Abstract
As a cell prepares to divide, its genetic material changes dramatically in both form and function. During interphase, a dynamic interplay between DNA compartmentalization and transcription functions to program cell identity. During mitosis, this purpose is put on hold and instead chromosomes function to facilitate their accurate segregation to daughter cells. Chromatin loops are rearranged, stacked and compressed to form X-shaped chromosomes that are neatly aligned at the center of the mitotic spindle and ready to withstand the forces of anaphase. Many factors that contribute to mitotic chromosome assembly have now been identified, but how the plethora of molecular mechanisms operate in concert to give rise to the distinct form and physical properties of mitotic chromosomes at the cellular scale remains under active investigation. In this review we discuss recent work that addresses a major challenge for the field: How to connect molecular level activities to large-scale changes in whole-chromosome architecture that determine mitotic chromosome size, shape and function.
Visualizing the spatial organization of chromosomes: Mapping DNA contacts and imaging chromatin
Understanding how mitotic chromosome properties emerge requires deep knowledge of their structure. Recent advances in genomics and imaging have allowed us visualize mitotic chromosomes in unprecedented detail and challenged the field to think beyond hierarchical models of mitotic chromosome structure depicted in textbooks.
The development of a chromosome conformation capture technique (Hi-C), which maps the frequency of inter- and intra-chromosomal contacts genome-wide, has revolutionized how we think about chromosome organization [1]. In interphase cells, a major feature that emerges are topologically associated domains (TADs). These megabase-sized regions of the chromosome contain DNA sequences that physically interact with each other more frequently than with regions outside the TAD, and are thought to regulate gene expression. A recent landmark study applied Hi-C to highly synchronized cultured cells to closely track any rearrangements of the genome at the onset of mitosis. The authors showed that during prophase, shorter-range interactions and TADs were replaced by a set of longer-range interactions that uniformly spanned the length of each chromosome [2]**. Combining these observations with extensive modeling, these long-range interactions were proposed to represent a “spiral staircase” of loops that form perpendicular to the central axis of each chromosome, and are broadly consistent with the radial loops observed decades ago by electron microscopy of salt-extracted chromosomes [3].
How do the fundamental units of chromatin, nucleosomes that consist of 2 copies each of histones H2A, H2B, H3 and H4, fit into this picture? A long-standing model in the field was based on hierarchical folding, in which nucleosomes are first organized into 30 nm fibers before being further packaged into the final condensed state of the mitotic chromosome. However, evidence for the formation of 30 nm chromatin fibers in vivo as examined by several techniques was lacking [4–6]. Recent studies made possible by advances in electron tomography have thoroughly dispelled the hierarchical folding model by showing that although nucleosomes are packed at a higher density in mitosis compared to interphase in both cultured vertebrate cells and fission yeast, they are irregularly arranged and lack a discernable higher order structure, appearing more like a molten polymer [7,8]. Moreover, depletion of core histones from Xenopus egg extracts revealed that nucleosomes are not required for creating the rod-like shape of mitotic chromosomes, although chromosomes lacking histones were fragile, appearing fuzzy and less compacted [9]*. Thus, nucleosome packing and higher order chromosome assembly may be largely independent processes [10].
Taken together, these studies present a new paradox: how do we reconcile the structured, large-scale DNA loop arrangement seen in Hi-C data with the apparent randomness of nucleosome-level organization in mitotic chromosomes? Below we discuss a number of recent mechanistic studies that provide new clues as to how many distinct molecular activities give rise to higher order chromosome morphology and physical properties.
Histone tail modifications and biomolecular condensates drive chromosome compaction
At the nanometer length scale, the current view is that core histones themselves influence higher order chromatin compaction by modulating nucleosome-nucleosome interactions during the cell cycle. Supporting this idea, it was shown that nucleosome arrays reconstituted using histones isolated either from interphase or mitosis resulted in either less or more compact structures, respectively [11]*.
Regulation at the level of the nucleosome appears to involve a series of post-translational modifications (PTMs) on histone tails that are either erased or added [12], and contribute to compaction of chromatin. The best understood example begins with Aurora B, the kinase subunit of the chromosomal passenger complex (CPC), which performs numerous roles in mitosis in all organisms [13] (Figure 1A). In budding yeast, it was shown that Aurora B phosphorylates serine 10 of histone H3 (H3S10) at mitotic onset, which then recruits Hst2, an enzyme that removes acetyl groups from lysine 16 of histone H4 (H4K16) and promotes inter-nucleosomal interactions between the positively charged histone H4 tail and an acidic patch on histone H2A [14]. This in vivo study agrees nicely with in vitro data using reconstituted nucleosome arrays [15].
Figure 1. Multiple molecular level activities cooperate to organize and shape mitotic chromosomes.
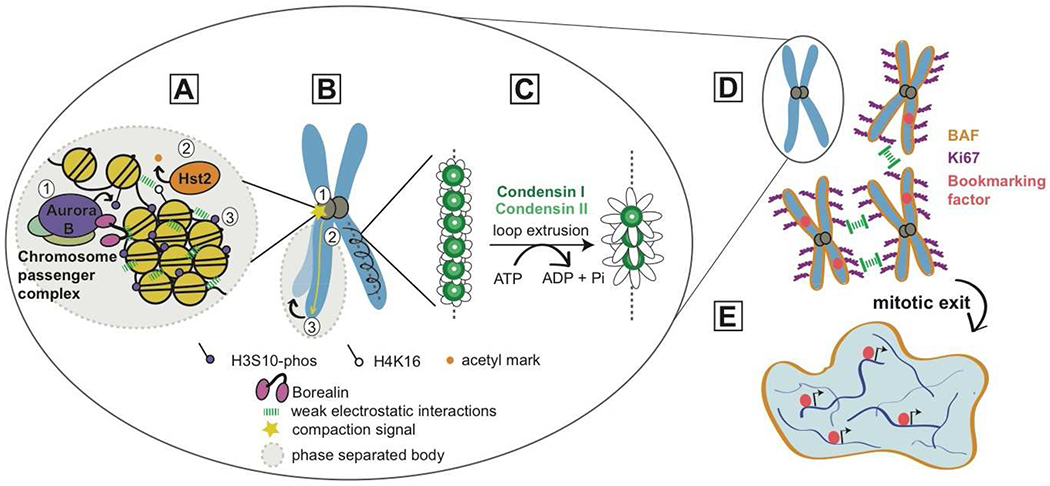
(A) Chromatin compaction at mitotic onset is driven by a cascade of histone post translational modifcations (PTMs) initiated by Aurora B, the kinase subunit of the chromosome passenger complex (CPC). Aurora B phosphorylates S10 of histone H3 (1), which recruits the deacetylase Hst2 that acts on histone H4 K16 (2). These changes result in increased multivalent interactions among neighboring nucleosomes, driven in part by an unstructured domain of Borealin, another subunit of the CPC (3). Ultimately, these interactions result in increased compaction of chromatin, which could be driven by phase separation.
(B) In the model proposed in Kruitwagen et al. [14], the compaction signal mediated by Aurora B initiates at the centromere (1) and spreads along the chromosome arm (2), resulting in a shortened, more compact chromosome (3).
(C) In the model proposed in Gibcus et al. [1], condensin I and II form loops of DNA perpendicular to the central axis. In this model, loop extrusion by the condensins creates larger loops of DNA, and thus axial shortening of the chromosome arm.
D-E) Interchromosomal organization is driven by two DNA binding molecules: Ki67 and BAF. Ki67 helps individualize chromosomes by acting as a “surfactant brush”, repelling chromosomes from one another. Upon mitotic exit, formation of a complete nucleus is aided by BAF, which crosslinks chromosomes at their periphery, preventing chromosome loss. Finally, mitotic bookmarking factors that remain on mitotic chromosomes help re-initiate transcriptional programs upon re-entry into interphase.
How does this molecular-scale mechanism play out in the context of an entire chromosome? An exciting study suggests that this signaling cascade may originate at the centromere in budding yeast [16]**. The authors observed that removing the centromere from a chromosome prevented centromeric phosphorylation of histone H3S10 and impaired condensation of that specific chromosome. Conversely, inserting a centromere sequence into an artificial mini-chromosome was sufficient to drive its compaction. Exactly how the compaction signal is transmitted along chromosome arms is unclear, but appears to involve histone deacetylation by Hst2 and regulation by the protein Shugoshin. It also remains to be seen whether or not the centromere is capable of playing a similar role in animal cells, where H3S10 phosphorylation, despite originating at the centromere [17], also occurs along the chromosome arms.
What features of the centromere could allow it to drive compaction of a chromosome? Intriguingly, a recent study indicates that the inner centromere has properties of a biomolecular condensate [18]**. The authors showed that an unstructured domain of the CPC subunit Borealin can drive phase separation of the complex in vitro, and provided evidence that the CPC exists in a phase separated state at the inner centromere in vivo. A separate study demonstrated that the unstructured domain of Borealin is also responsible for mediating multivalent interactions between the CPC and nucleosomes, suggesting that chromatin also participates in phase separation at the centromere [19]. Taken together, one appealing model is that a cascade of histone PTMs initiated by the CPC drives formation of a biomolecular condensate at the centromere that spreads and compacts chromatin along chromosome arms (Figure 1B). Another factor that could potentially initiate mitotic chromosome compaction in some systems is heterochromatin protein 1 (HP1), which maintains an active role at the centromere during mitosis [20,21], and has been shown to drive phase separation of chromatin in other contexts in vitro and in vivo [22,23].
Beyond the centromere, there is increasing evidence that phase separation, driven by weak multivalent interactions, plays a role in organizing interphase chromatin [24]. A recent study demonstrated that reconstituted nucleosome arrays were capable of phase separating into dense droplets both in vitro and when injected into nuclei of cultured cells. Decreasing their spacing increased the concentration of nucleosomes within the droplets, whereas acetylation of the arrays dissolved the droplets [25]**. These results suggest that tuning multivalent interactions among neighboring nucleosomes can affect the intrinsic compaction of chromatin. It will be of great interest to test these principles in the context of mitotic chromosomes, where it was shown that a transient increase in free magnesium during mitotic onset plays a role in chromatin compaction [26].
Taken together, these recent studies suggest that regulation at the nucleosome level can drive a global increase in chromatin compaction through two related pathways: the centromere-mediated initiation and spread of mitotic PTMs along the chromosome; and the promotion of multivalent interactions. Both mechanisms involve processes that occur at the nanometer length scale but manifest at the whole-chromosome scale, potentially through a process of phase separation.
ATPases drive whole chromosome organization and resolution
Although mitotic chromatin itself adopts a compacted conformation, other factors are required to produce structured and individually resolved chromosomes. Key to organization at larger scales is the DNA-based motor condensin, of which there are two types in vertebrates, condensin I and condensin II. When the ATPase subunit shared by both condensins was rapidly depleted from cultured cells, the volume of mitotic chromatin did not change, but chromosomes were misshapen and unresolved [27]. At the molecular level, condensins are ring-shaped protein complexes that change the topology of DNA by creating loops. This activity was directly demonstrated in single molecule experiments showing that condensin purified from yeast can create a loop on naked DNA and quickly “extrude” the DNA to form a progressively larger loop [28]**.
How are the smaller-scale DNA loops formed by condensins organized into higher order chromosome architecture? Based on Hi-C data from condensin-depleted vertebrate cells, it was proposed that condensin II forms large DNA loops during prophase, which are subsequently partitioned into smaller, nested loops by condensin I in prometaphase [2]**. The interplay between these two condensin-driven activities result in a series of loops arranged into a twisted helical structure perpendicular the central axis of a chromosome (Figure 1C). In the model proposed by the authors, axial shortening of the chromosome occurs as a result of loop extrusion by the two condensins.
Another open question is how the large-scale organization of DNA in chromosomes is directed by the spatial arrangement and temporal activity of condensins. One study addressed this question using quantitative, high resolution imaging and fluorescence correlation microscopy in single cultured cells to measure absolute numbers, spacing and dynamics of condensin molecules during mitotic progression [29]**. The authors then used direct measurements of chromosome length and published loop extrusion models [30] to derive DNA loop sizes as a function of mitotic progression, generating a comprehensive, quantitative model for condensin-driven mitotic chromosome compaction. Although many of the qualitative findings of this model are not new, its quantitative and predictive power is unprecedented and will undoubtedly aid future experimental design. Findings in other systems add further complexity. For example, in budding yeast, the binding kinetics of condensins on mitotic chromatin is not only dynamic, but also requires regulation by non-chromatin enzymes [31,32]. Additionally, a study in human cells showed the two ATPases in each condensin ring have different activities, with one promoting loop formation and the other stabilizing higher order connections between loops [33]. In the future it will be interesting to integrate these recently discovered parameters into models of loop extrusion-based mitotic chromosome formation.
In addition to condensin, the ATP-dependent DNA strand-passing enzyme topoisomerase II (topo2) is also required to resolve mitotic chromosomes from each other as they condense, which was beautifully demonstrated in a reconstitution experiment that identified core histones and their chaperones, condensin I, and topo2 as sufficient to remodel sperm chromatin into individualized chromatids [34]. Moreover, in a molecular dynamics simulation, loop extrusion by condensins and strand passing mediated by topo2 was sufficient to convert initially globular interphase chromosomes into elongated structures that resembled prophase chromosomes [35]. In vivo, however, interactions between topo2 and condensin activities appear to be more complex [36]. Acute inactivation of condensin I in Drosophila embryos in mitosis caused topo2-dependent chromosome concatenation and hypercompaction, thereby blocking the resolution and anaphase segregation of chromosomes [37]*.This result suggests that condensin is required continuously to drive decatenation rather than catenation of DNA molecules by topo2.
Taken together, these data suggest an exciting model in which the molecular-level ATP-dependent loop extrusion by condensin I and II, in cooperation with the DNA strand-passing activity by topo2, is the driving force that organizes mitotic chromosomes at a whole-chromosome scale.
BAF and Ki67 organize chromosomes at their periphery
In addition to being individually condensed and structured at a single-chromosome scale, mitotic chromosomes must also be organized relative to one another to facilitate their proper segregation. This level of organization is mediated by a set of proteins that act at the chromosome periphery. Promoting mitotic chromosome individualization is Ki67, a protein that acts as a brush-like surfactant on each mitotic chromosome, preventing coalescence of the genome into a single large mass [38]. At the end of mitosis, dimers of the barrier to autointegration (BAF) protein bind to chromatin. Containing two high affinity DNA binding sites, BAF functions as a cross-linker to connect chromosomes and promote the formation of a single nucleus [39]*. Thus, Ki67 and BAF use the simple molecular-scale activities such as DNA-binding and electrostatics to regulate the degree of whole chromosome resolution and clustering. This feature of whole chromosome organization is thought to be particularly important at the exit from mitosis, when the genome is re-compartmentalized inside the nuclear envelope and must exclude large cytoplasmic components that could not be exported through nuclear pores.
The emergence of whole chromosome mechanics and dynamics
As a result of the many mechanisms that generate higher order mitotic chromosome structure, distinct physical features emerge. One such property is the resistance to stretching forces, a parameter that can be directly measured by micromanipulating chromosomes isolated from human cells [40]. Interestingly, depleting condensin from chromosomes decreased their stiffness 10-fold, while increasing condensin levels had the opposite effect [41]. At the level of the nucleosome, increasing histone methylation, but not H3K9 acetylation, stiffened mitotic chromosomes [42]. Thus, factors that modulate organization over both shorter and longer length scales can affect physical properties of whole chromosomes.
Organization also occurs at an even higher level that determines how mitotic chromosomes are arranged within the cell. For example, a toroidal distribution of chromosomes at nuclear envelope breakdown facilitates their interaction with spindle microtubules [43]. Recently, it was observed that the two haploid sets of chromosomes are spatially segregated from one another throughout mitosis [44]. This “antipairing” behavior was observed in normal human and mouse somatic cells, but lost in a carcinoma cell line. The function of this organization is thought prevent recombination between homologues, which would result in genetic instability. It will be of great interest to discover the set of molecules that mediate this type of inter-chromosomal organization.
Emergent functions of mitotic chromosomes
The ultimate properties of condensed chromosomes including their size, shape, and stiffness facilitate their segregation by the mitotic spindle. These properties are dynamic and can be precisely tuned during a single cell division. For example, introducing an extra-long chromosome in budding yeast results in hyper-condensation of that chromosome during anaphase, a response that requires Aurora B phosphorylation of H3S10 [45]. Chromosome properties are also tuned across multiple cell divisions, for example to allow chromosome size to scale with cell size during early development, when many rapid cell divisions occurring in the absence of cell growth lead to progressively smaller cells [46]. The mechanisms tuning overall chromosome morphology, and the effects on downstream physical properties, remain poorly understood.
Other, more subtle functions are also programmed into mitotic chromosome structure and organization. A fascinating example is mitotic “bookmarking” in which regions of the genome are primed for transcription in the ensuing interphase [47,48]. Bookmarking factors can either reside within the chromatin itself, in the form of histone PTMs, or be mediated by transcription factors (TFs). A recent study in mouse ES cells showed that 100 out of 500 transcription factors tested were enriched on the mitotic chromosome relative to the cytoplasm [49]. These bookmarking TFs interact dynamically with mitotic chromatin, and in some cases, are able to preserve the local nucleosome architecture at the promoter [50]. Whether or not these bookmarking factors affect large-scale chromosome morphology, mechanics or dynamics remains to be seen.
Conclusions and future directions
In conclusion, multiple biochemical activities at the molecular level come together to mediate the large-scale chromosome organization and dynamics observed during mitosis. Despite recent advances in the field, two major challenges remain: 1) integrating the many sources of structural information (imaging, sequencing, biochemistry) into a comprehensive, multi-scale model for mitotic chromosome architecture, and 2) relating the structure and mechanics of mitotic chromosomes to their functions in cell division and epigenetic inheritance. In order to tackle these challenges, there is a great need in the field for reconstituted systems to study mitotic chromosome structure and function at the whole chromosome level. So far, Xenopus egg extracts is one of the few systems in which this is possible. A second approach that will undoubtedly push the field forward is to test our models of chromosome structure against the extremes that biology has to offer. For example, species with large genome sizes [51] or small chromosome numbers [52] could help us stretch our imaginations.
Acknowledgments
We would like to thank Hiro Funabiki, Pavan Choppakatla, Todd Stukenberg and Nathan Gamarra for their thoughtful comments on the manuscript. This work was supported in part by an R35 (GM118183) to R.H. and a Jane Coffin Childs Memorial Fund postdoctoral fellowship to C.Y.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement:
Nothing declared.
References
- 1.Mirny LA, Imakaev M, Abdennur N: Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol 2019, 58:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. : A pathway for mitotic chromosome formation. Science 2018, 5:eaao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]; **A combination of imaging, modeling and a Hi-C technique applied to cells progressing through mitosis revealed mechanisms by which DNA loop arrays are organized and remodeled to generate a helical arrangement around a central scaffold of condensins. Acute depletion of condensin I and II elucidated their different functions in loop formation and organization.
- 3.Earnshaw WC, Laemmli UK: Architecture of metaphase chromosomes and chromosome scaffolds. J. Cell Biol 1983, 96:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J: Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. U.S.A 2008, 105:19732–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risca VI, Denny SK, Straight AF, Greenleaf WJ: Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature 2017, 541:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T: Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. U.S.A 2016, 113:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC: ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357:eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai S, Chen C, Tan ZY, Huang Y, Shi J, Gan L: Cryo-ET reveals the macromolecular reorganization of S. pombe mitotic chromosomes in vivo. Proc. Natl. Acad. Sci. U.S.A 2018, 115:10977–10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shintomi K, Inoue F, Watanabe H, Ohsumi K, Ohsugi M, Hirano T: Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science 2017, 356:1284–1287. [DOI] [PubMed] [Google Scholar]; *Depletion of core histones from a cell-free reconstitution system revealed the dispensability of nucleosomes to generate higher order chromosome-like structures.
- 10.Takahashi M, Hirota T: Folding the genome into mitotic chromosomes. Curr. Opin. Cell Biol 2019, 60:19–26. [DOI] [PubMed] [Google Scholar]
- 11.Zhiteneva A, Bonfiglio JJ, Makarov A, Colby T, Vagnarelli P, Schirmer EC, Matic I, Earnshaw WC: Mitotic post-translational modifications of histones promote chromatin compaction in vitro. Open Biol 2017, 7:170076. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Core histones isolated under conditions that preserved their PTMs from interphase or mitotic cells and used to reconstitute chromatin displayed a less or greater propensity to induce chromatin aggregation, respectively.
- 12.Javasky E, Shamir I, Gandhi S, Egri S, Sandler O, Rothbart SB, Kaplan N, Jaffe JD, Goren A, Simon I: Study of mitotic chromatin supports a model of bookmarking by histone modifications and reveals nucleosome deposition patterns. Genome Res 2018, 28:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmena M, Wheelock M, Funabiki H, Earnshaw WC: The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol 2012, 13:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W, Neumann H: A cascade of histone modifications induces chromatin condensation in mitosis. Science 2014, 343:77–80. [DOI] [PubMed] [Google Scholar]
- 15.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu C-F, Nordenskiöld L: The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res 2011, 39:1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y: Centromeres License the Mitotic Condensation of Yeast Chromosome Arms. Cell 2018, 175:780–795. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Whereas chromosomes lacking a centromere failed to condense in mitosis in budding yeast, adding a centromere to a mini chromosome was sufficient to induce condensation. Shugoshin and the histone deacetylase Hst2 facilitated spreading of the condensation signal to chromosome arms.
- 17.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD: Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997, 106:348–360. [DOI] [PubMed] [Google Scholar]
- 18.Trivedi P, Palomba F, Niedzialkowska E, Digman MA, Gratton E, Stukenberg PT: The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol 2019, 21:1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The borealin subunit of the CPC was shown to phase separate the entire complex in vitro and contribute to phase separation of the CPC at the inner centromere in vivo.
- 19.Abad MA, Ruppert JG, Buzuk L, Wear M, Zou J, Webb KM, Kelly DA, Voigt P, Rappsilber J, Earnshaw WC, et al. : Borealin-nucleosome interaction secures chromosome association of the chromosomal passenger complex. J. Cell Biol 2019, 218:3912–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruppert JG, Samejima K, Platani M, Molina O, Kimura H, Jeyaprakash AA, Ohta S, Earnshaw WC: HP1α targets the chromosomal passenger complex for activation at heterochromatin before mitotic entry. EMBO J 2018, 37:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi Q, Chen Q, Liang C, Yan H, Zhang Z, Xiang X, Zhang M, Qi F, Zhou L, Wang F: HP1 links centromeric heterochromatin to centromere cohesion in mammals. EMBO Rep 2018, 19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH: Phase separation drives heterochromatin domain formation. Nature 2017, 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mir M, Bickmore W, Furlong EEM, Narlikar G: Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development 2019, 146:dev182766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK: Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179:470–484. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Histone tail-driven interactions were shown to drive phase separation of nucleosome arrays in vitro and when microinjected into nuclei. Phase separation was promoted by linker histone and inhibited by histone acetylation.
- 26.Maeshima K, Matsuda T, Shindo Y, Imamura H, Tamura S, Imai R, Kawakami S, Nagashima R, Soga T, Noji H, et al. : A Transient Rise in Free Mg2+ Ions Released from ATP-Mg Hydrolysis Contributes to Mitotic Chromosome Condensation. Curr. Biol 2018, 28:444–451. e6. [DOI] [PubMed] [Google Scholar]
- 27.Samejima K, Booth DG, Ogawa H, Paulson JR, Xie L, Watson CA, Platani M, Kanemaki MT, Earnshaw WC: Rapid degradation of condensins and 3D-EM reveal chromatin volume is uncoupled from chromosome architecture in mitosis. J. Cell. Sci 2018, doi: 10.1242/jcs.210187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C: Real-time imaging of DNA loop extrusion by condensin. Science 2018, 360:102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Single molecule experiments directly showed that condensin can act as an ATP-dependent motor that can extrude loops of tens of kilobase pairs of DNA.
- 29.Walther N, Hossain MJ, Politi AZ, Koch B, Kueblbeck M, Ødegård-Fougner Ø, Lampe M, Ellenberg J: A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J. Cell Biol 2018, 217:2309–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]; **High resolution fluorescence correlation spectroscopy of endogenously tagged condensin I and II revealed their dynamic localization during progression through mitosis, enabling a more comprehensive model of their different functions in loop formation and chromosome organization.
- 30.Goloborodko A, Marko JF, Mirny LA: Chromosome Compaction by Active Loop Extrusion. Biophys. J 2016, 110:2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thattikota Y, Tollis S, Palou R, Vinet J, Tyers M, D’Amours D: Cdc48/VCP Promotes Chromosome Morphogenesis by Releasing Condensin from Self-Entrapment in Chromatin. Mol. Cell 2018, 69:664–676. e5. [DOI] [PubMed] [Google Scholar]
- 32.Thadani R, Kamenz J, Heeger S, Muñoz S, Uhlmann F: Cell-Cycle Regulation of Dynamic Chromosome Association of the Condensin Complex. Cell Rep 2018. 23:2308–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elbatsh AMO, Kim E, Eeftens JM, Raaijmakers JA, van der Weide RH, García-Nieto A, Bravo S, Ganji M, Uit de Bos J, Teunissen H, et al. : Distinct Roles for Condensin’s Two ATPase Sites in Chromosome Condensation. Mol. Cell 2019. 76:724–737. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Acute inactivation of condensin I in Drosophila embryos caused disassembly of centromeric regions but hypercompaction of chromosome arms due to catenation by topoisomerase II.
- 34.Shintomi K, Takahashi TS, Hirano T: Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol 2015, 17:1014–1023. [DOI] [PubMed] [Google Scholar]
- 35.Goloborodko A, Imakaev MV, Marko JF, Mirny L: Compaction and segregation of sister chromatids via active loop extrusion. Elife 2016, 5:11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter J, Aragón L: A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet 2012, 28:110–117. [DOI] [PubMed] [Google Scholar]
- 37.Piskadlo E, Tavares A, Oliveira RA: Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. Elife 2017, 6:1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuylen S, Blaukopf C, Politi AZ, Müller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW: Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016, 535:308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW: DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170:956–972. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]; *The BAF protein was shown to promote the formation of a single nucleus by cross-linking DNA during nuclear assembly at the end of mitosis.
- 40.Marko JF: Micromechanical studies of mitotic chromosomes. Chromosome Res 2008, 16:469–497. [DOI] [PubMed] [Google Scholar]
- 41.Sun M, Biggs R, Hornick J, Marko JF: Condensin controls mitotic chromosome stiffness and stability without forming a structurally contiguous scaffold. Chromosome Res 2018, 26:277–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biggs R, Liu PZ, Stephens AD, Marko JF: Effects of altering histone posttranslational modifications on mitotic chromosome structure and mechanics. Mol. Biol. Cell 2019, 30:820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magidson V, O’Connell CB, Lončarek J, Paul R, Mogilner A, Khodjakov A: The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 2011, 146:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua LL, Mikawa T: Mitotic antipairing of homologous and sex chromosomes via spatial restriction of two haploid sets. Proc. Natl. Acad. Sci. U.S.A 2018, 115:E12235–E12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neurohr G, Naegeli A, Titos I, Theler D, Greber B, Díez J, Gabaldón T, Mendoza M, Barral Y: A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science 2011, 332:465–468. [DOI] [PubMed] [Google Scholar]
- 46.Heald R, Gibeaux R: Subcellular scaling: does size matter for cell division? Curr. Opin. Cell Biol 2018, 52:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palozola KC, Lerner J, Zaret KS: A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol 2019, 20:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behera V, Stonestrom AJ, Hamagami N, Hsiung CC, Keller CA, Giardine B, Sidoli S, Yuan Z-F, Bhanu NV, Werner MT, et al. : Interrogating Histone Acetylation and BRD4 as Mitotic Bookmarks of Transcription. Cell Rep 2019, 27:400–415. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raccaud M, Friman ET, Alber AB, Agarwal H, Deluz C, Kuhn T, Gebhardt JCM, Suter DM: Mitotic chromosome binding predicts transcription factor properties in interphase. Nat Commun 2019, 10:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Festuccia N, Owens N, Papadopoulou T, Gonzalez I, Tachtsidi A, Vandoermel-Pournin S, Gallego E, Gutierrez N, Dubois A, Cohen-Tannoudji M, et al. : Transcription factor activity and nucleosome organization in mitosis. Genome Res 2019, 29:250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuznetsova MA, Chaban IA, Sheval EV: Visualization of chromosome condensation in plants with large chromosomes. BMC Plant Biol 2017, 17:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drpic D, Almeida AC, Aguiar P, Renda F, Damas J, Lewin HA, Larkin DM, Khodjakov A, Maiato H: Chromosome Segregation Is Biased by Kinetochore Size. Curr. Biol 2018, 28:1344–1356. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]


