Abstract
The extensive genetic diversity of Ralstonia solanacearum , a serious soil-borne phytopathogen, has led to the concept that R. solanacearum encompasses a species complex [ R. solanacearum species complex (RSSC)]. Insertion sequences (ISs) are suggested to play an important role in the genome evolution of this pathogen. Here, we identified and analysed transposable elements (TEs), ISs and transposons, in 106 RSSC genomes and 15 Ralstonia spp. We mapped 10 259 IS elements in the complete genome of 62 representative RSSC strains and closely related Ralstonia spp. A unique set of 20 IS families was widespread across the strains, IS5 and IS3 being the most abundant. Our results showed six novel transposon sequences belonging to the Tn3 family carrying passenger genes encoding antibiotic resistance and avirulence proteins. In addition, internal rearrangement events associated with ISs were demonstrated in Ralstonia pseudosolanacearum strains. We also mapped IS elements interrupting avirulence genes, which provided evidence that ISs plays an important role in virulence evolution of RSSC. Additionally, the activity of ISs was demonstrated by transcriptome analysis and DNA hybridization in R. solanacearum isolates. Altogether, we have provided collective data of TEs in RSSC genomes, opening a new path for understanding their evolutionary impact on the genome evolution and diversity of this important plant pathogen.
Keywords: genome evolution, insertion sequence, mobile DNA, transposon
Data Summary
Genome data analysed in this work are available in the National Center for Biotechnology Information database. Individual accession numbers are listed in Table S1 (available with the online version of this article).
Impact Statement.
Ralstonia solanacearum is one of the most devastating plant pathogenic bacteria found worldwide. This soil-borne pathogen is composed of a large-scale group of strains varying in geographical distribution and pathogenic behaviour, known as the R. solanacearum species complex (RSSC). The observation of this heterogeneous group has led to the hypothesis that the mobile genetic elements (MGEs) may play an important role in shaping the genetic diversity of RSSC. The genome of R. solanacearum is organized into two circular replicons, a chromosome and a megaplasmid. Both replicons have a mosaic structure containing several MGEs, which may play relevant functions in the genome and virulence evolution of the pathogen. Here, we analysed a representative subset of 121 Ralstonia spp. genomes, including RSSC strains and Ralstonia pickettii , Ralstonia mannitolilytica and Ralstonia insidiosa , to investigate the repertoire of insertion sequences (ISs) and transposons. A great diversity of transposable elements (TEs) was found in the Ralstonia spp. genomes. A unique set of IS families was highly widespread across the strains. These findings have expanded our knowledge of the genetic basis of RSSC diversified adaptation based on its repertoire of TEs, and further studies are required to fully understand the evolutionary impact on genome evolution and pathogenicity of this important plant pathogen.
Introduction
Plant–pathogen interactions are intimate, complex and ancient, having developed from a never-ending war [1, 2]. Understanding how plant pathogenic bacteria are evolving to overcome plant resistance is crucial for designing disease control strategies. However, many evolutionary aspects of plan–pathogen interaction remain understudied. In order to form an association with hosts, some bacterial genomes undergo remarkable variations, such as insertions, duplications, inversions and translocations, until a stable long-term association is formed [3, 4]. To some extent, this process can be achieved by the accumulation of repetitive DNA, including transposable elements (TEs), prophages and paralogous genes; many of which have been recognized as non-functional sequences, which can play an important evolutionary role in specialized host adaptation [5].
TEs have garnered research interest as several pathogens possess a relatively high numbers of these mobile elements, which may be responsible for a bottlenecking relationship between pathogen and host [3]. The bacterial TEs, transposons and insertion sequences (ISs) are self-replicable intracellular mobile genetic elements (MGEs). Typically, ISs have single or multiple ORFs that encode a transposase protein, required for insertion into a new locus. In general, ISs have terminal inverted repeats (TIR) and are flanked by short direct repeats (DRs). These elements are distinguished from transposons because transposons carry cargo genes not involved in catalysing or regulating TE movement [6]. IS elements are typically the smallest TEs (<2 kb), and dramatically shape genome content by causing mutations, insertions, deletions, inversions of DNA and alterations of gene expression [7].
This process is believed to represent a great source of genomic diversification, allowing rapid evolution of pathogens or stimulating the emergence of new pathogenic races causing diseases in plants and animals [8]. ISs might play a crucial role in the genome evolution of the bacterium Ralstonia solanacearum , a serious soil-borne phytopathogen effecting agricultural production due to its extensive host range and aggressiveness [9]. However, a complete analysis of the TEs in the R. solanacearum genome has not been reported.
The genome of R. solanacearum is organized into two circular replicons, a chromosome and a megaplasmid; both encode housekeeping and accessory genes. They have similar genomic features (dinucleotide relative abundances, codon usage, and distribution and composition of simple sequence repeats), suggesting their co-evolution over a long time span [9, 10]. Genome comparisons of representative strains of R. solanacearum showed that genomic features, such as size, G+C content and number of genes, were conserved across the strains; however, many genomic rearrangements (e.g. inversion and translocation), as well as deletion and insertion of DNA, were also demonstrated among the strains [11, 12].
Owing to genome differentiation, R. solanacearum species complex (RSSC), which includes Ralstonia syzygii and blood disease bacteria (BDB), was proposed to encompasses three distinct species: Ralstonia pseudosolanacearum (formerly phylotypes I and III), R. solanacearum (IIA and IIB) and R. syzygii (formerly phylotypes IV and BDB) [13, 14]. To investigate the impact of TEs on the genome evolution of RSSC, we identified and analysed the MGEs present in the genomes of 106 RSSC strains and 15 Ralstonia spp. collected from diverse plant hosts and geographical origins.
Methods
Genome data and detection of TE sequences
The genomes of 106 RSSC and 15 Ralstonia spp. ( Ralstonia pickettii , Ralstonia mannitolilytica , Ralstonia insidiosa ) were downloaded from the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/genome) database in December 2018 (Table S1). Three different programs were used to identify IS elements. First, ISs were predicted by blastn [15] alignment against the ISfinder database, using default parameters (E value ≤10−5) [16], and a minimum alignment coverage of 50 % and with at least 70 % identity was considered. Next, two semi-automatic programs were used: ISsaga (insertion sequence semi-automatic genome annotation; http://issaga.biotoul.fr/ISsaga2/issaga_index.php) [17] and oasis (optimized annotation system for insertion sequences; https://github.com/dgrtwo/OASIS) [18]. All originally annotated IS elements were recovered from each program. The DRs, and TIR were manually identified and annotated using Geneious 11.1.5 (Biomatters) based blastn searches against ISfinder to identify known IS elements. An extensive survey of the IS elements within the Ralstonia ssp. genomes was analysed followed the Everyman’s Guide to Bacterial Insertion Sequences to identify partial IS copies and providing general features for each family [19]. Transposon sequences were identified by screening our local database of ISs to search for IS derivatives of transposons. We identified six sequences belonging to the Tn3 transposon family. Using the reference sequence, the predicted sequence was inspected for DR and TIR sequences that define the boundaries of the transposon. The complete nucleotide sequence was imported into Geneious in the GenBank format of corresponding records to help delimit genomic regions flanking the element. These six transposon sequences were registered in The Transposon Registry [20] as Tn6768, Tn6769, Tn6770, Tn6771, Tn6772 and Tn6773.
Virulence and antimicrobial-resistance-associated genes in TEs
Virulence and antimicrobial-resistance genes next to TEs were identified by performing a blastp search (using the following parameters: E value ≤10−5; amino acid identity >30 %; coverage >100 amino acids) on the Pathogen–Host Interactions database (PHI-base; www.phi-base.org) [21] and Ralsto TE3 [22], and by a standard blastn search against the Comprehensive Antibiotic Resistance Database (CARD; http://card.mcmaster.ca) [23]. To assess the impact of IS elements in the virulence genes, they were classified into three groups: IS insertions within a virulence ORF; impartial virulence ORF (less than 100 nt distant); and nearby ORF encoding a virulence genes.
Phylogenetic tree
The 16S rRNA gene sequences were obtained from the NCBI database and a distance matrix was constructed using ClustalW [24]. Subsequently, all the sequences were aligned and a phylogenetic tree was reconstructed in mega x [25] using maximum likelihood (1000 bootstrap replicates) and the substitution model Tamura–Nei+gamma distribution+invariable [25]. The generated output file (.tree) was visualized and annotated with the Interactive Tree of Life (iTOL) interface v4 (https://iTOL.embl.de/) [26].
Expression of ISs in the RSSC transcriptome
A transcriptome (61 Gbp) from R. solanacearum strain UW163 (accession numbers SRX1436103–SRX1436108, SRX1435115–SRX1435118, SRX1435038 and SRX1435071) [27] was retrieved in fastq format from the NCBI Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra) [28]. The expression profile of this strain was compared in basic minimal medium (BMM), casamino acid-peptone-glucose (CPG) liquid media (containing 1 g casein l−1, 10 g peptone l−1 and 5 g glucose l−1), and plant hosts (tomato, banana and melon) [27]. A quality check of the raw sequencing data was performed using the FastQC (v0.11.5) program and the reads were trimmed with Trimmomatic [29]. The alignment of quality trimmed data was performed using Bowtie2 version 2.2.8 [30]. The reads were mapped against reference genomes and the values were normalized with the edgeR 3.6.2 [31] library in RStudio and the gene fold change was calculated as log2(treatment/control – minimal medium). The expression of IS families found in the genome of UW163 was verified in the transcriptomic datasets.
Integration profile analysis
Seven R. solanacearum strains isolated from soil samples were selected, as detailed in Table S2. The isolates were cultured at 28 °C with a shaking speed of 150 r.p.m. in CPG medium. The genomic DNA was extracted using a Wizard genomic DNA purification kit (Promega) according to the manufacturer’s recommendations, checked for quality using a NanoDrop 2000 (Thermo Scientific) instrument and subjected to gel electrophoresis. Probes for IS1021 and ISRso10 were prepared and detected using a PCR DIG probe synthesis kit (Hoffmann–La Roche). For Southern hybridization, 10 µg genomic DNA was digested with EcoRI and incubated overnight at 37 °C. DNA denaturation, neutralization and transference were performed according to the Sambrook and Russel method [32].
Comparison of chromosomal rearrangements
Genome sequences of the strains KACC10722, T110 and SEPPX05 were obtained from NCBI in .gbk format, richness of IS copies in the chromosome being the major selection criteria. Multiple genome alignments were performed with Mauve software (version 2.3.1) [33], with the following parameters: alignment with progressive Mauve (aligner: Muscle 3.6); default seed weight (15); full alignment (minimum island size 50, maximum backbone gap size 50, minimum backbone size 50); use of seed families, yes; iterative refinement, yes; determination of locally collinear blocks (LCBs), yes).
Results
Great diversity of IS elements in the Ralstonia spp. genomes
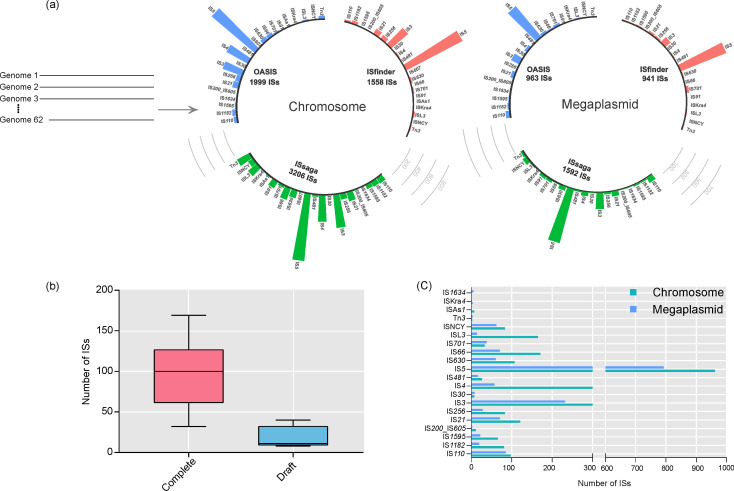
Our analysis showed 10 259 IS elements in the chromosome and megaplasmid of 62 Ralstonia spp. complete genomes using ISsaga [17], ISfinder [16] and oasis [18] (Fig. 1a). An overview of IS distributions in the 60 draft genomes revealed the mean number to be lower than in the complete genomes (Fig. 1b), indicating the effect of genome assembly bias. Therefore, to avoid bias in the analysis, we opted to work only with complete genomes. The IS numbers and families detected varied according to the computational tool, ISsaga being efficient for automated annotation of a total of 3206 ISs in the chromosome and 1592 in the megaplasmid (Tables S3a/S3b/S3c and S4a/S4b/S4c, Fig. 1a). ISsaga found the greatest number of ISs, and also encompasses a set of IS families identified by ISfinder and oasis; therefore, our further analysis was performed with the ISsaga dataset. Details for each IS annotation computational tool are listed in Tables S3a, S3b, S3c, S4a, S4b and S4c. Subsequently, we computed the IS family distribution in the replicons of the complete genomes. Our results showed a unique set of 20 IS families across the chromosome and megaplasmid of the Ralstonia spp. (Fig. 1c).
Fig. 1.
IS elements in Ralstonia spp. replicons. (a) A schematic diagram showing the IS numbers and families detected by the different computational tools. (b) Bar plot representing IS distributions in the complete and draft genomes. (c) Bar plot representing the distribution of IS families in the chromosome (green) and megaplasmid (blue).
Description of the major IS families in Ralstonia spp.
The IS5 family was the most abundant family found in the Ralstonia spp. genomes, followed by the IS3, IS4, IS110 and IS21 families (Table 1). A total of 1724 copies of IS5 were found, of which 962 copies were identified in the chromosome and 762 in the megaplasmid sequence. A total of 256 copies for this family were identified as partial. The element sizes ranged from 850 to 1200 nt in length and have been divided into four subgroups (Table 1). The IS3 family has 711 copies, of which 479 and 232 copies were found in the chromosome and megaplasmid, respectively. Within this family, 297 IS elements were identified as partial. The IS3 family encompasses five subgroups ranging from 1000 to 1750 nt in length. At least one to three different IS3 elements per genome were found. Ralstonia spp. genomes contain 436 copies of the IS4 family, 379 in the chromosome and 57 in the megaplasmid. Fifteen elements were identified as partial. The IS4 family encompasses only two subgroups (IS4 and IS50) ranging from 1110 to 1359 nt in length. One to three different ISs were found per genome. In total, 162 copies were identified as belonging to the IS110 family, of which 57 copies were in the chromosome and 105 in the megaplasmid. Also, 17 IS elements were identified as partial. IS110 members encoded a single ORF with size ranging from 1200 to 1253 nt. This family encompasses one subgroup (IS1111). At least one to two different IS elements per genome were found within this family. Moreover, 167 copies of IS21 family were identified, of which 121 were located in the chromosome and 46 in the megaplasmid. At least one to two different elements per genome were found in our dataset. Besides these five families describe here, another 13 families are listed in detail in Table 1.
Table 1.
Characteristics of IS elements found in the Ralstonia spp. genomes
|
Number of copies |
|||||||
|
Family |
Subgroup |
Size range (nt) |
Chromosome |
Megaplasmid |
Different IS(s) |
Partial |
Total |
|
IS21 |
– |
1700–1800 |
121 |
46 |
1–2 |
44 |
167 |
|
IS3 |
1000–1750 |
479 |
232 |
1–3 |
297 |
711 |
|
|
IS3 | |||||||
|
IS407 | |||||||
|
IS51 | |||||||
|
IS150 | |||||||
|
IS2 | |||||||
|
IS4 |
1100–1359 |
379 |
57 |
1–2 |
15 |
436 |
|
|
IS4 | |||||||
|
IS50 | |||||||
|
IS5 |
IS5 |
850–1200 |
962 |
793 |
1–3 |
226 |
1755 |
|
IS1031 | |||||||
|
IS427 | |||||||
|
IS903 | |||||||
|
IS110 |
IS1111 |
1200–1253 |
57 |
105 |
1–2 |
17 |
162 |
|
IS1182 |
IS1016 |
1330–1578 |
57 |
18 |
1–3 |
14 |
75 |
|
ISH4 | |||||||
|
IS1595 |
ISPna2 |
700–1287 |
49 |
39 |
1–2 |
22 |
88 |
|
|
ISSod11 |
|
|
|
|
|
|
|
IS256 |
– |
1200–1269 |
11 |
16 |
1–2 |
13 |
27 |
|
IS701 |
– |
1200–1500 |
33 |
38 |
1–2 |
7 |
71 |
|
ISL3 |
– |
1050–3000 |
165 |
14 |
1–2 |
35 |
179 |
|
ISNCY |
IS1202 |
1400–2000 |
83 |
62 |
1–2 |
53 |
145 |
|
Tn3 |
– |
1200–3000 |
201 |
30 |
1–2 |
225 |
231 |
|
ISAs1 |
– |
1200–1500 |
7 |
– |
1–3 |
– |
7 |
|
ISKra4 |
– |
1200–1500 |
3 |
1 |
1–3 |
2 |
4 |
|
IS30 |
– |
102–1071 |
7 |
9 |
1 |
2 |
16 |
|
IS481 |
– |
225–1968 |
19 |
17 |
1–2 |
7 |
36 |
|
IS630 |
– |
237–1128 |
59 |
61 |
1–2 |
48 |
120 |
|
IS66 |
– |
1515–2181 |
49 |
44 |
1–3 |
146 |
93 |
IS families are widespread throughout the RSSC strains
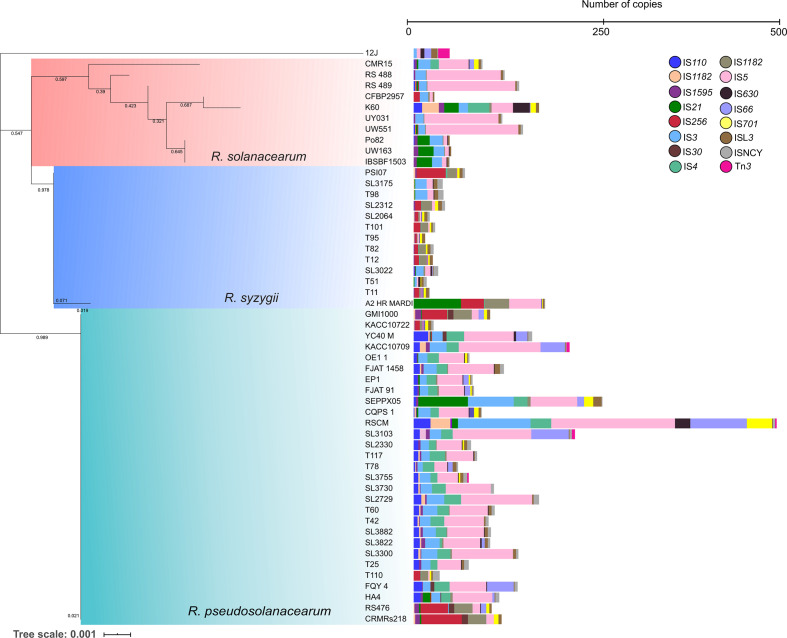
Comparisons of IS families between corresponding sets of Ralstonia ssp. complete genomes revealed the pattern of IS families among the RSSC strains (Fig. 2). The majority of IS families are widespread throughout the complex, the IS5 and IS3 families being shared by all RSSC genomes. Closely related strains tend to have similar patterns of ISs. However, several species-specific IS elements were noticed: such as IS30 only shared among six R. pseudosolanacearum strains; the IS4 and IS701 families mostly found in R. pseudosolanacearum strains, only shared by one R. solanacearum strain K60; most R. syzygii strains lack a set of IS110, IS256 and IS66 families, only found in one genome. Altogether, R. pseudosolanacearum strains shared numerous and diverse IS elements (n=3912), followed in number by R. solanacearum (n=855) and R. syzygii strains (n=559). A set of IS families found in 62 genomes of Ralstonia spp. were characterized in detail (Table S5).
Fig. 2.
Representation of IS family distribution in a RSSC phylogenetic context based on the 16S rRNA gene. The phylogenetic tree was generated with the maximum-likelihood method using mega x software (1000 bootstrap replications) and the substitution model Tamura–Nei+gamma distribution+invariable. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The tree was visualized and annotated using iTOL.
Characterization of six novel putative transposons in R. pseudosolanacearum genomes
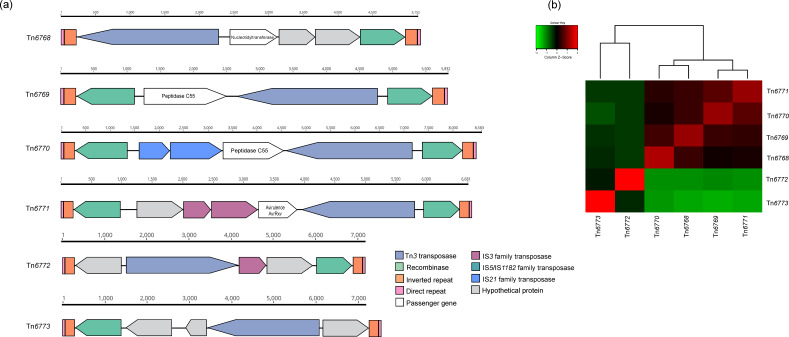
Tn6768, Tn6769, Tn6770, Tn6771, Tn6772 and Tn6773 are the novel putative transposons belonging to the Tn3 family identified in four R. pseudosolanacearum strains (RSCM, HA4-1, KACC10729 and SL3103) (Fig. 3a). Tn6768 and Tn6769 were found in the RSCM chromosome and megaplasmid, respectively; Tn6770 and Tn6771 were found in the megaplasmid and plasmid of the strain HA4-1, respectively. Tn6772 and Tn6773 were identified in the chromosome and megaplasmid sequences of strains SL3103 and KACC10709. The length of the transposon sequences ranged from 5.1 to 8.5 kb (Fig. 3a). Together the six transposons identified here shared 24 to 86 % of sequence identity, and were exclusively found in these three Chinese strains and one Korean strain (Fig. 3b). Commonly, all the transposons encode the Tn3 transposase family and recombinase proteins, which ensures the transposition process. Tn6768, Tn6769, Tn6770 and Tn6771 encode a serine recombinase family and they are flanked by a typical Tn3 family IRL sequence of 51 bp long (GGGGCCGTCTCAGAAAACGGAAAAAATCGTACGCTAAGCCCGGGTTGATGC), an IRR sequence of 42 bp (GGGGTCGTCTCAGAAAACGGAAAAAATCGTACGCTAAGCTCG) and an 8 bp long DR (CAAGATGG). However, Tn6772 and Tn6773 encode a putative tyrosine recombinase XerC-like and they are flanked by an IRL sequence of 51 bp long (AGCGTCTCGTGCAGCGCGGGATGGTCGCGATTAATCTGAAGGGGCGATCTT), an IRR sequence of 51 bp long (CATTGAGTCATGATTTTGACGAGTTTTATGCCTTG ATGGAATAAAGACCGA) and an 8 bp long DR (CCATAAGC). The Tn6768 includes two hypothetical proteins and nucleotidyltransferase genes as passengers. Tn6769 contains an additional recombinase and a passenger gene encoding peptidase C55. Tn6770, in addition to an extra recombinase and passenger genes encoding peptidase C55, also contains the IS21 family transposase. Tn6771 carries a pair of IS5 family transposases, a hypothetical protein gene and an additional passenger gene encoding avirulence effector protein, AviRxv (Fig. 3a). Tn6772 and Tn6773 include hypothetical proteins as passenger genes. We believe this is the first study reporting transposon elements in the RSSC genome.
Fig. 3.
Characterization of six novel transposons. (a) Schematic representation of six transposons belonging to the Tn3 family located in R. pseudosolanacearum strains RSSCM, HA4-1, KACC10709 and SL3103. Genes are indicated by coloured boxes, with the direction of transcription shown by the arrowheads. Transposition-related genes, passenger genes and terminal inverted repeats are as detailed in the key. (b) Heatmap of pairwise comparisons of the nucleotide sequences of the novel putative transposons. The colours represent the mean similarity values for the sequences, as shown in the key.
ISs mediate genomic rearrangements
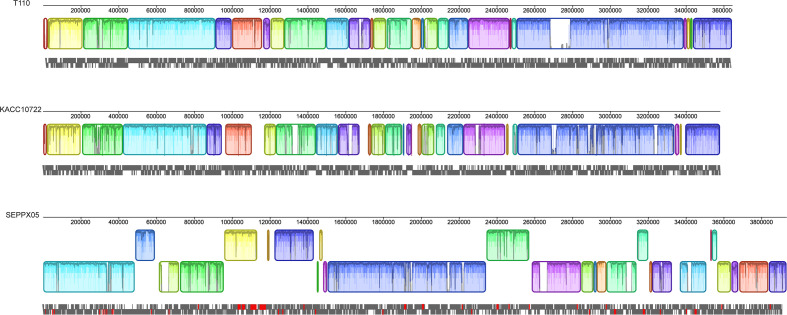
IS elements can shape genomic rearrangements by causing insertions, deletions and inversions [34]. Three R. pseudosolanacearum strains (SEPPX05, KACC10722 and T110) were selected. KACC10722 had 17 complete IS copies, while T110 had 57 IS copies on the chromosome sequence. Notably, these two strains share collinear syntenic blocks. R. pseudosolanacearum strain SEPPX05 had 156 IS copies. In genomes possessing a higher number of IS copies, these elements might have a larger impact on the sequence. Our analysis revealed numerous internal rearrangements in R. pseudosolanacearum strain SEPPX05, with a subset being mostly associated with repeated IS21 elements (Fig. 4), which is indicative that recombination between these ISs might be the cause of the rearrangements.
Fig. 4.
Mauve alignment of the three R. pseudosolanacearum genomes revealing numerous internal rearrangements in the strain SEPPX05. Coloured blocks represent co-linear blocks. Multiple genome alignments were performed by the Mauve software. IS21 family annotations are indicated by the red boxes, where available.
TEs linked to virulence-encoding regions
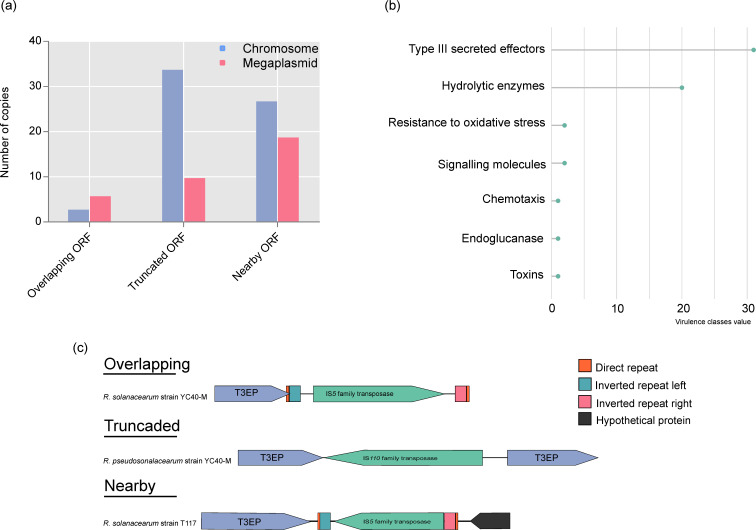
It is believed that for pathogens under a bottlenecking relationship with hosts, IS elements have a strong effect on their genome [35]. Therefore, we paid special attention to the genome context for each annotated IS in intergenic regions of RSSC virulence factors. Most of the elements were found to be truncated, inserted nearby or overlapping genes of virulence factors mainly found in the chromosome sequence (Fig. 5a). Most flanking genes were within type III secreted effectors, including a number of genes encoding type III effector proteins (T3EPs), hydrolytic enzymes (haemagglutinin-related genes), resistance to oxidative stress, signalling molecules, chemotaxis, endoglucanase gene and toxins (Fig. 5b). Details of the flanking genes are found in Table S6. Analysis revealed that 49 % (n=31) of the T3EP genes may be affected by ISs. Fig. 5c illustrates three examples, representing the three classes, mapped across the RSSC genomes. In R. pseudosolanacearum strain YC40-M, an IS5 is present within a T3EP gene, and an IS110 element disrupts another T3EP gene. An intergenic region is present upstream of a T3EP gene and downstream of gene encoding a hypothetical protein in R. pseudosolanacearum strain T117. A T3EP gene disrupted by ISs represented the most common flanking gene. Subsequently, we performed a blastx analysis of T3EP against the PHI-database [21] and Ralsto T3E database [22] to characterize the genes. More than half of the T3EP genes were identified as avirulence genes (avr).
Fig. 5.
ISs in the intergenic regions of virulence genes. (a) Dot plot showing the insertion of these elements for the three classes across the chromosome and megaplasmid sequence. (b) Lollipop chart showing the virulence classes value. (c) Schematic representation of three examples, representing the different classes, mapped in two strains of RSSC.
Comparative transcriptomics for expression of IS elements in planta for host-adapted R. solanacearum
TEs have been reported to play an important role during plant–pathogen interactions, as these elements increase microbial genetic variability and rapidly adapt to environmental changes [36]. However, little is known about the similarity or difference of the effect of these elements on the bacterial transcriptome under in vitro and in planta conditions. To address this question, we investigated transcriptome patterns of IS sequences using an in planta comparative RNA-seq dataset [27]. Gene expression for the R. solanacearum strain UW163 was studied in rich media and under in planta conditions during the colonization of banana, melon and tomato (Fig. S1). We observed that even though all IS elements were expressed under in vitro and in planta conditions, more genes were up-regulated for the in planta dataset than in the rich media dataset. Clearer host genotype effects were observed for ISs up-regulated during tomato and banana infection, which were down-regulated in melon plants, indicating the role of IS elements in the host adaptation of R. solanacearum (Fig. S1).
IS element activity in R. solanacearum isolates
In silico analysis indicated several IS copies. We searched for evidence of such elements in vitro in the R. solanacearum population. We selected seven R. solanacearum isolates from soil samples in Minas Gerais and Brasília, Brazil (Fig. S2a, Table S2), and performed Southern blotting using IS5 family transposase IS1021 and IS3 family transposase ISRso10 elements as probes (Fig. S2b). Most of the isolates showed the hybridization pattern for IS1021 and ISRso10 elements (Fig. S2c), suggesting the presence of these elements in the Brazilian R. solanacearum isolates. Within this population, we detected great polymorphism in the number of copies including isolates with no IS hybridization pattern.
Discussion
We report a curated TE identification method in 121 genomes of the RSSC and closely related Ralstonia spp. ( R. pickettii , R. insidiosa , R. mannitolilytica ). Our analysis found the majority of TEs in the complete genome sequences. However, these elements were also mapped in a low number in the draft genomes, which might be related to genome assembly artifacts that tend to occur near repetitive genomic regions resulting in only one contig with the elements collapsed [37]. Therefore, to avoid bias in the analysis, we opted to work only with complete genomes. In total, 10 259 IS elements were mapped using ISsaga, ISfinder and oasis. We showed that Ralstonia spp. shared a unique set of IS families, mainly IS5 and IS3. IS5 is a relatively heterogeneous group, the majority of its members having a single transposase (Tpase), but also some members may express Tpase by frameshifting [19]. The IS3 family has been found in 270 bacterial species, has over 554 members and is characterized by fusion ORF programmed translational frameshifting with 1200 and 1550 bp long sequences and inverted terminal repeats in the range of 20 to 40 bp [6, 19].
Especially for the RSSC, closely related strains tend to have similar patterns of IS elements. IS elements have demonstrated the ability to quickly multiply in genomes, resulting in a similar number of IS elements in closely related strains [37]. We noticed that R. pseudosolanacearum strains share numerous and diverse IS families. This study reflects on the coverage of these elements in their genome; for example, ISs constitute 3.9 % of the R. pseudosolanacearum strain SEPPX05 genome.
In addition to IS elements, we also report the presence of six novel transposon sequences that belonging to the Tn3 family. These transposons were found in three Chinese R. pseudosolanacearum strains (RSSCM, HA4-1 and KACC10709) and one Korean strain (SL3103). Interestingly, these transposons were only found in these strains. The transposon Tn6768 encodes the enzyme aminoglycoside nucleotidyltransferase in a passenger gene that confers resistance to a wide range of aminoglycosides, such as kanamycin A, and acts by transferring the nucleoside monophosphate group from a nucleotide to the 4′-hydroxyl group of kanamycin A [38, 39]. Although the wilt disease caused by R. solanacearum is not managed with antibiotics, our results showed acquisition of antibiotic resistance in this important phytopathogen. This is critical knowledge, because antibiotic-resistance genes are transferred by the mobile elements, which potentially might be acquired by other bacteria in the environment via horizontal gene transfer.
Putative avr genes were mapped as passengers in the sequence of Tn6771. The term ‘avr genes’ indicates an effector gene that encodes a determinant specifically to interact with the host [40]. Therefore, horizontal gene transfer of avr is recognized as a major epidemiological factor in new disease outbreaks [41], suggesting the role of these transposons in the pathogenicity of RSSC. Our findings demonstrated a collective data, showing the potential impact of these elements on Ralstonia host range. In this study, a large number of avr genes interrupted by ISs were found. As described by the gene-for-gene theory, avr genes are key determinants during plant–pathogen interactions [42]. The theory relies on the relationship between pathogen and host plant cultivars, this interaction occurs between an avr gene in the pathogen and an R (resistance) gene in the plant. When a pathogen possessing an avr attacks the plant that carries the corresponding R gene, resistance is induced in the plant, protecting it from the pathogen. Therefore, the inactivation of avr genes in bacteria can lead to virulence in a resistant host plant [40, 43]. Similarly, ISs have been found to interrupt avr genes in Pseudomonas syringae [44, 45] and be the mechanism of emergence of Fusarium oxysporum races as was demonstrated by Inami et al. [46]. In conclusion, these results provide evidence of MGEs as one of the driving forces for RSSC diversity.
As we demonstrated, in the genome of R. pseudosolanacearum strain SEPPX05, with a high number of IS copies, these elements have a strong influence on its organization, compared with two other R. pseudosolanacearum strains (KACC10722 and T110) with low numbers of copies. SEPPX05 had deletions, insertions and inversions, compared to most representative RSSC strains [47]. In addition, the strains KACC10722 and T110 are pathogenic to potato, and all three strains cause very high economic damage to crops in China [48].
Having shown the effect of IS elements in genome plasticity, we looked more closely at the impact of ISs in modulating RSSC virulence genes. Most IS elements were found in intergenic regions of genes encoding haemagglutinin-related protein, a class of adhesins produced by diverse pathogenic bacteria, responsible for the adhesion of bacteria during plant–pathogen interaction [49]. IS transposition is believed to activate the expression of a gene whose insertion creates an alternative promoter for the host gene or results in read-through transcription [34]. We mapped a few examples of insertions within virulence ORFs, such as ISs overlapping genes encoding haemolysin-like and type II secretion system family proteins, suggesting a possible interference of ISs in the transcription of these genes. Jeong and Timmis [50] reported transposition mediated by ISRso4 (IS5 family) in R. solanacearum , the inactivation of the global regulatory gene phcA modulated the expression of extracellular polysaccharides. Similarly, one IS was screened in the FJAT-1458 genome inserted into a phcA gene. FJAT-1458 is an avirulent strain and might be of use as a potential biocontrol agent as a plant vaccine for bacterial wilt [51].
Studies of the effect of IS transposition on phenotypic traits in bacteria have revealed a major modulation of IS expression under stress conditions [34, 37]. However, the mechanism of IS effects on pathogens under in planta conditions remains understudied. In this study, in planta bacteria RNA-seq data was used to link the expression of IS elements under in planta conditions. We found that while the IS elements were expressed both in vitro and in planta, the genes were up-regulated under in planta conditions compared with under rich media conditions. During plant–pathogen interactions, pathogens are challenged by abiotic and biotic stresses, such as reactive oxygen species, stress hormones, stress temperature, etc. [52]. A common feature of most ISs is that they are activated by stress and environmental factors [34]. Therefore, their transposition facilitates the establishment of the genetic variability that is required for adaptation [36]. This is the first evidence of IS activation in R. solanacearum under in planta conditions, suggesting the significant contribution of these elements to pathogen adaption.
The IS elements predicted in silico were assessed in vitro by analysing two IS elements in seven isolates of R. solanacearum from Brazil. The observed band polymorphism led to the hypothesis that these elements are involved in diversification [53]. Our analysis showed the widespread distribution of predicted IS elements in silico and in vitro, among R. solanacearum isolates. This might also indicate a recent activity of IS elements among the R. solanacearum population from Brazil. In conclusion, the research described here opens up new avenues for understanding the evolutionary impact of TEs on the genome evolution and diversity of the RSSC.
Data Bibliography
1. Xu J, Zheng H, Liu L et al. GenBank assembly no. GCA_000215325.1 (2011).
2. Guarischi-Sousa R, Puigvert M, Coll NS et al. GenBank assembly no. GCA_001299555.1 (2016).
3. Remenant B, Coupat-Goutaland B, Guidot A et al. GenBank assembly no. GCA_000197855.1, GCA_000427195.1 and GCA_000283475.1 (2010).
4. Bocsanczy AM, Huguet-Tapia JC, Norman DJ. GenBank assembly no. GCA_000525615.1 (2014).
5. Hayes MM, MacIntyre AM, Allen C. GenBank assembly no. GCA_001696875.1, GCA_002251695.1 and GCA_000285815.1 (2017).
6. Yuan K, Cullis J, Lévesque CA et al. GenBank assembly no. GCA_000710135.3 and GCA_000710695.1 (2015).
7. Patil VU, Girimalla V, Sagar V et al. GenBank assembly no. GCA_001373295.1 (2017).
8. Kotorashvili A, Meparishvili G, Gogoladze G et al. GenBank assembly no. GCA_002029865.1, GCA_002029885.1 and GCA_002029895.1 (2017).
9. Salanoubat M, Genin S, Artiguenave F et al. GenBank assembly no. GCA_000009125.1 (2002).
10. Chen D, Liu B, Zhu Y et al. GenBank assembly no. GCA_001887535.1 (2017).
11. Li P, Wang D, Yan J et al. GenBank assembly no. GCA_001891105.1 (2016).
12. Chen D, Liu B, Zhu Y et al. GenBank assembly no. GCA_002155245.1 (2017).
13. Liu Y, Tang Y, Qin X et al. GenBank assembly no. GCA_002220465.1 (2016).
14. Cao Y, Tian B, Liu Y et al. GenBank assembly no. GCA_000348545.1 (2013).
15. Ramesh R, Gaitonde S, Achari G et al. GenBank assembly no. GCA_000671315.1 and GCA_000671335.1 (2014).
16. Li X, Huang X, Chen G et al. GenBank assembly no. GCA_002162015.1 (2018).
17. Cho H, Song E-S, Heu S et al. GenBank assembly no. GCA_003515205.1, GCA_003515205.1, GCA_003515225.1, GCA_003515245.1, GCA_003515285.1, GCA_003515305.1, GCA_003515345.1, GCA_003515365.1, GCA_003515405.1, GCA_003515465.1, GCA_003515545.1, GCA_003515565.1, GCA_003515585.1, GCA_003515605.1, GCA_003515165.1, GCA_003515185.1, GCA_003515265.1, GCA_003515325.1, GCA_003515385.1, GCA_003515425.1, GCA_003515445.1, GCA_003515485.1, GCA_003515505.1 and GCA_003515525.1 (2019).
18. Tan X, Qiu H, Li F et al. GenBank assembly no. GCF_003999725.1 (2019).
19. Shan W, Yang X, Ma W et al. GenBank assembly no. GCA_000430925.2 and GCA_001876985.1 (2013).
20. Zou C, Wang K, Meng J et al. GenBank assembly no. GCA_001484095.1 (2016).
21. Albuquerque GMR, Souza EB, Silva AMF et al. GenBank assembly no. GCA_003595305.1 and GCF_003612975.1 (2017).
22. Remenant B, Coupat-Goutaland B, Guidot A. GenBank assembly no. GCF_000197855.1, GCF_000283475.1 and GCF_000427195.1(2010).
23. Badrun R, Abu Bakar N, Laboh R et al. GenBank assembly no. GCA_002012345.1 (2017).
24. Remenant B, de Cambiaire J-C, Cellier G et al. GenBank project no. PRJNA369602 (2011).
25. Daligault HE, Davenport KW, Minogue TD et al. GenBank assembly no. GCA_000743455.1 (2014).
26. Vaz-Moreira I, Tamames J, Martínez JL, Manaia CM. GenBank assembly no. GCA_001699815.1 and GCA_001699795.1 (2016).
27. Paterson J, Gross H. GenBank assembly no. GCA_002516395.2 (2018).
28. Ohtsubo Y, Fujita N, Nagata Y et al. GenBank assembly no. GCA_000471925.1 (2013).
29. Xu J, Zheng H, Liu L et al. GenBank assembly no. GCA_001663855.1 and GCA_001653935.1 (2016).
Supplementary Data
Funding information
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – finance code 001; Fundação Arthur Bernardes (Furnabe; Minas Gerais, Brazil) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Brasília, Brazil).
Acknowledgements
The authors thank Hilberty Lucas Nunes Correia for assistance with the transcriptome analysis.
Author contributions
M.F.S., conceived the study; O.S.G., K.F.C., J.C.S.A., A.S.F., T.S.S. and L.G.C.R., analysed the data and performed lab work; O.S.G., wrote the manuscript; M.F.S. and M.V.Q., critically reviewed the manuscript. All authors reviewed the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human nor animal experimentation is reported.
Footnotes
Abbreviations: DR, direct repeat; IS, insertion sequence; MGE, mobile genetic element; NCBI, National Center for Biotechnology Information; RSSC, Ralstonia solanacearum species complex; TE, transposable element; T3EP, type III effector protein; TIR, Terminal inverted repeat.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and two supplementary figures are available with the online version of this article.
References
- 1.Peyraud R, Dubiella U, Barbacci A, Genin S, Raffaele S, et al. Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J. 2017;90:720–737. doi: 10.1111/tpj.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider DJ, Collmer A. Studying plant-pathogen interactions in the genomics era: beyond molecular Koch's postulates to systems biology. Annu Rev Phytopathol. 2010;48:457–479. doi: 10.1146/annurev-phyto-073009-114411. [DOI] [PubMed] [Google Scholar]
- 3.Bobay L-M, Ochman H. The evolution of bacterial genome architecture. Front Genet. 2017;8:72. doi: 10.3389/fgene.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mira A, Klasson L, Andersson SGE. Microbial genome evolution: sources of variability. Curr Opin Microbiol. 2002;5:506–512. doi: 10.1016/S1369-5274(02)00358-2. [DOI] [PubMed] [Google Scholar]
- 5.Gil R, Latorre A. Factors behind junk DNA in bacteria. Genes. 2012;3:634–650. doi: 10.3390/genes3040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleckner N. Transposable elements in prokaryotes. Structure. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- 7.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold DL, Jackson RW. Bacterial genomes: evolution of pathogenicity. Curr Opin Plant Biol. 2011;14:385–391. doi: 10.1016/j.pbi.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, et al. Genome sequence of the plant pathogen Ralstonia solanacearum . Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 10.Castillo JA, Greenberg JT. Evolutionary dynamics of Ralstonia solanacearum . Appl Environ Microbiol. 2007;73:1225–1238. doi: 10.1128/AEM.01253-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li P, Wang D, Yan J, Zhou J, Deng Y, et al. Genomic analysis of phylotype I strain EP1 reveals substantial divergence from other strains in the Ralstonia solanacearum species complex. Front Microbiol. 2016;7:1719. doi: 10.3389/fmicb.2016.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remenant B, Coupat-Goutaland B, Guidot A, Cellier G, Wicker E, et al. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics. 2010;11:379. doi: 10.1186/1471-2164-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, et al. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics. 2016;17:90. doi: 10.1186/s12864-016-2413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safni I, Cleenwerck I, Vos PD, Fegan M, Sly L, et al. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. Indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int J Syst Evol Microbiol. 2014;64:3087–3103. doi: 10.1099/ijs.0.066712-0. [DOI] [PubMed] [Google Scholar]
- 15.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson DG, Lee M-C, Marx CJ. OASIS: an automated program for global investigation of bacterial and archaeal insertion sequences. Nucleic Acids Res. 2012;40:e174. doi: 10.1093/nar/gks778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. Everyman's guide to bacterial insertion sequences. Microbiol Spectr. 2015;3:550–590. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 20.Tansirichaiya S, Rahman MA, Roberts AP. The Transposon Registry. Mob DNA. 2019;10:40. doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urban M, Cuzick A, Rutherford K, Irvine A, Pedro H, et al. PHI-base: a new interface and further additions for the multi-species pathogen–host interactions database. Nucleic Acids Res. 2017;45:D604–D610. doi: 10.1093/nar/gkw1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabbagh CRR, Carrere S, Lonjon F, Vailleau F, Macho AP, et al. Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ. 2019;7:e7346. doi: 10.7717/peerj.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. Interactive tree of life (iTOL) V4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ailloud F, Lowe TM, Robène I, Cruveiller S, Allen C, et al. In planta comparative transcriptomics of host-adapted strains of Ralstonia solanacearum . PeerJ. 2016;4:e1549. doi: 10.7717/peerj.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russel DW. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 33.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol. 2017;43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 35.Stapley J, Santure AW, Dennis SR. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 2015;24:2241–2252. doi: 10.1111/mec.13089. [DOI] [PubMed] [Google Scholar]
- 36.Seidl MF, Thomma BPHJ. Transposable elements direct the coevolution between plants and microbes. Trends Genet. 2017;33:842–851. doi: 10.1016/j.tig.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Adams MD, Bishop B, Wright MS. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb Genom. 2016;2:e000062. doi: 10.1099/mgen.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fling ME, Kopf J, Richards C. Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycoside-modifying enzyme, 3"(9)-O-nucleotidyltransferase. Nucleic Acids Res. 1985;13:7095–7106. doi: 10.1093/nar/13.19.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krause KM, Serio AW, Kane TR, Connolly LE. Aminoglycosides: an overview. Cold Spring Harb Perspect Med. 2016;6:a027029. doi: 10.1101/cshperspect.a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leach JE, White FF. Bacterial avirulence genes. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel DW. Why do pathogens carry avirulence genes? Physiol Mol Plant Pathol. 1999;55:205–214. doi: 10.1006/pmpp.1999.0230. [DOI] [Google Scholar]
- 42.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. doi: 10.1146/annurev.py.09.090171.001423. [DOI] [Google Scholar]
- 43.Grennan AK. Plant response to bacterial pathogens: overlap between innate and gene-for-gene defense response. Plant Physiol. 2006;142:809–811. doi: 10.1104/pp.106.900207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deslandes L, Rivas S. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 2012;17:644–655. doi: 10.1016/j.tplants.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Kim JF, Charkowski AO, Alfano JR, Collmer A, Beer SV. Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae . Mol Plant Microbe Interact. 1998;11:1247–1252. doi: 10.1094/MPMI.1998.11.12.1247. [DOI] [Google Scholar]
- 46.Inami K, Yoshioka-Akiyama C, Morita Y, Yamasaki M, Teraoka T, et al. A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: inactivation of avirulence gene AVR1 by transposon insertion. PLoS One. 2012;7:e44101. doi: 10.1371/journal.pone.0044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Huang X, Chen G, Zou L, Wei L, et al. Complete genome sequence of the sesame pathogen Ralstonia solanacearum strain SEPPX 05. Genes Genomics. 2018;40:657–668. doi: 10.1007/s13258-018-0667-3. [DOI] [PubMed] [Google Scholar]
- 48.Jiang G, Wei Z, Xu J, Chen H, Zhang Y, et al. Bacterial wilt in China: history, current status, and future perspectives. Front Plant Sci. 2017;8:1549. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genin S, Denny TP. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol. 2012;50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 50.Jeong EL, Timmis JN. Novel insertion sequence elements associated with genetic heterogeneity and phenotype conversion in Ralstonia solanacearum . J Bacteriol. 2000;182:6541. doi: 10.1128/JB.182.22.6541-6541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen D, Liu B, Zhu Y, Zhang H, Chen Z, et al. Complete genome sequence of Ralstonia solanacearum FJAT-91, a high-virulence pathogen of tomato wilt. Genome Announc. 2017;5:e00900-17. doi: 10.1128/genomeA.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones P, Garcia BJ, Furches A, Tuskan GA, Jacobson D. Plant host-associated mechanisms for microbial selection. Front Plant Sci. 2019;10:862. doi: 10.3389/fpls.2019.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee H, Doak TG, Popodi E, Foster PL, Tang H. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli . Nucleic Acids Res. 2016;44:7109–7119. doi: 10.1093/nar/gkw647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.