Abstract
Purpose of Review
This review highlights the history, recent advances, and ongoing challenges of artificial intelligence (AI) technology in colonic polyp detection.
Recent findings
Hand-crafted AI algorithms have recently given way to convolutional neural networks with the ability to detect polyps in real-time. The first randomized-controlled trial comparing an AI system to standard colonoscopy found a 9% increase in adenoma detection rate, but the improvement was restricted to polyps smaller than 10 mm and the results need validation. As this field rapidly evolves, important issues to consider include standardization of outcomes, dataset availability, real-world applications, and regulatory approval.
Summary
AI has shown great potential for improving colonic polyp detection while requiring minimal training for endoscopists. The question of when AI will enter endoscopic practice depends on whether the technology can be integrated into existing hardware and an assessment of its added value for patient care.
Keywords: Artificial intelligence, machine learning, convolutional neural network, computer-aided detection, colonic neoplasm
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer death worldwide.[1] The decline in CRC mortality has been partly attributed to the increasing use of screening colonoscopy, [2, 3] but colonoscopy remains an imperfect test. Tandem colonoscopy studies show high polyp miss rates of up to 27% for diminutive polyps.[4, 5] The most widely accepted quality metric for screening colonoscopy is the adenoma detection rate (ADR), which is defined as the percentage of colonoscopies performed by an endoscopist in which at least one adenoma is detected. In a patient population with both women and men, the US Multi-Society Task Force on Colorectal Cancer recommends a target ADR ≥25%.[6] ADR is inversely correlated with the risk of interval cancer, such that every 1% increase in ADR is associated with a 3% decrease in the risk of CRC and 5% decrease in the risk of fatal CRC.[7]
Many technologies have been developed with the goal of increasing ADR. Both optical and mechanical approaches have shown promise in polyp detection.[8] Many of these devices share common shortcomings, such as the need to purchase specialized hardware or an initial learning curve required to use the device effectively.[9–11] On the other hand, high definition resolution—which is built into modern endoscopes and does not require additional equipment or training—has also been shown to improve ADR.[12, 13]
Artificial intelligence (AI) is a rapidly growing field with active investigation in many areas of medicine.[14, 15] Within gastroenterology, there has been keen interest in using AI as an adjunctive detection method in endoscopy. AI offers the promise of increasing polyp detection and even optical polyp diagnosis, all with minimal training on the part of the endoscopist. Although current clinical applications involve additional hardware, it is conceivable that future iterations of AI could be integrated directly into endoscope processors. In this review, we describe the history of AI in colonic polyp detection, recent research, and challenges ahead.
Hand-crafted Algorithms
Machine learning research for polyp detection began in the early 2000’s. The research was driven primarily by computer scientists and engineers using polyps as a basis for developing computer vision algorithms. Algorithms were “hand-crafted” to detect polyps based on certain features chosen by the designers. Commonly extracted features included color, shape, or textural information. While these algorithms typically showed high accuracy on carefully chosen datasets, they were limited in real-world applicability either due to slow processing time or variability in video quality. Additionally, because they were explicitly designed to detect certain features, lesions that did not have a typical appearance may not have been recognized, and non-polyp lesions that share characteristics with a polyp (e.g., stool or “suction polyp”) may result in a false positive result. Some notable work during this early period are summarized below and in Table 1.
Table 1.
Summary of hand-crafted algorithm studies.
| STUDY | ALGORITHM TYPE | POLYP FEATURES EXTRACTED | STUDY DESIGN | IMAGE TYPE | TRAINING SET | TESTING SET | PROCESSING TIME | OUTCOMES |
|---|---|---|---|---|---|---|---|---|
| KARKANIS 2003[46] | Hand-crafted | Texture | Ex-vivo | Video | 180 still images | 60 videos, 5–10 s each | 1.5 min per video | Sensitivity 94%, Specificity 99% |
| HWANG 2007[47] | Hand-crafted | Shape | Ex-vivo | Video | Not reported | 8621 frames | Not reported | Per-polyp sensitivity 96% |
| PARK 2012[48] | Hand-crafted | Combined spatial and temporal | Ex-vivo | Video | 364 video segments (300–1000 frames each) | 35 videos with >1 million frames | Not reported | AUROC 0.89 |
| BERNAL 2015[49] | Hand-crafted | Shape | Ex-vivo | Video | Not reported | CVC-ClinicDB - 612 frames | 10.5 s per frame | Accuracy (PPV) 70% |
| TAJBAKHSH 2015[50] | Hand-crafted | Context-shape | Ex-vivo | Video | Not reported | 19400 frames (proprietary), 300 frames in CVC-ColonDB | 2.6 s per frame | 48% sensitivity on proprietary database, 88% sensitivity in CVC-ColonDB |
| WANG 2015[20] | Hand-crafted | Texture | Ex-vivo | Video | 8 videos | 53 videos | 0.1 s per frame | Per-polyp sensitivity 97.7% |
| GEETHA 2016[51] | Hand-crafted | Combined color-texture | Ex-vivo | Still | Not reported | 703 frames | Not reported | Sensitivity 95%, Specificity 97% |
| ANGERMANN 2017[19] | Hand-crafted | Combined texture and spatio-temporal coherence | Ex-vivo | Video | Not reported | CVC-ClinicVideoDB - 18 videos with 10,924 frames | 20–185 ms with 0.3–1.8 s detection delay | 100% per-polyp sensitivity, 50% PPV |
Abbreviations: AUROC, Area Under the Receiver Operating Characteristic; PPV, positive predictive value
Karkanis et al. developed an algorithm using color wavelet covariance (CWC) to detect textural changes that could indicate adenomatous tissue.[16] The algorithm was developed and tested on 60 videos of 5–10 s each that contained polyps. From this dataset, a training set of 180 representative still images was chosen by an expert panel and a non-overlapping random set of 1200 images was chosen for testing. The algorithm performed well, with 93.6% sensitivity and 99.3% specificity for polyps that were confirmed to be adenomatous or hyperplastic on histology. The main limitation of this algorithm was the long processing time, which precluded real-time processing even in future iterations.[17, 18]
Recognizing the limitations of studying still images, Angermann et al. attempted to adapt algorithms designed for still-frame analysis to real-time video analysis.[19] They designed the algorithm to track a given polyp between frames, thus taking advantage of the additional temporal information of the video format as compared to still pictures. Applying different permutations of their algorithm to a publicly available dataset (CVC-ClinicVideoDB), the investigators were able to achieve 100% polyp detection but with variable processing time. Although they were able to process frames in as little as 20 ms, the total delay in detection was between 300 ms and 1.8 s, which again precluded real-time analysis.
Finally, in 2015 Wang et al. developed an algorithm approaching real-time analysis at 10 frames per second using edge detection.[20] The group reported results in terms of recall, defined as detection rate on a per-polyp basis rather than a per-frame basis. The system achieved 97.7% recall on 53 videos, missing only one polyp that was rejected by the algorithm for having blurred frames. This study is significant for being one of the first to apply an algorithm in a realistic test set running at near real-time speed.
Convolutional Neural Networks
A common limitation in earlier AI research was the available hardware. The bulk of data processing is performed by commercial graphics processing units (GPU’s). Researchers often conducted initial studies on the equivalent of personal computers, with plans to scale up using more robust systems in the future. In the last 3–4 years, the advent of a new generation of modern GPU’s has ushered in a second wave of innovation in AI. Modern algorithms have begun to use convolutional neural networks (CNN) and deep learning for computer-aided detection (CAD) of polyps. These algorithms are not designed to capture specific features of a polyp. Instead, the algorithm is trained to identify polyp and non-polyp features on a large dataset without explicit input from researchers.
CNN’s are modeled after the hierarchical visual processing system in the human brain. The basic idea is that neurons in the visual cortex process an image into progressively more complex shapes. An image is initially broken down into edge boundaries based on light vs. dark interfaces, then combined into simple shapes, and finally merged into recognizable complex features in subsequent layers (Figure 1).[21] A CNN seeks to replicate this idea with multiple layers of artificial neurons. A common architecture involves several “convolutional” layers, which segment the image into small chunks that can be easily processed. The outputs of these layers feed into “pooling” layers to reduce data size and reduce noise. Sequential layers feed into a neural network that ultimately produces a probability map that describes the likelihood of the image containing a desired target. The advantage of this system is that it can be trained to find any arbitrary feature in an image without the designer describing specific features. This avoids the primary limitation of prior hand-crafted algorithms that required hard-coding specific identifying features into the software. In the case of polyps, it may be difficult for an endoscopist to describe the features of a polyp in detail, but they may “know it when they see it”. Given an annotated dataset with sufficient sample size, CNN’s can be trained to recognize polyps or any other object.
Fig. 1.
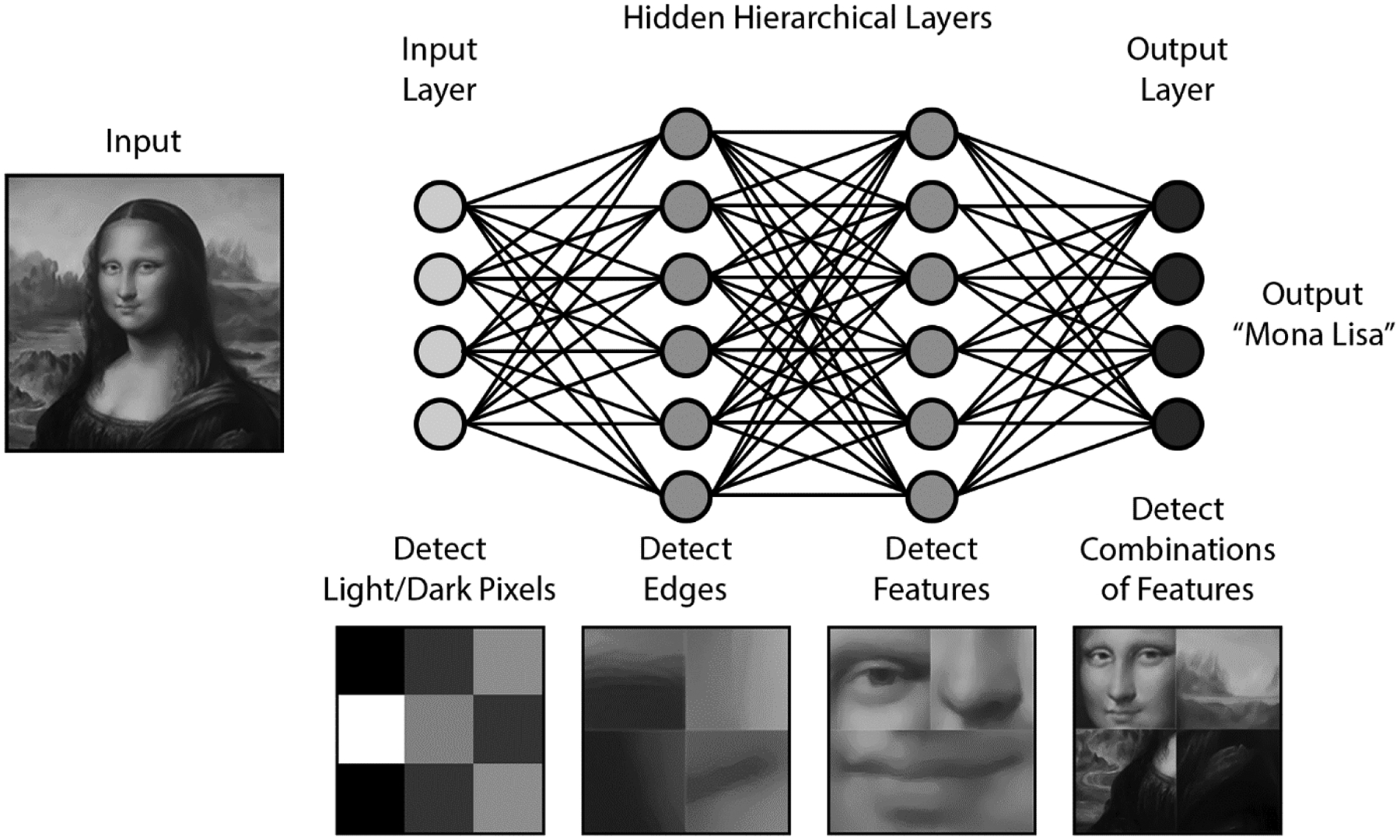
Convolutional network algorithm schematic as applied to facial recognition.
The image of the Mona Lisa is used under permission of Adobe Stock Services.
Early CNN Work
Billah et. al achieved 99% sensitivity and specificity on a public dataset by processing images using both hand-crafted features and a CNN.[22] The outputs from these algorithms were fed into a final algorithm (support vector machine) that reconciled the data to produce a single output. Although the reported sensitivity is high, the authors did not report processing time and therefore it is unclear if this method would be feasible in real-time.
Zhang et al. used a novel approach to solve the problem of limited training data.[23] They employed “transfer learning,” in which the algorithm initially trains on non-medical images before learning from polyp images. Using ImageNet, a database that includes natural images of objects such as cars, boats, and animals, the researchers increased the size of the training set to millions of images rather than the thousands typically used in polyp studies. This algorithm achieved 98% sensitivity and 1.00 area under the receiver operating characteristic (AUROC) curve. This study was also unique for comparing the performance of the algorithm to the performance of expert endoscopists. The algorithm outperformed the endoscopists in terms of accuracy (86% vs. 74%).
Real-time Analysis
Urban et al. reported one of the first algorithms applied in real-time.[24] They used ImageNet for pre-training and subsequently trained and tested the algorithm on multiple sets of colonoscopy images (see Table 2). They also tested the algorithm with a deliberately challenging set of 11 videos that showed quick withdrawal past polyps. The algorithm achieved 97% sensitivity at a specificity of 95%, with an overall accuracy of 96%.
Table 2.
Summary of convolutional neural network studies.
| STUDY | ALGORITHM TYPE | STUDY DESIGN | IMAGE TYPE | TRAINING SET | TESTING SET | PROCESSING TIME | OUTCOMES |
|---|---|---|---|---|---|---|---|
| BILLAH 2017[22] | CNN | Ex-vivo | Still | 14,000 combined training/testing | 14,000 combined training/testing | Not reported | 99% sensitivity, 99% specificity |
| YU 2017[36] | CNN | Ex-vivo | Video | ASU-Mayo 20 videos | ASU-Mayo 18 videos | 1.23 s per frame | 71% sensitivity, 88% PPV |
| MISAWA 2018[52] | CNN | Ex-vivo | Video | 411 video clips | 135 video clips | Not reported | 94% per-polyp sensitivity, 90% per-frame sensitivity with 60% false positive rate |
| PARK AND SARGENT 2016[53] | CNN | Ex-vivo | Still | 562 images | Same set as training | Not reported | 86% sensitivity, 85% specificity |
| POGORELOV 2018[54] | CNN | Ex-vivo | Still | Multiple sets from 1350 to 11954 frames | Multiple sets from 1350 to 11954 frames | Not reported | 75% sensitivity, 94% specificity |
| ZHANG 2017[23] | CNN | Ex-vivo | Still | Pretrained on ImageNet 1.2 mil and Places2015 2.5mil; trained on2262 | 150 random + 30 NBI images | Not reported | 98% sensitivity, 99% PPV, 1.00 AUROC |
| URBAN 2018[24] | CNN | Ex-vivo | Video | Pretrained on ImageNet 1.2 mil, trained on multiple sets: 8641 images, 1330 images, 9 videos, 53588 images from videos, 11 “challenging” videos | Multiple combinations of training datasets | 10 ms per frame (real-time) | 90% sensitivity with 0.5% false positive rate |
| KLARE 2019[25] | CNN | In-vivo cohort study | Live colonoscopy | Not available | 55 live colonoscopies | 50 ms latency | Per-polyp sensitivity 75%, ADR 29% (compared to endoscopist 31%) |
| WANG 2019[26] | CNN | Randomized controlled trial | Live colonoscopy | 5545 images | 1058 patients - 536 standard colonoscopy, 522 with CAD system | 25 fps with 77 ms latency | ADR increased in CAD group from 20% to 29%, polyps per procedure 0.97 vs. 0.51 |
Abbreviations: CNN, convolutional neural network; NBI, narrow-band imaging; PPV, positive predictive value; AUROC, Area Under the Receiver Operating Characteristic; ADR, adenoma detection rate; CAD, computer-aided detection
This study also demonstrated the real-world utility of CAD by showing the results of CNN-assisted video review by expert endoscopists. In 9 standard colonoscopy videos, 28 polyps were removed during the index colonoscopy. On expert review, without CNN assistance, 36 total polyps were identified. With the assistance of the CNN system, reviewers were able to identify 45 polyps, for a total of 17 polyps that were not seen on the index colonoscopy. However, none of the polyps detected with CNN assistance were larger than 10 mm. Perhaps most importantly, the algorithm ran at speeds faster than necessary for real-time analysis. Colonoscopy video runs at 25–30 fps or 33–40 ms per frame, and the algorithm was able to run at 10 ms per frame, which is more than 3 times the speed necessary for real-time analysis.
In-vivo Studies
Only two studies have applied CAD in-vivo during a live colonoscopy.
Klare et al. studied CAD prospectively on a series of 55 live colonoscopies.[25] Endoscopists in the study performed the colonoscopy as usual, while a second observer monitored the output of their polyp-detection system. The algorithm was not described in detail but used a combination of color, structure, and texture to identify polyps, with only a 50 ms delay. The group reported comparable polyp detection rate (PDR) (51% vs. 56%) and ADR (29% vs. 31%) between the CAD system and the endoscopists on a per-patient basis. However, on a per-polyp basis the system only detected 55 of 73 polyps identified on endoscopy. The system did not detect any additional polyps that were missed by the endoscopists. Therefore, the added value of this system in a real-world setting is questionable.
In 2019, Wang et al. published the only randomized clinical trial to date comparing a CAD system to standard colonoscopy for polyp detection.[26] The system is a CNN that processes data in real-time at 25 fps, with only a 77 ms delay on a secondary monitor. When a polyp is detected, a blue box appears on the screen over the area of the polyp and is accompanied by a sound cue (Figure 2).
Fig. 2.
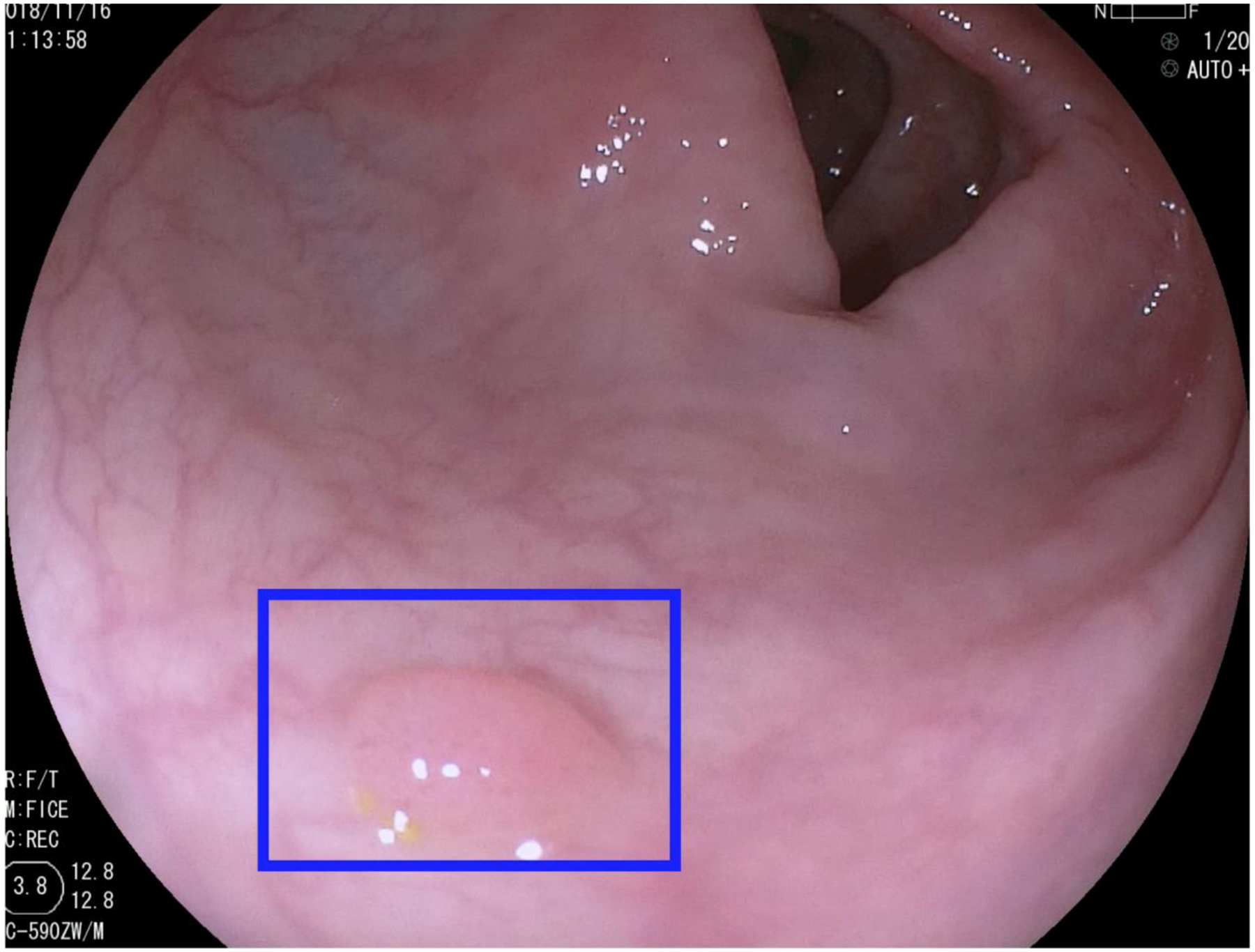
Sample output from a real-time AI system.[26] Image courtesy of Dr. Pu Wang and Dr. Tyler M. Berzin.
The trial included 1058 patients who were randomized to either standard colonoscopy or colonoscopy with a CAD system. The results showed increased ADR in the CAD group (29.1% vs.20.3%) and increased number of adenomas per patient (0.53 vs.0.31). The increased ADR was restricted to diminutive and small polyps, and there was no difference in polyps larger than 10 mm. In addition, a higher proportion of polyps detected by the CAD system were hyperplastic (43.6% vs.34.9%) and there was no difference in the proportion of advanced adenomas or sessile serrated polyps detected. There were only 39 reported non-polyp false positives in the CAD group, which is on average 0.075 false alarms per colonoscopy.
Total withdrawal time was slower in the CAD group (6.89 min vs.6.39 min), but the time difference was no longer statistically significant when time spent on removing polyps was excluded. This suggests that the additional withdrawal time was spent not on evaluating whether the system correctly identified a polyp but rather on resecting the detected polyps.
This study highlighted the full potential of AI in polyp detection, as well as its drawbacks. The system raised ADR with minimal increase in procedure time, but most of the adenomas detected were non-advanced and carry relatively low malignant potential.[27–30] The system also detected a larger number of hyperplastic polyps, which could increase procedural time and risk for unnecessary resection. Further development in real-time diagnosis of clinically relevant polyps could potentially counteract this issue. Another limitation of the study is that the ADR in the control group (20%) was lower than the quality benchmark recommended in the US.[31] This may be attributable to the fact that the study was conducted in China, where colorectal cancer incidence is lower than the US.[1] It remains to be seen whether these results will be confirmed in a Western population with a higher prevalence of adenomas.
Recent Abstracts
Within the last year, a number of conference abstracts on CAD have described some important and novel features. Many of these studies ran at real-time rates, and some used full-length, uncut colonoscopy videos. With real-world testing, these may develop into viable clinical AI systems. A summary of selected abstracts is shown in Table 3.
Table 3.
Recent abstracts on computer-aided polyp detection
| STUDY | ALGORITHM TYPE | STUDY DESIGN | STILL/VIDEO/LIVE | TRAINING SET | TESTING SET | PROCESSING TIME | OUTCOMES | NOVEL FEATURES |
|---|---|---|---|---|---|---|---|---|
| AHMAD 2019[55] | CNN | Ex-vivo | Video | 4664 frames | 24596 frames | Real-time | Sensitivity 85%, specificity 93% | Performance improved after additional training on new dataset |
| EELBODE 2019[56] | Recurrent CNN | Ex-vivo | Still | Not reported | 758 frames | Not reported | 92% sensitivity, 85% specificity | Improved performance with blue light imaging compared to white light |
| KA-LUEN LUI 2019[57] | CNN | Ex-vivo | Video | 8500 NBI images | 6 unedited videos | Real-time | Per-polyp sensitivity 100%, per-frame sensitivity 98.3%, specificity 99.7%, AUROC 0.99 | Utilizes NBI, mean polyp size was 2.6mm |
| MISAWA 2019[58] | CNN | Ex-vivo | Video | 3,017,088 frames | 64 videos | Not reported | 86% sensitivity with 26% false positive frames | Re-trained previously described algorithm on largest reported dataset, but performance was not significantly improved |
| OZAWA 2018 UEG[59] | CNN | Ex-vivo | Still | 16418 images | 7077 images | 20 ms per frame (real-time) | 92% sensitivity, 86% PPV, classification accuracy 83%, identified 97% of adenomas | Real-time detection and polyp classification combined in one system |
| OZAWA 2018 DDW[60] | CNN (same as above) | Ex-vivo | Still | 16418 images | 3533 images | Not reported (presumably real-time) | 92% sensitivity, 93% PPV, classification accurary 85% | Sensitivity and PPV were increased to 98% and 100% when only NBI images were used |
| REPICI 2019[61] | CNN | Ex-vivo | Video | Not reported | 338 video clips containing polyps | Real-time | Per-polyp sensitivity 99.7% (337/338) | Compared detection time to reaction time of endoscopists, algorithm was 1.27 s faster on average |
| SHICHIJO 2019[62] | CNN | Ex-vivo | Still | 9943 images | 1233 images | 30 ms per image | Per-polyp sensitivity 100%, per-image sensitivity 99%, 76% PPV | Dataset consisted entirely of laterally spreading tumors |
| YAMADA 2018[63] | CNN | Ex-vivo | Video | 139,983 images | 4840 images | 22 ms per image with 33 ms latency | Sensitivity 97%, specificity 99%, AUROC 0.975 | Compared to human endoscopists -had higher diagnostic yield in less time |
| ZHENG 2018[64] | CNN | Ex-vivo prospective cohort | Video | 50,000 images | 300 full colonoscopy videos | Not reported | 94.6% concordance with human endoscopist, detected 91 lesions that were not detected by endoscopists | Used algorithm to analyze full colonoscopy videos and had similar outcomes to human endoscopists -unclear outcome of additional lesions due to study design |
| ZHU 2018[65] | CNN | Ex-vivo | Still | Selection of 616 images | Selection of same 616 images | Not reported | Sensitivity 89% for SSA/P, 89% adenoma, 92% classification accuracy overall | Combined detection and classification including SSA/P and adenomas in a small curated dataset |
Abbreviations: CNN, convolutional neural network; NBI, narrow-band imaging; AUROC, Area Under the Receiver Operating Characteristic; PPV, positive predictive value; SSA/P, sessile serrated adenoma/polyp
Current Challenges
In this section, we highlight a number of challenges that remain in AI research for polyp detection.
Standardization of outcomes
Studies to date have used a wide range of study design and a number of primary outcomes. This can be partially attributed to the dual-specialty nature of the technology. Inherently, development of AI for use in colonoscopy requires the input of both engineers who design the software, as well as clinicians who can tailor it to clinical use. Earlier research frequently reported outcomes such as per-frame detection rate and recall, which are unfamiliar concepts to medical professionals. Instead, more clinically relevant outcomes include sensitivity, specificity, ADR, and polyp detection rate (PDR). These outcomes are seen more commonly in studies conducted by physicians.
In 2015, there was a notable attempt at a large-scale collaboration. The Medical Image Computing and Computer Assisted Intervention conference held an Automatic Polyp Detection challenge in which eight academic groups participated.[32] The competition tested these eight algorithms on three public databases, thus comparing the results in a standardized fashion. Although the performance of the algorithms was lackluster—the highest recall or per-polyp sensitivity was only 71%—the conference represented a significant and necessary attempt to standardize outcomes in this field.
Gastroenterologists could decide as a specialty what detection thresholds for which important outcomes (e.g., sensitivity or change in ADR) would be acceptable for use in clinical practice. For instance, the American Society for Gastrointestinal Endoscopy (ASGE) published a Preservation and Incorporation of Valuable endoscopic Innovations (PIVI) statement that recommends technologies that perform real-time polyp classification should have at least 90% negative predictive value for adenoma in order to be used in a “diagnose-and-leave” strategy.[33] While this PIVI document addressed polyp diagnosis rather than detection, similar guidance from gastroenterology societies or experts would help to focus the research on the most clinically meaningful questions and to standardize outcomes.
Training and testing datasets
The necessity of collecting a large number of images to train and test algorithms poses a logistic challenge. The ImageNet database has continued to grow, and it currently includes over 14 million annotated images of common objects such as appliances and animals.[34] By comparison, the datasets used in most polyp detection studies generally contain hundreds to thousands of images. Unfortunately, given the narrow scope of the research and the specialized knowledge required to annotate polyps, the work required to annotate millions of images is not feasible. There are several polyp datasets that are available either publicly or by request, but the largest of them is still less than 20,000 images.[35] As previously discussed, researchers have attempted to overcome this limitation by using transfer learning techniques that pre-trains an algorithm on a large generic dataset such as ImageNet prior to training the system on polyps.[23, 36] A collaborative effort to pool endoscopic images for research purposes would be another solution, and it may have the added benefit of serving as a gold standard test set for evaluating future algorithms.
Real-world application
Since the first clinical trial on this topic has already been published, all future studies should aim for real-world testing. Impressive outcomes on still images or pre-recorded videos in a lab setting may not translate to live testing. Real-time colonoscopy requires rapid processing time that may not be feasible with some algorithms, even with modern hardware. In addition, AI systems are often tested and trained on carefully curated high-quality images or pre-recorded videos. In actual practice, algorithms need to overcome the considerable challenges posed by blurry images, suboptimal bowel preparation, and variable light conditions.
Regulatory approval and reimbursement
As is the case for many areas of technology, research development in AI has far outpaced the formulation of government regulations. In the US, an executive order on AI was issued in February 2019, but this focused primarily on promoting research rather than highlighting legal considerations.[37] In contrast, the European Union General Data Protection Regulation was adopted in 2016 and stipulates that patients have a “right to explanation” of the logic behind an AI algorithm output. Some have cited the “right to explanation” as a potential barrier to AI application since the decisions made by neural networks are inherently difficult if not impossible to explain.[38] However, it is unclear if the regulation would apply in this case since the ultimate decision to act on the information remains with the endoscopist.
Current malpractice law protects physician decisions that are consistent with a reasonable standard of care. By this rule, physicians can expose themselves to liability only by following non-standard AI recommendations that may result in a negative outcome. If AI becomes standard of care in the future, situations may arise in which physicians could be penalized for appropriately rejecting questionable AI recommendations.[39]
In addition to regulatory approval and liability, insurance reimbursement may also be a future hurdle. The capital investment required to obtain this technology is unclear. Most current systems include separate hardware in addition to the standard colonoscopy tower, although in the future the software could be integrated as a standard feature in the colonoscope. Physicians will reasonably expect to be compensated for the additional investment in this new technology. Currently, most reimbursement is based on a fee-for-service model that does not reward physicians for improved outcomes. If quality-based reimbursement becomes the standard in the future, AI could be considered an added value for its potential improvement in ADR.
Future Directions
In order to adopt AI in clinical practice, more clinical trial data based on live colonoscopies will be needed. Physicians will expect consistent, validated data that shows AI increases ADR before the technology is widely accepted. Since the only trial to date was conducted in China, replication in other regions and populations will be a crucial next step.
In parallel with polyp detection, there has also been a large volume of research conducted on the use of AI for computer-aided diagnosis (CADx) or “optical biopsy.”[40–44] The goal of this technology is not only to identify but also to classify different types of polyps, such as adenomatous vs. hyperplastic. This would potentially allow for a “diagnose-and-leave” strategy in which hyperplastic polyps, which have no malignant potential, are left in place. Alternatively, for polyps determined to be adenomatous, a “resect-and-discard” strategy could help reduce pathology costs. Some current CADx systems are also capable of polyp detection, but many of the higher performance systems rely on the endoscopist to identify and capture high-quality images of polyps prior to classification.[42, 44, 45] Since a major potential pitfall of AI is time wasted on clinically non-significant polyps, a real-time combined detection and diagnosis system would provide the optimum efficiency in terms of time and cost.
Conclusions
AI technology for polyp detection remains in its infancy, but research is advancing rapidly. In a few years, the systems have progressed from still-frame analysis on carefully selected images to real-time analysis of uncut videos and even live colonoscopies. As the technology continues to mature, important questions about study design, regulatory approval, and possible integration with existing endoscopes need to be addressed. Notwithstanding these hurdles, it is more likely than not that the promise of AI-aided endoscopy in clinical practice will be realized in the near future.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest
Nicholas Hoerter declares no conflict of interest.
Peter Liang reports grants from Epigenomics, outside the submitted work.
Seth Gross reports personal fees from Olympus, outside the submitted work.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Nicholas Hoerter, NYU Langone Health, New York, NY, USA.
Seth A. Gross, Director of Clinical Care and Quality, Division of Gastroenterology, NYU Langone Health, New York, NY, USA.
Peter S. Liang, NYU Langone Health, VA New York Harbor Health Care System, New York, NY USA.
References
• Of importance
•• Of major importance
- 1.Bray F, Ferlay J, Soerjomataram I, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Cronin KA, Lake AJ, Scott S, et al. (2018) Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics: Annual Report National Cancer Statistics. Cancer 124:2785–2800. 10.1002/cncr.31551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fedewa SA, et al. (2017) Colorectal cancer statistics, 2017: Colorectal Cancer Statistics, 2017. CA Cancer J Clin 67:177–193. 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 4.Rex D, Cutler C, Lemmel G, et al. (1997) Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology 112:24–28. 10.1016/S0016-5085(97)70214-2 [DOI] [PubMed] [Google Scholar]
- 5.Heresbach D, Barrioz T, Lapalus MG, et al. (2008) Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy 40:284–290. 10.1055/s-2007-995618 [DOI] [PubMed] [Google Scholar]
- 6.Rex DK, Boland CR, Dominitz JA, et al. (2017) Colorectal Cancer Screening: Recommendations for Physicians and Patients From the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology 153:307–323. 10.1053/j.gastro.2017.05.013 [DOI] [PubMed] [Google Scholar]
- 7.Corley DA, Jensen CD, Marks AR, et al. (2014) Adenoma Detection Rate and Risk of Colorectal Cancer and Death. N Engl J Med 370:1298–1306. 10.1056/NEJMoa1309086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castaneda D, Popov VB, Verheyen E, et al. (2018) New technologies improve adenoma detection rate, adenoma miss rate, and polyp detection rate: a systematic review and meta-analysis. Gastrointest Endosc 88:209–222.e11. 10.1016/j.gie.2018.03.022 [DOI] [PubMed] [Google Scholar]
- 9.Wada Y, Fukuda M, Ohtsuka K, et al. (2018) Efficacy of Endocuff-assisted colonoscopy in the detection of colorectal polyps. Endosc Int Open 06:E425–E431. 10.1055/s-0044-101142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkinson NSS, Ket S, Bassett P, et al. (2019) Narrow-Band Imaging for Detection of Neoplasia at Colonoscopy: A Meta-analysis of Data From Individual Patients in Randomized Controlled Trials. Gastroenterology 157:462–471. 10.1053/j.gastro.2019.04.014 [DOI] [PubMed] [Google Scholar]
- 11.Hassan C, Senore C, Radaelli F, et al. (2017) Full-spectrum (FUSE) versus standard forward-viewing colonoscopy in an organised colorectal cancer screening programme. Gut 66:1949. 10.1136/gutjnl-2016-311906 [DOI] [PubMed] [Google Scholar]
- 12.Buchner AM, Shahid MW, Heckman MG, et al. (2010) High-Definition Colonoscopy Detects Colorectal Polyps at a Higher Rate Than Standard White-Light Colonoscopy. Clin Gastroenterol Hepatol 8:364–370. 10.1016/j.cgh.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 13.Subramanian V, Mannath J, Hawkey CJ, Ragunath K (2011) High definition colonoscopy vs. standard video endoscopy for the detection of colonic polyps: a meta-analysis. Endoscopy 43:499–505. 10.1055/s-0030-1256207 [DOI] [PubMed] [Google Scholar]
- 14.Esteva A, Kuprel B, Novoa RA, et al. (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thrall JH, Li X, Li Q, et al. (2018) Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J Am Coll Radiol 15:504–508. 10.1016/j.jacr.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 16.Karkanis SA, Iakovidis DK, Maroulis DE, et al. (2003) Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed 7:141–152. 10.1109/TITB.2003.813794 [DOI] [PubMed] [Google Scholar]
- 17.Maroulis DE, Iakovidis DK, Karkanis SA, Karras DA (2003) CoLD: a versatile detection system for colorectal lesions in endoscopy video-frames. Comput Methods Programs Biomed 70:151–166. 10.1016/S0169-2607(02)00007-X [DOI] [PubMed] [Google Scholar]
- 18.Iakovidis DK, Maroulis DE, Karkanis SA (2006) An intelligent system for automatic detection of gastrointestinal adenomas in video endoscopy. Comput Biol Med 36:1084–1103. 10.1016/j.compbiomed.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 19.Angermann Q, Bernal J, Sánchez-Montes C, et al. (2017) Towards real-time polyp detection in colonoscopy videos: Adapting still frame-based methodologies for video sequences analysis. In: Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures. Springer, pp 29–41 [Google Scholar]
- 20.Wang Y, Tavanapong W, Wong J, et al. (2015) Polyp-alert: Near real-time feedback during colonoscopy. Comput Methods Programs Biomed 120:164–179 [DOI] [PubMed] [Google Scholar]
- 21.Kaiser D, Haselhuhn T (2017) Facing a Regular World: How Spatial Object Structure Shapes Visual Processing. J Neurosci 37:1965. 10.1523/JNEUROSCI.3441-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Billah M, Waheed S, Rahman MM (2017) An Automatic Gastrointestinal Polyp Detection System in Video Endoscopy Using Fusion of Color Wavelet and Convolutional Neural Network Features. Int J Biomed Imaging 2017:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R, Zheng Y, Mak TWC, et al. (2017) Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features From Nonmedical Domain. IEEE J Biomed Health Inform 21:41–47. 10.1109/JBHI.2016.2635662 [DOI] [PubMed] [Google Scholar]
- • 24.Urban G, Tripathi P, Alkayali T, et al. (2018) Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 155:1069–1078.e8. 10.1053/j.gastro.2018.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]; A real-time polyp detection algorithm shown to improve polyp detection compared to expert review of colonoscopy video.
- •• 25.Klare P, Sander C, Prinzen M, et al. (2019) Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc 89:576–582.e1. 10.1016/j.gie.2018.09.042 [DOI] [PubMed] [Google Scholar]; A real-time algorithm that was tested in-vivo during live colonoscopies, its ADR was comparable to but slightly inferior to that of endoscopists.
- •• 26.Wang P, Berzin TM, Glissen Brown JR, et al. (2019) Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut gutjnl-2018–317500. 10.1136/gutjnl-2018-317500 [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the only randomized clinical trial using AI for polyp detection in live patients and found increased ADR compared to standard colonoscopy.
- 27.Atkin WS, Morson BC, Cuzick J (1992) Long-Term Risk of Colorectal Cancer after Excision of Rectosigmoid Adenomas. N Engl J Med 326:658–662. 10.1056/NEJM199203053261002 [DOI] [PubMed] [Google Scholar]
- 28.Noshirwani KC, van Stolk RU, Rybicki LA, Beck GJ (2000) Adenoma size and number are predictive of adenoma recurrence: implications for surveillance colonoscopy. Gastrointest Endosc 51:433–437. 10.1016/S0016-5107(00)70444-5 [DOI] [PubMed] [Google Scholar]
- 29.Martínez ME, Baron JA, Lieberman DA, et al. (2009) A Pooled Analysis of Advanced Colorectal Neoplasia Diagnoses After Colonoscopic Polypectomy. Gastroenterology 136:832–841. 10.1053/j.gastro.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini SD, Kim HM, Schoenfeld P (2006) Incidence of advanced adenomas at surveillance colonoscopy in patients with a personal history of colon adenomas: a meta-analysis and systematic review. Gastrointest Endosc 64:614–626. 10.1016/j.gie.2006.06.057 [DOI] [PubMed] [Google Scholar]
- 31.Rex DK, Schoenfeld PS, Cohen J, et al. (2015) Quality indicators for colonoscopy. Gastrointest Endosc 81:31–53. 10.1016/j.gie.2014.07.058 [DOI] [PubMed] [Google Scholar]
- • 32.Bernal J, Tajkbaksh N, Sánchez FJ, et al. (2017) Comparative validation of polyp detection methods in video colonoscopy: results from the MICCAI 2015 endoscopic vision challenge. IEEE Trans Med Imaging 36:1231–1249 [DOI] [PubMed] [Google Scholar]; Describes results of the first and only attempt to compare the performance of multiple algorithms directly in a standardized manner.
- • 33.Rex DK, Kahi C, O’Brien M, et al. (2011) The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc 73:419–422. 10.1016/j.gie.2011.01.023 [DOI] [PubMed] [Google Scholar]; ASGE statement establishing criteria for incorporation of polyp classification technology into practice.
- 34.Russakovsky O, Deng J, Su H, et al. (2015) ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis 115:211–252. 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 35.de Lange T, Halvorsen P, Riegler M (2018) Methodology to develop machine learning algorithms to improve performance in gastrointestinal endoscopy. World J Gastroenterol 24:5057–5062. 10.3748/wjg.v24.i45.5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L, Chen H, Dou Q, et al. (2017) Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J Biomed Health Inform 21:65–75. 10.1109/JBHI.2016.2637004 [DOI] [PubMed] [Google Scholar]
- 37.Artificial Intelligence for the American People. In: White House. https://www.whitehouse.gov/ai/. Accessed 12 Oct 2019
- 38.Edwards L, Veale M (2018) Enslaving the Algorithm: From a “Right to an Explanation” to a “Right to Better Decisions”? IEEE Secur Priv 16:46–54. 10.1109/MSP.2018.2701152 [DOI] [Google Scholar]
- 39.Price WN, Gerke S, Cohen IG (2019) Potential Liability for Physicians Using Artificial Intelligence. JAMA. 10.1001/jama.2019.15064 [DOI] [PubMed] [Google Scholar]
- 40.Chen P-J, Lin M-C, Lai M-J, et al. (2018) Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology 154:568–575 [DOI] [PubMed] [Google Scholar]
- 41.Byrne MF, Chapados N, Soudan F, et al. (2019) Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 68:94–100. 10.1136/gutjnl-2017-314547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross S, Trautwein C, Behrens A, et al. (2011) Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc 74:1354–1359. 10.1016/j.gie.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 43.Kominami Y, Yoshida S, Tanaka S, et al. (2016) Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc 83:643–649. 10.1016/j.gie.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 44.Mori Y, Kudo S, Misawa M, et al. (2018) Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med 169:357. 10.7326/M18-0249 [DOI] [PubMed] [Google Scholar]
- 45.Takemura Y, Yoshida S, Tanaka S, et al. (2010) Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest Endosc 72:1047–1051. 10.1016/j.gie.2010.07.037 [DOI] [PubMed] [Google Scholar]
- 46.Karkanis SA, Iakovidis DK, Maroulis DE, et al. (2003) Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed 7:141–152 [DOI] [PubMed] [Google Scholar]
- 47.Hwang S, Oh J, Tavanapong W, et al. (2007) Polyp detection in colonoscopy video using elliptical shape feature. In: 2007 IEEE International Conference on Image Processing. IEEE, pp II-465–II–468 [Google Scholar]
- 48.Park Sun Young, Sargent D, Spofford I, et al. (2012) A Colon Video Analysis Framework for Polyp Detection. IEEE Trans Biomed Eng 59:1408–1418. 10.1109/TBME.2012.2188397 [DOI] [PubMed] [Google Scholar]
- 49.Bernal J, Sánchez FJ, Fernández-Esparrach G, et al. (2015) WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111 [DOI] [PubMed] [Google Scholar]
- 50.Tajbakhsh N, Gurudu SR, Liang J (2015) Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imaging 35:630–644 [DOI] [PubMed] [Google Scholar]
- 51.Geetha K, Rajan C (2016) Automatic Colorectal Polyp Detection in Colonoscopy Video Frames. Asian Pac J Cancer Prev APJCP 17:4869–4873. 10.22034/APJCP.2016.17.11.4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Misawa M, Kudo S, Mori Y, et al. (2018) Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 154:2027–2029.e3. 10.1053/j.gastro.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 53.Park SY, Sargent D (2016) Colonoscopic polyp detection using convolutional neural networks. In: Medical Imaging 2016: Computer-Aided Diagnosis. International Society for Optics and Photonics, p 978528 [Google Scholar]
- 54.Pogorelov K, Ostroukhova O, Jeppsson M, et al. (2018) Deep Learning and Hand-Crafted Feature Based Approaches for Polyp Detection in Medical Videos. In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS). IEEE, Karlstad, pp 381–386 [Google Scholar]
- 55.Ahmad OF, Brandao P, Sami SS, et al. (2019) Tu1991 ARTIFICIAL INTELLIGENCE FOR REAL-TIME POLYP LOCALISATION IN COLONOSCOPY WITHDRAWAL VIDEOS. Gastrointest Endosc 89:AB647. 10.1016/j.gie.2019.03.1135 [DOI] [Google Scholar]
- 56.Eelbode T, Hassan C, Demedts I, et al. (2019) Tu1959 BLI AND LCI IMPROVE POLYP DETECTION AND DELINEATION ACCURACY FOR DEEP LEARNING NETWORKS. Gastrointest Endosc 89:AB632. 10.1016/j.gie.2019.03.1103 [DOI] [Google Scholar]
- 57.Ka-Luen Lui T, Yee K, Wong K, Leung WK (2019) 1062 USE OF ARTIFICIAL INTELLIGENCE IMAGE CLASSIFER FOR REAL-TIME DETECTION OF COLONIC POLYPS. Gastrointest Endosc 89:AB135. 10.1016/j.gie.2019.04.175 [DOI] [Google Scholar]
- 58.Misawa M, Kudo S, Mori Y, et al. (2019) Tu1990 ARTIFICIAL INTELLIGENCE-ASSISTED POLYP DETECTION SYSTEM FOR COLONOSCOPY, BASED ON THE LARGEST AVAILABLE COLLECTION OF CLINICAL VIDEO DATA FOR MACHINE LEARNING. Gastrointest Endosc 89:AB646–AB647. 10.1016/j.gie.2019.03.1134 [DOI] [Google Scholar]
- 59.Ozawa T, Ishihara S, Fujishiro M, et al. (2018) Novel computer-assisted system for the detection and classification of colorectal polyps using artificial intelligence.UEG Week 2018 Oral Presentations.pdf. United European Gastroenterology Journal, Austria, p A98 [Google Scholar]
- 60.Ozawa T, Ishihara S, Fujishiro M, et al. (2018) Sa1971 AUTOMATED ENDOSCOPIC DETECTION AND CLASSIFICATION OF COLORECTAL POLYPS USING CONVOLUTIONAL NEURAL NETWORKS. Gastrointest Endosc 87:AB271. 10.1016/j.gie.2018.04.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Repici A, Dinh NN, Cherubini A, et al. (2019) Su1716 ARTIFICIAL INTELLIGENCE FOR COLORECTAL POLYP DETECTION: HIGH ACCURACY AND DETECTION ANTICIPATION WITH CB-17–08 PERFORMANCE. Gastrointest Endosc 89:AB391–AB392. 10.1016/j.gie.2019.03.589 [DOI] [Google Scholar]
- 62.Shichijo S, Aoyama K, Ozawa T, et al. (2019) Tu2003 APPLICATION OF CONVOLUTIONAL NEURAL NETWORKS COULD DETECT ALL LATERALLY SPREADING TUMOR IN COLONOSCOPIC IMAGES. Gastrointest Endosc 89:AB653. 10.1016/j.gie.2019.03.1147 [DOI] [Google Scholar]
- 63.Yamada M, Saito Y, Imaoka H, et al. (2018) Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. United European Gastroenterology Journal, Austria, p A190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Y, Mak T, Jiang Y, et al. (2018) A study comparing colorectal polyp detection rates between endoscopists and artificial intelligence-doscopist. Colorectal Disease, France, p 22 [Google Scholar]
- 65.Zhu X, Nemoto D, Wang Y, et al. (2018) Sa1923 DETECTION AND DIAGNOSIS OF SESSILE SERRATED ADENOMA/POLYPS USING CONVOLUTIONAL NEURAL NETWORK (ARTIFICIAL INTELLIGENCE). Gastrointest Endosc 87:AB251. 10.1016/j.gie.2018.04.445 [DOI] [Google Scholar]


