Abstract
Background
Experimental murine models and human challenge studies of Salmonella Typhi infection have suggested that the gut microbiome plays an important protective role against the development of typhoid fever. Anaerobic bacterial communities have been hypothesized to mediate colonization resistance against Salmonella species by producing short-chain fatty acids, yet the composition and function of the intestinal microbiota in human patients with typhoid fever remain ill defined.
Methods
We prospectively collected fecal samples from 60 febrile patients admitted to Chittagong Medical College Hospital, Bangladesh, with typhoid fever or nontyphoidal febrile illness and from 36 healthy age-matched controls. The collected fecal samples were subjected to 16s rRNA sequencing followed by targeted metabolomics analysis.
Results
Patients with typhoid fever displayed compositional and functional disruption of the gut microbiota compared with patients with nontyphoidal febrile illness and healthy controls. Specifically, typhoid fever patients had lower microbiota richness and alpha diversity and a higher prevalence of potentially pathogenic bacterial taxa. In addition, a lower abundance of short-chain fatty acid–producing taxa was seen in typhoid fever patients. The differences between typhoid fever and nontyphoidal febrile illness could not be explained by a loss of colonization resistance after antibiotic treatment, as antibiotic exposure in both groups was similar.
Conclusions
his first report on the composition and function of the gut microbiota in patients with typhoid fever suggests that the restoration of these intestinal commensal microorganisms could be targeted using adjunctive, preventive, or therapeutic strategies.
Keywords: colonization resistance, microbiota, short-chain fatty acids, typhoid fever
Typhoid fever (TF) is a systemic infection caused by the gram-negative bacteria Salmonella enterica serovar Typhi (S. Typhi). This debilitating condition, characterized by prolonged fever and a wide range of other nonspecific clinical signs, is common in areas of Asia and Africa that lack clean water and sanitation [1, 2]. Recent increases in antimicrobial resistance among several S. Typhi haplotypes have complicated treatment options, and mortality rates are estimated to range between 128 000 to 161 000 deaths per year [3, 4]. Causes for the wide variety of clinical presentations as well as the severity of infection are not understood but are hypothesized to be driven by altered interactions between Salmonella serovars and the host response [5, 6].
Research interest into the microorganisms that reside in our gastrointestinal tract—the gut microbiome—has surged in the last 2 decades. With the advent and improvement of technical and computational modalities to chart the microbiome, researchers have uncovered that these communities have important functions in the protection against enteric and systemic infectious diseases [7–9]. In homeostasis, bacterial communities residing in the gut are able to impair the growth of opportunistic and enteric pathogens, a process termed colonization resistance [7]. As an example, bacterial species and their products can provide direct colonization resistance by nutrient depletion and antimicrobial peptide production within the intestine, as well as indirectly by enhancement of the intestinal epithelial barrier and the upregulation of mucosal T helper 17 (Th17) cells, group 3 innate lymphoid cells, and regulatory T cells [10].
Murine typhoid models with S. typhimurium underscore the protective role of gut commensals, as it has been shown that the presence of anaerobic microbiota-derived short-chain fatty acids, such as butyrate and propionate, reduces the growth of the pathogen by reducing both the oxygen availability and pH levels of the gut environment [11–13]. In addition, S. typhimurium has been shown to be capable of exploiting microbiome-derived nutrients and oligosaccharides, which allows for a growth advantage in the intestine over other potential pathogens, underscoring how these microbes are involved in an ongoing competitive colonization of the intestinal tract [14, 15]. The notion that the gut microbiota could be of importance in protection against typhoid fever has recently been observed in humans. A typhoid vaccine trial by Zhang and colleagues showed that differences in the composition and function of the gut microbiota in healthy adult volunteers were associated with altered host susceptibility to typhoid after challenge with wild-type S. Typhi [16].
Despite this evidence of a close relationship between S. Typhi and the microbiome, there remains a significant knowledge gap concerning the association between typhoid fever infection and human gut microbiome composition. This study aimed to investigate how the gut bacterial composition and function of patients with typhoid fever might differ from patients with other febrile illnesses or healthy controls and how these differences might be correlated with short-term outcome in a prospective cohort of patients admitted to Chittagong Medical College Hospital (CMCH), a 1000-bed teaching hospital in Chittagong Division of Bangladesh.
METHODS
Study Design and Participants
Patients admitted to CMCH between January 15, 2012, and July 5, 2012, were considered for enrollment, as described in previous studies conducted in this cohort [17–19]. Consecutive patients (≥16 years of age) who were admitted with an acute febrile illness (documented fever ≥38°C axillary, up to 48 hours after admission, history of fever <2 weeks) were recruited after giving informed written consent. Each study day, all the patients admitted to the wards were reviewed, and those with a history of fever of the appropriate duration had their temperature measured on at least 1 occasion. Eligible patients were then asked if they consented to participate in the study. Demographic and clinical information was recorded on a case record form at the time of admission and during the course of hospitalization.
A final diagnosis was made by the study team based on clinical presentation, basic laboratory results, and microbiology results, as described earlier [17–19]. Fecal samples were collected in plastic containers and were stored at −80°C within 24 hours [20, 21]. A group of local Bangladeshi healthy volunteers were recruited to donate stools, which were stored in a similar manner to the patient population. The study protocol was approved by the National Research Ethics Committee (NREC) of Bangladesh (BMRC/NREC/2010–2013/1543) and the Oxford Tropical Research Ethics Committee (OXTREC reference 25-11).
Microbiota Analysis
Fecal DNA was extracted and purified using a combination of bead-beating and the Maxwell 16 Tissue LEV Total RNA Purification Kit (Promega, Maddison, WI, USA), with Stool Transport And Recovery (STAR) buffer (Roche, Basel Switzerland) [22]. Polymerase chain reaction (PCR) products were purified using Ampure XP beads, and purified products were equimolar-pooled. The libraries were sequenced using an Illumina MiSeq platform (GATCBiotech, Konstanz, Germany) using V3 chemistry with 2×251 cycles [23]. Forward and reverse reads were truncated to 240 and 210 bases, respectively, and merged using USEARCH [24]. Merged reads that did not pass the Illumina chastity filter, had an expected error rate >2, or had <380 bases were filtered. Amplified sequence variants (ASVs) were inferred for each sample individually with a minimum abundance of 4 reads. Unfiltered reads were than mapped against the collective ASV set to determine the abundances. Taxonomy was assigned using the RDP classifier [25] and SILVA30 16S ribosomal database V132 [26]. Given prior observations demonstrating the potential benefits of butyrate as an immunomodulatory compound [11, 12], we assessed this feature in our cohort by measuring the abundance of bacterial taxa that are known to be drivers of butyrate production, in accordance with a study that analyzed butyrate-producing pathways from 15 publicly available data sets [27].
Targeted Measurement of Short-Chain Fatty Acids
Sample preparation of fecal extracts and nuclear magnetic resonance (NMR) spectroscopy for quantification of short-chain fatty acids (SCFAs) was performed as described in Kim et al., with some modifications [28]. Briefly, aqueous extracts of feces were prepared by mixing 50–100 mg of feces and 0.3 mL of deionized water, followed by mechanical homogenization in a Bullet Blender 24 (Next Avance Inc, Troy, NY, USA). The fecal slurry was centrifuged twice at 18 213×g for 10 minutes at 4°C, and 0.225 mL of the supernatant was mixed with 0.025 mL of 1.5-M potassium phosphate buffer (pH 7.4) containing 2 mM of sodium azide and 4 mM of sodium trimethylsilyl-propionate-d4 (TSP-d4) in D2O. One proton NMR spectrum was acquired for each sample in a 14.1 T Avance II NMR (Bruker Biospin Ltd, Billerica, MA, USA). Quantification of SCFAs from the NMR data was performed in ChenomX (Chenomx NMR suite 8.4) using the known concentration of TSP-d4.
Statistics
Statistical analysis was performed in the R statistical framework (version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria). To assess alpha diversity, we calculated the Shannon Diversity Index and Observed Taxa Richness index with the R phyloseq package [29]. Data were not normally distributed and are therefore presented as median (interquartile range [IQR]), while data were analyzed using a Wilcoxon matched-pairs signed-rank test for paired data or a Mann-Whitney test for unpaired data. Beta diversity metrics were calculated using weighted and unweighted UniFrac on a rarefied data set, after which principal coordinates analysis was performed to highlight the separation of microbiota composition based on antibiotic exposure and sampling time point. Differences in microbiota composition among groups and time points were tested for using permutational multivariate analysis of variance (PERMANOVA) on beta diversity matrices using the vegan R package. False discovery rate was adjusted for with Benjamini-Hochberg. To identify taxa that may be driving the significant differences detected between groups, differential abundance analysis was determined using DESeq2. Finally, potential predictors of outcome were assessed using linear regression models on log10-transformed relative abundances. Linear terms were confirmed by Wald tests in the rms R package (with P < .05 indicating nonlinearity).
RESULTS
Demographic and Clinical Data
Fecal samples of a total of 60 patients with a febrile illness and 36 healthy age-matched controls were collected. Fourteen of the 60 patients were diagnosed with typhoid fever, either by positive blood cultures for S. Typhi (n = 8; 57.1%) or positive S. Typhi PCR in blood, urine, and/or feces (n = 6; 42.8%). A total of 46 patients in the cohort were classified as having a nontyphoidal febrile illness (N-TF), of whom the majority were diagnosed with either a lower respiratory tract infection (36%) or urinary tract infection (27%). The baseline characteristics of all study participants are provided in Table 1. Patients with typhoid fever were slightly younger compared with nontyphoidal febrile illness patients and healthy controls (median age, 27 vs 35.5 vs 35.0 years, respectively; P = .044), while no differences in medical history were observed among the 3 groups. A comparison of clinical characteristics at admission showed that there were no apparent differences in clinical severity and subsequent length of hospital admission stay or mortality between TF and N-TF patients. TF patients had a longer duration of fever before admission, with more diarrhea, lower white blood cell counts, and higher aspartate transaminase (AST) and alanine transaminase (ALT) levels. There was more fluoroquinolone exposure in patients with TF before fecal sampling, but no differences in both exposure or duration of treatment with other antibiotic classes. An overview of infectious diagnoses, microbiological data, and antibiotic exposure before fecal sampling is depicted in Supplementary Table 1.
Table 1.
Baseline Demographics and Disease Characteristics at Admission
| Characteristic | Healthy Controls (n = 36) | Fever, No Typhoid (n = 46) | Typhoid (n = 14) | P Value |
|---|---|---|---|---|
| Age, median [IQR], y | 35.50 [27.00–40.00] | 35.00 [23.00–54.50] | 27.00 [22.00–31.00] | .044 |
| Male sex, No. (%) | 18 (50.0) | 25 (54.3) | 10 (71.4) | .387 |
| Past history of typhoid fever, No. (%) | 3 (8.3) | 8 (17.4) | 2 (14.1) | .513 |
| Diabetes, No. (%) | 2 (5.6) | 6 (13.0) | 0 (0.0) | .189 |
| Signs and symptoms | ||||
| Headache, No. (%) | 25 (54.3) | 8 (57.1) | 1.000 | |
| Chest pain, No. (%) | 10 (21.7) | 0 (0.0) | .088 | |
| Cough, No. (%) | 25 (54.3) | 1 (7.1) | .006 | |
| Anorexia, No. (%) | 21 (45.7) | 8 (57.1) | .818 | |
| Abdominal pain, No. (%) | 14 (30.4) | 7 (50.0) | .306 | |
| Diarrhea, No. (%) | 3 (6.5) | 6 (42.9) | .004 | |
| Vomiting, No. (%) | 17 (37.0) | 8 (57.1) | .302 | |
| Constipation, No. (%) | 6 (13.0) | 2 (14.3) | 1.000 | |
| Dysuria, No. (%) | 6 (13.0) | 1 (7.1) | .899 | |
| Days of fever before admission, median [IQR] | 4.00 [2.25–6.75] | 6.00 [5.00–9.50] | .048 | |
| Vital signs | ||||
| Temperature, median [IQR] | 38.90 [38.32–39.18] | 38.55 [38.31–39.27] | .888 | |
| Pulse, median [IQR] | 106 [92–116] | 106 [96–110] | .909 | |
| Systolic blood pressure, median [IQR] | 109 [100–120] | 113 [101–120] | .478 | |
| Respiratory rate, median [IQR] | 24 [20–36] | 24 [21–32] | .713 | |
| Glasgow Coma Scale, median (IQR) | 15 [15–15] | 15 [15–15] | .258 | |
| White blood cell count, median [IQR] | 12 800 [8200–16 000] | 5850 [4300–9825] | .002 | |
| Hemoglobin, median [IQR] | 11.60 [10.75–13.25] | 12.60 [9.90–13.33] | .431 | |
| Platelets, median [IQR] | 200 000 [170 000–250 000] | 180 000 [162 500–230 000] | .481 | |
| Creatinine, median [IQR], mg/dL | 1.00 [0.80–1.20] | 0.90 [0.90–1.00] | .263 | |
| ALT, median [IQR] | 27.50 [19.75–46.50] | 57.5 [30.00–126.25] | .003 | |
| AST, median [IQR] | 65.00 [48.25–84.75] | 700.00 [245.25–2220.00] | <.001 | |
| Outcome | ||||
| Length of hospital stay, median [IQR], d | 4.00 [3.00–5.00] | 5.50 [4.00–8.50] | .059 | |
| Mortality, No. (%) | 1 (2.2) | 0 (0.0) | 1.000 |
Abbreviations: ALT, alanine transaminase; AST, aspartate transaminase; IQR, interquartile range.
Typhoid Fever Is Characterized by Significantly Altered Patterns of Microbiota Disruption
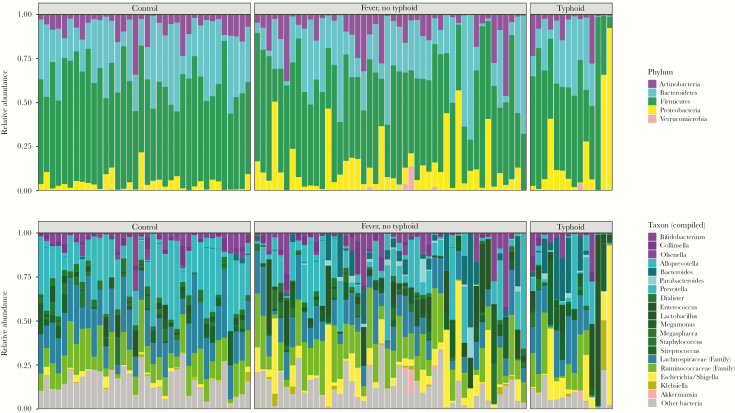
Sequencing of fecal samples taken at the time of hospital admission yielded a total of 3 552 949 high-quality 16S rRNA gene sequences (average 37 010 per sample). We observed significantly altered states of gut microbiota composition between TF- and N-TF patients compared with controls. On the microbiota phylum level, TF and N-TF patients showed a significant reduction in Actinobacteria (P = .04), while Proteobacteria were significantly enriched (P = .0001) (Figure 1A). In addition, major differences were observed between both patient groups and controls on a microbiota genus level. Importantly, potentially invasive bacterial genera were significantly more abundant in patients, including Enterococcus species (P < .0001) and Escherichia/Shigella species (P < .0001) (Figure 1).
Figure 1.
Fecal microbiota composition (phylum and genus levels) among patients with nontyphoidal febrile illness (n = 46), typhoid fever (n = 14), and controls (n = 36). Each bar represents 1 sample; phyla (A) and compiled taxa (B) are indicated with colors and expressed as percentage of the total DNA reads. Ruminococcaceae and Lachnospiraceae families and genera, which made up ≥5% of the total microbiota in at least 1 sample, are included in Figure B; other genera are pooled within the category “Other bacteria.”
Supplementary Figure 1 shows an overview of microbiota composition linked to a timeline of hospital stay and antibiotic exposure for each TF and N-TF patient. An unbiased comparison of microbial taxa in gut communities showed that typhoid fever is characterized by a distinct composition of the microbiome when compared with patients with a nontyphoidal febrile illness. On the genus level, TF subjects were shown to be significantly more colonized by Streptococcus, with a decrease in “beneficial” obligate anaerobic taxa, such as Ruminococcus species, Faecalibacterium species, and other Lachnospiraceae when compared with N-TF patients (Supplementary Figure 2).
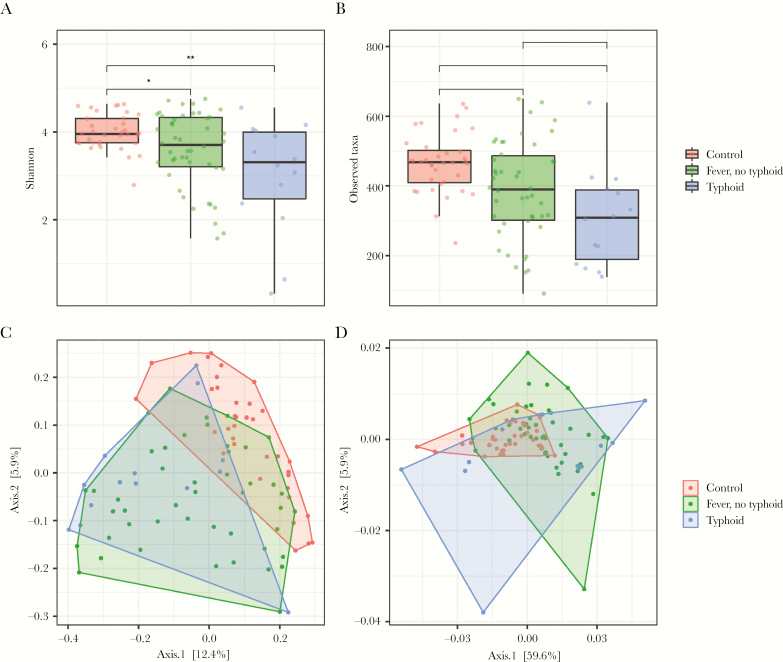
In tandem with the differences in community composition, strong alterations were observed in microbiota alpha diversity and richness between patients with typhoid fever and nontyphoidal febrile illness (N-TF vs control vs TF: median Shannon, 3.71 vs 3.96 vs 3.31; Wilcoxon, P = .048 and P = .004, respectively; median Observed Taxa, 390 vs 468 vs 308; Wilcoxon, P = .008 and P < .001, respectively) (Figure 2A and B). In addition, community richness was significantly lower in TF patients compared with N-TF patients (Wilcoxon, P = .040). Similar patterns were observed in beta diversity metrics of the microbiota (Figure 2C and D), as samples that were collected from TF and N-TF patients differed from controls in both weighted and unweighted UniFrac analyses (PERMANOVA, P < .0001 and P < .0001) (Supplementary Table 2). Notably, beta diversity between TF and N-TF patients was comparable in the weighted UniFrac model, but was significantly altered with an unweighted UniFrac approach (PERMANOVA, P = .014) (Supplementary Table 2), which suggests that the taxa that drive the separation between TF and N-TF patients are of relatively low abundance.
Figure 2.
Overview of differences in alpha and beta diversity among patients with nontyphoidal febrile illness (n = 46), typhoid fever (n = 14), and controls (n = 36). The Shannon index and the Observed Taxa (OT) index were used to calculate the diversity community (A) and richness (B) within each individual microbiota sample. Data are presented as a dot plot with a line at the median. Principal coordinates analysis of unweighted (C) and weighted (D) UniFrac distances of samples of patients with typhoid fever, nontyphoidal febrile illness, or healthy controls. *P < .05; **P < .01; ***P < .001.
Lower Abundance of Short-Chain Fatty Acids Producing Taxa in Typhoid Fever, With Diminished Fecal Butyrate, Acetate, and Propionate Levels
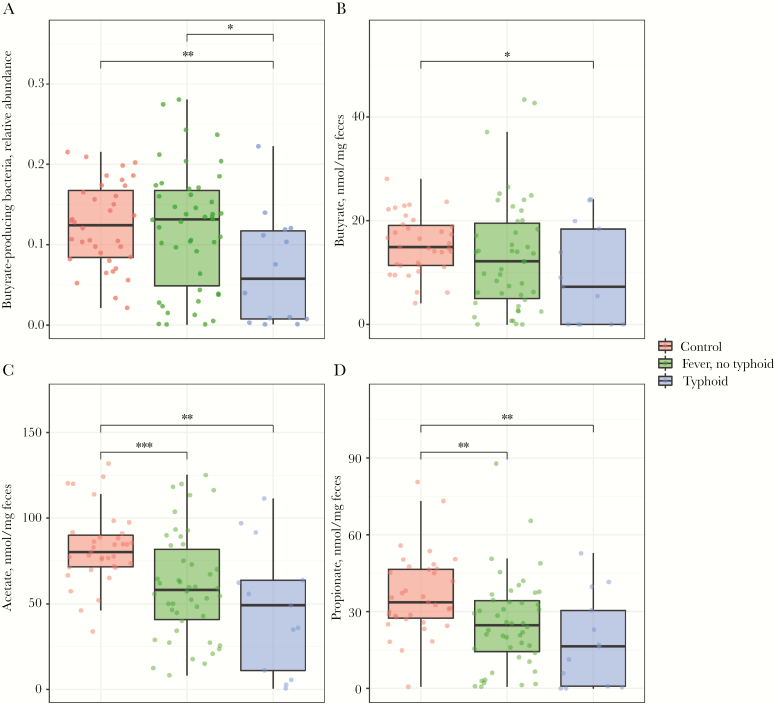
TF patients had a profoundly lowered abundance of butyrate-producing genera, compared with both N-TF patients and healthy controls (N-TF vs TF vs control; median % of total sequences, 12.0 vs 6.9 vs 12.4; Wilcoxon, P = .020 and P = .009, respectively) (Figure 3A). We subsequently performed targeted NMR analysis to measure volatile short-chain fatty acids on a different aliquot of the same fecal samples and found that the measured fecal concentrations of butyrate, acetate, and propionate were significantly diminished in TF patients compared with controls (Figure 3B–D). Of note, the fecal concentration of butyrate strongly correlated with the abundance of butyrate-producing bacteria (R = .76; P < .0001) (Supplementary Figure 3). Finally, linear regression analysis indicated that low diversity and low abundance of butyrate-producing bacteria were not associated with length of hospital stay in both TF and N-TF patients (Supplementary Figure 4A and B).
Figure 3.
Lower amount of anaerobic bacteria with equally low abundance of fecal short-chain fatty acids in fecal samples of typhoid fever patients. Relative abundance of intestinal butyrate-producing bacteria (A), absolute butyrate concentration (B), absolute acetate concentration (C), and absolute propionate concentration (D) in fecal samples of patients with nontyphoidal febrile illness (n = 46), typhoid fever (n = 14), or healthy controls (n = 36). Data are presented as a dot plot with a line at the median. *P < .05; **P < .01; ***P < .001. Abbreviation: Ns, nonsignificant.
DISCUSSION
In this prospective observational study conducted at Chittagong Medical College Hospital, Bangladesh, we found that patients with typhoid fever had significantly altered patterns of compositional and functional disruption of the gut microbiota compared with patients with nontyphoidal febrile illness and healthy local controls. Specifically, TF patients had lower microbiota richness and alpha diversity, lower amounts of obligately anaerobic bacteria and fecal short-chain fatty acids, and a higher prevalence of potentially pathogenic bacterial taxa in the gut when compared with nontyphoidal febrile patients. The marked differences we observed between TF and N-TF patients cannot be fully explained by an altered exposure to antibiotics. Typhoid fever patients were exposed to more fluoroquinolones, which are known to have little effect on the butyrate-producing compartment of the microbiota [30, 31], whereas exposure to beta-lactam antibiotics and metronidazole was comparable between groups. In addition, the total amount of antibiotic exposure between groups before hospital admission did not differ.
We identify 2 potential reasons that could explain the typical characteristics of the typhoid fever microbiome as observed in our cohort of patients. First, S. Typhi infection itself will most likely alter the intestinal environment, leading to an aerobic expansion and a loss of obligate anaerobic bacteria. This has been observed in preclinical models with S. typhimurium by the Bäumler lab [11, 12]. Second, patients with TF could have a disrupted microbiome before the development of TF, potentially increasing their risk of initial infection due to reduced colonization resistance. For example, the ultra-low amounts of short-chain fatty acids observed in this cohort with TF—and their potential relationship with acidification-mediated resistance in the gut environment [13]—could benefit expansion of S. Typhi bacteria in the intestine and increase the a priori risk of developing TF.
An important limitation of this cohort study is the small sample size and limited follow-up. Given these limitations, no significant association was detected between either low diversity or low abundance of butyrate-producing bacteria and length of hospital admission. However, there is evidence that dysbiosis could have a negative impact on long-term outcomes, as it has been shown that some of the bacterial species constituting the intestinal microbiota contribute to protection against both enteric and systemic infections. For example, systemic exposure of microbiota-derived ligands, such as flagellin and lipopolysaccharide, increases the activity of alveolar macrophages and bone marrow–derived neutrophils, which enhances the killing of gram-positive and gram-negative pathogens in the lung [32–36]. In addition, studies in mice have implicated that microbiota-derived butyrate restores IL-10 levels in the lung and the intestine by inhibiting histone deacetylase 3 (HDAC3), which contributes to microbiota-induced immunomodulation [37, 38]. We observed a similar importance of these microbial communities in a cohort study of patients receiving allogeneic hematopoietic stem cell transplant, as patients who harbored a high abundance of butyrate-producing bacteria were less likely to develop viral lower respiratory tract infections compared with patients who lost these bacterial species [30].
The consequences of typhoid fever infection on altered immunomodulation have also been described previously; transcriptional profiling of peripheral blood to investigate the host response of 29 individuals who contracted the disease in Vietnam showed that the infection induced a distinct immunosuppressive signature in peripheral blood that persisted for 1–9 months after acute infection [39]. Patients who retained this immunosuppressive signature were thought to be more susceptible to reinfection or relapse [40, 41], and one could hypothesize that TF-induced dysbiosis could potentially play a role in this phenomenon.
Given the aforementioned evidence of the role of the microbiota in systemic priming of immune responses and the extensive amount of disruption of the microbiota observed in our cohort, it is vital to investigate new treatment strategies that preserve these microorganisms during infection. For example, overuse of antibiotics and their associated transient alteration of the microbiota have implications for outcome, which was supported by a recent systematic review that found a statistically significant association between antibiotic exposure and subsequent risk of community-acquired infections [42]. In addition, our study highlights a new opportunity for the development of next-generation pre- or probiotics to enhance the abundance of obligate anaerobic bacteria as a potential adjuvant therapy during or following the treatment of TF and other infectious diseases.
Other limitations of this study include the cross-sectional setup, which limited our ability to provide clear insights on both the causes and long-term consequences of TF-related dysbiosis. In addition, the single-center setup of this cohort might not be fully comparable to other study settings, where exposure to microbial pathogens, antimicrobial treatment, and sanitation measures differ. Therefore, both large longitudinal cohort studies and additional human-controlled S. Typhi challenge models are warranted to further uncover the intricate crosstalk between S. Typhi, the intestinal microbiota, and its host.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank all patients and their families who participated in the study, and we are grateful to the valuable contributions of past and present group members for their work on contributing to the understanding of the pathogenesis of typhoid fever. We also thank Mark Davids for his aid in preprocessing the 16S rRNA sequences, Jorn Hartman for his help with the workup of the intestinal microbiota samples, and Marieke Heijink for her work on the setup of the targeted SCFA measurements.
Financial support. This work was supported by the Netherlands Organization for Scientific Research (VIDI grant number 91716475 to W.J.W.), the Wellcome Trust of Great Britain (106158/Z/14/Z) and the European Society of Paediatric Infectious Diseases. Rapeephan Maude was a Wellcome Trust Masters Fellow in Public Health and Tropical Medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Meiring JE, Gibani M; TyVAC Consortium Meeting Group The Typhoid Vaccine Acceleration Consortium (TyVAC): vaccine effectiveness study designs: accelerating the introduction of typhoid conjugate vaccines and reducing the global burden of enteric fever. Report from a meeting held on 26–27 October 2016, Oxford, UK. Vaccine 2017; 35:5081–88. [DOI] [PubMed] [Google Scholar]
- 2. Wain J, Hendriksen RS, Mikoleit ML, et al. . Typhoid fever. Lancet 2015; 385:1136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Britto CD, Wong VK, Dougan G, Pollard AJ. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl Trop Dis 2018; 12:e0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen J. ‘Frightening’ typhoid fever outbreak spreads in Pakistan. Science 2018; 361:214. [DOI] [PubMed] [Google Scholar]
- 5. Tsolis RM, Young GM, Solnick JV, Bäumler AJ. From bench to bedside: stealth of enteroinvasive pathogens. Nat Rev Microbiol 2008; 6:883–92. [DOI] [PubMed] [Google Scholar]
- 6. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog 2012; 8:e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 2016; 352:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol 2017; 2:135–43. [DOI] [PubMed] [Google Scholar]
- 9. Harris VC, Haak BW, Boele van Hensbroek M, Wiersinga WJ. The intestinal microbiome in infectious diseases: the clinical relevance of a rapidly emerging field. Open Forum Infect Dis 2017; 4:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 2017; 279:90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bronner DN, Faber F, Olsan EE, et al. . Genetic ablation of butyrate utilization attenuates gastrointestinal salmonella disease. Cell Host Microbe 2018; 23: 266–273.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rivera-Chávez F, Zhang LF, Faber F, et al. . Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe 2016; 19:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jacobson A, Lam L, Rajendram M, et al. . A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 2018; 24:296–307.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thiennimitr P, Winter SE, Bäumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol 2012; 15:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schieber AM, Lee YM, Chang MW, et al. . Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 2015; 350:558–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y, Brady A, Jones C, et al. . Compositional and functional differences in the human gut microbiome correlate with clinical outcome following infection with wild-type Salmonella enterica serovar Typhi. MBio 2018; 9:e00686-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Jong HK, Parry CM, van der Vaart TW, et al. . Activation of coagulation and endothelium with concurrent impairment of anticoagulant mechanisms in patients with typhoid fever. J Infect 2018; 77:60–7. [DOI] [PubMed] [Google Scholar]
- 18. Maude RR, Ghose A, Samad R, et al. . A prospective study of the importance of enteric fever as a cause of non-malarial febrile illness in patients admitted to Chittagong Medical College Hospital, Bangladesh. BMC Infect Dis 2016; 16:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Jong HK, Garcia-Laorden MI, Hoogendijk AJ, et al. . Expression of intra- and extracellular granzymes in patients with typhoid fever. PLoS Negl Trop Dis 2017; 11:e0005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vandeputte D, Tito RY, Vanleeuwen R, et al. . Practical considerations for large-scale gut microbiome studies. FEMS Microbiol Rev 2017; 41:154–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carruthers LV, Moses A, Adriko M, et al. . The impact of storage conditions on human stool 16S rRNA microbiome composition and diversity. PeerJ 2019; 7:e8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Costea PI, Zeller G, Sunagawa S, et al. . Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 2017; 35:1069–76. [DOI] [PubMed] [Google Scholar]
- 23. Kozich JJ, Westcott SL, Baxter NT, et al. . Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 25. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quast C, Pruesse E, Yilmaz P, et al. . The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems 2017; 2:e00130–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim HK, Kostidis S, Choi YH. . NMR analysis of fecal samples. In: Giera M, ed. Clinical Metabolomics: Methods and Protocols. Methods in Molecular Biology. Vol. 1730 New York, NY: Springer Science + Business Media, LLC; 2018; 317–28. [DOI] [PubMed] [Google Scholar]
- 29. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haak BW, Littmann ER, Chaubard JL, et al. . Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 2018; 131:2978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhalodi AA, van Engelen TSR, Virk HS, Wiersinga WJ. Impact of antimicrobial therapy on the gut microbiome. J Antimicrob Chemother 2019; 74:i6–i15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gray J, Oehrle K, Worthen G, et al. . Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med 2017; 9:aaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clarke TB, Davis KM, Lysenko ES, et al. . Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuijt TJ, Lankelma JM, Scicluna BP, et al. . The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016; 65:575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lankelma JM, Birnie E, Weehuizen TAF, et al. . The gut microbiota as a modulator of innate immunity during melioidosis. PLoS Negl Trop Dis 2017; 11:e0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abt MC, Osborne LC, Monticelli LA, et al. . Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014; 111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chakraborty K, Raundhal M, Chen BB, et al. . The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat Commun 2017; 8:13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson LJ, Dunstan SJ, Dolecek C, et al. . Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc Natl Acad Sci U S A 2009; 106:22433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Croswell A, Amir E, Teggatz P, et al. . Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun 2009; 77:2741–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gopinath S, Carden S, Monack D. Shedding light on Salmonella carriers. Trends Microbiol 2012; 20:320–7. [DOI] [PubMed] [Google Scholar]
- 42. Malik U, Armstrong D, Ashworth M, et al. . Association between prior antibiotic therapy and subsequent risk of community-acquired infections: a systematic review. J Antimicrob Chemother 2018; 73:287–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.