Abstract
Several aggregation prone RNA-binding proteins, including FUS, EWS, TAF15, hnRNP A1, hnRNP A2 and TDP-43, are mutated in neurodegenerative diseases. The nuclear-cytoplasmic distribution of these proteins is controlled by proteins in the Karyopherin family of nuclear transport factors (Kaps). Recent studies have shown that Kaps not only transport these proteins, but also inhibit their self-association/aggregation, acting as molecular chaperones. This chaperone activity is impaired for disease-causing mutants of the RNA binding proteins. Here we review physical data on the mechanisms of self-association of several disease-associated RNA binding proteins, through liquid-liquid phase separation and amyloid fiber formation. In each case we relate these data to biophysical, biochemical and cell biological data on the inhibition of self-association by Kaps. Our analyses suggest that Kaps may be effective chaperones because they contain large surfaces with diverse physical properties that enable them to engage multiple different regions of their cargo proteins, blocking self-association.
Introduction
Karyopherins transport macromolecular cargos between the nucleus and the cytoplasm through the nuclear pore complex (NPC), controlling the nuclear-cytoplasmic distribution of thousands of molecules [1–6]. Kaps bind cargos and phenylalanine-glycine (FG) motifs in NPC subunits simultaneously to move traffic across the NPC [7,8]. Kaps are divided into importins, which bring cargo into the nucleus, exportins, which take cargo out of the nucleus, and bidirectional transporters, which can move cargo both directions. Many Importins bind nuclear localization signals (NLSs) in their cargos. For example, Karyopherin-β2 (Kapβ2/Transportin-1) binds proline-tyrosine NLSs (PY-NLSs) of many cargos [9,10]. Importin-β (Impβ/Kapβ1) binds some cargos directly, but binds many others through its complex with the adaptor protein, Importin-α (Impα/Kapα), which binds the lysine-rich classical-NLS (cNLS) [11–13]. Several importins also bind directly to folded domains of cargos [14–17]. Analogous to importins, some exportins bind linear motifs in their cargos and others bind to folded domains [2,6]. Bidirectional Kaps appear to recognize folded domains [18–20]. The mechanisms of cargo recognition by Kaps have been reviewed extensively elsewhere [6,10].
Many Kaps, including Kapβ2 and Impα/β1, act to control the nuclear-cytoplasmic distribution of RNA binding proteins [11,21–23]. These cargos function in diverse processes, ranging from RNA splicing to DNA repair to transcription. Some of these cargos, including FUS, hnRNP A1 and TDP-43, are mutated in the neurodegenerative diseases amyotropic lateral sclerosis (ALS), frontotemporal lobe dementia (FTD) and multi-system proteinopathy (MSP) [24,25]. The wild type versions of these proteins are predominantly nuclear in healthy cells. In response to stresses, they translocate to the cytoplasm and enter stress granules (SGs), biomolecular condensates that form through self-assembly of stalled translation pre-initiation complexes and associated factors [23,26,27]. Disease-causing variants of the proteins constitutively mislocalize to the cytoplasm, either because of impaired NLSs or physical changes that increase self-association, with concomitant localization in aberrant SGs [27–31]. Kaps co-localize with these aberrant stress granules in patient tissues, animal models and transfected cells [32]. There is therefore substantial interest in understanding the physical mechanisms that drive RNA binding proteins to self-associate, and whether and how Kaps modulate self-association and cellular localization. Here we review recent biophysical, biochemical, cell biological and organismic data that have revealed molecular mechanisms underlying these processes, and their regulation by Kaps, and have suggested potential new strategies for treatment of neurodegenerative diseases.
FET-Kapβ2 binding
The three FET proteins, FUS, EWS and TAF15 (or TAFII68), are ubiquitously expressed in human tissues, mostly localized to the nucleus, bind nucleic acids and function in RNA processing, transcription, splicing, and DNA repair [33]. FUS, TAF15, and to a lesser extent EWS, also localize to SGs in response to stresses [27]. Wildtype and mutant proteins are mislocalized and aggregated in the cytoplasm of neurons in ALS and FTD patients [34–38].
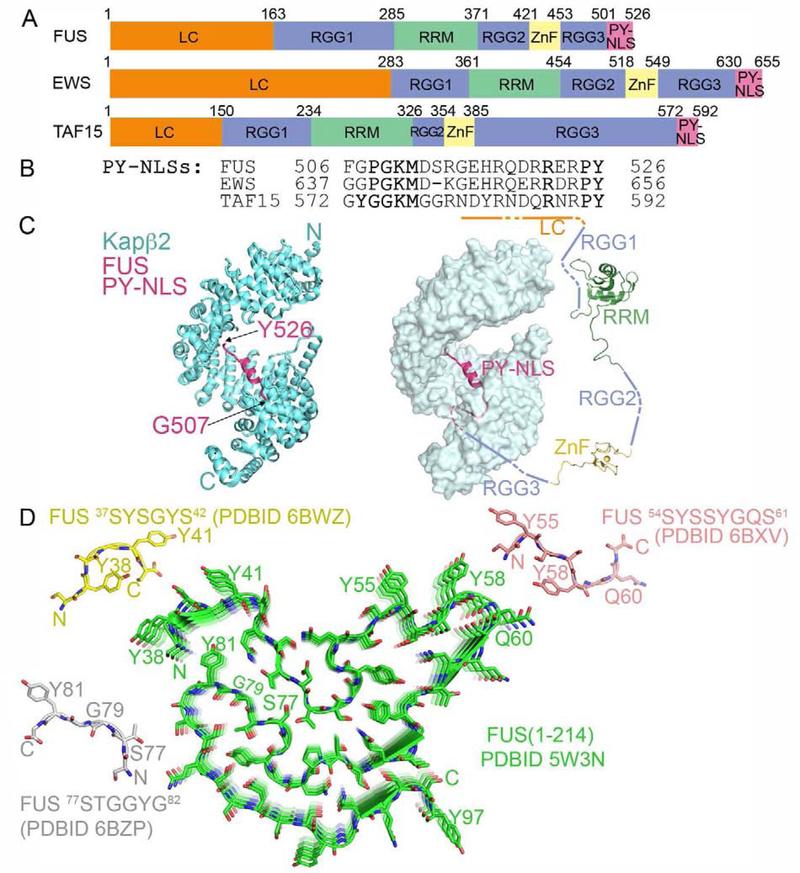
Each FET protein has an N-terminal low complexity (LC) serine-, tyrosine-, glycine- and glutamine-rich region followed by an arginine-glycine-glycine region (RGG1), an RNA recognition motif (RRM) domain, a second RGG region (RGG2), a zinc finger domain (ZnF), a third RGG region (RGG3) and a PY-NLS (Figure 1A). The LC, RGG1, RGG2, RGG3 and PY-NLS are all intrinsically disordered; the 90-residue RRM and the 30-residue ZnF are small globular domains. The LC and RGG regions self-associate, both individually and with each other, to mediate liquid-liquid phase separation (LLPS) and fibrillization [27,39–44]. RRM, ZnF and the RGGs bind RNA and the PY-NLS binds Kapβ2 for transport into the nucleus [45–49]. [9,21,50].
Figure 1. The FET proteins are nuclear import cargos of Kapβ2.
A. Domain organization. B. PY-NLS sequences with Kapβ2-binding epitopes underlined. C. Structure of Kapβ2 (cyan) bound to FUS PY-NLS (magenta; 4FDD). Left, cartoon representation of Kapβ2. Right, surface representation with a schematic of FUS domains. RRM (2LCW) and ZnF (6G99) domains. D. Protomers of short FUS LC segments (crystal structures of 37SYSGYS42 (yellow), 54SYSSYGQS61 (pink) and 77STGGYG81 (grey)) and solid-state NMR structure of FUS LC (residues 1–214; green). All four segments form cross-β structures.
All FET PY-NLSs are similar in sequence (Figure 1B) and structures of the FUS PY-NLS (residues 507–526) bound to HEAT repeats 9–17 of Kapβ2 are likely representative of the family (Figure 1C) [50,51]. The PY-NLS consists of a structurally conserved N-terminal hydrophobic motif (508PGKM511), a central arginine-rich α-helix and a C-terminal PY motif, all of which make numerous interactions with Kapβ2, affording high affinity (KD ~ 10–70 nM) [44,50].
ALS mutations are frequently found in the FUS LC and PY-NLS, and slightly less often in the RGGs [23,31,34,52–54]. Mutated sites in the PY-NLS contact Kapβ2 in the cargo complex, explaining why the alterations weaken Kapβ2 binding, leading to aberrant localization and aggregation (see below) [32,42,50]. In EWS and TAF15, ALS mutations are located primarily in the LC and RGGs [55,56], although disease mutations are occasionally found in the PY-NLS of EWS [55].
Self-association of the FET proteins
Upon concentration at low temperature, the FUS LC region forms hydrogels, which contain cross-β fibrils [41]. The solid-state NMR structure of the FUS LC fibrils showed a 57-residue core, which stacks through cross-β interactions (Figure 1D) [57]. Crystal structures of short, 6–8 residue FUS LC peptides show bent coils termed LARKS (low-complexity aromatic-rich kinked segments) stacking into cross-β structures that make non-hydrophobic and water-mediated contacts with neighbors. These structures are distinct from the extremely stable steric zippers of amyloid fibrils, which are canonical β-strands with hydrophobic inter-strand contacts and water-less interfaces [58–61]. These differences were proposed to explain the extreme stability of steric zippers as compared to the sensitivity of LARKS-based hydrogels to changes in temperature and solution conditions. The conformations of the FUS LARKS peptides are different from the same segments in the larger FUS LC core, possibly due to different constructs and experimental conditions (Figure 1D). It remains unknown which aspects of these structures will track to full-length FUS.
Solution NMR and Raman spectroscopy studies of dilute/monomeric and phase-separated FUS LC (residues 1–163) provide yet a different set of information [62,63]. Extensive biophysical studies found no evidence of stable secondary structural elements, and all major residue types seem to participate in transient intra- and intermolecular FUS-FUS contacts [63]. Averaged over time, individual residues appear to contact many residues across the protein to produce a conformationally heterogeneous, dynamically rearranging ensemble of molecules in the condensed phase. Distributed interactions are consistent with observations that all groups of five sequential tyrosine residues tiled across the sequence contribute similarly to phase separation of a FUS LC fusion with a tandem array of SH3 domains [64]. The different constructs, buffer conditions and temperatures used by various labs likely explain the differences in observed FUS LC behavior [57,60–63]. Further research is necessary to determine which of these behaviors are relevant to full-length FUS, where both LC and RGG contribute to self-association.
FUS RGG2 and RGG3 can also phase separate weakly on their own, but greatly promote phase separation of the LC region [42–44,65]. These effects do not require the PY-NLS [44]. Cation-π arginine-tyrosine interactions and the distributed charge in the arginine guanidinium group are important to LLPS [44]. Thus, in full length FUS, LC-RGG, LC-LC and RGG-RGG interactions contribute to self-association and phase separation. Lessons learned from FUS likely apply to the similarly organized EWS and TAF15.
Kapβ2 inhibits self-association of the FET proteins
Stoichiometric Kapβ2 inhibits LLPS, hydrogel formation or fibrilization of purified FUS [32,42–44]. Kapβ2 also solubilizes phase-separated FUS and preformed hydrogels or solid aggregates that contain fibrils. Kapβ2 has similar effects on EWS and TAF15, all dependent on tight binding to the PY-NLSs [32]. Interference with Kapβ2-PY-NLS interactions by a peptide inhibitor, RanGTP or ALS mutations in the FUS PY-NLS, all diminish the chaperone activity of Kapβ2. FUS without its PY-NLS is solubilized only with >50-fold higher concentrations of Kapβ2 [32,42,44].
Kapβ2 also inhibits FUS condensates in cells [32,42,43]. Co-expression in yeast of human Kapβ2 with FUS eliminated FUS cytoplasmic aggregates (which form since yeast Kapβ2 does not bind FUS) [32]. Kapβ2 overexpression decreased cytoplasmic FUS-positive SGs in HeLa, neural progenitor, HEK293T cells and ALS-patient derived fibroblasts [32]. High affinity binding of Kapβ2 to FUS, rather than nuclear import, is key; a mutant KapTL, which binds FUS but cannot import it, effectively suppressed FUS-positive SGs in HeLa cells [32], and Kapβ2 can suppress FUS-positive SGs even when nuclear import is blocked with wheat germ agglutinin [42]. Thus, the chaperone function of Kapβ2, rather than the ability to decrease cytoplasmic FUS, is essential to SG disruption.
In patient derived fibroblasts, SG depletion by Kapβ2 coincides with restored expression of FUS mRNA targets that had been depleted by a FUS R521H mutant [32]. In an ALS fly model, decreasing Kapβ2 expression enhanced neurodegeneration caused by FUS, FUS R521H and FUS R518K (bind Kapβ2 slightly weaker) but not FUS P525L (binds Kapβ2 10-fold weaker). Oppositely, increasing Kapβ2 expression in FUS R521H flies increased lifespan [32]. These results suggest therapeutic potential for modulating aggregation of FUS.
How Kapβ2 prevents FUS-FUS interactions
How does Kapβ2 binding to the PY-NLS inhibit FUS-FUS interactions mediated by the LC and RGG regions? It does not act simply as a large object anchored to the FUS C-terminus that blocks interactions through non-specific steric and/or electrostatic effects. Other proteins that bind FUS, such as an antibody raised against the FUS PY-NLS, HDAC1 and PRMT1, do not inhibit FUS self-association [32,42]. When the FUS PY-NLS is replaced with a cNLS or an NES, neither of the respective cognate Kaps, Impα or the 123 kDa CRM1, inhibited LLPS [44]. Dynamic light scattering analysis also showed that Kapβ2 has significant self-adhesion and therefore cannot confer repulsive tendencies to the Kapβ2-FUS complex [44]. Therefore, simply binding of Kapβ2 to FUS is insufficient to inhibit FUS-FUS self-association.
A variety of data rather suggest that Kapβ2, when anchored to the PY-NLS, acts by weakly binding a number of regions distributed across FUS that are involved in self-association. Small angle X-ray scattering (SAXS) analyses showed that FUS compacts upon binding Kapβ2, indicating global changes that extend well beyond the PY-NLS [44]. Consistent with this idea, when Kapβ2 is added to FUS lacking the PY-NLS, NMR analyses showed weak dynamic interactions with the LC, RGG, RRM, and ZnF [44]. Anchoring of FUS PY-NLS to Kapβ2 should enable these interactions to effectively occur in cis, strengthening them entropically. Together, the FUS-Kapβ2 interactions constrain the FUS chain and interrupt the myriad FUS-FUS interactions (LC-LC, LC-RGG, RGG-RGG) to inhibit and reverse FUS LLPS and fibrillization. Kapβ2 may engage all FUS sites simultaneously, or more likely dynamically samples different collections of contacts that rapidly interconvert. Further physical analyses are necessary to identify the regions of Kapb2 that bind FUS, and understand disruption of FUS self-association in atomic detail.
Kapβ2 chaperone function, methylation state and RNA-binding of FUS
FUS is normally heavily methylated on arginines in healthy cells [66,67], but FUS in neurons of FUS-FTD patients is hypomethylated and accumulates with EWS, TAF15 and Kapβ2 in the cytoplasm [36,67–70]. Arginine methylation weakens LC-RGG and RGG-RGG interactions [42]. Hypomethylated FUS forms solid-like condensates while hypermethylated FUS forms liquid-like condensates; both can be dissolved by Kapβ2 [43].
The FUS RRM and ZnF bind RNAs weakly [45,71] while the RGGs bind with nanomolar affinity [71,72]. Low concentrations of RNA can promote FUS self-association through multivalent interactions that enhance intermolecular networks [47,62]. In contrast, high concentrations of RNA disrupt FET proteins LLPS as they saturate binding sites on individual FUS molecules and prevent intermolecular networks [73]. High RNA concentrations seem important to keep the proteins soluble in the nucleus, while low RNA concentrations in the cytoplasm may cause aberrant assembly in disease. Kapβ2- and RNA-binding to FUS seem mutually exclusive, suggesting that Kapβ2 may inhibit FET self-association in the cytoplasm through blocking interactions with RNA [44]. In the nucleus, RanGTP prevents Kapβ2 from competing with FET-RNA interactions. The regulatory logic of Ran-Kapβ2 interactions and differential concentrations of RNA may act synergistically to maintain FET solubility in the cell.
Kapβ2 and the hnRNP A/B proteins
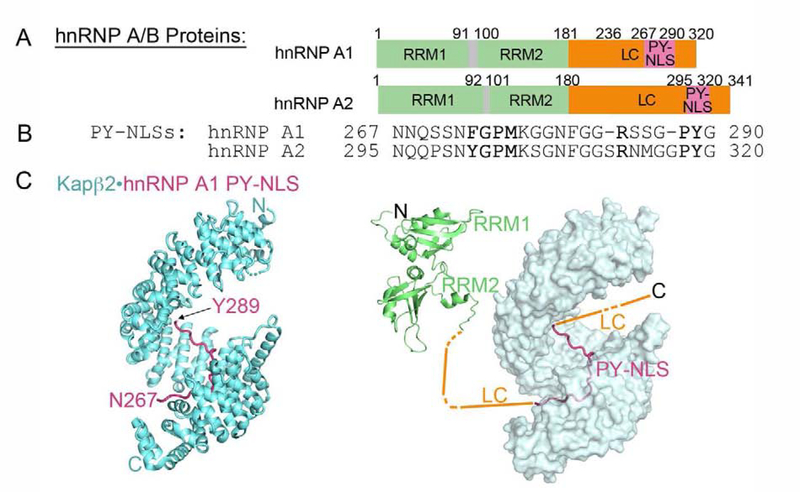
The Kapβ2 cargos, hnRNP A/B, are splicing factors that bind RNA. They localize mostly to the nucleus in healthy cells, but translocate to SGs upon stress [49,74–76]. Family members hnRNP A1 and hnRNP A2 are mutated in patients with MSP and ALS, where they aggregate into cytoplasmic inclusions, often partially co-localizing with TDP-43 [77]. Both proteins have a similar domain organization: two RRM domains followed by a LC region, which also houses the PY-NLS (Figure 2A). The hnRNP A1/A2 PY-NLSs (residues 273–289 and 301–319, respectively) bind the same site in Kapβ2 that contacts the FUS PY-NLS (Figure 2B, C) [9]. Residues immediately N-terminal to the PY-NLSs (D262V in hnRNP A1, D290V and P298L in hnRNP A2) are mutated in families with MSP, ALS and Paget’s disease [77–79]. PY-NLSs of hnRNP A1 and A2 enable both nuclear import and also chaperoning by Kapβ2.
Figure 2. The hnRNP A/B family of proteins are nuclear import cargos of Kapβ2. A, B.
Domain organization (A) and PY-NLS sequences (B) of hnRNP A1 and hnRNP A2. The Kapβ2 binding epitopes of the PY-NLSs are underlined. C. Structure of Kapβ2 (cyan) bound to the PY-NLS of hnRNP A1 (magenta); 2H4M. Kapβ2 is shown with a cartoon representation on the left, and with a surface representation on the right. A schematic of other hnRNP A1 domains are shown in the right panel. Structures of the folded tandem RRM1-RRM2 (2LYV) domains are shown.
Self-association of the hnRNP A/B proteins
hnRNP A/B proteins can phase separate and form amyloid fibers in vitro both alone and in the presence of RNA [41,49,77]. Self-association is driven largely by the LC region, but the RRM domains also likely contribute in the presence of RNA [49,78,80]. Crystal structures and fibril-diffraction guided structural models of hnRNP A1 peptides (209GFGGNDNFG217, 234GGGYGGS240 , 243GYNGFG248, 246GFGNDGSNF254, 260YNDFGNY266) showed bent coils/β-strands that stack in-register into cross-β structures [60,81]. The aspartate residues in some of these peptides stack in-register down the β-sheet, generating a column of negative charges and electrostatic repulsion that could attenuate cross-β fibril stability and afford greater dynamics. One of these aspartates, Asp 262, when mutated to valine or asparagine in disease, could decrease electrostatic repulsion and explain increased fiber formation by the mutant [81].
Solid-state NMR analyses of fibrils of the entire hnRNP A2 LC suggested a 44-residue core within residues 280–341 that forms cross-β structures [82]. The D290V disease variant produces the same cross-β structure but with more stable fibrils, likely through decreased electrostatic repulsion, as in the hnRNP A1 peptide with D262V mutation. Solution NMR analyses of a similar fragment at low concentrations unfavorable to fiber formation showed disordered random coils that sample dynamic and transient intra- and intermolecular interactions [78]. As in FUS, individual residues contact many others, with little apparent specificity or structural preference. Analogous analyses of phase-separated hnRNP A2 LC also showed structurally disordered and dynamic protein [78]. Finally, a very recent solution NMR study analyzed an hnRNP A1 LC fragment (residues 186–320) prevented from aggregation through deletion of a six-residue element (residues 259–264) containing a predicted steric zipper. This fragment does not have persistent secondary structure, but compacts and undergoes LLPS through contacts between aromatic residues distributed across the sequence [83]. The relatively uniform spacing of the aromatics appears to disfavor aggregation to produce a dynamic liquid phase.
Kapβ2 inhibits self-association of hnRNP A/B proteins
Kapβ2 inhibits hnRNPs A1 and A2 from forming hydrogels/fibrils by binding to their PY-NLSs [32]. In cells, Kapβ2 overexpression decreases the number of sodium arsenite-induced hnRNP A1-and hnRNP A2-positive SGs [32]. Expressing Kapβ2 reduced cytoplasmic aggregation of hnRNP A2(D290V), restored it to the nucleus and rescued the fly flight muscles from degeneration caused by the mutation [32]. The mechanism by which Kapβ2 disrupts hnRNP A1/A2 self-association has not been examined. The location of the PY-NLS adjacent to the fiber-forming LC sequences suggests that Kapβ2 may simply sterically block proximal LC-LC interactions. Alternatively, or in addition, Kapβ2 may interact more extensively through weak and distributed contacts, as it does with FUS.
Importinα/β and TDP-43
TDP-43 is mostly localized to the nucleus and functions in mRNA processing, stability, transport and translation [22,84,85]. TDP-43 in neuronal cells under stress also accumulates in SGs [26,86]. Wildtype TDP-43 accumulates into aberrant cytoplasmic aggregates in 90% of ALS patients, ~50% of FTD patients and in Alzheimer’s disease patients, regardless of the genetic lesion causing disease [87]. TDP-43 is also mutated in ALS and FTD [87].
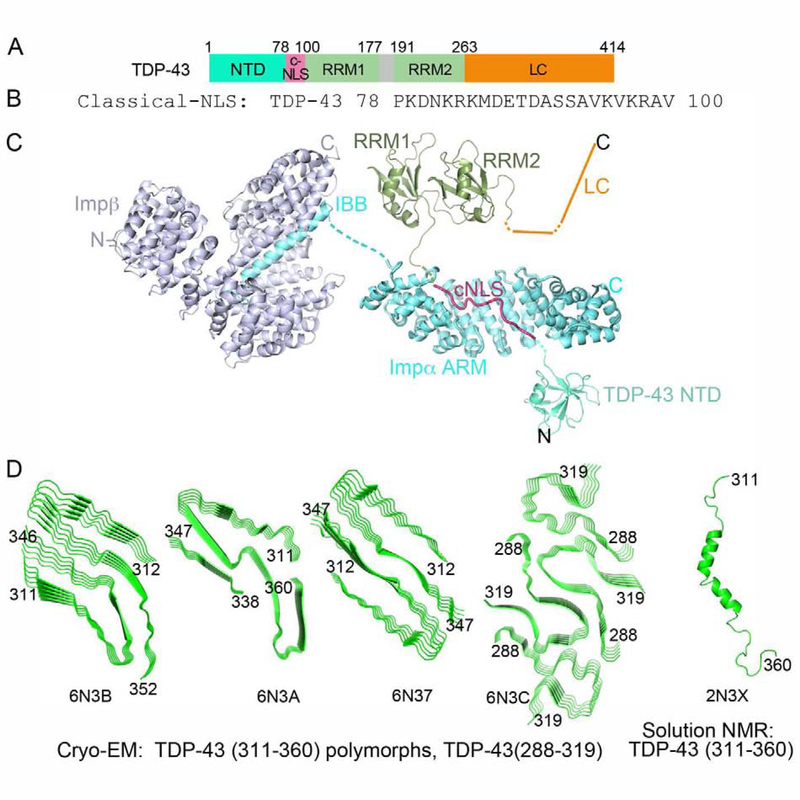
TDP-43 contains a globular Dix-like N-terminal domain (NTD) followed by two tandem RRM domains and an LC region (Figure 3A). A bipartite cNLS that binds Impα/β is located in the NTD-RRM1 linker (Figure 3B, C) [22,85]. A putative NES (residues 239–250) was previously reported, but the motif is buried in the RRM2 fold and does not bind CRM1 [85,88,89]. Instead, TDP-43 exits the nucleus via passive diffusion. The 150-residue disordered LC region has only one tyrosine residue but contains eight FG, FS or FN dipeptide motifs.
Figure 3. TDP-43 is a nuclear import cargo of the Impα/β heterodimer. A, B.
Domain organization (A) and the bipartite cNLS sequence (B) of TDP-43 (Impα-binding epitopes underlined). C. Structures of Impα (cyan) bound to the bipartite cNLS of Nucleoplasmin (magenta) (1EJY) and of Impβ (blue) bound to the IBB of Impα (cyan) (1QGK), along with the schematic of TDP-43 domains: 1) NTD (5MDI) connected to the the Nucleoplasmin cNLS (a model for the TDP-43 cNLS), followed by RRM1-RRM2 (4BS2) and a schematic of the LC domain. D. Left to right: Cryo-EM structures of TDP-43 segment spanning residues 311–360 (three polymorphic cross-β structures), residues 286–331 which self-associates into one cross-β structure, and the solution NMR structure of monomeric TDP-43 residues 311–360.
Self-association of TDP-43
Several elements of TDP-43 contribute to self-association [49,90]. Its LC region readily phase separates and many fragments within it can form fibrils [90–92]. Like other DIX domains, the NTD oligomerizes in a phosphorylation-sensitive manner, enhancing phase separation of the full-length protein probably by increasing the effective valency of the LC region [93]. RRM1-RRM2 do not self-assemble but may facilitate LLPS in the presence of RNA [90,94,95].
X-ray structures of short LC peptides show either steric zipper or LARKS fibrils, depending on the fragment examined [92]. Cryo-electron microscopy structures of fibrils generated by larger (40–50 residue) LC segments show cores that stack into cross-β arrays (Figure 3D) [96]. Several groups reported solution NMR analyses of the LC region [90,91,97]. Studies of LC segments under conditions favoring either monomer or phase separation both found helical elements flanked by random coils (Figure 3D) [90,91]. Another study of a monomeric 26-residue LC segment found a flexible ensemble with few hydrophobic contacts or β-structure [97]. CD analyses of some LC segments showed initial partial α-helical or coil structure that transformed over time into β-sheet rich aggregates [91,97].
Impα/β inhibits self-association of TDP-43
Impα/β inhibits TDP-43 fibrilization and can disassemble pre-formed fibrils [32]. Tight binding to the cNLS is essential to these effects. The consequences of these activities have not been studied in cells, and no structural or biochemical analyses of the Impα/β-TDP-43 interaction have been reported. Future mechanistic work should examine whether Impα alone can chaperone TDP-43, whether importin binding to the cNLS affects NTD oligomerization and/or RNA-binding to the RRMs, and whether importin makes contacts with other TDP-43 regions outside of the cNLS.
Karyopherins as chaperones
Impα/β and Kap121 can inhibit phase separation of FUS when its PY-NLS is replaced with the cognate ligands of the two importins, the cNLS and IK-NLS, respectively [44]. As described above, Impα/β also blocks TDP-43 fibrillization [32]. It had long been suggested that negatively charged importins could act as chaperones for abundant, positively charged cargos like histones and ribosomal proteins [98]. Structural evidence supporting this suggestion was published recently - Importin-9 wraps around the H2A-H2B histone dimer core to shield it from aggregation [15]. Collectively, these findings suggest that β-Importins may act generally to modulate assembly of aggregation-prone transport cargos. Why are Importins effective chaperones?
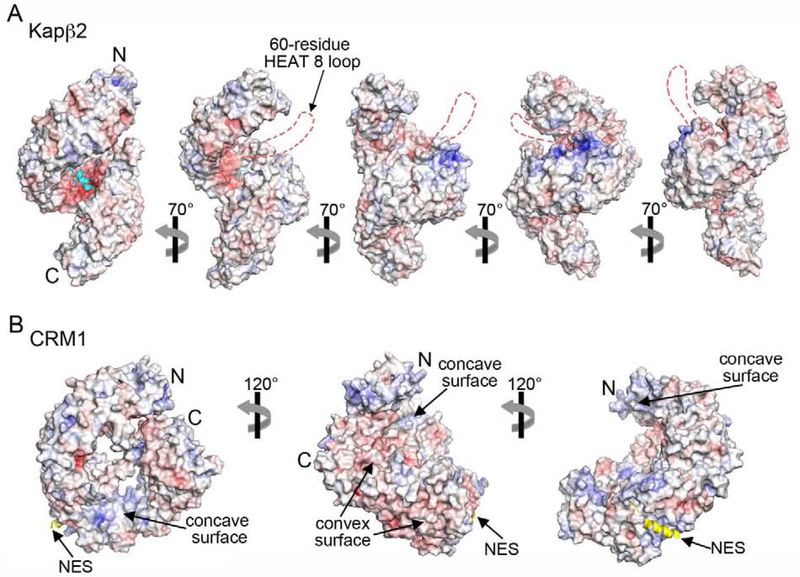
β-Importins are large proteins (~100–150 kDa) composed of 20–21 tandem HEAT repeats that form superhelical shapes (Figure 4) [6,99]. Their non-globular architectures produce large surface areas for diverse interactions with many binding partners or cargos. Importins are highly acidic, with long acidic loops and large negatively-charged surfaces on their concave sides, suitable to contact basic elements of RNA-binding proteins such as RGG regions and RRM domains, which often contribute to higher order assembly (Figure 4A). All Kaps also have many hydrophobic patches on their convex spines that bind FG repeats of nucleoporins. These same patches may be involved in analogous interactions with tyrosine or phenylalanine residues in the LC regions of RNA-binding proteins that mediate self-association. Finally, importins typically bind the NLS sequences of their cargos with nanomolar affinity, enhancing other interactions with the HEAT repeat surfaces that would be weak if they occurred in trans. Together, these properties may enable Importins in general to prevent self-assembly of their RNA-binding cargo proteins.
Figure 4. Comparison of charge distributions for an importin versus an exportin.
A. Electrostatic surface potential of Kapβ2 (4FDD) in a series of orientations. The bound FUS PY-NLS (cyan) is drawn as cartoon, and the negatively charged disordered 60-residue loop at HEAT repeat 8 is represented with dashed red lines. B. Electrostatic surface potential of the exportin CRM1 (3GB8) in three orientations. The bound NES of cargo Snurportin-1 is shown in yellow.
Many RNA-binding proteins, including FUS, hnRNP A1 nor TDP-43, shuttle between the nucleus and the cytoplasm. All three proteins exit the nucleus via passive diffusion or with bound mRNAs; none are transported by exportins [44,88,89]. When the PY-NLS of FUS was replaced with an NES that binds CRM1, the exportin failed to inhibit LLPS of FUS [44,89]. Thus, it appears that importins but not exportins can chaperone FUS. Differences in physical characteristics between importins and exportins may explain this specificity [44]. Exportins are less negatively charged (isoelectric points 5.0–5.5) and have rather different charge distributions (Figure 4B). While the concave sides of importins have large contiguous negatively charged surfaces, the analogous surfaces of exportins have basic patches. The spatial relationships between NES- and FG-binding sites in exportins are also quite different from those between NLS- and FG-binding sites in importins [100–102]. NLSs bind acidic patches on concave surfaces of importins while NESs bind to a hydrophobic groove on the convex surface of CRM1. These differences may account for the different chaperone activities of importins and exportins.
Future perspectives
Many questions remain about the mechanism of the importins’ chaperone function. How general is the mechanism of Kapβ2 chaperoning of Fus, which occurs through distributed, dynamic contacts with many regions of the cargo protein? Is this mode of action used by other importin-RNA-binding protein pairs? How else could importins prevent and reverse protein aggregation? Are importins other than Kapβ2, Impα/β and Importin-9 capable of chaperoning and what proteins do they act on? Do exportins also have chaperone functions, perhaps in the nucleus where they bind protein cargos in the presence of RanGTP? Finally, given the ability of Kapβ2 to abolish cytoplasmic aggregates of FUS, hnRNP A1 and hnRNP A2 in cells and rescue degeneration in fly disease models, could importins be therapeutic targets for neurodegenerative diseases?
Acknowledgement
This work was funded by NIGMS of NIH under Awards R01GM069909 (Y.M.C.), R01GM56322 (M.K.R.) and T32 GM131963 (CS), the Welch Foundation Grants I-1532 (Y.M.C.) and I-1544 (M.K.R.) and the University of Texas Southwestern Endowed Scholars Program (Y.M.C.). Work in M.K.R’s lab was supported by the Howard Hughes Medical Institute.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chook YM, Blobel G: Karyopherins and nuclear import. Curr Opin Struct Biol 2001, 11:703–715. [DOI] [PubMed] [Google Scholar]
- 2.Cook A, Bono F, Jinek M, Conti E: Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 2007, 76:647–671. [DOI] [PubMed] [Google Scholar]
- 3.Gorlich D, Kutay U: Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 1999, 15:607–660. [DOI] [PubMed] [Google Scholar]
- 4.Twyffels L, Gueydan C, Kruys V: Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett 2014, 588:1857–1868. [DOI] [PubMed] [Google Scholar]
- 5.Weis K: Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 2003, 112:441–451. [DOI] [PubMed] [Google Scholar]
- 6.Xu D, Farmer A, Chook YM: Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol 2010, 20:782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D: The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, et al. : Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 2015, 163:734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM: Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soniat M, Chook YM: Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J 2015, 468:353–362. [DOI] [PubMed] [Google Scholar]
- 11.Chook YM, Suel KE: Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta 2011, 1813:1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalderon D, Richardson WD, Markham AF, Smith AE: Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984, 311:33–38. [DOI] [PubMed] [Google Scholar]
- 13.Dingwall C, Sharnick SV, Laskey RA: A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1982, 30:449–458. [DOI] [PubMed] [Google Scholar]
- 14.Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, Chook YM, Borden KL: Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci U S A 2016, 113:5263–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padavannil A, Sarkar P, Kim SJ, Cagatay T, Jiou J, Brautigam CA, Tomchick DR, Sali A, D’Arcy S, Chook YM: Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An S, Yoon J, Kim H, Song JJ, Cho US: Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon J, Kim SJ, An S, Cho S, Leitner A, Jung T, Aebersold R, Hebert H, Cho US, Song JJ: Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding. J Mol Biol 2018, 430:822–841. [DOI] [PubMed] [Google Scholar]
- 18.Aksu M, Trakhanov S, Gorlich D: Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat Commun 2016, 7:11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aksu M, Trakhanov S, Vera Rodriguez A, Görlich D: Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122. The Journal of Cell Biology 2019, 218:1839–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunwald M, Bono F: Structure of Importin13-Ubc9 complex: nuclear import and release of a key regulator of sumoylation. Embo j 2011, 30:427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suel KE, Gu H, Chook YM: Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol 2008, 6:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE: Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 2008, 121:3778–3785. [DOI] [PubMed] [Google Scholar]
- 23.Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. : ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 2010, 29:2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison AF, Shorter J: RNA-binding proteins with prion-like domains in health and disease. Biochem J 2017, 474:1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King OD, Gitler AD, Shorter J: The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 2012, 1462:61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li YR, King OD, Shorter J, Gitler AD: Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013, 201:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. : A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 28.Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. : ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 2015, 88:678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shorter J, Taylor JP: Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis 2013, 1:e25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S: Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci 2010, 30:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dormann D, Haass C: TDP-43 and FUS: a nuclear affair. Trends Neurosci 2011, 34:339–348. [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. : Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 2018, 173:677–692 e620.•• This study reveals that nuclear-import receptors or importins prevent and reverse phase transitions of NLS-containing RNA-binding proteins. Importin overexpression can prevent RNA-binding proteins from accumulating in stress granules, restore them to the nucleus, and rescue degeneration caused by disease-linked FUS and hnRNP A2.
- 33.Ederle H, Dormann D: TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett 2017, 591:1489–1507. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. : Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009, 323:1205–1208. [DOI] [PubMed] [Google Scholar]
- 35.Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR: FUS pathology in basophilic inclusion body disease. Acta Neuropathol 2009, 118:617–627. [DOI] [PubMed] [Google Scholar]
- 36.Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, et al. : FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain 2011, 134:2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IR: Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol 2009, 118:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, et al. : Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol 2010, 119:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RH, Jr., Lindquist S, et al. : A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol 2011, 9:e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD: Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 2011, 9:e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. : Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, et al. : Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 2018, 173:706–719 e713.•• This study shows that RGG domains drive FUS phase separation, and nuclear import receptor TNPO1 prevents RGG-driven FUS phase separation and stress granule accumulation. Arginine methylation of FUS reduces its phase separation, and loss of arginine methylation (as in FTD-FUS) increases FUS phase separation and stress granule accumulation.
- 43.Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. : FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 2018, 173:720–734 e715.•• This study shows that intermolecular hydrogen bonding and cation-pi interactions between the LC domain and arginines in the C-terminal regions of FUS contribute its phase separation. Arginine methylation affect these interactions, hence gelation, phase separation of FUS; hypomethylation of arginines in FUS (as in FTLD) increases phase separation and gelation. The study also shows that Transportin (or Karyopherinβ2) chaperones FUS in neuron terminals and reduces phase separation/gelation of methylated and hypomethylated FUS.
- 44.Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, Lin Y, Peeples WB, Saltzberg D, Soniat M, et al. : Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173:693–705 e622.•• This study shows that nuclear import receptor Karyopherinβ2 or TNPO1 inhibits FUS phase separation. Karyopherinβ2 is anchored to FUS through high affinity interactions with its PY-NLS, and NMR analysis shows that Karyopherinβ2 also makes weak and dynamic interactions across all domains of FUS including those that participate in FUS-FUS interactions that lead to phase separation. Karyopherinβ2-FUS interactions compete with FUS-FUS interactions to inhibit phase separation.
- 45.Iko Y, Kodama TS, Kasai N, Oyama T, Morita EH, Muto T, Okumura M, Fujii R, Takumi T, Tate S, et al. : Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem 2004, 279:44834–44840. [DOI] [PubMed] [Google Scholar]
- 46.Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB: RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet 2013, 22:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz JC, Wang X, Podell ER, Cech TR: RNA seeds higher-order assembly of FUS protein. Cell Rep 2013, 5:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thandapani P, O’Connor TR, Bailey TL, Richard S: Defining the RGG/RG motif. Mol Cell 2013, 50:613–623. [DOI] [PubMed] [Google Scholar]
- 49.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP: Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang ZC, Chook YM: Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc Natl Acad Sci U S A 2012, 109:12017–12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu C, Zhang J, Gao F, Yang L, Jia M, Zhu H, Gong W: FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS One 2012, 7:e47056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. : Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009, 323:1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kino Y, Washizu C, Aquilanti E, Okuno M, Kurosawa M, Yamada M, Doi H, Nukina N: Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Research 2010, 39:2781–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H: Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging 2011, 32:2323.e2327–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. : Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 2012, 21:2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. : A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 2011, 108:20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R: Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 2017, 171:615–627 e616.• Solid-state NMR analysis of the 214-residue FUS LC fibrils reveals a 57-residue fibril core that is not polymorphic and lacks hydrophobic interactions. Phosphorylation of residues in the core abolishes FUS LC phase separation and binding to FUS LC hydrogels.
- 58.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. : Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007, 447:453–457. [DOI] [PubMed] [Google Scholar]
- 59.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D: Structure of the cross-beta spine of amyloid-like fibrils. Nature 2005, 435:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS: Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science 2018, 359:698–701.• This paper reports atomic structures of five different protein segments of low complexity domains that are known to be part of membraneless assemblies/organelles. They all form similar kinked β-sheets, with aromatic residues stacking down the sheet and interacting between sheets. These structures, with relatively weak inter-sheets interactions are named LARKS for low-complexity aromatic-rich kinked segments to distinguished them from steric zippers of amyloid fibrils, which have strong interdigitating side chain interactions between sheets.
- 61.Luo F, Gui X, Zhou H, Gu J, Li Y, Liu X, Zhao M, Li D, Li X, Liu C: Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat Struct Mol Biol 2018, 25:341–346.• This paper reports microED and X-ray crystallography structures of two tandem (S/G)Y(S/G) motifs in the FUS LC named reversible amyloid cores (RACs) that form reversible fibrils. The RACs form an ordered-coil fibril spine and a labile fibril spine with wet interface, respectively.
- 62.Burke KA, Janke AM, Rhine CL, Fawzi NL: Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 2015, 60:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL: Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol 2019, 26:637–648.• This paper studies interactions within the FUS LC that stabilize its condensed phase-separated state using NMR and Raman spectroscopy, mutagenesis and molecular simulation. The studies show conformational heterogeneity of FUS with no evidence of formation of secondary structures. Hydrogen bond, π/sp2 and hydrophobic interactions especially involving tyrosine and glutamine residues stabilize FUS-FUS interactions in the phase-separated liquid state.
- 64.Lin Y, Currie SL, Rosen MK: Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem 2017, 292:19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. : A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174:688–699 e616.• This study uses mutagenesis to understand phase separation of proteins in the FUS family. Phase separation is driven by mutivalent interactions between tyrosine residues in the N-terminal LC domain and arginine residues in the C-terminal RNA-binding RGG domains. Negatively charged residues, glycine, glutamine and serine residues affect phase separation.
- 66.Du K, Arai S, Kawamura T, Matsushita A, Kurokawa R: TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem Biophys Res Commun 2011, 404:991–996. [DOI] [PubMed] [Google Scholar]
- 67.Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, et al. : Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 2012, 31:4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neumann M, Valori CF, Ansorge O, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ishihara K, Ang LC, Bilbao JM, et al. : Transportin 1 accumulates specifically with FET proteins but no other transportin cargos in FTLD-FUS and is absent in FUS inclusions in ALS with FUS mutations. Acta Neuropathol 2012, 124:705–716. [DOI] [PubMed] [Google Scholar]
- 69.Brelstaff J, Lashley T, Holton JL, Lees AJ, Rossor MN, Bandopadhyay R, Revesz T: Transportin1: a marker of FTLD-FUS. Acta Neuropathol 2011, 122:591–600. [DOI] [PubMed] [Google Scholar]
- 70.Troakes C, Hortobagyi T, Vance C, Al-Sarraj S, Rogelj B, Shaw CE: Transportin 1 colocalization with Fused in Sarcoma (FUS) inclusions is not characteristic for amyotrophic lateral sclerosis-FUS confirming disrupted nuclear import of mutant FUS and distinguishing it from frontotemporal lobar degeneration with FUS inclusions. Neuropathol Appl Neurobiol 2013, 39:553–561. [DOI] [PubMed] [Google Scholar]
- 71.Ozdilek BA, Thompson VF, Ahmed NS, White CI, Batey RT, Schwartz JC: Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res 2017, 45:7984–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahama K, Oyoshi T: Specific binding of modified RGG domain in TLS/FUS to G-quadruplex RNA: tyrosines in RGG domain recognize 2’-OH of the riboses of loops in G-quadruplex. J Am Chem Soc 2013, 135:18016–18019. [DOI] [PubMed] [Google Scholar]
- 73.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, et al. : RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360:918–921.• The paper reports that RNA regulates phase separation of prion-like RNA-binding proteins; low RNA/protein ratios promote phase separation whereas high RNA/protein ratios prevent it. The authors suggest that high concentrations of RNA in the nucleus keeps RNA-binding proteins soluble in that compartment.
- 74.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G: A novel receptor-mediated nuclear protein import pathway. Cell 1996, 86:985–994. [DOI] [PubMed] [Google Scholar]
- 75.Bonifaci N, Moroianu J, Radu A, Blobel G: Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci U S A 1997, 94:5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brumwell C, Antolik C, Carson JH, Barbarese E: Intracellular trafficking of hnRNP A2 in oligodendrocytes. Exp Cell Res 2002, 279:310–320. [DOI] [PubMed] [Google Scholar]
- 77.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. : Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013, 495:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, Fawzi NL: Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol Cell 2018, 69:465–479 e467.• Characterization of the structure and interactions of the hnRNP A2 LC by NMR spectroscopy, molecular simulation and microscopy to understand liquid-liquid phase separation and granule formation. hnRNP A2 LC is disordered as monomer and when it is phase-separated, with transient interactions distributed across the entire LC. Methylation of arginines in the RGG motifs of the hnRNP A2 LC decreases phase separation.
- 79.Qi X, Pang Q, Wang J, Zhao Z, Wang O, Xu L, Mao J, Jiang Y, Li M, Xing X, et al. : Familial Early-Onset Paget’s Disease of Bone Associated with a Novel hnRNPA2B1 Mutation. Calcif Tissue Int 2017, 101:159–169. [DOI] [PubMed] [Google Scholar]
- 80.Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL: The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 2015, 163:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, et al. : Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 2019, 10:2006.• This paper reports that hnRNPA1 forms a reversible amyloid. It also reports atomic structures of the reversible amyloid-forming cores, which revealed stacking aspartate residues. This distinct structural characteristic contributes to fibril reversibility and explains the role of aspartate mutations in familial ALS.
- 82.Murray DT, Zhou X, Kato M, Xiang S, Tycko R, McKnight SL: Structural characterization of the D290V mutation site in hnRNPA2 low-complexity-domain polymers. Proc Natl Acad Sci U S A 2018, 115:E9782–E9791.• Solid state NMR analyses of the hnRNP A2 LC fibrils, from the wild type protein and one with the disease D290V mutation, show the LC domain forming in-register cross-β structures. is part of a rigid parallel cross-β structure. Residue D290 is immobilized in the polymer, and the D290V stabilizes the polymer.
- 83.Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T: Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020, 367:694–699.• This paper uses nuclear magnetic resonance spectroscopy, small-angle x-ray scattering, and multiscale simulations to show that the valency of aromatic residues in prion-like domains determines the extent of temperature-dependent compaction of individual molecules in dilute solutions and the full binodals that quantify concentrations of PLDs within coexisting dilute and dense phases. It also shows that uniform patterning of aromatic residues promotes phase separation while inhibiting aggregation.
- 84.Geuens T, Bouhy D, Timmerman V: The hnRNP family: insights into their role in health and disease. Hum Genet 2016, 135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM: Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 2008, 283:13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C: Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J Biol Chem 2012, 287:23079–23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cook C, Zhang YJ, Xu YF, Dickson DW, Petrucelli L: TDP-43 in neurodegenerative disorders. Expert Opin Biol Ther 2008, 8:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pinarbasi ES, Cagatay T, Fung HYJ, Li YC, Chook YM, Thomas PJ: Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci Rep 2018, 8:7083.• The previously identified NES in TDP-43 does not bind XPO1 and cannot direct nuclear export. TDP-43 nuclear export of TDP-43 is not dependent upon XPO1 recognition of a putative NES, but likely mediated by passive diffusion.
- 89.Ederle H, Funk C, Abou-Ajram C, Hutten S, Funk EBE, Kehlenbach RH, Bailer SM, Dormann D: Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci Rep 2018, 8:7084.• • The predicted NESs of TDP-43 and FUS are not functional. Bot proteins exported independently of CRM1/Exportin-1, and are likely to leave the nucleus by passive diffusion. The study also shows that transcription can control localiziation of TDP-43.
- 90.Conicella AE, Zerze GH, Mittal J, Fawzi NL: ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 2016, 24:1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang LL, Che MX, Zhao J, Zhou CJ, Xie MY, Li HY, He JH, Hu HY: Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J Biol Chem 2013, 288:19614–19624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guenther EL, Cao Q, Trinh H, Lu J, Sawaya MR, Cascio D, Boyer DR, Rodriguez JA, Hughes MP, Eisenberg DS: Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat Struct Mol Biol 2018, 25:463–471.• This paper reports crystal structures of 10 different peptide segments from the LCD of TDP-43. Six segments form steric-zipper β-sheet structures characteristic of pathogenic aggregation, and four others form the more labile LARKS structures. Familial ALS mutations convert LARKS to irreversible aggregates.
- 93.Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, et al. : A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J 2018, 37.• This study shows that the TDP-43 NTD forms heat-to-tail polymers that are disruped by a phosphomimetic substitution at S48, which also decreases phase separation and RNA splicing activity. NMR analysis of a TDP-43 NTD mutant that prevents polymerization results in a structure of a head-to-tail NTD dimer.
- 94.Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE: Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 2005, 348:575–588. [DOI] [PubMed] [Google Scholar]
- 95.Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FH: Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol 2013, 20:1443–1449. [DOI] [PubMed] [Google Scholar]
- 96.Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS: Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat Struct Mol Biol 2019, 26:619–627.• This paper reports Cryo-EM structures of two segments, SegA (residues 311–360) and SegB (residues 286–331) of TDP-43 that form pathogenic cores. SegA forms three polymorphs of similar shape folds while SegB produced one structure. Energetic analysis suggests that the SegA polymorphs form irreversible fibrils while the SegB forms both reversible and irreversible fibrils.
- 97.Mompean M, Buratti E, Guarnaccia C, Brito RM, Chakrabartty A, Baralle FE, Laurents DV: “Structural characterization of the minimal segment of TDP-43 competent for aggregation”. Arch Biochem Biophys 2014, 545:53–62. [DOI] [PubMed] [Google Scholar]
- 98.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D: Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 2002, 21:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lott K, Cingolani G: The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta 2011, 1813:1578–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM: Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 2009, 458:1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fung HY, Chook YM: Atomic basis of CRM1-cargo recognition, release and inhibition. Semin Cancer Biol 2014, 27:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Port SA, Monecke T, Dickmanns A, Spillner C, Hofele R, Urlaub H, Ficner R, Kehlenbach RH: Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export. Cell Rep 2015, 13:690–702. [DOI] [PubMed] [Google Scholar]