Abstract
NIPP1 is a ubiquitously expressed nuclear protein that regulates functions of protein Ser/Thr phosphatase-1 in cell proliferation and lineage specification. The role of NIPP1 in tissue homeostasis is not fully understood. Here we show that the selective deletion of NIPP1 in mouse epidermis resulted in epidermal hyperproliferation, a reduced adherence of basal keratinocytes and a gradual decrease in the stemness of hair follicle stem cells, culminating in hair loss. This complex phenotype was associated with chronic sterile skin inflammation and could be partially rescued by dexamethasone treatment. NIPP1-deficient keratinocytes massively expressed pro-inflammatory chemokines and immunomodulatory proteins in a cell-autonomous manner. Chemokines subsequently induced the recruitment and activation of immune cells, in particular conventional dendritic cells and Langerhans cells, accounting for the chronic inflammation phenotype. Our data identify NIPP1 as a key regulator of epidermal homeostasis and as a potential target for the treatment of inflammatory skin diseases.
Keywords: chemokines, cytokines, inflammation, hair-follicle stem cells, NIPP1, protein phosphatase-1
INTRODUCTION
NIPP1, for nuclear inhibitor of protein phosphatase 1 (PP1), is encoded by Ppp1r8 and is ubiquitously expressed in the nucleus of metazoan and plant cells (Ceulemans et al. 2002). The heterodimeric PP1:NIPP1 holoenzyme is inactive under steady-state but can be activated by the phosphorylation-induced allosteric removal of the C-terminal PP1-inhibitory domain of NIPP1 (Beullens et al. 2000). Activated PP1:NIPP1 selectively dephosphorylates phosphoproteins that are recruited via the N-terminal forkhead-associated (FHA) domain of NIPP1 and include such diverse proteins as protein methyltransferase EZH2, pre-mRNA splicing factors SF3B1/SAP155 and CDC5L, and protein kinase MELK (Boudrez et al. 2000; Boudrez et al. 2002; Vulsteke et al. 2004; Nuytten et al. 2008). NIPP1 has been identified as a key regulator of proliferation and lineage specification (Van Eynde et al. 2004; Boens et al. 2016; Ferreira et al. 2017). Indeed, Ppp1r8−/− embryos suffered from a reduced cell proliferation and died at the gastrulation stage (Van Eynde et al. 2004). Likewise, the postnatal deletion of NIPP1 in testis resulted in the progressive loss of germ cells (Ferreira et al. 2017). In contrast, the deletion of NIPP1 in liver epithelial cells caused a slow-onset hyperproliferation of biliary epithelial cells (Boens et al. 2016).
Skin epidermis forms a stratified epithelium, known as interfollicular epidermis (IFE), and is associated with different appendages, including hair follicles (HFs), sebaceous glands and sweat glands (Belokhvostova et al. 2018). The epidermis and its appendages have a high cellular turnover and are maintained by different stem-cell populations (Gonzales and Fuchs 2017; Belokhvostova et al. 2018). In the IFE, stem cells give rise to proliferating keratinocytes in the basal layer. When basal keratinocytes detach, they move upwards to the suprabasal layers where they withdraw from the cell cycle and differentiate into spinous and granular keratinocytes and finally into dead corneocytes (Hänel et al. 2013). In the bulge region of the HFs, hair follicle-stem cells (HFSCs) generate progenitor cells that differentiate into the various cell lineages of the hair shaft and inner root sheath (Gonzales and Fuchs 2017). The fate of HFSCs and progenitor cells is tightly controlled by their microenvironment or niche. Skin homeostasis depends on a complex interaction between keratinocytes and other skin-resident cells, including immune cells (Heath and Carbone 2013; Kumari and Pasparakis 2017; Kabashima et al. 2019). Keratinocytes themselves can also act as innate immune cells as they can release chemokines that recruit and activate immune cells (Fritz et al. 2017; Otsuka et al. 2017; Eyerich et al. 2018). The chronic release of chemokines by keratinocytes and immune cells can cause severe inflammatory skin diseases, such as atopic dermatitis and psoriasis (Li et al. 2018).
We have generated a mouse model for the inactivation of both Ppp1r8 alleles in keratinocytes. We report here that adult mice lacking NIPP1 in the epidermis develop a stress response that culminates in a massive release of cytokines, the recruitment of immune cells and chronic sterile skin inflammation.
RESULTS
Epidermal-barrier dysfunction and hair loss in Ppp1r8−/− mouse skin
We selectively inactivated Ppp1r8 in keratinocytes using CRE recombinase under control of the Keratin-14 promoter (see Methods section). Tg(Krt14-Cre)/Ppp1r8fl/+and Tg(Krt14-Cre)/Ppp1r8fl/− were used as controls (CTR) and skin-specific NIPP1 knockout (SKO) mice, respectively (Fig. S1a). Immunostaining, immunoblotting and qRT-PCR revealed efficient removal of NIPP1 from the IFE and HFs in SKO tail and back (dorsal) skin (Figs. 1a–d).
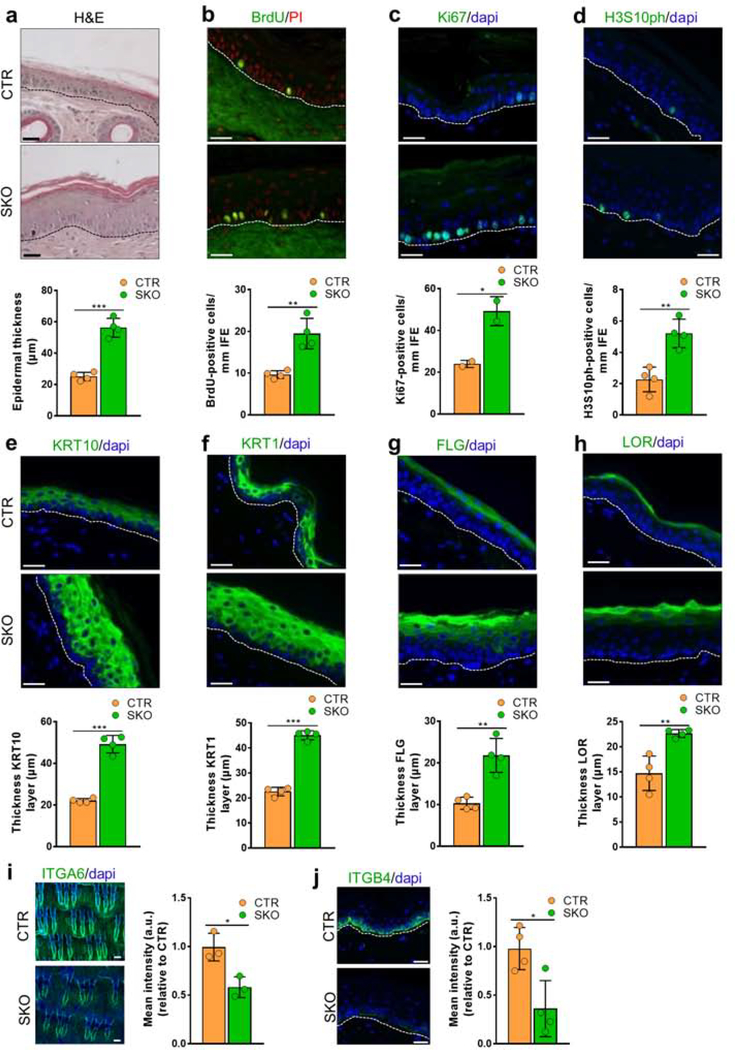
Figure 1. Macroscopic phenotype associated with the inactivation of Ppp1r8 in mouse skin.
(a) NIPP1 immunostains (green) in longitudinal sections of tail and back skin from CTRs and SKOs at 8 weeks (tail) and 4 weeks (back). Dapi (blue) was used as a nuclear counterstain. Left panels show stainings for the interfollicular epidermis, while the middle and right panels show stainings for hair follicles. Scale bars, 50 μm.
(b) The level of NIPP1 in extracts of tail epidermis from 4 CTRs and SKOs of 8 weeks was visualized by immunoblotting. GAPDH was used as a loading control.
(c) Quantification of the data in panel b (n = 4).
(d) qRT-PCR analysis of the Ppp1r8 transcript in tail and back epidermis from CTRs and SKOs of 8 weeks (n=4). Hprt was used as a housekeeping gene for normalization.
(e) Representative pictures of the ears and tail of CTRs and SKOs at 8 weeks.
(f) Representative pictures of back skin and tail of CTRs and SKOs at 8–12 months.
(g) Ulcerated skin lesions in SKOs of 8 months.
(h) Quantification of transepidermal water loss (TEWL) from the back of CTR and SKO mice of 8 weeks (n=6).
The bar graphs are represented as means ± SD. **, p<0.01; ***, p<0.001 (unpaired student’s t-test).
Adult SKOs could be macroscopically distinguished from CTR mice at the age of 8 weeks by their thickened ears and a dry, flaky tail that showed patches with lost hair and coloration (Fig. 1e). At later ages the SKOs also showed a more global body-hair loss (alopecia) (Fig. 1f), and visibly suffered from pruritus and associated scratching (Movie S1; Figs. S1b–c). Moreover, SKOs of > 6 months often developed chronic lesions around the ears, snout and throat (Fig. 1g). These lesions did not appear to be caused by a deficiency in wound-healing as no differences were noted between CTRs and SKOs in the healing efficiency of 5-mm punch biopsies on the back skin (Figs. S1d–e). Both CTR and SKO embryos stained with toluidine blue at E16.5, but not at E18.5 (Fig. S1f), demonstrating that deletion of NIPP1 did not affect the establishment of the embryonic epidermal barrier. Interestingly, transepidermal-water-loss (TEWL) assays in adult mice revealed a nearly twofold increased water loss in SKOs (Fig. 1h), indicating epidermal-barrier dysfunction.
Epidermal hyperplasia in the SKOs
Histological analysis identified increased epidermal thickness (acanthosis) in both tail (Fig. 2a) and back skin (Fig. S2a) of SKO mice at the age of 8 weeks and 9 months, respectively. Acanthosis in the SKO tails was associated with hyperproliferation of cells in the basal layer, as demonstrated by a twofold higher incorporation of bromo-deoxyuridine (BrdU) during a 4h-pulse labeling (Fig. 2b), and similarly elevated levels of Ki67 (Fig. 2c) and histone H3 phosphorylated at Ser10 (H3S10ph) (Fig. 2d). The back skin of 8-week-old SKOs also contained more BrdU-positive cells in the IFE (Fig. S2b). In further agreement with a hyperproliferation phenotype (Liakath-Ali et al. 2014), the basal-cell marker Keratin-14 (KRT14) was also expressed in the suprabasal layers (Fig. S2c). Moreover, the suprabasal layers were expanded, as demonstrated by stainings for the spinous-layer specific markers Keratin-10 (KRT10) and Keratin-1 (KRT1) (Figs. 2e–f), and the granular-layer specific markers Filaggrin (FLG) and Loricrin (LOR) (Figs. 2g–h). These observations were validated by qRT-PCR analysis (Figs. S2d–g). In back skin, the thickness of the KRT14-layer, but not the FLG layer, was increased in SKO mice of 8 weeks (Fig. S2h). Since hyperproliferation is generally associated with reduced adherence of keratinocytes to the basement membrane, we quantified the expression of integrins, which link the keratin intermediate filaments to the extracellular matrix. The level of integrins ITGA6 and ITGB4 in tail skin was decreased by 40% and 60%, respectively, in the SKOs, as deduced from immunostainings (Figs. 2i–j) and qRT-PCRs (Fig. S2i), suggesting a reduced engagement of NIPP1-null keratinocytes to laminins in the extracellular matrix. This finding is consistent with the presence of basal cells in the suprabasal layers (Fig. S2c)and an increased TEWL (Fig. 1h). In addition to these in vivo experiments, we also examined the effect of NIPP1 depletion on the proliferation of cultured human keratinocytes (HaCaT). Both MTT and viability assays (IncuCyte) showed a decreased growth rate after the siRNA-mediated knockdown of NIPP1 (Figs. S2j–k). The IncuCyte scratch wound assay showed that NIPP1 depletion delayed the migration of HaCaT cells into the created wound but, eventually, the wound healed completely (Figs. S2l–m). We conclude that NIPP1 deletion reduces the proliferation and migratory capacity of HaCaT cells. This is opposite to what we observed for keratinocytes in the SKOs and suggests that the response to a depletion of NIPP1 is context-dependent and affected, for example, by other (epi)dermal cell types.
Figure 2. Epidermal hyperproliferation in SKOs.
(a) Hematoxylin-Eosin (H&E) staining of tail skin sections of CTR and SKO mice at the age of 8 weeks. Hematoxylin was used as a nuclear counterstain. The bar diagram shows quantification of the epidermal thickness (n=4).
(b-d) Tail skin sections from 8-week-old CTR and SKO mice were immunostained for incorporated BrdU after 4h-pulse (b) and for the proliferation markers Ki67 (c) and H3S10ph (d). Propidium iodide (PI) (b) or dapi (c, d) were used as nuclear counterstain. Bar diagrams show the quantifications of the corresponding immunostainings (a, b and d, n=4; c, n=2). Scale bars, 25 μm.
(e-h) Tail skin sections from 8-week-old CTR and SKO mice were immunostained for Keratin-10 (KRT10) (e), Keratin-1 (KRT1) (f), Filaggrin (FLG) (g) and Loricrin (LOR) (h). Dapi was used as a nuclear counterstain. Bar diagrams show the quantifications of layer thickness (n=4). Scale bars, 25 μm.
(i) Tail epidermal wholemounts of 8-week-old CTR and SKO mice were immunostained for the integrin ITGA6 (green) and quantified by measuring the mean intensity (n=3). Dapi was used as a nuclear counterstain. Scale bars, 100 μm.
(j) Tail skin sections from 8-week-old CTR and SKO mice were immunostained for the integrin ITGB4 (green) and quantified (n=4). Dapi was used as a nuclear counterstain. Scale bars, 50 μm.
Bar data are represented as means ± SD. *, p<0.05; **, p<0.01; ***, p<0.001 (unpaired student’s t-test).
Reduced stemness and adherence of HFSCs in the SKOs
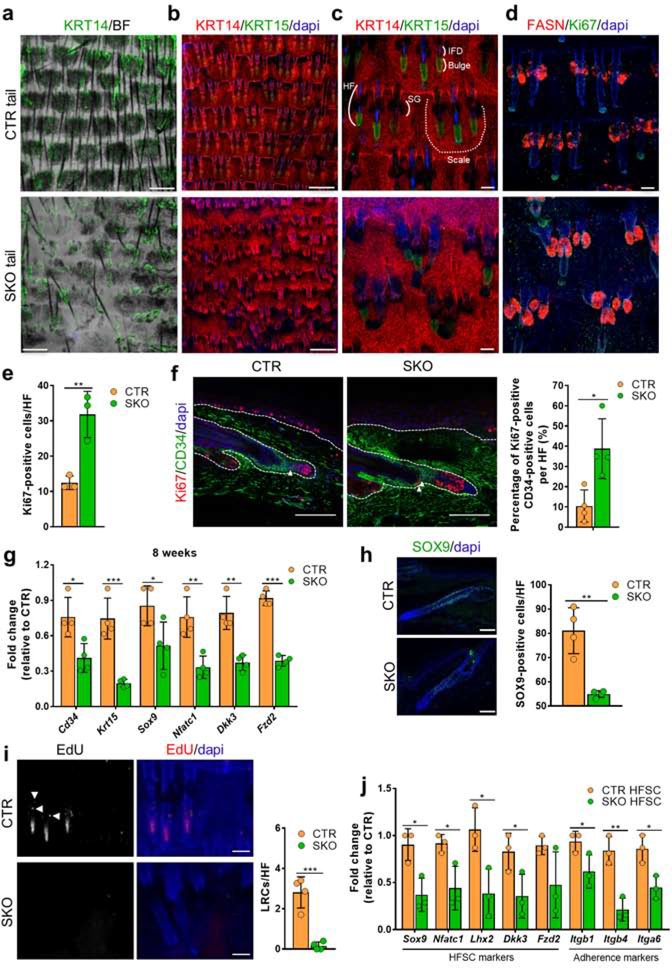
Tail epidermal wholemounts of SKOs showed loss of scales (Fig. 3a), aberrations in HF arrangement (Fig. 3b) and alterations in HF-bulge morphology (Fig. 3c). Indeed, the arrangement of HFs in triplets was distorted in SKO tails (Fig. 3b), but the total number of HFs was not significantly altered (Fig. S3a). Immunostaining of tail wholemounts revealed that bulges (KRT15-positive) were absent or less organized in NIPP1-deficient HFs (Fig. 3c). However, the infundibulum (Fig. 3c) and associated sebaceous glands (FASN-positive) were always visible in the SKO tail (Fig. 3d). Quantification of Ki67-staining in the wholemounts also showed an increased proliferation of cells in SKO HFs (Fig. 3e). In addition, co-immunostaining of CD34 and Ki67 demonstrated hat HFSCs (CD34-positive) in the SKOs were more proliferative in the telogen phase of the hair cycle (Fig. 3f).The bulge HFSCs are required to maintain and renew HFs and also produce the cells of the keratinized hair shaft. We hypothesized that the hair phenotype in the SKOs is (partially) explained by a loss or dysfunction of HFSCs. Consistent with this notion, we found that the expression of various HFSC markers was substantially decreased in SKO tail epidermis (Fig. 3g), and these differences further increased with age (Fig. S3b). Furthermore, immunostaining for the HFSC markers SOX9 and CD34 confirmed a reduced number of HFSCs in SKO tail (Figs. 3h and S3c) and back skin (Fig. S3d). Also, the number of EdU label-retaining-cells (LRCs) following a chase period of 77 days (Fig. S3e)was severely reduced in HFs of the SKO tail (Fig. 3i) and back skin (Fig. S3f). This is indicative for a loss of slow-cycling stem cells (Fuchs 2009; Kretzschmar and Watt 2014), which likely stems from hyperproliferation-induced exhaustion of HFSCs (Fig. 3f). Finally, we used fluorescence-activated cell sorting (FACS) to isolate HFSCs (CD34high/ITGA6high) from CTR and SKO tail epidermis (Fig. S3g). The number of HFSCs in the SKOs was reduced by some 40%, as quantified by cell-sorting analysis (Fig. S3h). Moreover, qRT-PCR analysis of the CD34high/ITGA6high cell population (HFSCs) revealed that HFSC markers, such as Sox9, Nfatc1, Lhx2, Dkk3 and Fzd2, and the cell-adhesion markers Itgb1, Itgb4 and Itga6 were significantly reduced in purified HFSCs of the SKOs, suggesting that NIPP1-deficient HFSCs had lost factors essential for stemness and displayed a reduced matrix adherence (Fig. 3j). Collectively, these data demonstrated that NIPP1 deletion resulted in a reduction of HFSCs with the remaining HFSCs displaying reduced stemness and cell adherence, consistent with the observed hair-loss phenotype.
Figure 3. Hyperproliferation, partial loss and reduced stemness of HFSCs in SKOs.
(a-d) Tail epidermis wholemounts of 8-week-old CTR and SKO mice were immunostained for the basal layer marker Keratin-14 (KRT14) (a-c), the bulge marker Keratin-15 (KRT15) (b, c), the sebaceous gland marker Fatty acid synthase (FASN) (d), and the proliferation maker Ki67 (d). Dapi was used as a nuclear counterstain. BF, bright field. Scale bars, 500 μm (a, b) and 100 μm (c, d). SG, sebaceous gland; IFD, infundibulum; HF, hair follicle.
(e) Quantification of Ki67-positive cells in telogen HFs as shown in (d) (bar diagram, n=3). Ki67-positive cells were counted in the total HF except for the hair germ.
(f) Tail skin sections from CTR and SKO mice of 10 weeks were immunostained for the bulge HFSC marker CD34 and the proliferation marker Ki67. Double-positive cells are indicated by white arrowheads. The number of Ki67-positive, CD34-positive cells in the HF in tail skin was quantified (bar diagrams, n=4). Dapi was used as a nuclear counterstain. Scale bars, 100 μm.
(g) qRT-PCR analysis of the indicated HFSC markers in tail epidermis of CTR and SKO mice of 8 weeks (n=4). Hprt was used as housekeeping gene for normalization.
(h) Tail skin sections from CTR and SKO mice of 8 weeks were immunostained for the HFSC marker SOX9. The number of positive cells per HF was quantified (bar diagrams, n=4). Dapi was used for nuclear staining. Scale bars, 75 μm.
(i) Tail epidermal wholemounts from EdU-injected CTR and SKO mice were stained for EdU. White dots are EdU-positive cells (i.e. label retaining cells (LRCs)), while the white signal in the lowest part of the HF is caused by autofluorescence of the hair shaft. LRCs are indicated by white arrowheads. Dapi was used as a nuclear counterstain. Scale bars, 100 μm. Bar diagrams (n=4) show the quantification of the number of EdU-positive cells in the hair follicle (HF).
(j) qRT-PCR analysis of the indicated genes in FACs purified HFSCs (n=3). Hprt was used as a housekeeping gene for normalization.
Bar data are represented as means ± SD. *, p<0.05; **, p<0.01; ***, p<0.001 (unpaired student’s t-test).
The overt SKO phenotype stems from a chemokine-driven sterile inflammatory response
SKO tail and back (epi)dermis showed an increased infiltration of leucocytes, as demonstrated by staining for CD45+ cells (Figs. 4a and S4a) and flow cytometry (Fig. 4b). CD117-staining disclosed an increased number of mast cells in SKO tail skin (Fig. S4b). We quantified immune-cell types by flow cytometry and found that SKO tail skin contained more antigen-presenting cells, including conventional dendritic cells and Langerhans cells, but not macrophages (Fig. 4c). FACS analysis did not reveal biologically meaningful differences in T-cell development and homeostasis in thymus and spleen at the age of 4 weeks (Fig. S4c) and 8–12 weeks (Fig. S4d), making a contribution of these organs to the observed SKO immune response unlikely. Also, Gram and Periodic Acid Schiff (PAS) stainings were negative (data not shown), indicating that the immune response in the SKOs was not elicited by bacterial or fungal infections, respectively, but caused by pathogen-free or sterile inflammation.
Figure 4. Sterile inflammation in the SKOs.
(a) Tail-skin sections from CTR and SKO mice of 8 weeks were immunostained for the pan-immune cell marker CD45. Dapi was used as a nuclear counterstain. Bar diagrams show the quantifications (n=4). Area = 100.000 μm2. For each mouse, 15 pictures were analysed and the area was extrapolated to 100.000 μm2. Scale bars, 50 μm.
(b) Flow-cytometric quantification of CD45+ cells in the tail skin of 8-week-old CTRs and SKOs (n=6).
(c) Representative flow-cytometric profiles and quantifications of the indicated types of CD45+ immune cells in the tail skin of 8-week-old CTR and SKO mice (n=6).
The bar data are represented as means ± SD. *, p<0.05; **, p<0.01 (unpaired student’s t-test).
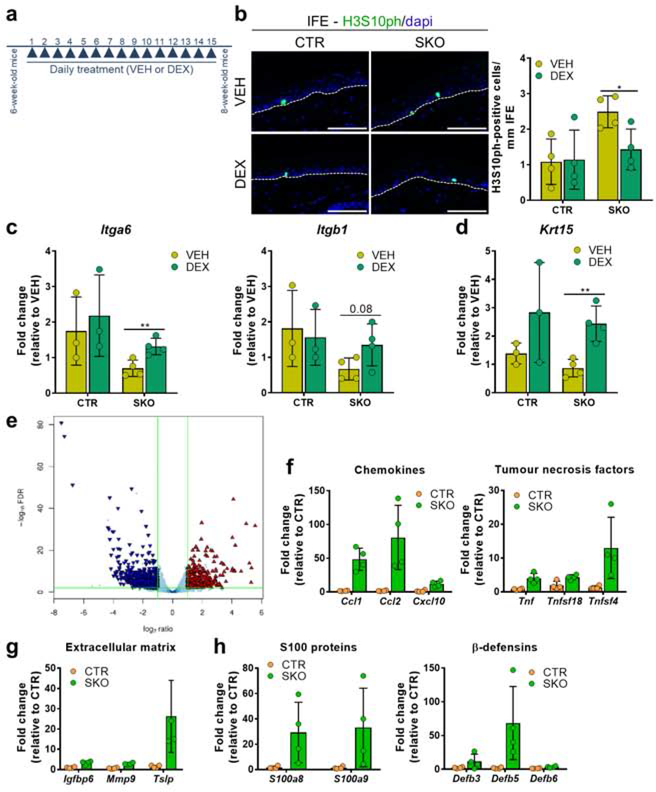
To delineate the contribution of leucocytes to the SKO phenotype, we treated 6-week-old CTR and SKO mice daily, for two weeks, with either vehicle or the immune-suppressant dexamethasone (Fig. 5a). Strikingly, the hyperproliferation of keratinocytes in the IFE of the SKOs was reversed by this treatment, as deduced from immunostainings for H3S10ph (Fig. 5b). Furthermore, the reduced expression of the integrin Itga6 (Fig. 5c) and the HFSC marker Krt15 (Fig. 5d) were also alleviated by the dexamethasone treatment. The reduced expression of the integrin Itgb1 was not significantly rescued (Fig. 5c). Together, these data suggest that epidermal hyperproliferation in the SKOs as well as the loss and reduced adherence of the HFSCs are, at least partially, caused by chronic sterile skin inflammation.
Figure 5. Contribution of chemokine-driven sterile inflammation to the SKO phenotype.
(a) Scheme for the treatment of CTRs and SKOs with vehicle (VEH) or dexamethasone (DEX). Skin samples were isolated 24h after the last treatment.
(b) Immunostaining of tail skin sections from vehicle/dexamethasone-treated CTRs and SKOs for H3S10ph. Bar diagrams show quantifications of H3S10ph immunostainings in the interfollicular epidermis (IFE; n=4). Scale bars, 75 μm.
(c,d) qRT-PCR analysis for the expression of the adherence markers Itga6 and Itgb1 (c) and the HFSC marker Krt15 (d) in vehicle/dexamethasone-treated skin samples.
(e) Volcano plot of differentially expressed genes between CTR and SKO mice of 8 weeks (n=4). The triangles are colored blue and red if they are classified as down- or upregulated genes, respectively, based on the corrected p-values (FDR<0.01, denoted by the horizontal green-colored line) and an absolute log2-ratio larger than 1 (denoted by the vertical green-colored lines.
(f-h) Bars charts of expression data of the indicated genes (FDR<0.01). The indicated genes belong to the families of chemokines and tumor necrosis factors (f), extracellular matrix proteins (g), or antimicrobial peptides (h). Data are expressed as a fold change ± SD (n=4) in the SKOs as compared to CTRs. The expression of all described genes (f-h) is significantly different between CTRs and SKOs based on the corrected p-value (FDR<0.01) of the RNA-seq analysis.
The bar data in b-d are represented as means ± SD (n≥3). *, p<0.05; **, p<0.01 (unpaired student’s t-test). The bar data in f-h are represented as means ± SD.
To obtain insights into the molecular signaling pathway(s) that are affected by the deletion of NIPP1, we performed a comparative transcriptome analysis (RNA-seq) on tail epidermis of CTR and SKO mice (Fig. 5e). In total, 846 protein-encoding genes were differentially expressed (FDR < 0.01, two-fold cut-off) (Table S4). The 61% differentially expressed genes (DEGs) that were downregulated in the SKOs were strongly enriched for genes encoding keratins or keratin-associated proteins (Figs. S5a–c;Table S5), consistent with aberrant HF development and associated hair loss. The 39% DEGs that were upregulated in the SKOs were mostly enriched for the Gene Ontology entries ‘Immune_System_Process’, ‘Extracellular_Space’ and ‘Cytokine_Activity’ (Fig. S5d; Table S6). We noted a strong upregulation of different types of cytokines in tail epidermis (Fig. 5f), which was confirmed by qRT-PCR (Fig. S5e). In fact, the chemokine Ccl2 was the most upregulated gene(> 40-fold), and the chemokines Ccl1, Ccl20, Cxcl10 and Cxcl9 were at least 8 times more expressed in SKO epidermis (Table S4 and S6). Other immunomodulatory genes, including extracellular matrix proteins (Fig. 5g) or antimicrobial peptides, such as S100 proteins and β-defensins (Fig. 5h), were also upregulated in the SKOs. The chemokines Ccl1 and Ccl2 were also upregulated in back epidermis (8 weeks), albeit to a lesser extent (Fig. S5f).
Differential RNA-seq analysis also disclosed prominent changes in the expression of many genes of the Epidermal-Differentiation-Complex (EDC) locus, which encodes proteins that are essential for the terminal differentiation of keratinocytes (Fig. S5g) (Poterlowicz et al. 2017). The transcript level of 17 EDC genes was increased in the SKOs, while 5 genes were less expressed (Fig. S5h). Interestingly, genes that contribute to terminal epidermal differentiation (e.g. S100a8/a9, various Lce genes) generally showed an increased expression. In contrast, genes that are expressed in HFs (S100a3, Tchh, Tchhl1, Crnn), and contribute to terminal hair differentiation, were downregulated in the SKOs. Collectively, the DEGs independently confirmed an increased epidermal differentiation and diminished development of the hair lineage in the SKOs, and suggested that the observed inflammatory phenotype in the SKOs is chemokine-driven.
Inflammation in the SKOs is initiated by keratinocyte-derived chemokines
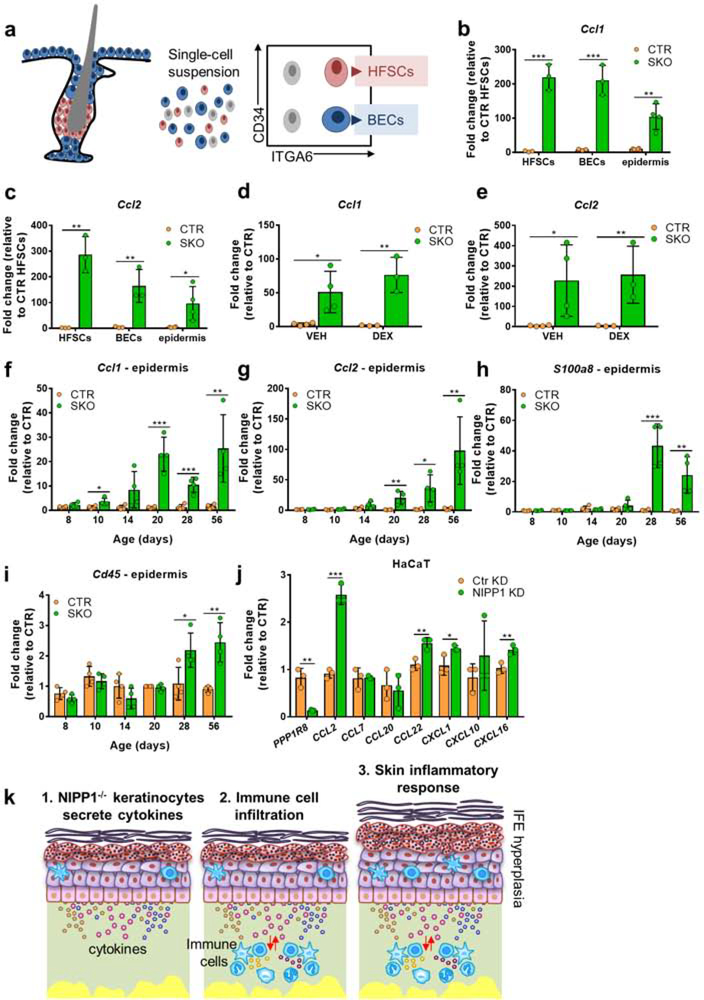
Since keratinocytes as well as resident and recruited immune cells in the skin can produce cytokines, we explored the cellular source of increased cytokine expression in the SKOs. For this purpose, we isolated basal epidermal cells (BECs; CD34-neg/ITGA6high) and HFSCs (CD34high/ITGA6high) from the epidermis of CTR and SKO tails by FACS (Fig. 6a). qRT-PCR analysis revealed a massive increase in the expression of cytokines, including Ccl1, Ccl2, Cxcl1, Cxcl2 and Tnf, in the purified SKO cell populations (Figs. 6b–c; Fig. S6a–c). In general, the relative expression of these cytokines was higher in SKO HFSCs and BECs than in SKO epidermis. Interestingly, cytokine expression by Ppp1r8−/− keratinocytes was not affected by a prolonged treatment with dexamethasone (Figs. 6d–e) which did, however, partially suppress the global SKO phenotype (Figs. 5b–d). It should be taken into account that the dexamethasone treatment was started at 6 weeks when the phenotype was largely established, including the recruitment of immune cells. Together, these data suggested that the expression of chemokines in NIPP1-deficient keratinocytes was not caused by the observed sterile inflammatory response but the consequence of endogenous stress signaling. We proceeded to analyze cytokine expression and immune infiltration in tail epidermis of SKO mice of different ages. The deletion of NIPP1 from pre-adult SKO epidermis was confirmed by qRT-PCR (Fig. S6d). We already detected a small but significant increase in the expression of the chemokinesCcl1 and Ccl2 in SKOs of 10 and 20 days,respectively (Figs. 6f–g). In contrast, the induction of other immunoregulatory genes, such as S100a8 and Mmp9,wasonly detected from 28 days onwards (Figs. 6h and S6e). The immune infiltration was evaluated in both epidermis and dermis by examining the level of Cd45 transcripts. The increased level of Cd45 transcripts was only detected from 20 days onwards in the dermis (Fig. S6f) and from 28 days in epidermis (Fig. 6i). Finally, the cell-autonomous character of cytokine secretion by NIPP1-depleted keratinocytes was independently validated by our observation that cultured human HaCaT keratinocytes also secrete cytokines after the siRNA-mediated knockdown of NIPP1 (Fig. 6j), albeit to a lesser extent than observed for Ppp1r8−/− mouse keratinocytes. Altogether, these data suggested that NIPP1-null keratinocytes release cytokines, including CCL1/2, in a cell-autonomous manner from an age of ~10 days onwards (Fig. 6k). CCL1/2, and other cytokines, act as chemoattractant for immune cells, in particular dendritic cells that express the corresponding chemokine receptors. Activated immune cells subsequently release additional cytokines that contribute to the observed sterile inflammatory response and SKO phenotype at later ages.
Figure 6. Chemokine release from NIPP1-depleted keratinocytes.
(a) Schematic representation of FACS purification of HFSCs and basal epidermal cells (BECs).
(b,c) qRT-PCR analysis of the expression of Ccl1 (b) and Ccl2 (c) in total epidermis and FACS-purified HFSCs and BECs.
(d,e) qRT-PCR analysis of the expression ofCcl1 (d) and Ccl2 (e) in vehicle/dexamethasone-treated mice, as described in Fig. 5a.
(f-i) qRT-PCR analysis of the expression of Ccl1 (f), Ccl2 (g), S100a8 (h), and Cd45 (i) in tail epidermis of CTR and SKOs at the indicated ages.
(j) qRT-PCR analysis of the indicated transcripts in the human keratinocyte cell line HaCaT after treatment with NIPP1-specific siRNA (NIPP1 KD) or nontargeted siRNA (Ctr KD) for 24 h.
(k) Model of chemokine-induced inflammation in the SKOs. NIPP1-null keratinocytes secrete pro-inflammatory chemokines, which attract and activate immune cells that secrete additional cytokines and thereby contribute to the inflammatory response.
For all qRT-PCR data, Hprt was used as a housekeeping gene for normalization. All data are represented as means ± SD (n≥3). *, p<0.05; **, p<0.01; ***, p<0.001 (unpaired student’s t-test).
DISCUSSION
We have shown that Ppp1r8−/− keratinocytes spontaneously and constitutively release chemokines that attract immune cells and induce a chronic sterile inflammatory response. Both HFSCs and BECs purified from SKOs showed a vastly increased expression of proinflammatory chemokines (Fig. 6). This increased chemokine expression was not affected by a prolonged treatment with an immunosuppressant and was also detected in cultured HaCaT keratinocytes, following the siRNA-mediated knockdown of NIPP1, confirming the cell-autonomous and inflammation-independent nature of this response. Importantly, the increased expression of chemokines in the SKO epidermis preceded the recruitment of immune cells to the skin by 1–2 weeks, demonstrating that the initial trigger for the inflammatory response was not provided by recruited immune cells (Figs. 6 and S6f).
Inflammation is a protective response aimed at removing the causes of cell stress (e.g. infection), clearing damaged cells and initiating tissue repair (Karin and Clevers 2016). However, when the cause of stress is not well-resolved, the inflammation becomes chronic and can lead to tissue destruction and pathologies such as psoriasis and atopic dermatitis (Rotty and Coulombe 2012; Lessard et al. 2013). Consistent with a chronic-inflammation phenotype the skin of the SKOs displayed properties of both tissue repair and destruction. On the one hand the epidermis of SKOs was hyperplastic (Fig. 2) and showed an increased expression of epithelial keratins and terminal-epidermal-differentiation genes (Fig. S5). On the other hand, the HFSCs of the SKOs were hyperproliferative and showed a decreased stemness and adherence, accounting for the degeneration of HFs (Fig. 3), hair loss and the gradual appearance of skin wounds (Fig. 1). Importantly, the SKO phenotype was partially rescued by the repeated administration of an immunosuppressant (Fig. 5), indicating that it was caused by chronic inflammation. At first glance, the SKO phenotype is reminiscent of the hepatic response to the deletion of NIPP1 in hepatoblast-derived epithelial cells, which also involved both a hyperplastic and inflammatory response (Boens et al. 2016). However, inflammation in Ppp1r8−/− livers developed much later than the hyperproliferation of bile-duct progenitor cells, arguing against a causal relationship.
Inflammation-induced epidermal hyperproliferation, as elicited in the NIPP1 SKOs, is well understood and is mainly mediated by increased signaling through the NF-κB pathway (Liu et al. 2017). However, the mechanism underlying the degeneration of HFs in the SKOs is less clear and can involve multiple mechanisms. HFs normally escape immune surveillance but the deletion of NIPP1 somehow results in the loss of their ‘immune privilege’ status, making the HSFCs accessible to removal by immune cells (Azzawi et al. 2018). HFSCs are also known to be extremely sensitive to stress-signaling, which causes their differentiation and migration out of the stem-cell niche (Schuler et al. 2018). It is therefore possible that the activation of a stress-signaling pathway, induced by the absence of NIPP1 (see below), accounts for the differentiation and loss of HFSCs in the SKOs. Importantly, HFs can also degenerate in an inflammation-independent manner, for example by depletion of core components of the Polycomb Repressive Complex 2, including EZH1/2 (Ezhkova et al. 2011), an established substrate of PP1:NIPP1 (Minnebo et al. 2013). Since HF degeneration in the SKOs was only partially rescued by a dexamethasone treatment (Fig. 5), it cannot be excluded that HF degeneration in the SKOs is (partially) inflammation-independent and due to decreased PRC2-mediated signaling.
It is not yet clear how NIPP1 regulates the expression of cytokines in keratinocytes. Since NIPP1 recruits phosphoproteins for regulated dephosphorylation by associated PP1, we speculate that NIPP1 controls the phosphorylation of transcription factors and/or transcriptional co-repressors/activators that regulate the expression of cytokine genes. Consistent with this view, it has been shown that NIPP1 controls the phosphorylation status and activity of the transcriptional co-repressors Enhancer of Zeste Homolog 2 (EZH2) and Krüppel-associated box domain-associated protein 1 (KAP1) (Ferreira et al. 2018; Minnebo et al. 2013; Smith-Moore et al. 2018). Since many of cytokine genes with an altered expression in the SKOs have NF-κB binding sites in their promoters, NIPP1 may affect cytokine expression through PP1-mediated dephosphorylation of NF-κB (Richmond 2002; Taniguchi and Karin 2018). In addition, NIPP1 may function itself as a transcription factor, as suggested by a genome-wide promoter binding profiling of NIPP1 in HeLa cells using the DamID technique (Verheyen et al. 2015). Interestingly, the latter approach identified the Cxcl1 promoter, as well as the Krtap19–4 and Krtap19–5 promoters, as putative binding sites for NIPP1.
Currently it is not clear which stress pathway is deregulated in keratinocytes following the depletion of NIPP1 and leads to an increased expression of cytokines. The stable expression of a PP1-NIPP1 fusion in HeLa cells resulted in the accumulation of DNA damage, R-loops and hypercondensed chromatin, and was associated with a reduced DNA-repair capacity (Winkler et al. 2018). Conversely, the deletion of NIPP1 in skin or liver increased the repaircapacity of DNA damage induced by mutagens, which correlated with an increased expression of key DNA-repair proteins (unpublished data). The exact role of PP1:NIPP1 in DNA-damage and -repair signaling is not yet understood but we speculate that it involves the (de)phosphorylation of substrates since the effects of a PP1-NIPP1 fusion on DNA damage were dependent on a functional substrate-binding FHA-domain and an active PP1 moiety (Winkler et al. 2018). Interestingly, all known substrates of PP1:NIPP1 have been implicated in DNA damage or repair signaling (Maréchal et al. 2014; Savage et al. 2014; Beke et al. 2015; Rondinelli et al. 2017), suggesting that the phenotype of the NIPP1 SKOs may be caused by an altered phosphorylation status of PP1:NIPP1 substrates.
In conclusion, we have shown that deletion of NIPP1 in keratinocytes triggers the release of cytokines, which unleashes a chronic inflammatory response and culminates in a pathological phenotype that is reminiscent of human inflammatory skin diseases such as atopic dermatitis and psoriasis. It will be interesting to investigate whether the activation of PP1:NIPP1, for example with compounds that disrupt the interaction of the C-terminal inhibitory domain of NIPP1 with PP1, alleviate the development of these inflammatory skin diseases in various models.
MATERIALS AND METHODS
Animals
Mice were housed in a pathogen-free animal facility under standard 12h-light/dark cycles with water and chow ad libitum. The generation of skin-specific NIPP1 KO and CTR mice is described in Supplementary Materials and Methods. All experimental protocols were in accordance with and approved by the Guide of Care of Experimental Animals of the KU Leuven Ethical Committee (license number 053/2018).
Mouse treatments
Pups of 10 days were intraperitoneally injected with 100 μl of 6.25 mg/ml 5-ethynyl-2’-deoxyuridine (EdU) every 12 hours for a total of 4 injections and imaged using the Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Thermo Fisher Scientific). Skin samples were collected 77 days after the last injection. Mice were intraperitoneally injected daily, for 15 consecutive days, with 4 mg/kg rapidexon/dexamethasone (Eurovet Animal Health, Dechra) or vehicle (saline). The skin-barrier assays, i.e. embryo whole-mount dye-penetration assays and transepidermal water loss; the assessment of scratching behavior and the wound-healing assays are detailed described in Supplementary Material and Methods.
Flow cytometry
HFSCs and BECs, immune cells in the skin, and T cell maturation and activation in spleens and thymi were analysed by flow cytometry as detailed in Supplementary Materials and Methods.
RNA-sequencing and qRT-PCR
Total RNA was isolated from snap-frozen mouse epidermis, dermis, FACS-purified HFSCs and BECs or from HaCaT cells (stored at −80°C). For RNA-sequencing, total RNA was isolated from snap-frozen tail epidermis from 4 CTR and 4 SKO mice (age of 8 weeks). qRT-PCR and RNA sequencing is further described in Supplementary Materials and Methods.
Biochemical procedures and (immuno)histochemical analysis
Total lysates were made from tail epidermis and subjected to Western blot analyses as detailed in Supplementary Materials and Methods. Skin tissue was fixed and used for (immuno)histochemical analysis according to standard protocols. All antibodies are listed in Supplementary Table S2. See also Supplementary Materials and Methods.
Cell culture
HaCaT cells were cultured in DMEM medium with 4.5 g/L glucose and 2 mM L-glutamine (Sigma-Aldrich), supplemented with 1 mM sodium pyruvate (Sigma-Aldrich), 10% FCS and 1% Penicillin/Streptomycin. The siRNA transfections were performed using RNAiMAX transfection reagent (Thermo Fisher Scientific). Knockdowns were verified by qPCR analysis of Ppp1r8 expression. See also Supplementary Materials and Methods.
Data analysis
The results are expressed as means ± standard deviation (SD) and analyzed with the unpaired Student t-test. Statistical analysis was performed in GraphPad Prism version 7 (GraphPad software, Inc). See also Supplementary Materials and Methods.
DATA AVAILABILITY
Gene expression data are available at GEO under accession number at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116844).
Supplementary Material
ACKNOWLEDGEMENTS
Maud De Meyer, Fabienne Withof, Annemie Hoogmartens and Lies Pauwels provided expert technical assistance. Mieke Anthonissen is acknowledged for expert advice on TEWL assays, and Pier Andrée Pentilla, Christèle Nkama and Pradeep Kumar (https://gbiomed.kuleuven.be/english/corefacilities/facs) for FACS sorting. We thank Dr. Fiona M. Watt (King’s College London) for allowing the wholemount stainings to be performed in her laboratory and for giving feedback on the data. Prof. W. Birchmeier (Max-Delbrueck-Center for Molecular Medicine, Germany) is acknowledged for the donation of Tg (Krt14-Cre) mice. Rekin’s Janky of the Nucleomics Core at KU Leuven analyzed the RNA-seq data.
FUNDING
This work was supported by the Belgian Foundation Against Cancer, FWO grant G078717N, a KU Leuven BOF-grant (GOA/15/016), and NIH grant 1R01CA188382-01. Iris Verbinnen was a research fellow of the FWO Flanders.
Abbreviations
- NIPP1
nuclear inhibitor of protein phosphatase 1 (PP1)
- FHA
forkhead associated
- IFE
interfollicular epidermis
- HF
hair follicle
- HFSCs
hair follicle stem cells
- CTR
control
- SKO
skin-specific NIPP1 knockout
- TEWL
transepidermal water loss
- BrdU
bromo-deoxyuridine
- H3S10ph
histone H3 phosphorylated at Ser10
- KRT
keratin
- FLG
filaggrin
- LOR
loricrin
- ITG
integrin
- FASN
fatty acid synthase
- EdU
5-ethynyl-2’-deoxyuridine
- LRC
label-retaining-cell
- FACS
fluorescence-activated-cell-sorting
- DEGs
differentially expressed genes
- EDC
epidermal differentiation complex
- BEC
basal epidermal cell
- PRC2
polycomb repressive complex 2
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Azzawi S, Penzi LR, Senna MM. Immune Privilege Collapse and Alopecia Development: Is Stress a Factor. Ski. Appendage Disord. 2018. p. 236–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beke L, Kig C, Linders JTM, Boens S, Boeckx A, van Heerde E, et al. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci. Rep 2015;35(6):e00267–e00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belokhvostova D, Berzanskyte I, Cujba AM, Jowett G, Marshall L, Prueller J, et al. Homeostasis, regeneration and tumour formation in the mammalian epidermis. Int. J. Dev. Biol 2018;62(6–8):571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens M, Vulsteke V, Van Eynde A, Jagiello I, Stalmans W, Bollen M. The C-terminus of NIPP1 (nuclear inhibitor of protein phosphatase-1) contains a novel binding site for protein phosphatase-1 that is controlled by tyrosine phosphorylation and RNA binding. Biochem. J 2000;352 Pt 3:651–8 [PMC free article] [PubMed] [Google Scholar]
- Boens S, Verbinnen I, Verhulst S, Szekér K, Ferreira M, Gevaert T, et al. Brief Report: The Deletion of the Phosphatase Regulator NIPP1 Causes Progenitor Cell Expansion in the Adult Liver. Stem Cells. 2016;34(8):2256–62 [DOI] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Groenen P, Van Eynde A, Vulsteke V, Jagiello I, et al. NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem 2000;275(33):25411–7 [DOI] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Waelkens E, Stalmans W, Bollen M. Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J. Biol. Chem 2002;277(35):31834–41 [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. BioEssays. 2002. p. 371–81 [DOI] [PubMed] [Google Scholar]
- Eyerich S, Eyerich K, Traidl-Hoffmann C, Biedermann T. Cutaneous Barriers and Skin Immunity: Differentiating A Connected Network. Trends Immunol. 2018. p. 315–27 [DOI] [PubMed] [Google Scholar]
- Van Eynde A, Nuytten M, Dewerchin M, Schoonjans L, Keppens S, Beullens M, et al. The Nuclear Scaffold Protein NIPP1 Is Essential for Early Embryonic Development and Cell Proliferation. Mol Cell Biol. 2004;24:5863–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25(5):485–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M, Boens S, Winkler C, Szekér K, Verbinnen I, Van Eynde A, et al. The protein phosphatase 1 regulator NIPP1 is essential for mammalian spermatogenesis. Sci. Rep 2017;7(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M, Verbinnen I, Fardilha M, Van Eynde A, Bollen M. The deletion of the protein phosphatase 1 regulator NIPP1 in testis causes hyperphosphorylation and degradation of the histone methyltransferase EZH2. J. Biol. Chem. American Society for Biochemistry and Molecular Biology; 2018;293(47):18031–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz Y, Klenotic PA, Swindell WR, Yin ZQ, Groft SG, Zhang L, et al. Induction of Alternative Proinflammatory Cytokines Accounts for Sustained Psoriasiform Skin Inflammation in IL-17C+IL-6KO Mice. J. Invest. Dermatol 2017;137(3):696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E The Tortoise and the Hair: Slow-Cycling Cells in the Stem Cell Race. Cell. 2009. p. 811–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU, Fuchs E. Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev. Cell. 2017;43(4):387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänel KH, Cornelissen C, Lüscher B, Baron JM. Cytokines and the skin barrier. Int. J. Mol. Sci 2013. p. 6720–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath WR, Carbone FR. The skin-resident and migratory immune system in steady state and memory: Innate lymphocytes, dendritic cells and T cells. Nat. Immunol 2013. p. 978–85 [DOI] [PubMed] [Google Scholar]
- Kabashima K, Honda T, Ginhoux F, Egawa G. The immunological anatomy of the skin. Nat. Rev. Immunol 2019. [DOI] [PubMed] [Google Scholar]
- Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016. p. 307–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. Markers of epidermal stem cell subpopulations in adult mammalian skin. Cold Spring Harb. Perspect. Med 2014;4(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Pasparakis M. Epithelial cell death and inflammation in skin. Curr. Top. Microbiol. Immunol 2017. p. 77–93 [DOI] [PubMed] [Google Scholar]
- Lessard JC, Pina-Paz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci 2013;110(48):19537–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakath-Ali K, Vancollie VE, Heath E, Smedley DP, Estabel J, Sunter D, et al. Novel skin phenotypes revealed by a genome-wide mouse reverse genetic screen. Nat. Commun 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yao Q, Mariscal AG, Wu X, Hülse J, Pedersen E, et al. Epigenetic control of IL-23 expression in keratinocytes is important for chronic skin inflammation. Nat. Commun 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct. Target. Ther 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, et al. PRP19 Transforms into a Sensor of RPA-ssDNA after DNA Damage and Drives ATR Activation via a Ubiquitin-Mediated Circuitry. Mol. Cell 2014;53(2):235–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnebo N, Görnemann J, O’Connell N, Van Dessel N, Derua R, Vermunt MW, et al. NIPP1 maintains EZH2 phosphorylation and promoter occupancy at proliferation-related target genes. Nucleic Acids Res. 2013;41(2):842–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytten M, Beke L, Van Eynde A, Ceulemans H, Beullens M, Van Hummelen P, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27(10):1449–60 [DOI] [PubMed] [Google Scholar]
- Otsuka A, Nomura T, Rerknimitr P, Seidel JA, Honda T, Kabashima K. The interplay between genetic and environmental factors in the pathogenesis of atopic dermatitis. Immunol. Rev 2017. p. 246–62 [DOI] [PubMed] [Google Scholar]
- Poterlowicz K, Yarker JL, Malashchuk I, Lajoie BR, Mardaryev AN, Gdula MR, et al. 5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells. PLoS Genet. 2017;13(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond A NF-κB, chemokine gene transcription and tumour growth. Nat. Rev. Immunol 2002. p. 664–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B, Gogola E, Yücel H, Duarte AA, Van De Ven M, Van Der Sluijs R, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol 2017;19(11):1371–8 [DOI] [PubMed] [Google Scholar]
- Rotty JD, Coulombe PA. A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol 2012;197(3):381–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, et al. Identification of a BRCA1-mRNA Splicing Complex Required for Efficient DNA Repair and Maintenance of Genomic Stability. Mol. Cell 2014;54(3):445–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler N, Timm S, Rübe CE. Hair Follicle Stem Cell Faith Is Dependent on Chromatin Remodeling Capacity Following Low-Dose Radiation. Stem Cells. 2018;36(4):574–88 [DOI] [PubMed] [Google Scholar]
- Smith-Moore S, Neil SJD, Fraefel C, Linden RM, Bollen M, Rowe HM, et al. Adenoassociated virus Rep proteins antagonize phosphatase PP1 to counteract KAP1 repression of the latent viral genome. Proc. Natl. Acad. Sci 2018;115(15):E3529–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Karin M. NF-B, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol 2018. p. 309–24 [DOI] [PubMed] [Google Scholar]
- Verheyen T, Görnemann J, Verbinnen I, Boens S, Beullens M, Van Eynde A, et al. Genomewide promoter binding profiling of protein phosphatase-1 and its major nuclear targeting subunits. Nucleic Acids Res. 2015;43(12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulsteke V, Beullens M, Boudrez A, Keppens S, Van Eynde A, Rider MH, et al. Inhibition of Spliceosome Assembly by the Cell Cycle-regulated Protein Kinase MELK and Involvement of Splicing Factor NIPP1. J. Biol. Chem 2004;279(10):8642–7 [DOI] [PubMed] [Google Scholar]
- Winkler C, Rouget R, Wu D, Beullens M, Van Eynde A, Bollen M. Overexpression of PP1–NIPP1 limits the capacity of cells to repair DNA double-strand breaks. J. Cell Sci 2018;131(13):jcs214932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data are available at GEO under accession number at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116844).