Abstract
The US Environmental Protection Agency’s ToxCast program has generated toxicity data for thousands of chemicals but does not adequately assess potential neurotoxicity. Networks of neurons grown on microelectrode arrays (MEAs) offer an efficient approach to screen compounds for neuroactivity and distinguish between compound effects on firing, bursting, and connectivity patterns. Previously, single concentrations of the ToxCast Phase II library were screened for effects on mean firing rate (MFR) in rat primary cortical networks. Here, we expand this approach by retesting 384 of those compounds (including 222 active in the previous screen) in concentration-response across 43 network activity parameters to evaluate neural network function. Using hierarchical clustering and machine learning methods on the full suite of chemical-parameter response data, we identified 15 network activity parameters crucial in characterizing activity of 237 compounds that were response actives (“hits”). Recognized neurotoxic compounds in this network function assay were often more potent compared to other ToxCast assays. Of these chemical-parameter responses, we identified three k-means clusters of chemical-parameter activity (i.e., multivariate MEA response patterns). Next, we evaluated the MEA clusters for enrichment of chemical features using a subset of ToxPrint chemotypes, revealing chemical structural features that distinguished the MEA clusters. Finally, we assessed distribution of neurotoxicants with known pharmacology within the clusters and found that compounds segregated differentially. Collectively, these results demonstrate that multivariate MEA activity patterns can efficiently screen for diverse chemical activities relevant to neurotoxicity, and that response patterns may have predictive value related to chemical structural features.
Keywords: Neurotoxicity, Screening, ToxCast
Introduction
It is now well-established that humans are exposed to far more compounds than can be evaluated for hazard potential using animal-based approaches. Because the need for information regarding the potential toxicity of thousands of compounds exists, significant efforts have been applied to characterize biological activity of environmentally relevant compounds using in vitro assays. Combined with information on chemical structure, exposure and metabolism, this approach can be useful for prioritizing chemicals for further testing or for screening-level risk decisions. The US Environmental Protection Agency’s Toxicity Forecaster (ToxCast) program (Dix et al. 2007; Judson et al. 2010; Kavlock et al. 2012; Richard et al. 2016) provides hazard, exposure, structure, and other information on thousands of compounds. A major component of the ToxCast program is data from over 1000 in vitro assay endpoints that examine a wide range of cellular responses mapped to ~ 300 signaling pathways [https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data]. Currently, over 3800 unique compounds have or are undergoing screening in some or all of these assays, with the most complete assay coverage, to date, available for ~ 1100 compounds tested in the earliest Phases I and II of the program. Compound categories represented within this ~ 1100 chemical portion of the Phase II test set (i.e., the ph1_v2 and ph2 subsets) include pesticides, pharmaceuticals, antimicrobials, flame retardants, food-additives, and “green” chemicals, among others (Richard et al. 2016).
Nervous system function is sensitive to disruption by a wide variety of natural toxins, pharmaceuticals, pesticides and industrial chemicals. Therefore, it is important that screening approaches include assays that identify compounds with potential neurotoxicity hazard. The ToxCast assay battery includes assays that evaluate interactions of compounds with ion channels and second messengers, membrane bound, and/or nuclear receptors, as well as other potential targets that may mediate neurotoxicity. However, these assays do not capture the breadth of potential targets that may be linked to neurotoxicity, nor does interaction of a chemical at the receptor level necessarily result in neuroac- tivity/neurotoxicity. Thus, incorporation of functional, cell- based assays into screening approaches such as the ToxCast program can provide additional information that is useful for decision-making. Previously, we demonstrated that assessment of compound effects on neural network function using microelectrode arrays (MEAs) can identify neurotoxic and neuroactive compounds with high sensitivity and selectivity (McConnell et al. 2012; Valdivia et al. 2014). Subsequently, we used this assay to screen ToxCast PhII compounds at a single concentration (Strickland et al. 2018).
Primary cortical cultures grown on multi-well MEAs (mwMEAs) exhibit spontaneous electrical spikes and groups of spikes (bursts) that are associated with neuronal action potentials (Wheeler and Nam 2011). Further, the activity of neurons in these cultures demonstrates synchronous and oscillatory activity, which are related to synaptogenesis and the balance of excitatory and inhibitory synapses (Muramoto et al. 1993; Maeda et al. 1995; Chiappalone et al. 2006). These cortical networks are sensitive to modulation by compounds with a wide variety of modes of action (Johnstone et al. 2010), including those that modulate voltage-gated sodium channels (Meyer et al. 2008; Shafer et al. 2008; Scelfo et al. 2012; Mohana Krishnan and Prakhya 2016; Baskar and Murthy 2018), and glutamatergic (Frega et al. 2012; Lantz et al. 2014) and GABAergic (Mack et al. 2014; Bradley et al. 2018) agonists and antagonists. In addition, MEAs are sensitive to a broad range of chemicals and have been used to screen or examine the effects of illicit (Honde- brink et al. 2016) and antiepileptic drugs (Colombi et al. 2013), components of harmful algae (Nicolas et al. 2014; Alloisio et al. 2016), neuroactive toxins (Pancrazio et al. 2014; Kasteel and Westerink 2017), and metals (Dingemans et al. 2016; Huang et al. 2016). Results with MEAs are also reproducible across laboratories (Novellino et al. 2011; Vassallo et al. 2017). This work demonstrates that measuring changes in firing rates and patterns of extracellularly- recorded action potentials from neural networks using MEAs is a rapid, cost effective, and accurate approach to screen compounds for neurotoxicity hazard.
We previously screened ToxCast compounds (Strickland et al. 2018) using cortical networks grown on MEAs and, based on testing a single concentration, identified a subset of 326 compounds that altered the mean firing rate (MFR) of cortical networks. However, this approach does not establish potency, nor does it provide hit confirmation. In addition, the complex pattern of neural activity measured by MEAs can be described by multiple parameters that evaluate different aspects of network activity, such as bursting, coordinated bursting, synchrony and oscillatory behavior; these parameters were not examined in our previous analysis. For small numbers (~ 10–50) of chemicals, effects on bursting and other network parameters have been used to group or identify compounds with similar modes of action (Gramowski et al. 2004, 2006; Mack et al. 2014; Gramowski-Voß et al. 2015; Bader et al. 2017; Bradley et al. 2018). In our previous work, we also demonstrated that application of chemotype analysis identified chemical sub-structures, or features enriched in the hit subset, which provided initial structure-activity relationship inferences. In the present study, we sought to: (1) rescreen a subset of hits from the Strickland et al. 2018 study in a concentration-response mode to confirm and further characterize the potency of those chemicals while expanding the parameters that were evaluated beyond the mean firing rate (MFR) to include other parameters of network function; (2) utilize machine learning and k-means clustering analysis to determine the parameter subset most informative in characterizing neuroactivity and compare these MEA parameter potencies to other ToxCast assays; and (3) potentially characterize the active compounds using chemotype enrichment analysis and identify putative parameter patterns that may define specific modes of action.
Materials and methods
The overall process for our analysis is as follows: 384 compounds were screened in concentration-response across 43 MEA parameters. These compound-parameter associations were evaluated using the ToxCast Analysis Pipeline (tcpl) to identify active compound-parameter associations (Filer 2016; Filer et al. 2017). Of the parameters these compounds were active in, the parameters most informative in distinguishing between neuroactive and non-neuroactive compounds were identified by reducing the full, 43-parameter set using (1) hierarchical clustering, and (2) machine learning methods. From the final, reduced set of compoundparameter associations, we used k-means clustering to characterize bioactivity patterns. Finally, we identified enriched chemotypes within these k-means bioactivity clusters.
Chemical selection and preparation
A total of 384 compounds from EPA’s ToxCast chemical library were selected for this analysis from the previous, single concentration (40 pM) screen of 1055 ToxCast compounds (Strickland et al. 2018). These included 222 compounds that were active in the initial MEA assay that were selected for concentration response follow-up testing to determine potency, and 162 compounds that did not alter spontaneous neural activity beyond the criterion for a “hit” [exceeded 2× the activity threshold for DMSO-treated wells (see Strickland et al. 2018)] following the initial single-concentration screen. A complete list of all compounds screened in concentration response can be found in the supplemental materials.
Stock solutions of each compound used in the present study (made to a target concentration of 20 mM in dimethyl sulfoxide, DMSO) were received from EPA’s ToxCast Chemical Contractor (Evotec, Branford, CT) in sealed round-bottom 96-well plates. Upon arrival, compounds were transferred from round-bottom 96-well plates to individual micro-centrifuge tubes and stored at - 80 °C until use. Prior to each experiment, compounds were thawed, vortexed, and immediately diluted in DMSO (5–0.015 mM) in a v-bottom 96-well plate. Compounds were subsequently diluted 1:10 in NB/B27 media in a second v-bottom 96-well plate for dosing. To dose, 10 pl from the dosing plate was transferred to the designated well on the 48-well MEA plate. Each well of the mwMEA contained 500 pl of media (1:50 dilution), resulting in a final concentration range of 0.03–40 pM for each compound, spaced in half-log increments. DMSO (0.2% by volume; n = 3 wells/ plate), GABAa antagonist bicuculline (25 pM BIC; n = 2 wells/plate), and Lysis buffer solution (~ 4% by volume; n = 1 well/plate) were included on each mwMEA as controls to assess concentration-related changes in neural function and cell health following compound exposure.
To complete the concentration response determination for all 384 compounds from the Phase II library, ~ 6–12 compounds were screened per week in triplicate over ~ 52 weeks.
Primary cortical culture on MEAs
Prior to plating rat primary cortical cultures, multi-well microelectrode (mwMEA) plates from Axion Biosystems Inc. (Atlanta, GA) were prepared for culture by coating with polyethyleneamine as previously described (Valdivia et al. 2014; Strickland et al. 2018). Each 48-well MEA plate consisted of a total of 768 nano-textured gold platinum microelectrodes (~ 40–50 pm diameter, 350 pm center-to-center spacing) with 16 electrodes/well plus four integrated ground electrodes (M768-KAP Kapton, Axion Biosystems Inc., Atlanta, Georgia).
All procedures involving laboratory animals were approved by the National Health and Environmental Effects Research Laboratory’s institutional laboratory animal health care and use committee and complied with applicable federal guidelines for laboratory animal experimentation. Primary cortical neural cultures were prepared as previously described in Strickland et al. (2018) from Long-Evans rat pups on postnatal days 0–1, with minor modifications. Full media changes occurred on day in vitro (DIV) 5, 9 and 12, or, if the experiment was planned for DIV 13, cells received a ½ media change on DIV 12 (24 h prior to experiment).
Multiplexed screening approach
Previous experiments established a multiplexed approach that allows for simple and rapid characterization of compound effects on both neurophysiological and cellular viability parameters from within the same well of the mwMEA plate, allowing for these data to be obtained for each compound from the same network (Wallace et al. 2015). First, baseline and treated data are collected for effects of compounds on neurophysiology using MEA recordings. Effects on cell health were determined immediately following the conclusion of the mwMEA recording in the presence of the compound. Cellular viability was determined using two commercially available assays: Lactate Dehydrogenase (LDH) release and CellTiter Blue (CTB).
MEA recordings
Spontaneous activity in the cortical cultures was recorded using an Axion Biosystems Maestro 768 channel amplifier and Axion Integrated Studios (AxIS) v1.8 (or later) software. The amplifier recorded from all channels simultaneously (gain = 1200 ×; sampling rate = 12.5 kHz/channel): raw signals were filtered with a Butterworth band-pass filter (300–5000 Hz), which filters out slower local field potentials leaving only fast potentials, i.e., “spikes”, arising from extracellular currents associated with action potentials (Pine 2006; Wheeler and Nam 2011). On-line spike detection of filtered signals was conducted with the AxIS adaptive spike detector, using a threshold of 8 × the root mean squared (rms) noise on each channel. Any electrodes with rms noise levels greater than 5 pV were grounded (e.g., no data were recorded). Once grounded, an electrode was grounded for the duration of the experiment. All recordings were conducted at 37 °C. Wells were deemed usable if on the day of the exposure > 10 electrodes were active (defined as > 5 spikes/min). On DIV 13 or 15, a minimum of three wells from one cortical culture preparation were treated with each compound (0.03–40 pM). Prior to recording baseline activity, each mwMEA plate was placed in the Maestro at 37 °C and allowed to sit for 20 min while firing rates stabilized. Baseline activity (40 min) was recorded before the addition of each compound. An additional 40 min of spontaneous activity was recorded in the presence of each compound. Changes in network parameters relative to baseline were assessed following compound treatment.
Cytotoxicity assays
All compounds were examined for cytotoxicity using the mwMEA multiplexed approach outlined in Wallace et al (2015) with the following modifications. Immediately following the 40 min recording in the presence of compounds, 50 pl of media was removed from each well and transferred to a sterile 96-well plate. This was used to determine LDH released from cells during compound exposure using a kit from Promega where absorbance was determined in a Molecular Devices VersaMax plate reader at 490 nm.
Metabolic activity was determined using the CellTiter Blue (CTB) assay (Promega Cat. #8081), following the manufacturer’s instructions with the following modifications. Following the removal of the 50 pl sample for the LDH assay, 450 pl of media was removed from each well and replaced with 200 pl of fresh media containing a 1:6 dilution of CTB reagent for the determination of metabolic activity. Fluorescence was measured in a Fluorostar Optima fluorimeter using an excitation wavelength of 544 nm and an emission wavelength of 590 nm.
Data analysis
Raw recordings were re-played and analyzed with the AxIS 2.3 Neural Statistics Compiler. The Burst Detector Settings used the Inter-Spike Interval Threshold Algorithm with a maximum Inter-Spike Interval of 100 ms and had at least 5 spikes. Network burst detection was enabled with a maximum inter-spike interval of 100 ms, a minimum of 10 spikes, 25% of the electrodes participating and a Synchrony Window of 20 ms. This provided 41 different well-level (e.g., averaged across all electrodes in the well) parameters of neural network function, each of which represented an average activity over the 40 min recording period, to which weighted mean firing rate and number of active electrodes were added, for a total of 43 parameters. Data for each of these parameters were analyzed using the ToxCast Analysis Pipeline (tcpl, version 1.2.2) (Filer et al. 2017), which is available publicly as an R package (Filer 2016). Based on the low variability in DMSO activity, no transformation or normalization was assigned using tcpl. Outliers were determined for individual endpoints as follows: sd = standard deviation, x = (dose - baseline) for each sample, and removed if x < mean(x)—6*sd(x) or x > mean(x) + 6*sd(x). The responses for assay parameters that indicated a decrease or loss of function were multiplied by - 1. The difference between each sample response and baseline value was used to generate concentration-response curves. A chemical was considered active in an assay if at least one median response value was greater than three times the baseline median absolute deviation (BMAD), a measure of median response values for all compounds across the lowest two concentrations for that assay that is indicative of baseline variability. The curves for each chemical-assay combination were fit with a constant model, a constrained Hill model, and a constrained gain-loss model, as described in tcpl. The model with the lowest Akaike Information Criterion (AIC) value was determined to be the best fit (Filer et al. 2017). Analyses were carried out using R version 3.5.1 (R Core Team 2018). All data, R-code and other associated files are available at the following DOI (https://doi.org/10.23719/1504294).
Neuroactive and non-neuroactive chemical classifications
A list of neuroactive and non-neuroactive chemicals within the set of 384 compounds tested here was developed to serve as the “neurotoxicity training set” to classify compounds from our chemical set and to aid in identifying the parameters most crucial in characterizing neuroactivity. Neuroactive chemicals were identified from the active chemical set based on generally well-established activity towards neurotransmitter receptors, ion channels or other neurobiological targets yielding a list of 41 neuroactive chemicals within our chemical set (Table S1). Non-neuroactive chemicals were identified for this study by selecting chemicals negative in this concentration-response screen as well as the previous single-point screen (Strickland et al. 2018). This chemical list was then evaluated for potential neurotoxicity by searching *chemical name* and *neurotoxicity* in PubMed. Chemicals with evidence of neurotoxicity in the literature were removed from our list giving a total of 32 non-neuroactive chemicals (Table S1).
Clustering methods
Complete hierarchical clustering of chemical-parameter results was conducted to reduce redundant parameters. This was done using a Euclidian distance function weighted to account for differences in each chemical-parameter AC50 and winning (lowest AIC) model. K-means clustering of chemical-parameter results to identify similar chemical- parameter patterns was conducted using JMP version 14.0 (SAS Institute Inc.). The input data for the potency-driven k-means clustering analysis were AC50 values (in pM) for chemical-parameter hits and an assignment of 1 mM for non-hits (about 8 times the highest AC50 value). The input data for the direction of activity-driven k-means clustering were a binary value distinguishing between increased and decreased activity. Validation of k-means clusters was conducted by comparing the percentage of variance explained as a function of the number of clusters (the elbow method), and using NbClust (version 3.0), an R package that uses nearly 30 indices to select the optimal number of clusters (Charrad et al. 2014). These k-means clusters were used to characterize the chemical-parameter results.
Machine learning methods
WEKA version 3.8.1 (Frank et al. 2016) was used to rank parameters with information gain ranking and Pearson’s correlation and to perform sequential minimal optimization (SMO) Support Vector Machines (SVMs), Artificial Neural Networks (ANNs) and Random Forest machine learning classification algorithms. These algorithms were used to classify neuroactive and non-neuroactive compounds from the neurotoxicity training set and select the ranked parameters most crucial for classification using the AC50s from the chemical-parameter hits (in pM) as the parameter value. Inactive values were set to 1 mM, about eightfold larger than the highest AC50. All classifications were conducted using tenfold cross-validation. The percent correct assignment of the compounds using these machine learning methods was the basis for evaluating classifier performance. A plot of correlations between chemical parameters was developed using R/corrplot (version 0.84) (Wei and Simko 2017).
Chemical potency comparisons
For each active chemical in the MEA data set, the AC50 of the most potent MEA parameter was compared to the potencies of all other active (ToxCast hit call = 1) ToxCast and Tox21 assays for that chemical. The most-recent, full release of the invitrodb_v2 collection of ToxCast and Tox21 assay information (October 2015) was obtained from the ToxCast website (https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data ) (U.S. EPA 2015). Figures were generated using R/ggplot2 package (version 3.1.0) (Wickham 2016).
ToxPrint chemotype enrichment analysis
Chemical feature analysis was conducted using the Chem- otyper application (https://chemotyper.org/), with the latest ToxPrint feature set (V2.0_r711; https://toxprint.org/ ), developed by Altamira (Altamira, Columbus, OH, USA) and Molecular Networks (Molecular Networks, Erlangen, GmbH) under contract from the US Food and Drug Administration (FDA) (Yang et al. 2015). The feature set contains 66 different categories of substructures (e.g., C#N and AlkaneCyclic, which contain nitrile groups and cyclic alkanes, respectively), or chemotypes (CTs), with 729 more specific chemotypes within them (e.g., C#N contains C#N_ cyano_cyanohydrin; a nitrile group bonded to a CO, and C#N_nitrile_generic; indicating any chemical that contains a nitrile group). We used the 66 broader, un-nested categories for analysis. Chemical hits within the potency-driven k-means clusters were given structural fingerprints based on the presence of a given chemotype within that chemical’s structure.
Chemotype enrichment was conducted using the hypergeometric test. Enrichment was calculated based on the proportion of each CT within a k-means cluster relative to the proportion of that CT within all k-means clusters. Chem- otypes were considered significantly enriched if p values were < 0.05. Figures were generated using R/ggplot2 package (version 3.1.0) (Wickham 2016) and R/pheatmap package (version 1.0.12) (Kolde 2018).
Results
Characterization of chemical potency
Of the 384 chemicals included in the experiment, 375 chemicals were active on at least one network parameter, for a total of 5566 chemical-parameter hits. The LDH assay exhibited small BMAD values, resulting in responses for many chemicals being greater than the 3*BMAD threshold, even though they did not demonstrate a concentration response. This decreased the reliability of this parameter for detecting truly cytotoxic compounds and therefore cytotoxicity was characterized using only the CTB assay. Nineteen of these chemicals were also active in the CTB assay indicating potential cytotoxicity. To avoid counting cytotoxic chemical activity as activity in a parameter, the AC50 values of these chemicals in the CTB assay were compared to the AC50 values of the chemicals in any other parameter and only those chemical-parameter hits with AC50s less than the chemical- CTB AC50 were counted as hits. This reduced the number of “active” chemicals from 375 to 374 and the number of positive chemical-parameter hits from 5566 to 5423.
Characterization of the most informative parameters
Many of the parameters extracted from the MEA recordings in the analysis were highly correlated. To reduce the parameter set to the most informative parameters, we conducted hierarchical clustering on the 43 parameters with a distance function adjusted to include differences between chemical- parameter AC50s and winning AIC models. Within these 43 parameters, 14 pairs clustered together closely and were narrowed down to one parameter within each of these redundant pairs (Figure S1). Many of these parameters were variants of the other (e.g., “mean firing rate” vs. “weighted mean firing rate”) or evaluated the same characteristic using different statistics (e.g., “mean” vs. “median ISI within burst”). In selecting between the two closest parameters to keep in the analysis, preference was given to the parameter that was not a variant of the other (e.g., “mean firing rate”) and calculation of averages over standard deviations (e.g., “average network burst duration”). The “Number of Active Electrodes” and “Number of Bursting Electrodes” parameters were both excluded due to promiscuity among several chemicals. This reduced the number of parameters from 43 to 27 and the number of active chemicals from 374 to 323.
To reduce the parameter set further, machine-learning methods were employed to rank the remaining 27 parameters and identify those most informative in distinguishing between neuroactive and non-neuroactive chemicals in the neurotoxicity training set. Parameters were ranked using an information gain ranking approach and Pearson’s correlation approach with WEKA (Table S2). These 27 parameters were further narrowed down to the top 20, 15, 10, and 5 parameters and used to classify neuroactive and non-neuroactive chemicals in the neurotoxicity training set using SVM, ANN, and Random Forest machine learning methods with WEKA. To determine the best method for classifying active chemicals as hits, this process was repeated three times with the neurotoxicity chemical training set included in the machine learning process narrowed down to those active in 1 or more parameters, 2 or more parameters, or 3 or more parameters.
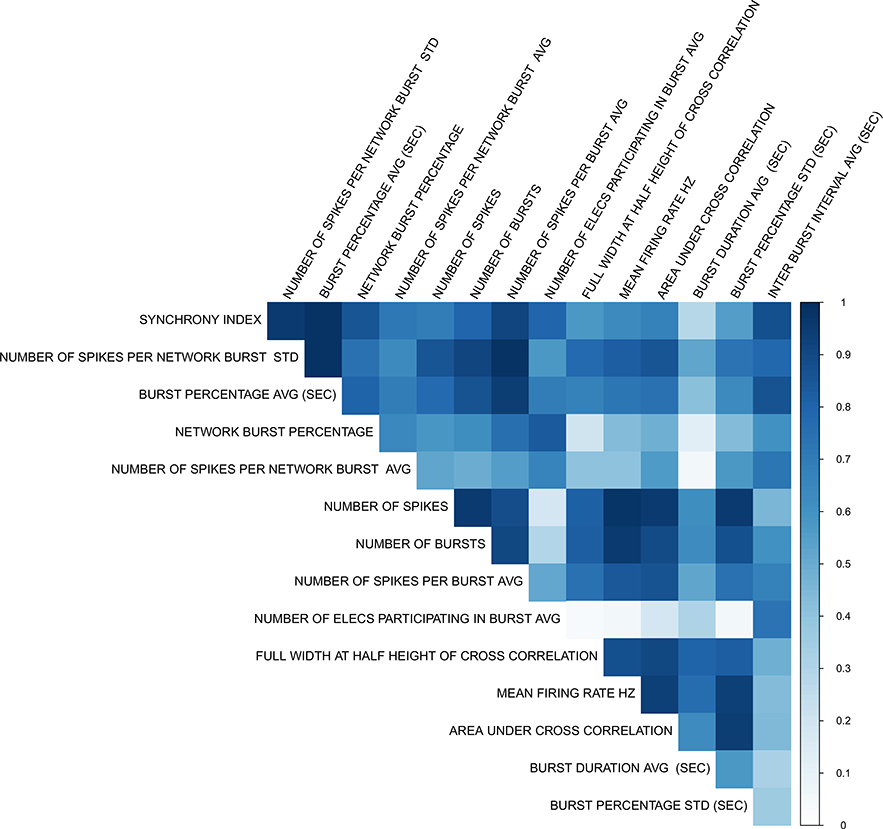
Across all three machine learning models, requiring a minimum of three parameter hits to count a chemical as active gave the greatest classification accuracy between neuroactive and non-neuroactive chemicals as did reducing the number of parameters to the top 15. Although the order differed slightly, the top 15 parameters were the same when ranked using both an information gain ranking approach and Pearson’s correlation. As described in Table 1, these parameters cover different general activity, bursting, and connectivity measures of neuroactivity. The pairwise correlation between these parameters is shown in Fig. 1. Between the three machine-learning models, the Random Forest model had the highest classification accuracy overall. For the Random Forest model, with chemicals requiring a minimum of three parameter hits to be counted as active, 98.4% of neuroactive versus non-neuroactive chemicals in the training set were correctly classified with the 15 parameter set, and this dropped to 96.9% with all 27 parameters and 93.8% with the top 5 parameters. Based on this high correct classification accuracy, we reduced the 27 parameters to the top 15 parameters and only counted chemicals as active if they were active in at least three of these 15 parameters. This reduced the number of chemicals labeled “active” across all chemicals tested from 323 to 237. If a chemical was active in both the up and down direction for a parameter, then the direction with the lower AIC was selected. These processes reduced the total number of chemical-parameter hits from 3112 to 2060. These data are summarized in Fig. 2 with more in-depth data available in Table S3.
Table 1.
Top 15 parameters
| Category | Parameter | Description/definition |
|---|---|---|
| General activity (“Firing”) | Mean firing rate (Hz) | Total number of spikes over the duration of the analysis |
| Number of spikes | Total number of spikes over the duration of the analysis | |
| Number of bursts | Total number of single-electrode bursts over the duration of the analysis | |
| Burst structure | Burst duration (average) | Average time from the first to the last spike in a burst |
| Interburst interval (average) | Average time between the start of bursts | |
| Burst percentage-avg | The number of spikes in a burst divided by the total number of bursts, times 100 | |
| Burst percentage-std | Standard deviation of the burst percentage | |
| Number of spikes/burst (avg) | Average number of spikes in a burst | |
| Connectivity | Network burst percentage | The number of spikes in network bursts divided by the total number of spikes, times 100 |
| #Spikes/network burst-avg | The average number of spikes in a network burst | |
| #Spikes/network burst-std | The standard deviation of the number of spikes in a network burst | |
| #Electrodes participating in Burst-avg | The average number of electrodes participating in a network burst | |
| Area under cross-correlation | Area under the well-wide pooled inter-electrode cross-correlation | |
| Full-width at half-height of cross-correlation | Distance along the x-axis from the left half height to the center half height of the normalized cross correlogram | |
| Synchrony Index | A unit measure of synchrony between 0 and 1 | |
Definitions are summarized from Axion Biosystems neural metric tool metric definitions (v2.4)
Fig. 1.
Parameter correlations of chemical-parameter AC50s for final 15 MEA parameters
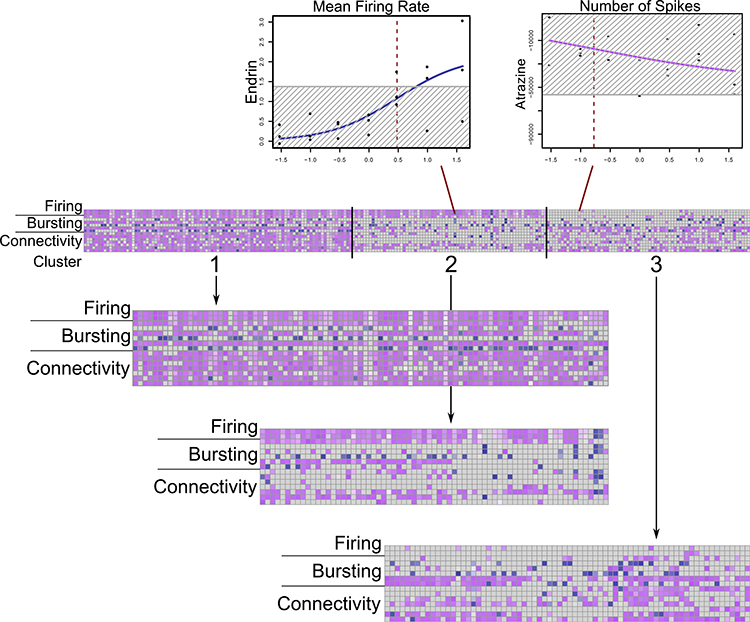
Fig. 2.
Heatmap of three potency-driven activity patterns in k-means clusters of chemical-parameter hits. Blue = increased activity, purple = decreased activity. Darker colors = lower chemical AC50s. Sample dose-response curves shown for an active and inactive chemical-parameter. Red lines = log10(AC50), grey box = 3*BMAD
Among the final chemical-parameter data, five times more chemical-parameters had decreased activity than increased activity. The “burst percentage average” parameter was the most promiscuous of our final parameter set with 193 chemicals active. “Number of spikes” and “mean firing rate” were the next two most active parameters with 170 and 163 chemical hits, respectively. Seven chemicals were active in all 15 parameters: cyfluthrin, fluazinam, pyridaben, ema- mectin benzoate, nitrofen, dichlorodiphenyldichloroethane, and PharmaGSID_48509.
Comparison of MEA potencies to ToxCast assays
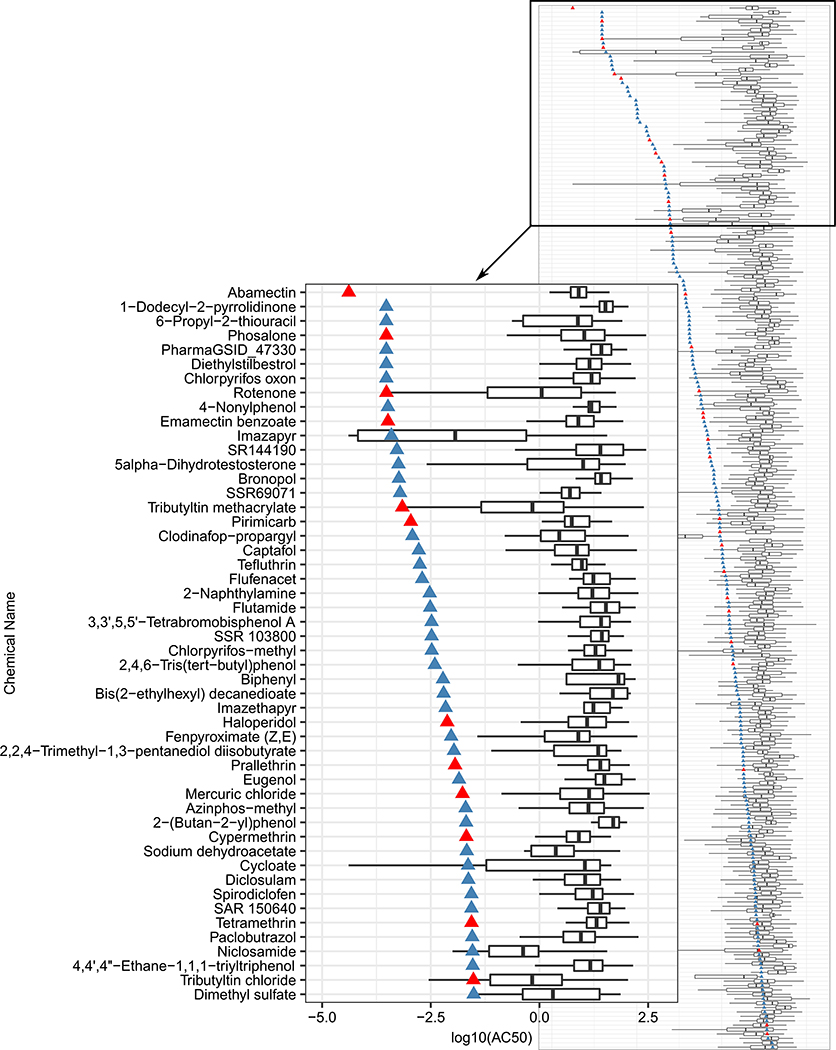
To characterize the relative potency of the MEA parameters to other ToxCast assays, we plotted the AC50 of the most potent MEA parameter for each chemical against the EC50s of active ToxCast assays for that same chemical (Fig. 3, SI Fig. 2, Table S4). Of the 1192 assay endpoints in the original ToxCast data set, 852 were active in our final set of 237 chemicals. Among the known neuroactive compounds included in the chemical training set, the MEA parameter was consistently more sensitive than other ToxCast assays (Fig. 3, SI Fig. 2). Abamectin was the most potent chemical with an AC50 of 4.1 × 10−5 in the area under cross-correlation MEA parameter. All 15 of the MEA parameters were among the most potent for the full chemical-parameter set.
Fig. 3.
Comparison of ToxCast assay potencies to most potent MEA parameter for each compound. Triangle = log10(AC50) for the most potent MEA parameter for each chemical, red = known neuroactive compounds (Table S1), blue = other compounds. Inset figure = compounds with log10(AC50) ≤ — 1.5. Full figure=Figure S2
Characterization of chemical activity patterns
The chemical-parameter hits, when clustered, could suggest that different clusters reflect different modes of action for neurotoxicity (Fig. 2). We used k-means clustering with a range of 1–15 clusters to determine the best grouping for acute effects on network activity measured with MEAs. After validation using both the elbow method and the NBClust R package (Charrad et al. 2014), we determined that three clusters formed the best grouping for the chemical activity patterns based on the potency of effects. The first cluster (Cluster 1), contained 95 compounds, while the second (Cluster 2) and third (Cluster 3) contained 69 and 72 chemicals, respectively. Closer examination of these potency-driven “bioactivity clusters” indicates that compounds in Cluster 1 (Fig. 2) alter nearly all of the 15 network parameters, whereas Cluster 2 compounds, which also altered firing parameters, had much more limited actions on bursting parameters and actions on connectivity were generally more limited to synchrony measures. By contrast, Cluster 3 was differentiated from Cluster 2 by an almost complete lack of effect on firing parameters, and more robust effects on bursting and connectivity parameters, especially connectivity parameters. Further, by considering the directionality of changes within each cluster, sub-clusters of activity could be determined. Compounds within these 3 clusters were grouped by the chemical-parameter direction of change (up versus down) to determine if including this information clarified differing patterns of activity among the compounds. The results indicated that, within each of the three main clusters, up to 5 sub-clusters could be identified based on the directionality of the changes in MEA parameters. For example, Cluster 2 was divided into three sub-clusters with different firing and bursting parameter activity. Sub-Clusters 1 and 2 consistently decreased firing and connectivity parameter activity, but Sub-Cluster 1 had more bursting parameter activity than Sub-Cluster 2. In contrast, Sub-Cluster 3 consistently increased firing, bursting, and connectivity parameters. These chemical clusters and sub-clusters are described in Table S5. Whereas these sub-clusters further clarified different biological activity patterns within the main clusters, the number of chemicals within each sub-cluster was variable. To make direct comparisons between clusters easier, only the original set of three potency-driven clusters was used for additional analysis.
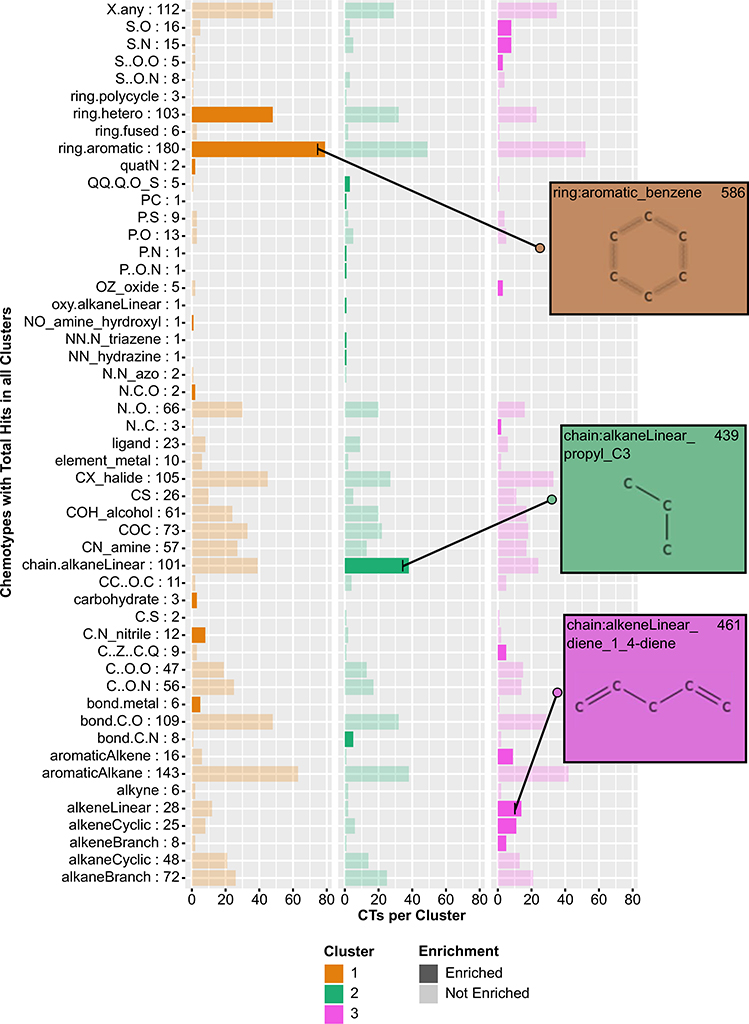
An analysis of the sub-structural features of each chemical within the three clusters was conducted to identify any features that could be contributing to the original, potency-driven chemical clustering pattern. We used the ToxPrint CTs broad, un-nested structure categories to identify chemical structural fingerprints based on the presence of a given CT within that chemical’s structure. Of the 237 chemicals within k-means clusters, 51 of the 66 CTs were represented at least once, and only one chemical, SSR 103800, a mixture, did not have any assigned CTs. The 236 chemicals with corresponding CTs are given in Table S6. To determine if structure could explain the parameter activity pattern among these potency-driven k-means clusters, we calculated enrichment of each chem- otype within one of the three k-means clusters relative to all three clusters. Of the 51 different CT categories analyzed, 27 were enriched in one of the three clusters (Fig. 4, Table S7). Cluster 1, 2, and 3 were enriched for 8, 9, and 10 different CTs, respectively. No single CT was enriched in multiple clusters. In addition to the distinct CT patterns within each cluster, particular motifs tended to be enriched within the same cluster. For example, ring structures (e.g., aromatic and hetero rings) were consistently enriched in Cluster 1, whereas sulfur-containing compounds (e.g., S-N and S-O) were consistently enriched in Cluster 3. The aromatic rings CT enriched in Cluster 1 were present in the most chemicals, with 79 chemicals in Cluster 1 containing the motif. Compared to the 147 chemicals now labeled as inactive, the active chemical set has more aromatic structures and carbon chains. Unlike the active set, the inactive set contains nucleobase and amino acid CTs as well as different nitrogen structures, such as N = [N+] (data not shown).
Fig. 4.
Barplot describing enriched chemotypes within three potency-driven k-means clusters. Number of CTs within all clusters in row label. Darker bars = enriched CTs within cluster. Inset images depict structures within respective CT. More complete CT descriptions can be found in the chemotyper application
Discussion
The present study provided concentration-response characterization of positives identified by single concentration screening of 1055 compounds conducted in Strickland et al. (2018). Of the 222 compounds that were active on the MFR parameter at the highest concentration in the previous single concentration screen analysis, 189 compounds (85%) were active in the final chemical set in the present multi-concentration-response screening follow-up analysis, and 136 were active on the MFR parameter. The direction of chemical activity on the MFR parameter for these 136 compounds was in agreement for both the previous single concentration screen and the current multiconcentration screen, with 135 compounds decreasing MFR activity and 1 compound increasing MFR activity. In the previous work (Strickland et al. 2018), hit calls were based on changes in mean firing rate relative to changes caused by DMSO for a single concentration of compound. In the current analysis, a “neurotoxicity training set” of chemicals that were contained within the original Tox- Cast test set was used as a benchmark for training and validation of the models to determine active compounds following exposure to multiple concentrations. This work provides AC50 values for effects of these compounds on neural function, a parameter that is not well represented in the current panel of ToxCast assays. Further, the present work demonstrated that multiple parameters describing neural network activity in MEAs provide a more robust assessment of chemical neuroactivity than MFR alone, as no individual parameter fully predicted the positive or negative status of all other parameters in the set, and 15 parameters best distinguished between active and inactive chemicals in our neurotoxicity training set. Finally, the present analysis demonstrated that chemical expression patterns vary across these 15 parameters and the results reveal three patterns of bioactivity that could indicate distinct chemical mechanisms of action.
The complex spiking, bursting and coordinated activity of neural networks grown on MEAs can be described by over 200 parameters (Gramowski-Voß et al. 2015; Bader et al. 2017). To date, assessment of neuroactivity/ neurotoxicity using MEAs has focused largely on compound actions on mean firing rate (MFR) (Ylä-Outinen et al. 2010; Defranchi et al. 2011; McConnell et al. 2012; Scelfo et al. 2012; Nicolas et al. 2014; Valdivia et al. 2014; Hondebrink et al. 2016) and/or burst rate (Keefer et al. 2001b, a; Pancrazio et al. 2001). Previous studies have also demonstrated that consideration of multiple parameters of network activity can provide additional information to help characterize the actions of compounds on network activity and can be used to identify groups of compounds with similar modes of action (Gramowski et al. 2004, 2006; Gramowski-Voß et al. 2015; Bader et al. 2017; Bradley et al. 2018). These groups have used multiple parameters of network activity and grouped them into classifications such as “general activity”, “burst structure”, “oscillatory behavior” (not considered here) and “synchrony” (similar to “connectivity”) to examine functional relationships between smaller sets of compounds (Gramowski et al. 2004, 2006; Gramowski-Voß et al. 2015; Bader et al. 2017). Interestingly, in these cases, the authors selected a set of ~ 12 parameters to use for characterization of compound effects. However, these previous studies generally examined small sets of compounds with a limited chemical space, and the methods/rationale for selecting parameters were not clearly stated. Thus, the extent to which this approach could be applied to a set of compounds with diverse structures is unknown. The present study examined a set of 384 compounds drawn from a more diverse chemical space to consider the information provided by what initially was a set of 43 parameters of network function. Using unsupervised analysis approaches, this was reduced to a set of 15 parameters that separated neuroactive from non-neuroactive compounds in a neurotoxicity training set with high accuracy. Interestingly, although functional domains represented by these parameters were not a consideration in the selection of the final 15 parameters used here, this final set reflects 3 major domains of network activity (overall activity rate, bursting structure, and connectivity), as outlined in Table 1, Fig. 2, and Table S3. That these domains of activity emerged without a priori selection indicates that effects on firing, bursting and network activity in vitro are indeed important in describing how different compounds disrupt neural network activity following acute exposure. Interestingly, our results identifying these functional domains as important are consistent with the studies mentioned above, despite our unbiased approach using a neuroactive training set of compounds compared to the unknown methods utilized in the other studies.
The present results indicated that the actions of compounds on network activity could be described by three different patterns of bioactivity. By examining the chemotypes enriched within each chemical cluster, we found that chemical structure can help explain the pattern of neuroactivity in our chemical-parameter results. Different chemotypes were enriched in different clusters, demonstrating the importance of chemical structure in influencing different patterns of bioactivity, which could in turn be surrogates for different mechanisms of activity. Further, several examples illustrate how chemical structure and biological activity are interrelated and influence how network activity is affected by different chemical classes. Triazole fungicides were grouped in Cluster 1 (flusilazole, hexaconazole, propiconazole, difenoconazole) and Cluster 2 (tebuconazole, paclobutra- zole), whereas the strobulirun fungicides were all found in Cluster 1. Pyrethroid insecticides were distributed among Clusters 1 and 3 whereas organochlorine insecticides that act on GABAa receptors were distributed more evenly across all three clusters. Further, the sub-clustering with respect to the direction of chemical-parameter activity improved the separation of these compounds within the potency-driven clusters. For example, the triazole fungicides in Cluster 2 were in Sub-Cluster 3, whereas the GABAA agonists and antagonists in Cluster 2 were in Sub-Clusters 1 and 2. The GABAA compounds in Cluster 1 were in a different subcluster than the pyrethroids in Cluster 1. In general, segregation of compounds into different clusters and sub-clusters may in part reflect differences in mechanisms of action. For example, the triazole fungicides may interfere with voltagegated calcium (Heusinkveld et al. 2013) and potassium channel (Sung et al. 2012) function, while pyrethroids modify kinetics of voltage-gated sodium channel function (Narahashi 2002). This demonstrates the utility of this assay to assess and distinguish different neuroactivity patterns for compounds with varied effects on neural network function. While certain compound classes tended to group into these different neuroactivity patterns, the neuroactivity patterns did not consist of entire compound classes. Two factors may contribute to this. First the promiscuity of many of the compounds in this chemical set towards multiple targets likely invokes different patterns of response; these promiscuous actions likely increase as concentrations of compounds increase. Second, beyond a few categories of compounds (pyrethroids, triazoles, organochlorines), most compound classes had five or fewer representative members in the current test set. Both of these issues make it hard to distinguish truly unique patterns of response based on chemical class.
These results, demonstrating that evaluation of multiple neural network parameters in conjunction with consideration of chemical structure results in predictable outcomes within the neuroactive space, are important from the standpoint of evaluating unknown compounds. The relationship between structure and function will provide increased confidence that compounds for which toxicological information is lacking may have relevant effects if they have network parameter activity and structural profiles similar to known neuroactive compounds that have been more thoroughly characterized. Further, these different structure/activity inferences represent different chemical classes, providing an approach to predict neuroactivity for different chemical exposures. This information will be useful in the context of prioritization for additional screening/testing and for designing more focused follow-up studies.
We previously demonstrated enrichment of certain chemotypes among the active compounds (compared to inactive compounds) in the single concentration screen (Strickland et al. 2018). The approach taken here differs from the previous approach in two important respects. First, we used the 66 broad chemotype classifications rather than the specific chemotypes used in Strickland et al. (2018). For example, the enriched “chain:aromaticAlkane_ Ph-C1-Ph” and “chain:aromaticAlkane_Ph-C6” which were enriched in Strickland et al. (2018) would both be part of the “aromaticAlkane” classification for chemotypes in the present data set. Second, we also calculated enrichment from one cluster to the next (meaning we were relating active compounds to each other) rather than relating actives to inactives as was done in the single-point screen. Although direct comparisons between the enrichment of chemotypes in the two studies are therefore difficult, there were examples of consistency across the two studies. For example, of the overlapping chemicals between the single-point screen (Strickland et al. 2018) and the present analysis, 4/5 “ring:hetero_[5]_N_pyrazole” and 2/4 “ring:hetero_[5]_O_dioxolane_(1_3-)” chemicals were in the “ring:hetero” enriched cluster. A comparison of chem- otype enrichment in the active vs inactive compounds in the present data set will be conducted in the future.
These data have other uses as well. By identifying those compounds with more potent AC50 values in the MEA assay than in other ToxCast assays, potential neurotoxicity could be identified as an endpoint of concern, particularly when compared to assays that do not measure endpoints of relevance to the nervous system. Additionally, by comparing activity for these compounds in other ToxCast assays to activity in the MEA assay, the relationship between neuroactivity and broader biological activity can be elucidated. We found that, for the majority of known neuroactive compounds in the chemical training set, the MEA parameters were more sensitive than or had potencies similar to other ToxCast assays. This was also the case with other active compounds in our data set with unknown neuroactivity. For example, 4-nonylphenol, a common environmental contaminant, has an MEA AC50 similar to known neuroactive compounds like rotenone and phosalone, and the MEA parameter AC50 was more sensitive than other ToxCast assays. While this compound’s neuroactive potential is not well- understood, there are other indications that 4-nonylphenol can impact nervous system function. For example, 4-nonyl- phenol has been found to inhibit acetylcholinesterase activity across organisms (Li 2008a, b; Vidal-Liñán et al. 2015), and to augment neurotoxic behavioral effects with co-exposure to diazinon in Daphnia (Zein et al. 2015). Other endocrine- disrupting chemicals have been implicated in neurological disorders (Kajta and Wójtowicz 2013; Cano-Nicolau et al. 2016), and, while the mechanism(s) of 4-nonylphenol’e effects on network activity have yet to be elucidated, our analysis highlights the potential for 4-nonylphenol to also affect neuroactivity.
Within the ToxCast assay suite are assays that measure interactions of compounds with ion channels, G-protein and membrane-bound receptors that are of relevance to nervous system function. Comparing the results of the present study to those assays could serve to help develop putative adverse outcome pathways (AOPs) related to neurotoxicity. Whereas many different mechanisms of neurotoxicity are well-established in the literature, few well-documented AOPs for neurotoxicity have been described to date (Gong et al. 2015; Bal-Price et al. 2015, 2017; Sachana et al. 2018; Li et al. 2019). These data therefore can serve as a resource to either strengthen existing AOPs or support the development of new ones.
While the data and analysis presented here have a number of different uses for neurotoxicity screening, some limitations of the model should be considered when interpreting the results of those analyses. The approach taken here was designed to optimize rapid and efficient testing of large numbers of compounds. Thus, a minimal number of wells (3) per treatment were evaluated, and our combined analysis approach demonstrated a high accuracy (98.4%) of separating neuroactive from non-neuroactive compounds. However, increasing the number of wells/treatment may be valuable for further characterization of hits, or testing of a smaller number of compounds, as this may provide a more robust data set for traditional statistical analyses (e.g., ANOVA followed by post-hoc testing). The primary cortical model used here has been widely utilized for the study of neuroactive/ neurotoxic effects of chemicals. However, it is a single brain region, and activity of compounds that have preferential effects on other brain regions [e.g., striatum (dopaminergic) or brainstem (glycinergic)] may not be captured or reflected accurately by the cortical model. The lack of activity of cholinergic compounds in this particular model has been previously established (McConnell et al. 2012; Valdivia et al. 2014), although other laboratories have reported effects of cholinergic compounds in similar models (Defranchi et al. 2011; Hondebrink et al. 2016). Other factors that could also influence chemical responses are the lack of a blood-brain barrier in the MEA system as well as a limited metabolic capability of the primary cortical cells compared to the liver.
Conclusion
Primary cortical cultures grown on MEAs respond to a broad range of structurally diverse compounds with changes in electrical activity. We evaluated neuroactivity for ToxCast chemicals in multiple network activity parameters to develop a robust assessment of neurotoxicity, and these MEA parameters were more sensitive overall than other ToxCast assays. By characterizing the concentration-response of literature- identified active compounds using a set of metrics that describes different aspects of network function, it was demonstrated that compounds could be grouped by profiles of biological activity, and that these groups were consistent with the underlying chemotypes present in the compounds tested here. The current model resulted in a reduced set of network parameters recommended for processing the MEA multi-concentration screen potency data to better identify true neurotoxic-actives. With such modifications, this assay will be increasingly useful for identifying and characterizing the potential neurotoxicity of compounds for which toxicity data are lacking.
Supplementary Material
Acknowledgements
The authors acknowledge the outstanding tissue culture and general laboratory support of Ms Theresa Freuden- rich at the US Environmental Protection Agency (EPA). In addition, the authors thank Drs Katie Paul-Friedman and Holly Mortensen at the US EPA for their useful comments on a previous version of this manuscript.
Funding This work was supported in part by the National Health and Environmental Effects Research Laboratory (NHEERL) and in part by CRADA 644–11 between the US EPA and Axion Biosystems. This work was supported by the National Institutes of Health [ES025128, ES030007] and the US Environmental Protection Agency [STAR R835802].
Footnotes
Compliance with ethical standards
Conflict of interest JDS was an employee of Axion Biosystems when the data collection for this work was conducted. Axion is a manufacturer of microelectrode array recording equipment and supplies. All other authors declare that they have no conflicts of interest.
This document has been subjected to review by US EPA Office of Research and Development and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. This work was supported in part by CRADA 644–11 between the US EPA and Axion Biosystems.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00204-019-02636-x) contains supplementary material, which is available to authorized users.
References
- Alloisio S, Giussani V, Nobile M et al. (2016) Microelectrode array (MEA) platform as a sensitive tool to detect and evaluate Ostre- opsis cf. ovata toxicity. Harmful Algae 55:230–237. 10.1016/j.hal.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Bader BM, Steder A, Klein AB et al. (2017) Functional characterization of GABAA receptor-mediated modulation of cortical neuron network activity in microelectrode array recordings. PLoS ONE 12:e0186147 10.1371/journal.pone.0186147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal-Price A, Crofton KM, Sachana M et al. (2015) Putative adverse outcome pathways relevant to neurotoxicity. Crit Rev Toxicol 45:83–91. 10.3109/10408444.2014.981331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal-Price A, Lein PJ, Keil KP et al. (2017) Developing and applying the adverse outcome pathway concept for understanding and predicting neurotoxicity. Neurotoxicology 59:240–255. 10.1016/j.neuro.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar MK, Murthy PB (2018) Acute in vitro neurotoxicity of some pyrethroids using microelectrode arrays. Toxicol Vitr 47:165–177. 10.1016/j.tiv.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Bradley JA, Luithardt HH, Metea MR, Strock CJ (2018) In vitro screening for seizure liability using microelectrode array technology. Toxicol Sci 163:240–253. 10.1093/toxsci/kfy029 [DOI] [PubMed] [Google Scholar]
- Cano-Nicolau J et al. (2016) Estrogenic effects of several BPA analogs in the developing zebrafish brain. Front Neurosci 10:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrad M,Ghazzali N, Boiteau V, Niknafs A (2014) An examination of indices for determining the number of clusters: NbClust Package R topics documented. J Stat Softw 61:1–36 [Google Scholar]
- Chiappalone M, Bove M, Vato A et al. (2006) Dissociated cortical networks show spontaneously correlated activity patterns during in vitro development. Brain Res 1093:41–53. 10.1016/j.brainres.2006.03.049 [DOI] [PubMed] [Google Scholar]
- Colombi I, Mahajani S, Frega M et al. (2013) Effects of antiepileptic drugs on hippocampal neurons coupled to micro-electrode arrays. Front Neuroeng 6:1–11. 10.3389/fneng.2013.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defranchi E, Vogel S, van Ravenzwaay B et al. (2011) Feasibility assessment of micro-electrode chip assay as a method of detecting neurotoxicity in vitro. Front Neuroeng 4:1–12. 10.3389/fneng.2011.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemans MML, Schütte MG,Wiersma DMM et al. (2016) Chronic 14-day exposure to insecticides or methylmercury modulates neuronal activity in primary rat cortical cultures. Neurotoxicology 57:194–202. 10.1016/j.neuro.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT et al. (2007) The toxcast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 95:5–12. 10.1093/toxsci/kfl103 [DOI] [PubMed] [Google Scholar]
- Filer DL (2016) tcpl: ToxCast Data Analysis Pipeline. R package version 1.2.2. https://CRAN.R-project.org/package=tcpl [Google Scholar]
- Filer DL, Kothiya P, Woodrow Setzer R et al. (2017) Tcpl: The Tox- Cast pipeline for high-throughput screening data. Bioinformatics 33:618–620. 10.1093/bioinformatics/btw680 [DOI] [PubMed] [Google Scholar]
- Frank E, Hall MA, Witten IH (2016) The WEKA Workbench. Online Appendix[dummy] for “Data Mining: Practical Machine Learning Tools and Techniques, Fourth Edi. Morgan Kaufman [Google Scholar]
- Frega M, Pasquale V, Tedesco M et al. (2012) Cortical cultures coupled to micro-electrode arrays: a novel approach to perform in vitro excitotoxicity testing. Neurotoxicol Teratol 34:116–127. 10.1016/j.ntt.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Gong P, Hong H, Perkins EJ (2015) Ionotropic GABA receptor antagonism-induced adverse outcome pathways for potential neurotoxicity biomarkers. Biomark Med 9:1225–1239. 10.2217/bmm.15.58 [DOI] [PubMed] [Google Scholar]
- Gramowski A, Jugelt K, Weiss DG, Gross GW (2004) Substance identification by quantitative characterization of oscillatory activity in murine spinal cord networks on microelectrode arrays. Eur J Neurosci 19:2815–2825. 10.1111/j.0953-816X.2004.03373.x [DOI] [PubMed] [Google Scholar]
- Gramowski A, Jügelt K, Stüwe S et al. (2006) Functional screening of traditional antidepressants with primary cortical neuronal networks grown on multielectrode neurochips. Eur J Neurosci 24:455–465. 10.1111/j.1460-9568.2006.04892.x [DOI] [PubMed] [Google Scholar]
- Gramowski-Voß A, Schwertle HJ, Pielka AM et al. (2015) Enhancement of cortical network activity in vitro and promotion of GABAergic neurogenesis by stimulation with an electromagnetic field with a 150 MHz carrier wave pulsed with an alternating 10 and 16 Hz modulation. Front Neurol 6:1–12. 10.3389/fneur.2015.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusinkveld HJ, Molendijk J, Van den Berg M, Westerink RHS (2013) Azole fungicides disturb intracellular Ca2+ in an additive manner in dopaminergic PC12 cells. Toxicol Sci 134:374–381. 10.1093/toxsci/kft119 [DOI] [PubMed] [Google Scholar]
- Hondebrink L, Verboven AHA, Drega WS et al. (2016) Neurotoxicity screening of (illicit) drugs using novel methods for analysis of microelectrode array (MEA) recordings. Neurotoxicology 55:1–9. 10.1016/j.neuro.2016.04.020 [DOI] [PubMed] [Google Scholar]
- Huang T, Wang Z, Wei L et al. (2016) Microelectrode array-evaluation of neurotoxic effects of magnesium as an implantable biomaterial. J Mater Sci Technol 32:89–96. 10.1016/).jmst.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone AFM, Gross GW, Weiss DG et al. (2010) Microelectrode arrays: a physiologically based neurotoxicity testing platform for the 21st century. Neurotoxicology 31:331–350. 10.1016/j.neuro.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ et al. (2010) In vitro screening of environmental chemicals for targeted testing prioritization: The toxcast project. Environ Health Perspect 118:485–492. 10.1289/ehp.0901392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajta M, Wójtowicz AK (2013) Impact of endocrine-disrupting chemicals on neural development and the onset of neurological disorders. Pharmacol. Reports 65:1632–1639 [DOI] [PubMed] [Google Scholar]
- Kasteel EEJ, Westerink RHS (2017) Comparison of the acute inhibitory effects of Tetrodotoxin (TTX) in rat and human neuronal networks for risk assessment purposes. Toxicol Lett 270:12–16. 10.1016/j.toxlet.2017.02.014 [DOI] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K et al. (2012) Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25:1287–1302. 10.1021/tx3000939 [DOI] [PubMed] [Google Scholar]
- Keefer EW, Gramowski A, Gross GW (2001a) NMDA receptor- dependent periodic oscillations in cultured spinal cord networks. J Neurophysiol 86:3030–3042. 10.1152/jn.2001.86.6.3030 [DOI] [PubMed] [Google Scholar]
- Keefer EW, Gramowski A, Stenger DA et al. (2001b) Characterization of acute neurotoxic effects of trimethylolpropane phosphate via neuronal network biosensors. Biosens Bioelectron 16:513–525. 10.1016/S0956-5663(01)00165-8 [DOI] [PubMed] [Google Scholar]
- Kolde R (2018) pheatmap: Pretty Heatmaps. R package version 1:10 [Google Scholar]
- Lantz SR, Mack CM, Wallace K et al. (2014) Glufosinate binds V-methyl-D-aspartate receptors and increases neuronal network activity in vitro. Neurotoxicology 45:38–47. 10.1016/j.neuro.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Li MH (2008a) Effects of nonionic and ionic surfactants on survival, oxidative stress, and cholinesterase activity of planarian. Che- mosphere 70:1796–1803 [DOI] [PubMed] [Google Scholar]
- Li MH (2008b) Effects of nonylphenol on cholinesterase and car- boxylesterase activities in male guppies (Poecilia reticulata). Ecotoxicol. Environ Saf 71:781–786 [DOI] [PubMed] [Google Scholar]
- Li J, Settivari R, LeBaron MJ, Marty MS (2019) An industry perspective: a streamlined screening strategy using alternative models for chemical assessment of developmental neurotoxicity. Neurotoxicology 73:17–30. 10.1016/j.neuro.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Mack CM, Lin BJ, Turner JD et al. (2014) Burst and principal components analyses of MEA data for 16 chemicals describe at least three effects classes. Neurotoxicology 40:75–85. 10.1016/j.neuro.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Maeda E, Robinson H, Kawana A (1995) The mechanisms of generation and propagation of synchronized bursting in developing networks of cortical neurons. J Neurosci 15:6834–6845. 10.1523/JNEUROSCI.15-10-06834.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell ER, McClain MA, Ross J et al. (2012) Evaluation of multiwell microelectrode arrays for neurotoxicity screening using a chemical training set. Neurotoxicology 33:1048–1057. 10.1016/j.neuro.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DA, Carter JM, Johnstone AFM, Shafer TJ (2008) Pyrethroid modulation of spontaneous neuronal excitability and neurotransmission in hippocampal neurons in culture. Neurotoxicology 29:213–225. 10.1016/j.neuro.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Mohana Krishnan B, Prakhya BM (2016) In vitro evaluation of pyrethroid-mediated changes on neuronal burst parameters using microelectrode arrays. Neurotoxicology 57:270–281. 10.1016/j.neuro.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Muramoto K, Ichikawa M, Kawahara M et al. (1993) Frequency of synchronous oscillations of neuronal activity increases during development and is correlated to the number of synapses in cultured cortical neuron networks. Neurosci Lett 163:163–165. 10.1016/0304-3940(93)90372-R [DOI] [PubMed] [Google Scholar]
- Narahashi T (2002) Nerve membrane ion channels as the target site of insecticides. Mini Rev Med Chem 2:419–432 [DOI] [PubMed] [Google Scholar]
- Nicolas J, Hendriksen PJM, van Kleef RGDM et al. (2014) Detection of marine neurotoxins in food safety testing using a multielectrode array. Mol Nutr Food Res 58:2369–2378. 10.1002/mnfr.201400479 [DOI] [PubMed] [Google Scholar]
- Novellino A, Scelfo B, Palosaari T et al. (2011) Development of microelectrode array based tests for neurotoxicity: assessment of interlaboratory reproducibility with neuroactive chemicals. Front Neuroeng 4:1–14. 10.1016/0014-4827(72)90159-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancrazio JJ, Keefer EW, Ma W et al. (2001) Neurophysiologic effects of chemical agent hydrolysis products on cortical neurons in vitro. Neurotoxicology 22:393–400. 10.1016/S0161-813X(01)00028-6 [DOI] [PubMed] [Google Scholar]
- Pancrazio JJ, Gopal K, Keefer EW, Gross GW (2014) Botulinum toxin suppression of CNS network activity in vitro. J Toxicol 10.1155/2014/732913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine J (2006) A history of MEA development In: Taketani M, Baudry M (eds) Advances in network electrophysiology. Springer, pp 3–23 [Google Scholar]
- R Core Team (2018) R: a language and environment for statistical computing. Austria, Vienna [Google Scholar]
- Richard AM, Judson RS, Houck KA et al. (2016) ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29:1225–1251. 10.1021/acs.chemrestox.6b00135 [DOI] [PubMed] [Google Scholar]
- Sachana M, Rolaki A, Bal-Price A (2018) Development of the adverse outcome pathway (AOP): chronic binding of antagonist to V-methyl-D-aspartate receptors (NMDARs) during brain development induces impairment of learning and memory abilities of children. Toxicol Appl Pharmacol 354:153–175. 10.1016/j.taap.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc . JMP Version 14.0. Cary, NC, 1989–2007 [Google Scholar]
- Scelfo B, Politi M,Reniero F et al. (2012) Application of multielectrode array (MEA) chips for the evaluation of mixtures neurotoxicity. Toxicology 299:172–183. 10.1016/j.tox.2012.05.020 [DOI] [PubMed] [Google Scholar]
- Shafer TJ, Rijal SO, Gross GW (2008) Complete inhibition of spontaneous activity in neuronal networks in vitro by deltamethrin and permethrin. Neurotoxicology 29:203–212. 10.1016/j.neuro.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Strickland JD, Martin MT, Richard AM et al. (2018) Screening the ToxCast phase II libraries for alterations in network function using cortical neurons grown on multi-well microelectrode array (mwMEA) plates. Arch Toxicol 92:487–500. 10.1007/s00204-017-2035-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D-J, Kim J-G, Won KJ et al. (2012) Blockade of K and Ca2 channels by azole antifungal agents in neonatal rat ventricular myocytes. Biol Pharm Bull 35:1469–1475 U.S. EPA (2015) ToxCast and Tox21 Summary Files from invitrodb_v2 [DOI] [PubMed] [Google Scholar]
- Valdivia P, Martin M, LeFew WR et al. (2014) Multi-well microelectrode array recordings detect neuroactivity of ToxCast compounds. Neurotoxicology 44:204–217. 10.1016/j.neuro.2014.06.012 [DOI] [PubMed] [Google Scholar]
- Vassallo A, Chiappalone M, De Camargos LR et al. (2017) A multi-laboratory evaluation of microelectrode array-based measurements of neural network activity for acute neurotoxicity testing. Neurotoxicology 60:280–292. 10.1016/j.neuro.2016.03.019 [DOI] [PubMed] [Google Scholar]
- Vidal-Liñán L, Bellas J, Salgueiro-Gonzalez N, Muniategui S, Beiras R (2015) Bioaccumulation of 4-nonylphenol and effects on biomarkers, acetylcholinesterase, glutathione-S-transferase and glutathione peroxidase, in Mytilus galloprovincialis mussel gilla. Environ Pollut. 200:133–139. 10.1016/j.envpol.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Wallace K, Strickland JD, Valdivia P et al. (2015) A multiplexed assay for determination of neurotoxicant effects on spontaneous network activity and viability from microelectrode arrays. Neurotoxicology 49:79–85. [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V (2017) R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84) [Google Scholar]
- Wheeler BC, Nam Y (2011) In vitro microelectrode array technology and neural recordings. Crit Rev Biomed Eng 39:45–61. 10.1615/CritRevBiomedEng.v39.i1.40 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York [Google Scholar]
- Yang C, Tarkhov A, Marusczyk J et al. (2015) New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model 55:510–528. 10.1021/ci500667v [DOI] [PubMed] [Google Scholar]
- Ylä-Outinen L, Heikkila J, Skottman H et al. (2010) Human cell-based micro electrode array platform for studying neurotoxicity. Front Neuroeng 3:1–9. 10.3389/fneng.2010.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.