.6 %) and broiler meat (50.6 %) in Europe [1] and its prevalence is increasing in breeding hens in some European Member States [2]. In humans, S. Infantis has been classified as the fourth most prevalent serovar in non-typhoidal Salmonella (NTS) human infections in Europe [1].
In Italy, for instance, data obtained in 2014 and 2016 from cross-sectional studies in the broiler sector at slaughter, according to the harmonized AMR Monitoring sampling scheme (Commission Implementing Decision 2013/652/EU, http://data.europa.eu/eli/dec_impl/2013/652/oj), revealed that S. Infantis accounted for 75 and 90 % of all isolates detected, respectively. Among flock, prevalences of 9.6 % (68/709) in 2014 and 8.7 % (70/807) in 2016 were reported [3]. Moreover, over the last decade, multi-drug resistant (MDR) S. Infantis has increasingly been reported from food-producing animals and humans [3]. For instance, among flock prevalence of Italian extended-spectrum cephalosporin (ESC)-resistance (R) S. Infantis isolates in broilers has worryingly increased from 0.4 % (3/709) in 2014 to 2.0 % (16/807) in 2016.
In Italy, we detected an emerging clone of extended spectrum beta-lactamases (ESBL)-producing S. Infantis harbouring an IncI/IncP chimeric megaplasmid of around 300 kbp, carrying virulence, fitness and MDR genes/traits and named p-ESI-like [4], since it was similar to that described in ESC-susceptible (S) S. Infantis in Israel (pESI) in 2014 [5].
Later on, pESI-like-positive S. Infantis isolates were also observed in the USA and Switzerland [6, 7].
Specifically, pESI-like is characterized by the presence of an IncP replication origin, IncI genes as ardA, pilL, sogS, trbA and specific genetic markers as pESI backbone, the gene conferring increased bacterial tolerance to environmental mercury (mer operon), yersiniabactin and K88-like fimbria fim, as already described in Aviv et al. [5]. Furthermore, pESI-like-borne AMR genetic determinants encoding resistance to different antimicrobial classes, as ESBL (e.g. bla CTX-M), tetracyclines (e.g. tetA), trimethoprim (e.g. dfrA), sulphonamides (e.g. sul1) or aminoglycosides (e.g. aadA) have been previously reported [4]. Recent studies in Italy have also demonstrated the plasticity of the pESI-like positive S. Infantis clone, having the capacity to acquire additional transferable resistance against another highest priority critically important antimicrobial (HPCIA) such as colistin, mediated by mcr-1 [8], carried by a different conjugative plasmid (IncX4).
Moreover, additional traits of resistance and enhanced fitness such as the yersiniabactin biosynthetic protein gene irp2, the Infantis plasmid-encoded fimbria gene ipf, toxin/antitoxin (T/AT) elements (e.g. ccdB/A; pemK/I), and the genes conferring resistance to quaternary ammonium compounds (e.g. qacEΔ1), have been also previously associated to pESI-like [4, 5, 8]. The presence of such plasmids in bacteria harbouring these accessory genetic elements, has been previously described as an infection of the bacterial cell, also defining the host/pathogen relationship as symbiotic or parasitic depending on the advantages offered by the plasmid-borne genes [9].
Whole-genome sequencing (WGS) is an easy, thorough and efficient approach to share detailed genetic information about micro-organisms isolated from different countries, without the need of sharing biological samples. WGS analysis provides a large set of data from which all the relevant information about a bacterial isolate can be mined using bioinformatics pipelines instead of ‘classical’ typing methods, as prediction of sequence types (STs) by multilocus sequence typing (MLST) and plasmid (p)STs by pMLST. Moreover, genotyping methods, as SNPs, core-genome MLST (cgMLST) and hierarchical clustering (HC) of cgMLST, have demonstrated a high-resolution capacity in cluster analysis to assess population structure or source attribution of outbreak-related isolates or even isolates with unknown epidemiological relationships [10, 11].
The aim of this work was to characterize S. Infantis and pESI-like plasmid from different European countries by using WGS and bioinformatics analysis, and to investigate the genetic relatedness of S. Infantis clones and plasmids by phylogenetic analysis across 12 European countries and across animal production sectors, human and food sources.
Methods
Genome sequences of Salmonella Infantis
Raw reads from a total of 382 genomes of S. Infantis isolates were studied in depth. Raw reads were collected in the frame of the ENGAGE project, a collaboration between eight institutions across Europe with the objective to improve the cooperation between European institutions in food safety and public health protection using WGS [12]. Partners from ENGAGE and National Reference Laboratories for Antimicrobial Resistance (NRL-ARs) from the European Union (EU) were invited to participate, providing the paired-ends raw reads of already sequenced S. Infantis genomes or by sending isolates to be sequenced by the Italian NRL-AR, Istituto Zooprofilattico Sperimentale del Lazio e della Toscana (IZSLT) ‘M. Aleandri’. Nine different countries participated in the study providing the following number of isolates/genomes: Denmark (n=56), Finland (n=3), Germany (n=38), Ireland (n=19), Italy (n=166), Luxembourg (n=17), Netherlands (n=26), Poland (n=16), UK (n=31). The country of origin of the samples investigated did not always correspond with the country of sampling/isolation. In this regard, 14 samples had been imported from other EU or non-EU countries, in particular four isolates from Hungary, five from Ukraine, one from Romania, and three had a travel history (America, Ecuador and Peru) (Table S1, available in the online version of this article). In addition, ten S. Infantis genomes previously isolated and sequenced in USA were downloaded from ENA database [7] and included as an outgroup. Each of the 382 studied isolates were detected from representative samples of different Epidemiological Units collected from 2001 to 2017 in the frame of various sampling contexts and from animal (n=128), human (n=79), food (n=95), feed (n=12) and food/farm environmental (n=66) sources. Additionally, two samples (n=1 meat/bone meal and n=1 compost samples) were classified as ‘other’. Detailed information can be found in Tables 1 and Table S1.
Table 1.
Number of isolates classified by country of origin and sample type
|
Sample type |
|||||||
|---|---|---|---|---|---|---|---|
|
Country of origin |
Animal |
Environment |
Feed |
Food |
Human |
Other |
Total |
|
Denmark |
24 |
12 |
1 |
16 |
|
|
53 |
|
Finland |
1 |
|
|
|
|
|
1 |
|
Germany |
9 |
10 |
4 |
16 |
|
|
39 |
|
Hungary |
2 |
2 |
|
|
|
|
4 |
|
Ireland |
2 |
5 |
|
2 |
|
2 |
11 |
|
Italy |
80 |
13 |
|
22 |
50 |
|
165 |
|
Luxembourg |
|
1 |
|
5 |
11 |
|
17 |
|
N.A. |
3 |
|
3 |
8 |
|
|
14 |
|
Netherlands |
|
|
|
5 |
11 |
|
16 |
|
Poland |
2 |
9 |
|
7 |
|
|
18 |
|
Probably Imported from Ukraine |
|
|
|
1 |
|
|
1 |
|
Romania |
|
|
|
1 |
|
|
1 |
|
Travel history: America |
|
|
|
|
1 |
|
1 |
|
Travel history: Ecuador |
|
|
|
|
1 |
|
1 |
|
Travel history: Peru |
|
|
|
|
1 |
|
1 |
|
Ukraine |
|
|
|
5 |
|
|
5 |
|
UK |
3 |
14 |
4 |
3 |
|
|
24 |
|
USA |
2 |
|
|
4 |
4 |
|
10 |
|
Total |
128 |
66 |
12 |
95 |
79 |
2 |
382 |
Molecular characterization using WGS
For WGS, all S. Infantis isolates were sequenced using Illumina Technology. Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession numbers PRJEB23728 and PRJEB34620. Individual genome accession numbers are reported in Table S1. All raw reads were analysed using a common pipeline.
Briefly, the quality of raw reads was evaluated using FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and trimmed with TrimmomaticPE v0.22 [13] with the following parameters: Q30 as the minimum quality required for maintaining a base from the beginning and from the end of the read and a window size of 10 with Q20 as average quality. Processed reads were ‘de novo’ assembled using SPAdes v3.11.0 [14] with the default parameters. Quality of the obtained assemblies was assessed with quast v4.6.3 [15].
The assemblies obtained were analysed using on-line SISTR (https://lfz.corefacility.ca/sistr-app/) [16] in order to confirm the serotype ‘in silico’.
SNP-based phylogenetic analysis
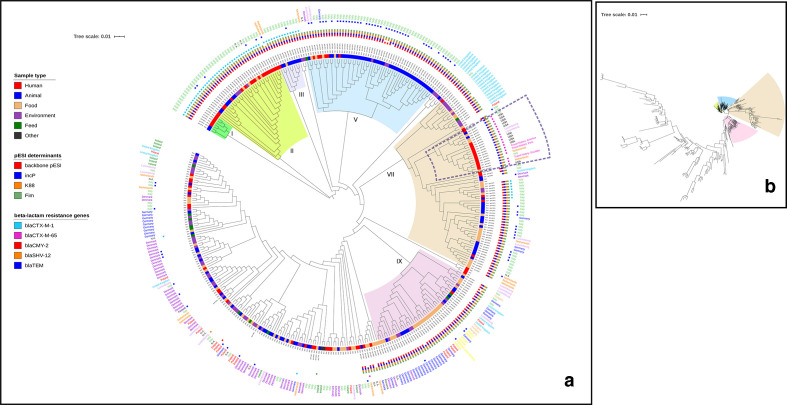
A SNP tree was built with CSI Phylogeny 1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) [17] using as reference S. Infantis SINFA (LN649235), according to the quality parameters previously reported [8]. Raw reads from all the 382 S. Infantis involved in the study were aligned against the reference genome S. Infantis SINFA (LN649235), using BWA v. 0.7.2 [18]. The depth at each mapped position was calculated using genomeCoverageBed (BEDTools v. 2.16.2) [19]. SNPs were called using ‘mpileup’ (SAMTools v. 0.1.18) [20]. SNPs were filtered out if the depth at the SNP position was not at least 10× or at least 10 % of the average depth for the particular genome mapping and if the mapping quality was below 25 or the SNP quality was below 30, calculated by BWA and SAMTools, respectively. The pruning distance was set at 10 bp. Then, all genome mappings were compared and all positions where SNPs were called in at least one mapping were validated in all mappings and ignored if fails validation. The validation includes both depth and Z-score for the SNP filtering. Maximum-likelihood tree was created using FastTree [21], based on a total of 8444 informative SNPs (Fig. 1) and edited with iTOL (http://itol.embl.de/) [22]. Clusters separation was performed based on the topology and the branch length of the tree.
Fig. 1.
Maximum-likelihood SNP-based phylogeny of 382 S. Infantis genomes using as reference S. Infantis SINFA (LN649235) and based on a total of 8444 informative SNPs. (a) Circular representation of the phylogeny, obtained using iTOL (http://itol.embl.de/) ignoring branch length. Dotted lines mark the subcluster containing isolates from human clinical cases, harbouring mostly bla CTX-M-65 (study outgroup). Colours of the isolate ID indicate the different country of origin. Colour squares indicate, from the innermost to the outermost: sample type, pESI determinants, ESBL/AmpC resistance genes, blaTEM gene type 1A, 1B or 216 (blue square). Year of isolation and the presence of gyrA chromosomal point mutations are also indicated for each genome. (b) Circular representation of the phylogeny showing the branch lengths.
Two more trees were built as previously described, only including the raw reads of pESI-like positive isolates (n=245), using as reference S. Infantis SINFA (LN649235) and the Italian pESI-like plasmid reference [4], based on 1375 and 1326 informative SNPs, respectively. Both trees were compared using the ‘cophylo’ function of R [23].
The Italian pESI-like plasmid reference sequence was obtained as follows: the raw reads from the sequence of the transconjugant [4] were mapped against the reference genome of E. coli K12, used as recipient in the conjugation experiment. The unmapped reads were assembled ‘de novo’ using Spades v3.11.0. The final assembly was considered as the plasmid sequence.
cgMLSTs and MST
cgMLSTs were determined using the Enterobase (http://enterobase.warwick.ac.uk/) web based on the 3002 alleles scheme [24] and the HC classification of Enterobase.
For each genome the following different parameters, were considered:
All the 3002 alleles were different among the strains. This threshold is usually applied for a limited number of isolates, as for those outbreak-related.
HC20 (the clusters included all strains with links no more than 20 alleles apart, representing 0.66 % of the alleles) and HC50 (the clusters included all strains with links no more than 50 alleles apart, representing 1.66 % of the alleles).
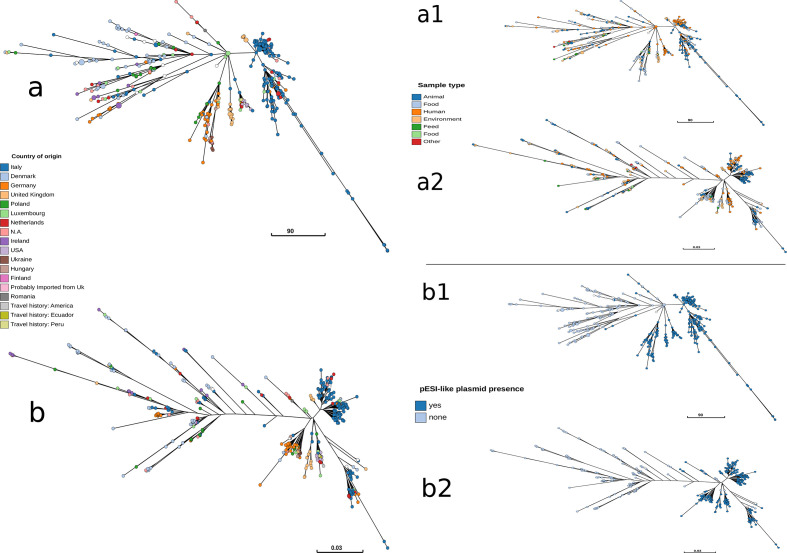
The MST of the allelic profiles obtained from EnteroBase was built using a global optimal eBURST method based on Kruskal’s algorithm. The analysis and the visualization was performed by using a stand-alone version of GrapeTree [25]. GrapeTree was also used for a topology comparison between the obtained SNP tree and the core-genome MST (Fig. 2a, b).
Fig. 2.
Representation as MSTs of the cgMLST (a) and the SNP tree (b), coloured according to the country of origin. Representation as MSTs of the cgMLST(a1,b1) and the SNP tree (a2,b2), coloured according respectively to the sample type (a1,a2) and the presence of pESI-like (b1,b2).
Megaplasmid molecular typing and characterization
The presence of the pESI-like plasmid was determined using blast v2.2.31 [26] by detecting the presence of specific pESI markers, including the IncP origin of replication and the specific genes of IncI plasmid, pESI backbone, K88, Fim, fitness and virulence genes [4, 8]. The presence of the genes was determined only for those obtained with an identity and coverage value >99 % with the reference sequence.
Plasmid incompatibility groups and pMLST [8, 27] were assigned using the Center for Genomic Epidemiology (CGE) tools (https://cge.cbs.dtu.dk/services/pMLST/) by using the default parameters.
Antimicrobial resistance phenotypes and genotypes
The genomes confirmed as S. Infantis were molecular characterized using different bioinformatics tools to assign STs by MLST analysis (https://cge.cbs.dtu.dk/services/MLST/) [28], and to determine the genetic basis of AMR (https://cge.cbs.dtu.dk/services/ResFinder/) [29] using the default parameters.
The results of the genomic analysis on antimicrobial resistance determinants were further compared with phenotypic data of antimicrobial resistance. For this purpose, a minimum panel of antimicrobial drugs was selected based on test and result data provided by the partners and common to all the isolates included in the collection (Table S1). The minimum panel included ampicillin (AMP), cefotaxime (FOT), gentamicin (GEN), ciprofloxacin (CIP) (nalidixic acid; NAL), tetracycline (TET), trimethoprim (TMP), sulfamethoxazole (SMX) and chloramphenicol (CHL). MIC data collected were interpreted according to epidemiological cut-off values included in the Annex A of the EU Decision 2013/652/EU (http://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32013D0652&from=IT). A minority of quantitative data were provided as disk diffusion (DD) inhibition diameters (Table S1), and were interpreted according to epidemiological cut-off values or, when these have not been made available (CIP, NAL and SMX), according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) Clinical Breakpoints.
Results
Phylogenetic trees
Chromosome SNP-based phylogenetic tree
In the phylogenetic tree built with the genomic sequences of the 382 isolates studied, the minimum and maximum SNPs differences between isolates were seven and 1098, respectively. Nine main clusters (I–IX) including isolates mostly pESI-like positive, were identified when considering a branch length <0.05. Clusters I, II and III consisted, respectively, of seven, 34 and nine pESI-like positive isolates harbouring bla CTX-M-1 that were collected from different sources, mainly from Italy. Cluster IV contained 58 pESI-like positive isolates collected from different sources obtained from five countries (Netherlands, Denmark, Luxembourg, Germany and Italy) and most of them (40/58, 68.9 %) characterized by the presence of the bla TEM-1 gene (Fig. 1).
Clusters VII, VIII and IX contained S. Infantis isolated from different sources from 11 countries. Within the cluster VII, there was a subcluster (branch length <0.02) containing isolates from human clinical cases, harbouring bla CTX-M-65, including those isolated from the USA or with travel history to America (Fig. 1a).
Plasmid SNP-based phylogenetic tree
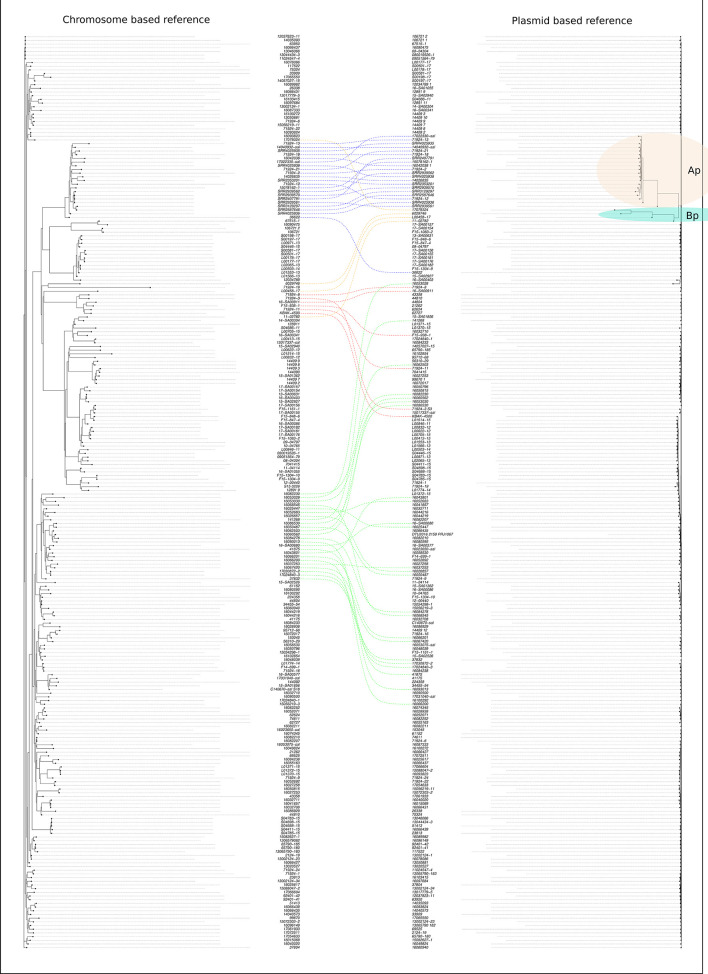
In the phylogenetic tree built using the genomes of 245 isolates presenting pESI-like plasmid (~300 kb), and the plasmid sequence as reference (~286 kb), SNP variation ranged from 0 to 827 different SNPs detected between the sequences. In this SNPs plasmid-based tree, just two small well-differentiated clusters were identified (cluster Ap and cluster Bp) containing 20 and 4 isolates, respectively, that differ from the other plasmid sequences by 106 up to 827 SNPs. The larger cluster (cluster Ap) contained the ten isolates collected from different sources in the USA [7] and ten isolates of human origin collected from the Netherlands (n=5, two with a known travel history to South America), Italy (n=3, one with a known travel history to America) and Luxembourg (n=2). Interestingly, all but one isolates of cluster Ap were ESBL producers (CTX-M-65 type) and the majority of them (12/20) also harboured the fosA3 gene, associated to fosfomycin resistance.
The sequences of the plasmids, not included in clusters Ap and Bp, differed from each other by 1 to 178 SNPs. Cluster Ap was the only one whose group correlates with the chromosome reference-based phylogenetic tree (Fig. 3, blue dotted line).
Fig. 3.
Comparison of chromosome and plasmid-reference-based trees built with S. Infantis pESI-like positive isolates. Blotted lines of the same colour link the same isolate IDs included in the two trees. Two small well-differentiated clusters were identified as clusters Ap and Bp. Cluster Ap was the only one whose group correlates with the chromosome reference-based phylogenetic tree (blue dotted line). The other three groups were chosen, as an example, to evince the genetic relatedness of isolates belonging to different clusters in the two trees.
From the comparison of both plasmid and chromosome-reference-based pESI-like positive trees, a different path in the chromosome (different tree clustering and SNP differences) and plasmid evolution, was observed (Fig. 3).
MLST and cgMLST analysis
Overall, the majority of the S. Infantis isolates (356/382) belonged to ST32. The other STs represented were two different single locus variant (SLV) of ST32, ST2283 (17/382) and ST1032 (5/382), differing in the sucA (alpha ketoglutarate dehydrogenase) or the dnaN (DNA polymerase III beta subunit) alleles, respectively; ST479 (1/382) and ST580 (1/382). Two genomes from the USA, retrieved from ENA, had low quality to be assigned to a ST filtered out due to the internal quality parameters implemented by Enterobase (Table S1).
The results of cgMLST obtained considering all the 3002 different alleles, indicated, as expected, a large variability of cgSTs with almost a different cgST assigned for each isolate, while cgSTs 93 308, 102 164 and 140 061 were assigned to three isolates each (Table S1). Three isolates were not assigned to any cgST (Table S1) according to Enterobase. Interestingly, the cgST-93308 was assigned to three Italian S. Infantis (IDs: 12037823/11, 13 044 434–3 and 14 075 324) isolated in different years (2012, 2013, 2014) from three different sources: a chicken holding environment, a human clinical sample and a caecal content of a broiler chicken, respectively. The cgST-102164 was assigned to three Danish isolates (IDs: F15-1092-1, F14-1239-1, F14-1262-4), all isolated in 2014 from environmental samples of different broiler chicken farms. Similarly, cgST-140061 was assigned to three isolates from Ireland (S16-002738, S16-003238, S16-003532), all collected from water treatment facilities in 2016 (Table S1).
Moreover, when the HC20 parameter was used, 63 different groups were identified, with the HC20-7828 group being the most populated, containing 150/382 isolates (38.3 %). When applying the HC50 parameters, only six different groups were identified with the most populated being the HC50-36, which included 323/382 isolates (84.5 %) (Table S1).
MSTs
The MST built based on the allelic profiles (cgMLST) of a total of 377 isolates, indicated a distinction between isolates with and without the megaplasmid, with a few exceptions (Fig. 2b). No clear clustering based on sample type (Fig. 2b) or geographical location (Fig. 2a) was observed.
The topology of the SNP-based MST was comparable to the topology of the cgMLST-based MST (Fig. 2a, b).
Molecular typing and characterization of pESI-like plasmid
Overall, 245 of 382 S. Infantis analysed harboured a pESI-like plasmid. Among these, 232 isolates collected from samples of ten European countries of origin (Denmark, Germany, Hungary, Italy, Luxembourg, the Netherlands, Poland, Romania, the UK and Ukraine). In particular, 220/245 isolates contained four pESI plasmid markers: pESI backbone, oriV from IncP (AM261769), K88 and fim genes (Fig. 1, Table S1). Regarding the alleles of the IncI1 pMLST profiles, 91.8 % of the genomes containing the pESI backbone sequence (n=213) presented the same profile (ardA2, pilL3, sogS9, trbA21 and rep absent, designated as profile A) (Table S1). Moreover, 25/245 isolates originating from different countries were also considered pESI-like positive since they tested positive for oriV from incP, K88 and Fim but tested negative for the pESI backbone gene (Table S1). As for the pMLST profiles, most of the pESI backbone-negative isolates (n=18) contained a different combination of alleles (ardA11, pilL3, sogS14, trbA8 and rep absent; designated profile B), while only 3/25 isolates, presented the IncI1 pMLST profile A (Table S1).
Fitness determinants
The genes coding for the toxin/antitoxin (T/AT) system PemI/K were present in all the isolates harbouring pESI-like, whereas those coding for the T/AT system CcdA/B were present in all but one pESI-like-positive S. Infantis (Table S1). In most of the isolates, these four genes were found in the same contig (Fig. S1).
Regarding the T/AT system HicA/B, it was present in 43/245 S. Infantis harbouring pESI-like, carried by the IncX4 plasmid and associated to the presence of the bla TEM-1 gene.
Moreover, 236/245 isolates (96,3 %) harbouring the megaplasmid, contained the merA [mercury (II) reductase] gene (Table S1), usually located in the same contig of the pESI backbone and the origin of replication (Fig. S1). Overall, 198/245 isolates also contained the qacEΔ gene, a defective and possibly attenuated form [30] of the qacE (quaternary ammonium compounds and disinfectant resistance gene) as previously detected in isolates from Italy [4].
AMR phenotypes and genotypes
According to Resfinder 3.0 results, the study of accessory resistance genes investigated in the 382 S. Infantis, revealed the presence of a specific AMR genes pattern in pESI-like-positive isolates, compared with pESI-like-negative ones (Fig. 4).
Fig. 4.
Graphical representation of the antimicrobial resistance genotypes (accessory resistance genes) of the 382 S. Infantis isolates clustered according to the chromosome SNP-based phylogeny, (linear representation). Coloured squares indicate the presence of resistance genes for the following antimicrobial classes: green squares: trimethoprim; black squares: tetracyclines; light blue squares: sulphonamides; orange squares: phenicols; pink squares: macrolides; yellow squares: polymyxins; red squares: ESBL/AmpC genes; blue squares: aminoglycosides.
The minimum common panel of the antimicrobial resistance phenotypes (AMP, FOT, GEN, CIP, (NAL), TET, TMP, SMX and CHL), provided by the different partners (n=353) or previously published [4, 7, 8], are reported in Table S1. Overall, the concomitant phenotypical resistance to tetracycline, sulfamethoxazole and trimethoprim in MDR pESI-like-positive isolates (n=177) was mediated by the pESI-like borne tet(A), sul1 and dfrA14 genes, respectively. These above mentioned three genes were all located in a contig belonging to the megaplasmid [4].
Extended-spectrum cephalosporin resistant (ESC-R) isolates (n=82 resistant to cefotaxime) harboured bla CTX-M-1 (n=61), bla CTX-M-65 (n=19), bla CMY-2(n=1) and bla SHV-12 (n=1) genes.
In addition, 68 isolates (17.8 %) harboured at least one gene encoding macrolide resistance (mph, mef, erm(B) (Table S1). In particular, 2 Danish isolates (F15_1093_1 and F15-1093-4) presented the erm(B) gene (98 % identity) in combination with mph. For these two isolates, also susceptibility tests to the macrolide antimicrobial azithromycin were available, and they proved to be resistant, with a MIC value>=64 mg/L .
Four different known chromosomal point mutations in the gyrA gene associated with phenotypical (fluoro)quinolones resistance were found in 251 S. Infantis of 382 isolates analysed: 167 isolates presented the D87G, 28 the D87Y, 55 the S83Y and one the S83F point mutations. 131 isolates did not present any known mutation in the gyrA gene, nor phenotypical (fluoro)quinolone resistance. Six isolates presented plasmid mediated (fluoro)quinolone resistance qnrB genes (five qnrB19 and one qnrB81 genes), all of them being positive also for chromosomal point mutations. One (fluoro)quinolone resistant isolate (ID: 51564/33) tested negative for any known mutation in the gyrA gene but presented a point mutation in the parC gene (T57S), also involved in (fluoro)quinolone resistance. All these results were in agreement with the phenotypical resistance patterns provided (Table S1).
Discussion
S. Infantis is the serovar most frequently isolated in broiler flocks and the fourth most frequent in breeding flocks and laying hens in the European Union [31]. In the present study, we carried out an in-depth molecular characterization of S. Infantis and pESI-like circulating in 12 European countries (with five of them considered as major producers of poultry meat) and from different sources, including 79 clinical isolates from human sources collected in three European countries (with isolates from three human cases with travel-history to America).
Additionally, recent publications indicated that, besides Italy [4], an increasing prevalence of MDR S. Infantis has been reported in European countries such as Switzerland, Slovenia, or outside Europe, e.g. in Egypt, Iran and Ecuador [6, 32–35]. Moreover, a S. Infantis strain carrying a megaplasmid named pSI54/04 and sharing some common features with pESI, such as resistance and virulence determinants [5], has been recently described in Hungary [36].
As previously demonstrated [4, 8], in Italy there is a stable population of S. Infantis carrying the pESI-like megaplasmid. It has been proven that this plasmid frequently harbours AMR genes against at least three antimicrobial classes (sulphonamides, tetracyclines and trimethoprim), with variable presence of the aminoglycoside resistance gene aadA1, along with resistances to heavy metals and several toxin/anti-toxin systems fostering its vertical transmission [4, 5]. The minimum pESI-borne sul1, dfrA, qacEΔ gene pattern indicates the presence of class I integron [37, 38].
Besides the presence of pESI-like-borne tet(A), sul1 and dfrA14 genes, a wide diversity of resistance genes has been observed in the studied S. Infantis population. For example, it is worth to mention the presence of acquired macrolide resistance genes in 17.8 % of the isolates tested, including not only mph and mef genes, but also the erm(B) gene. This latter one in combination with mph, was found in two Danish isolates displaying azithromycin resistance, with MICs>=64 mg/L . These findings reveal the need of continuous monitoring of acquired macrolide resistance in major zoonotic pathogens, in order to take action towards its spread in animal productions and along the food chain. As for ESBL genes, two variants of pESI-like megaplasmids have been described in the present study, one associated with European isolates carrying bla CTX-M-1 and the other associated with American isolates, an outgroup carrying mostly bla CTX-M-65. However, both variants share the resistance genes for at least four antimicrobial classes (trimethoprim, tetracyclines, sulphonamides, aminoglycosides) the toxin-antitoxin systems and the genes enhancing Salmonella fitness. Differently from what has been previously observed in the pig industry in Germany [39], carbapenemase-encoding genes were not found in the dataset of this study, suggesting a very limited occurrence of this AMR feature so far, even considering that the use of third- and fourth-generation cephalosporins can co-select for carbapenem resistance. Noteworthy, although third- and fourth-generation cephalosporins have been never licensed for poultry in Europe, it is well known they have been routinely used ‘off-label’ in poultry breeding hatcheries until 2012 [40].
Similarly to what has been previously reported [41, 42], we have observed that at the core-genome level (chromosome), the studied European S. Infantis population is composed by different clones showing no clear correlation of clustering with source or geographical location of the isolates, with few exceptions (clusters I–III), probably due to the dominance of the Italian isolates within our dataset (Figs 1, 2a b). Conversely, the pESI-like plasmids from the European populations are more genetically homogeneous. Yet, as we already discussed in previous studies [4, 8], we suggest that once the megaplasmid is acquired by S. Infantis, it would likely quickly spread in the local S. Infantis population because of the presence of the specific plasmid-borne genes, which enhance the colonization, virulence and fitness behaviour of the strain. An example is the locus harbouring the genes coding for multiple type II toxin/antitoxin modules PemI/K and CcdA/B, detected in all but one pESI-like-positive isolates herein described. These genetic elements play a central role in bacterial adaptability in response to stress conditions and in the maintenance of plasmids or genomic islands [43], also promoting the ecological success of certain clones, such as the pESI-like-positive-ESBL-producing S. Infantis clone [4]. In particular, the CcdA-CcdB complex has been reported to contribute to the maintenance of plasmids or genomic islands by activation of post-segregational killing mechanisms of the cell [43].
According to the chromosomal SNPs phylogenetic tree (Fig. 1), pESI-like plasmid could potentially colonize all the different European populations of S. Infantis studied, regardless the source of isolation. As can be observed in Fig. 1, in at least ten European countries from which S. Infantis positive-samples originated, the megaplasmid has been spreading in different S. Infantis clonal lineages. These findings are remarkable, since extending to other European countries what we have previously demonstrated in Italy, where the presence of pESI-like in S. Infantis dates back to 2011 [4].
This study helps explaining how a ‘parasitic or symbiotic’ plasmid, carrying accessory and useful genes for bacterial fitness, spreads regardless the national borders, by colonizing animal primary production systems (i.e. the broiler chicken industry) by certain bacterial clones(s) and contributing to human illness through contamination of the food chain. For instance, in the UK, MDR S. Infantis is very rare in poultry and is eradicated whenever it occurs because of the high risk of further spread. The UK isolates in this study included strains from poultry meat that had been imported from Eastern Europe. One of these strains subsequently spread at a broiler farm within the same UK company that processed the meat and persisted for several flock cycles, including spreading to two other farms in the company that used the same thinning team, before it was successfully controlled (Rob Davies, personal communication). Similar isolates have also been obtained in UK from raw meat pet food containing imported chicken meat and associated animal by-products and dogs [44]. Such lateral dissemination, along with infection of parent breeding flocks in some countries, is considered to be an important route for national and international dissemination of S. Infantis [31]. However, the spread of different S. Infantis lineages containing the pESI-like plasmid may also have been favoured by the EU trade of broiler chicken grandparent breeding stock, and by the pyramidal structure of the poultry industry, with only a few primary breeding companies at the top of the pyramid that produce broilers for the whole world. If a biological agent, e.g. a bacterial agent with its plasmidic and core and accessory genome content (including AMR determinants) enter the broiler production chain, it may be transferred globally, as previously observed and discussed in the case of ESBL/AmpC-producing E. coli [45]. This issue should be monitored and considered for risk-management purposes, and its investigation would also benefit of ad hoc studies targeting breeding stock at import stage. As for the cgMLST results, a large number of different cgSTs (Table S1) were identified within the European S. Infantis population, and this is the likely consequence of the dataset analysed. Indeed, the isolates included in our study are the result of surveillance/monitoring activities (2001–2017), and are not known to be outbreak-related (Table S1). However, some isolates presented the same cgST. This is the case of the cgST-93308 assigned to three Italian S. Infantis isolated from human, animal and environmental sources. This feature is in agreement with what has been previously demonstrated for zoonotic transmission events of the pESI-like-positive S. Infantis in Italy [4, 8].
As for the comparison of the chromosome SNPs-based analysis and MST trees, they showed a substantial agreement in cluster separation based on the presence of pESI-like (Figs 1, 2b). A clear correlation of clustering of the isolates was not seen neither with geographical location nor with sample types, with few exceptions probably related to the dominance of the Italian isolates compared with the isolates from other countries (Fig. 2a, b). Considering the inherent technical differences between the two methodologies, the different algorithms behind the two approaches and the nature of the dataset (not-outbreak related isolates), the topology of both trees was very similar, especially for clusters with closely related isolates. However, similarly to what has been previously reported for other serovars [11], there were some differences between the cgMLST and SNP analyses when more distant relationships among the clusters were examined (Fig. 2a, b).
According to the expansion characteristics of the plasmid observed among the countries investigated and the population structure of S. Infantis revealed in this study using WGS, both variants of the pESI-like plasmid (pESI-CTX-M-1 positive and pESI-CTX-M-65 positive) act as parasitic plasmids that colonize a clone, with the apparent potential to spread to other clones, and to ‘infect’ other local S. Infantis populations.
Interestingly, all isolates containing bla CTX-M-65 were all located in a well-defined cluster, both in the chromosome-based and in the plasmid-based phylogenetic trees, as well as in the MST approach (Figs 1, 2a and 3). This cluster contained isolates from the USA and from the Italian and Dutch human patients with travel history to America. Most importantly, this subpopulation seems not to be related to the European S. Infantis population in the food chain, neither at chromosomal nor at plasmid level and there is no evidence that this cluster circulates in food-animal productions in EU countries. Indeed, to the best of our knowledge, a single unreferenced ‘food sample’ in Switzerland with a CTX-M-65-positive S. Infantis has been reported so far [6].
It is worth noticing that the capacity of transmission of this plasmid, despite of its size, has been demonstrated both by ‘in vitro’ experiments of transconjugation with E. coli [4] and ‘in vivo’ among the microbiota of a laboratory animal or to other Salmonella enterica serovars, such as S. Typhimurium [46]. These experiments suggest that the pESI transfer from S. Infantis to other Salmonella serovars or to other bacteria from the normal microbiota acting as reservoirs of virulence and antimicrobial resistance genes may occur also in ‘field conditions’.
The acquisition of the megaplasmid by a permissive bacterial host, such as Salmonella in poultry breeding stock, could be a decisive factor for a quicker transmission of the plasmid within European countries and across Europe. Furthermore, the extensive use of antibiotics, or disinfectants such as quaternary ammonium compounds or even heavy metals like mercury compounds in agriculture, could favour the selection of pESI-like positive S. Infantis. Once the parasitic plasmid has ‘infected’ the bacterial cells, it is hard to cure because of the presence of accessory genetic elements harboured by the mega plasmid such as T/AT systems and the post-segregational killing mechanisms that prevent the plasmid loss, as already described for the emerging S. Infantis clone in Italy [4, 8]. It remains to be seen whether the plasmid ‘infection’ will persist at population level even after the elimination of the selection pressure and the consequent reduction of the fitness of the bacterium, which still carries a plasmid that now may be considered ‘useless’, or providing advantages at present not clearly understood [9, 47]. Taken together, our findings underline the importance of applying a global One Health approach, integrating human and animal surveillance systems and comparing genomic data from different sources and different geographic areas, to help control the spread of potentially MDR zoonotic pathogens, such as S. Infantis. Proper intervention strategies are urgently needed to prevent further dissemination of MDR S. Infantis within animal productions and human disease. Insights into plasmids such as the pESI-like megaplasmid and other mobile genetic elements across and beyond Europe, and into predictors of their success in animal productions, are considered of further help in the surveillance of zoonotic disease agents. Indeed, these genetic elements often provide to zoonotic bacterial hosts additional traits of virulence, fitness and resistance to various antimicrobial drugs, including those classified as highest priority critically important to humans.
Data Bibliography
1. Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession numbers PRJEB23728 and PRJEB34620. Individual genome accession numbers are reported in Table S1.
2. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014 Jul;58(7):3895–903. doi: 10.1128/AAC.02412–14. Epub 2014 Apr 28.
3. Carfora V, Alba P, Leekitcharoenphon P, Ballarò D, Cordaro G, Di Matteo P, Donati V, Ianzano A, Iurescia M, Stravino F, Tagliaferri T, Battisti A, Franco A. Colistin Resistance Mediated by mcr-1 in ESBL-Producing, Multidrug Resistant Salmonella Infantis in Broiler Chicken Industry, Italy (2016–2017). Front Microbiol 2018 Aug 17;9:1880. doi: 10.3389/fmicb.2018.01880. eCollection 2018. Erratum in: Front Microbiol. 2018 Oct 08;9 : 2395.
4. Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, Tolli R, D'Incau M, Staffolani M, Di Giannatale E, Hendriksen RS, Battisti A. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and and 2014. PLoS One Dec 30;10(12):e0144802. doi: 10.1371/journal.pone.0144802. eCollection 2015.
5. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM and Lund O. Multilocus Sequence Typing of Total Genome Sequenced Bacteria. J Clin Micobiol 2012. 50(4): 1355–1361. doi: 10.12.0/JCM.06094–11
6. Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukherjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, Zhao S. Comparative Analysis of Extended-Spectrum-β-Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States. Antimicrob Agents Chemother 2017 Jun 27;61(7). pii: e00488-17. doi: 10.1128/AAC.00488–17. Print 2017 Jul.
7. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes J Antimicrob Chemother. 2012 Nov; 67(11): 2640–4. doi: 10.1093/jac/dks261. Epub 2012 Jul 10.
Supplementary Data
Funding information
This work was supported by the project Establishing Next Generation sequencing Ability for Genomic analysis in Europe (ENGAGE) co-funded by the European Food Safety Authority (EFSA, GP/EFSA/AFSCO/2015/01/CT1).
Acknowledgements
The ENGAGE-EURL-AR Network Study Group consortium is represented by: Burkhard Malorny and Maria Borowiak, German Federal Institute for Risk Assessment (BfR), Department for Biological Safety, Max-Dohrn Str. 8-10, 10589 Berlin, Germany; Rosemarie Slowey, Department of Agriculture, Food and the Marine, National Reference Laboratory for Antimicrobial Resistance (food, feed and animal health), Backweston Laboratory Campus, Young’s Cross Celbridge, Co Kildare, Ireland; Joël Mossong and Monique Perrin, Laboratoire National de Santé, Microbiology Dept, Luxembourg; Kees T. Veldman, Wageningen Bioveterinary Research, Department of Bacteriology and Epidemiology, National Reference Laboratory for Antimicrobial Resistance, Netherlands; Eelco Franz, National Institute for Public Health and the Environment (RIVM), Centre for Infectious Disease Control (CIb), Bilthoven, Netherlands; Dariusz Wasyl, National Veterinary Research Institute, Department of Microbiology, Department of Omics Analyses, National Reference Laboratory for Salmonellosis and Antimicrobial Resistance, Puławy, Poland; Magdalena Zajac, National Veterinary Research Institute, Department of Microbiology, National Reference Laboratory for Salmonellosis and Antimicrobial Resistance, Puławy, Poland; Liljana Petrovska, and Rob Davies, Animal and Plant Health Agency, Addlestone, Surrey, UK; Suvi Nykäsenoja, Finnish Food Authority, Microbiology Unit, Helsinki, Finland; Henry Kuronen, Finnish Food Authority, Veterinary Bacteriology and Pathology Unit, Kuopio, Finland. The authors wish to thank Fabiola Feltrin, Fiorentino Stravino, Valentina Donati, Luigi Sorbara, Roberta Onorati and Carmela Buccella for outstanding technical assistance. We wish to acknowledge Enterobase for its commitment and effort in maintaining and curating the Salmonella (cg/wg) MLST databases.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The conclusions, findings and opinions expressed in this scientific paper reflect only the view of the authors and not the official position of the European Food Safety Authority.
Footnotes
Abbreviations: AMP, ampicillin; cgMLST, core genome multiLocus sequence typing; CHL, chloramphenicol; CIP, ciprofloxacin; DD, disk diffusion; ESBL, extended-spectrum beta-lactame; ESC-R, extended-spectrum cephalosporin resistance; EU, European Union; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FOT, cefotaxime; GEN, gentamicin; HC, hierarchical clustering; HPCIA, highest priority critically important antimicrobial; IZSLT, istituto zooprofilattico sperimentale del lazio e della toscana; MDR, multidrug resistant; MIC, minimun inhibitory concentration; MLST, multiLocus sequence typing; MST, minimum spanning tree; NAL, nalidixic acid; NRL-ARs, National Reference Laboratories for antimicrobial resistance; NTS, non-typhoidal Salmonella; pST, plasmid sequence type; SMX, sulfamethoxazole; SNP, single-nucleotide polymorphism; STs, sequence types; T/AT, toxin/antitoxin system; TET, tetracycline; TMP, trimethoprim; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table and one supplementary figure are available with the online version of this article.
Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession numbers PRJEB23728 and PRJEB34620.
References
- 1.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018 The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA Journal. 2018;16:5500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2017 The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA Journal. 2017;15:5077–5228. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2018 The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA Journal. 2018;6:5182. doi: 10.2903/j.efsa.2018.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One. 2015;10:e0144802. doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, et al. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol. 2014;16:977–994. doi: 10.1111/1462-2920.12351. [DOI] [PubMed] [Google Scholar]
- 6.Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, et al. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010-2015: poultry-related multidrug resistant clones and an emerging ESBL-producing clonal lineage. Front Microbiol. 2017;8:1322. doi: 10.3389/fmicb.2017.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate H, Folster JP, Hsu C-H, Chen J, Hoffmann M, et al. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother. 2017;61:e00488-17. doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carfora V, Alba P, Leekitcharoenphon P, Ballarò D, Cordaro G, et al. Colistin resistance mediated by mcr-1 in ESBL-producing, multidrug resistant Salmonella Infantis in broiler chicken industry, Italy (2016-2017) Front Microbiol. 2018;9:1880. doi: 10.3389/fmicb.2018.01880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLean RC, San Millan A. Microbial evolution: towards resolving the plasmid paradox. Curr Biol. 2015;25:R764–R767. doi: 10.1016/j.cub.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Janowicz A, De Massis F, Ancora M, Cammà C, Patavino C, et al. Core genome multilocus sequence typing and single nucleotide polymorphism analysis in the epidemiology of Brucella melitensis infections. J Clin Microbiol. 2018;56:pii: e00517-18. doi: 10.1128/JCM.00517-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearce ME, Alikhan N-F, Dallman TJ, Zhou Z, Grant K, et al. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar enteritidis outbreak. Int J Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendriksen RS, Pedersen SK, Leekitcharoenphon P, Malorny B, Borowiak M, et al. Final report of engage ‐ establishing next generation sequencing ability for genomic analysis in Europe. EFS3. 2018;15 doi: 10.2903/sp.efsa.2018.EN-1431. [DOI] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella in silico typing resource (SISTR): an open Web-Accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. 1000 genome project data processing subgroup. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Bork P. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 24.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, et al. "GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens". Genome Res. 2018 doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 27.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, et al. The 3' conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/AAC.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bolton D, et al. EFSA Panel on Biological Hazards (EFSA BIOHAZ Panel) Salmonella control in poultry flocks and its public health impact. EFSA Journal. 2019;17:e05596. doi: 10.2903/j.efsa.2019.5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pate M, Mičunovič J, Golob M, Vestby LK, Ocepek M. Salmonella Infantis in broiler flocks in slovenia: the prevalence of multidrug resistant strains with high genetic homogeneity and low biofilm-forming Ability. Biomed Res Int. 2019;2019:4981463–13. doi: 10.1155/2019/4981463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ammar AM, Abdeen EE, Abo-Shama UH, Fekry E, Kotb Elmahallawy E. Molecular characterization of virulence and antibiotic resistance genes among Salmonella serovars isolated from broilers in Egypt. Lett Appl Microbiol. 2019;68:188–195. doi: 10.1111/lam.13106. [DOI] [PubMed] [Google Scholar]
- 34.Ranjbar R, Rahmati H, Shokoohizadeh L. Detection of common clones of Salmonella enterica serotype Infantis from human sources in Tehran hospitals. Gastroenterol Hepatol Bed Bench. 2018;11:54–59. [PMC free article] [PubMed] [Google Scholar]
- 35.Vinueza-Burgos C, Baquero M, Medina J, De Zutter L, Occurrence DZL. Occurrence, genotypes and antimicrobial susceptibility of Salmonella collected from the broiler production chain within an integrated poultry company. Int J Food Microbiol. 2019;299:1–7. doi: 10.1016/j.ijfoodmicro.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Szmolka A, Szabó M, Kiss J, Pászti J, Adrián E, et al. Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella infantis in broiler and human population in Hungary. Food Microbiol. 2018;71:25–31. doi: 10.1016/j.fm.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Kücken D, Feucht H, Kaulfers P. Association of qacE and qacEDelta1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol Lett. 2000;183:95–98. doi: 10.1016/S0378-1097(99)00636-9. [DOI] [PubMed] [Google Scholar]
- 38.Guerra B, Soto S, Cal S, Mendoza MC. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob Agents Chemother. 2000;44:2166–2169. doi: 10.1128/AAC.44.8.2166-2169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borowiak M, Szabo I, Baumann B, Junker E, Hammerl JA, et al. Vim-1-Producing Salmonella Infantis isolated from swine and minced pork meat in Germany. J Antimicrob Chemother. 2017;72:2131–2133. doi: 10.1093/jac/dkx101. [DOI] [PubMed] [Google Scholar]
- 40.EMA (European Medicines Agency) 2018 Reflection paper on off-label use of antimicrobials in veterinary medicine in the European Union. https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-label-use-antimicrobials-veterinary-medicine-european-union-first-version_en.pdf
- 41.Gymoese P, Kiil K, Torpdahl M, Østerlund MT, Sørensen G, et al. WGS based study of the population structure of Salmonella enterica serovar Infantis. BMC Genomics. 2019;20:870. doi: 10.1186/s12864-019-6260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokoyama E, Murakami K, Shiwa Y, Ishige T, Ando N, et al. Phylogenetic and population genetic analysis of Salmonella enterica subsp. enterica serovar Infantis strains isolated in Japan using whole genome sequence data. Infect Genet Evol. 2014;27:62–68. doi: 10.1016/j.meegid.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Goeders N, Van Melderen L. Toxin-Antitoxin systems as multilevel interaction systems. Toxins. 2014;6:304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anonymous Salmonella in livestock production 2018 data – AphA 2019 – to be published on line OCT 2019. 2019.
- 45.Dierikx CM, van der Goot JA, Smith HE, Kant A, Mevius DJ. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One. 2013;8:e79005. doi: 10.1371/journal.pone.0079005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aviv G, Rahav G, Gal-Mor O. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of Warm-Blooded hosts. mBio. 2016;7:e01395–16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iranzo J, Puigbò P, Lobkovsky AE, Wolf YI, Koonin EV. Inevitability of genetic parasites. Genome Biol Evol. 2016;8:2856–2869. doi: 10.1093/gbe/evw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.