Abstract
Background
Since February 2020, Italian hospitals registered COVID-19 (COronaVIrus Disease 19) cases more often than the rest of the Europe. During this epidemic, health authorities requested swab tests, while seeking new patient paths.
Methods
A dual laboratory approach was evaluated, consisting of patient care reports for viral RNA detection on swabs and rapid serological tests in 516 patients (192 symptomatic or paucisymptomatic and 324 asymptomatic).
Results
We found the molecular positive fraction equal to 12% (23/192) among symptomatic/paucisymptomatic (S/P) and 15.4% (50/324) in asymptomatic (As) sets. Among subsets, we observed serologically positive results, corresponding to 35% (8/23) for S/P and 38% (19/50) for As. Among molecular negative cases, we detected specific Immunoglobulin G or M (Ig G or Ig M) positivity in the S/P cohort equal to 6.6% (11/167) and 6% (15/246) in As cases. For indeterminate molecular results, we found S/P serological positivity equal to 100% (1/1) and 54% (13/24) in As patients. We found higher (p < 0.05) seropositivity in older patients (n = 8) among symptomatic and positives for viral RNA (n.23).
Conclusions
It has been observed that a dual approach of serological and molecular tests detects a higher absolute number of disease cases in a pandemic context,which could improve monitoring and health surveillance efficacy. The age-related seropositivity frequency in this study, if confirmed, could enhance the validity of serological tests, especially in older patients.In these subjects, molecular positivity accompanied by serological positivity (distinct for M and G immunoglobulins) should help determine disease status and support decisions related to patient management.
Keywords: COronaVIrus disease 19, Symptomatic/paucisymptomaticpatients, Asymptomatic patients, Serological tests, Swabs
1. Introduction
At the time of this writing in Tuscany, a central district of Italy, an outbreak of coronavirus disease 2019 (COVID-19) has been ongoing, caused by the 2019 novel coronavirus, and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; previously called 2019-nCoV). Evidence suggests rapid local spread of the virus from Northwest regions (from Lombardy and Veneto). The first Italian patient was registered at the end of February and the Italian Civil Protection Department recorded the number of infected SARS-CoV-2 patients, consisting of 69,176. In Tuscany, there were 2,699 infected people. Given the experience of recent coronavirus contacts, we learned that the health care system must consider the risk of contagion between health workers and the spread of the virus within hospital environments.
Numerous scientific publications [1], [2], [3] report studies on viral infection in hospital settings, which leads to the epidemiological problem; this is due to lower response effectiveness of the treatments for infected and non-infected patients. This problem could be resolved through hospital structure organization efficiency and valid diagnostic approaches for the suspicious infectious cases.
The Tuscany District (TD), following the Coronavirus Regional Task Force (issuedFebruary 24, 2020) provisions [4], planned diagnostic-therapeutic strategies for the SARS COV2 pandemic [5],consisting of nasopharyngeal swab execution for all individuals with medium and high-risk contacts.
In Italian areas, it is difficult to carry out swab tests forthewhole population, with the number of certified and authorized laboratories for research of viral RNA; above all, there is the procurement difficulty for the necessary resources to assist in the analytical process (reagents, instruments, and Personal Protective Equipment, PPE). As such, it is critical to secure more immediate and faster diagnostic strategies.
In the Republic of China, the first nation afflicted with Covid-19, other diagnostic tools based on the qualitative determination of IgG and IgM antibodies against SARS COV2 were used. Qualitative antibody determinations have been performed with rapid response devices and applied in hospital triage for symptomatic patients [6].
The Italian Ministry of Health has not yet recommended Ig G and Ig M antibody detection for the diagnostic routine with COVID-19. In this study, the San Donato Hospital in Arezzo, in the Southeast Tuscany Local Health Unit (USL Toscana Sud-Est), evaluated the effectiveness of the combined use of antibody tests and molecular investigation for medium and high-risk contacts in asymptomatic, paucisymptomatic,or symptomatic patients. This validated approach could help with diagnosis, for increased clinical safety management as well as to optimize the infrastructural resources for eligible patients, who must be placed in dedicated hospital sectors. This procedure, to screen suspicious infectious cases, aims to reduce intrahospital spread of COVID-19 and to properly treat all patients.
There is a great amount of evidence-based data in the literature, demonstrating the detection value of immunoglobulin, which can identify cycles of the infectious virology sector [7], [8], [9]. This study proposes to link serological information to molecular data and to resolve diagnosis difficulties in a global emergency scenario.
2. Materials and methods
2.1. Analysis samples and population
From March 17 to 21, 2020, we analyzed 516 nasopharyngeal swabs from patients in the Emergency Room (ER) and from subjects undergoing health surveillance by territorial and hospital prevention departments. All patients provided informed consent in accordance with the Declaration of Helsinki. At the triage phase, every patient has a clinical evaluation and is classified as symptomatic/paucisymptomatic in all or one basal symptom (fever, cough,shortness of breath). The analyzed population was classified by age, gender, and symptomatology, in Table 1 .
Table 1.
Population distribution by age, gender, and symptomatology.
| Symptomatic / Paucisymptomatic Set | Asymptomatic Set | Total | ||
|---|---|---|---|---|
| Age (years) | Average | 66.8 | 46.0 | 53.7 |
| Median value | 73.0 | 45.1 | 52.9 | |
| Gender count | Females | 85 | 196 | 281 |
| Males | 107 | 128 | 235 |
2.2. Molecular analysis
In these study samples, we looked for the presence of SARS COV2 viruses, using a molecular biology method approved by the World Health Organization (WHO). The molecular kit is a CE-IVD (European Conformity, In Vitro Diagnostics, Mountain View, CA, USA) device; this method (Seegene Inc., Seoul, Korea) uses a PCR design-based, multitarget detection of genetic viral regions for the envelope protein (EP), RNA-dependent RNA polymerase (RdRp) and Nucleocapsid (N) [10].
This kit also contains an exogenous reaction control (IC), added during the first step in the pre-extraction phase, to determine the use of 10 ng in each individual sample for analysis. This allows us to verify the correct conditions for reverse transcription and subsequent PCR.
The analytical positivity determination system uses following approach:samples with valid amplification for all three targets, or only two, are considered positive. Samples without any viral signal fluorescence and valid amplification (within the 40th cycle) for IC, are considered negative. The ones that do not present a fluorescence signal for viral targets and IC are defined as invalid, are instead indeterminate in the case of single amplification of one viral target with a valid IC signal. Automation Platform was used for sample processing: Nimbus Platform (Hamilton, Italy), locally distributed from Arrow Diagnostics (San Marino and Vatican City, Italy).
2.3. Serological analysis
After the swab collection, a blood sample is taken (purple cap tube, Ethylenediaminetetraacetic acid -EDTA- additive) to detect Immunoglobulins G and M (IgG and IgM) with a fast-determining kit (Acro Biotech, Inc.,Rancho Cucamonga, CA, USA). This Rapid Test consists of a membrane-based qualitative immunoassay for detection of IgG and IgM antibodies to the 2019-nCoV in whole blood, serum, or plasma. The kit has two components, IgG and IgM. Human IgG and IgM are recognized by the chromatographic method for anti-human IgG and IgM antibodies. A goat anti-mouse IgG is used for the control line system. As stated, results without a valid signal are considered invalid.All statistical analyses are assessed by GraphPad Instat by Graph Pad Software, v. 3.05,while applying One-way ANOVA, repeated measures ANOVA, the Kruskal-Wallis test, the Friedman test, and a comparison posttest Bonferroni.
3. Results
3.1. Results for the global population and a serological test for analytical performance
Among 516 analyzed nasopharyngeal swabs, we identified 413 SARS COV2 negative, 73 positive, 25 undetermined, the remaining 5 defined as invalid. Immunoglobulin serological results for these patients consisted of 83 Ig positive (17 for Ig M, 34 for Ig G, and 32 simultaneously positive for Ig M/ Ig G). Table 2 shows complete results with Ig G and Ig M frequency, distributed among different molecular types.
Table 2.
Results distribution of molecular and serological analysis.
| ImmunoglobulinResults | ||||||
|---|---|---|---|---|---|---|
| MolecularResults | Count | Ig M positive | Ig G positive | Ig M and IgG pos | Total amountseropositivity among different molecularresponses | Total Ig positive /Molecular Type |
| NEGATIVE | 413 | 14 | 8 | 4 | 26 | 26/413 (6.3%) |
| POSITIVE | 73 | 3 | 15 | 9 | 27 | 27/73 (37%) |
| INDETERMINATE | 25 | 0 | 11 | 3 | 14 | 14/25 (56%) |
| INVALID | 5 | 0 | 0 | 0 | 0 | 0/5 (0%) |
The use of the molecular test, the gold standard to detect infection, and the study of the basis data in Table 2, allow to calculate the analytical limit value for serological testing: specificity (0.94), sensitivity (0.37), positive predictive value (PPV) (0.51), and negative predictive value (NPV) (0.94).
3.2. Results among gender
Gender distribution in an analyzed cohort had 235 males and 281 females; among 73 SARS COV2 positives, there were 39 males (53%) and 34 females (47%), while negative cases were comprised of 229 females (55%) and 184 males (45%). Indeterminate cases had15 females (60%) and 10 males (40%). The gender-based incidence study does not reveal statistically significant differences (p = 0.5), Table S1 and Graph S1 in Supplementary.
3.3. Results distribution by symptomatology
The analyzed cohort has 324 asymptomatic and 192 symptomatic or paucisymptomatic patients (S/P). These two sets are stratified based on molecular and immunological results (Table 3 ).
Table 3.
Molecular and serological characteristic distribution in the symptomatology profile.S/P set is composed of 23 (12%) molecular positive tests (MOL POS), 167 (87%) molecular negatives (MOL NEG), 1 (0.5%) molecular indeterminate (MOL IND), and 1 (0.5%) molecular invalid (MOL INV). The As set is composed of 50 (15.4%) molecular positive tests (MOL POS), 246 (76%), molecular negatives (MOL NEG), 25 (7.4%), molecular indeterminates (MOL IND), and 4 (1.2%) molecular invalids (MOL INV). Both sets present a MOL POS fraction of immunologically positive responses (IG POS), in particular the S/P set is composed of 8/23 (35%) and the As set of 19/50 (38%). S/P and As MOL NEGs are constituted by a similar percentage of IG POS (6.6 and 6%, respectively);S/P MOL IND was registered as a single case that was IG POS, while in the As set, the detection percentage of IG POS was 7.4%. No IG POS was detected among MOL INV samples.
| Symptomatic / Paucisymptomatic Set (S/P set) | Asymptomatic Set (As set) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases number | IG POS | IG NEG | IG POS % | IG NEG % | Cases number | IG POS | IG NEG | IG POS % | IG NEG % | |
| MOL POS | 23 (12%) | 8 | 15 | 35% | 75% | 50 (15.4%) | 19 | 31 | 38% | 62% |
| MOL NEG | 167 (87%) | 11 | 156 | 6.6% | 93.4% | 246 (76%) | 15 | 231 | 6% | 94% |
| MOL IND | 1 (0.5%) | 1 | 0 | 100% | 0% | 24 (7.4%) | 13 | 11 | 54% | 46% |
| MOL INV | 1 (0.5%) | 0 | 1 | 0% | 100% | 4 (1.2%) | 0 | 4 | 0% | 100% |
| Total Amount | 192 | 20 | 172 | 10.4% | 89.6% | 324 | 47 | 277 | 14.5% | 85.5% |
3.4. Results as age-related
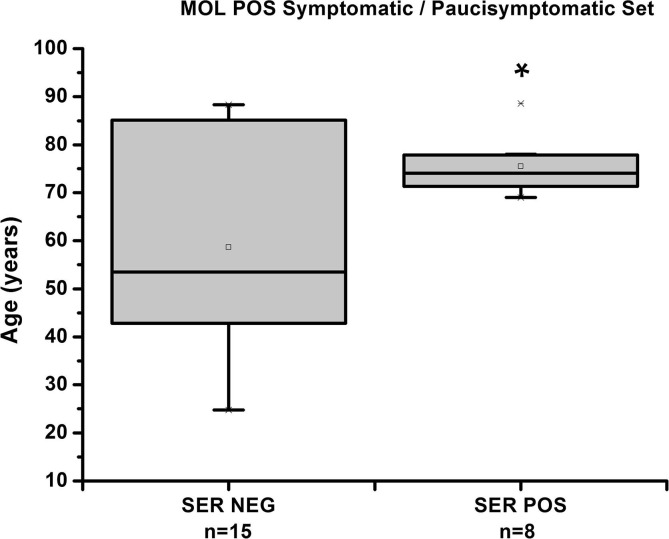
Positivity incidence for patient age shows no statistical difference among cohorts of S/P and As (Table S2 database, Supplementary) except for the subset of SER POS (n = 8) vs. SER NEG (n = 15) of the MOL POS in the P/S set. So we observe a significant difference in patient age for SER POS vs.SER NEG (p < 0.05). This last set shows an age average of 58.6 years vs. 75.5 years for the SER POS (Graph 1 , complete statistical analysis in Table S3, Supplementary).
Graph 1.
Distribution based on age of SER POS and SER NEG subsets of the MOL POS in S/P set (C.I. 95%). The line through the box indicates the median value.
4. Discussion
In Italy, there are state directives from the Ministry of Health, which do not impose immunological screening fortheCOVID-19 emergency [11]. However, there is global interest from the scientific community on a combined approach for detecting infected patients and evaluating their clinical state [12], [13]. This study aimed to evaluate the multiple approach, based on molecular analysis of swabs and serological screening for Ig G and M, in order to decrease improper diagnosis and viral spread in the general territory and the hospital structures.
The serological results distribution among molecularly screened patients (Table 2) yields an idea of how important it is to use Ig detection in this situation. A consistent fraction of MOL IND (56%) is positive for Ig (G and/or M), with this data indicating that 3/11 (~30%) are probably referred for disease onset, or the molecular indeterminate result, while the concomitant presence of Ig M suggests a relatively low viral burden,despite that virus contact occurred. The 26/413 (6.3%) serological positives among MOL NEG indicate concrete contact with the virus, so we observe more than 50% Ig M positivity (18/26) in this set. This provides value for serological exams; exclusive use of the molecular test with swabs does not provide a diagnosis for an infected patient in the hospital context.
Serological investigation is a simple method for the laboratory; these tests, based on the chromatographic technique, are fast (about 15 min to execute) [14] and do not require additional instrumentation. The investigation of the presence of Ig G (8/26 of serologically positive responses in MOL NEG) can determine disease status, but this aspect is not well-delineated by molecular investigation alone. The most commonly used techniques of RNA analysis do not exploit the quantitative aspect, based on Real Time Polymerase Chain Reaction (RT-PCR) for qualitative use. This cannot evaluate the patient's disease phase (incremental viral load in disease progress or a decrease in the remission phase are not yet distinguishable). Only an analysis with reference standards and frequent patient monitoring allows the healthcare system to diagnose the actual state of the disease [15], [16], [17].
The incidence of Ig positive in a MOL POS cohort (27/73, 37%) indicates that the molecular method is the gold standard to screen patients, but reveals that a consistent fraction are probably in the serologically undetectable set (onset of disease), or that Ig kits must be developed for a major specificity [18].
The problem of indeterminate cases regarding molecular response is becoming critical: in the screening phase, it raises doubts about specificity of the kits used, as the monitoring context is an insidious aspect of the interpretation of infectious patients in clinical remission. The cross reactivity of the oligonucleotide’s sequences in molecular kits [10] and the viral fluctuation known to make up these pathologies [19], represent a fundamental problem for patient monitoring. The systematic use of serological assessment could be a valid method of framing the patient's clinical status, in cases in which the molecular response is not exhaustive, or in which it provides a doubtful result. In this study, more than 50% (14/25, 56%) of the MOL IND cases were characterized by a positive response to immunoglobulins (Table 2). This data allows us to increase the clinical utility of the laboratory data.
We found 1.9% (8/413) of Ig G positivity in MOL NEG (Table 2). The incidence of only IgG positivity in the population has been an important parameter for evaluating herd immunity. This type of investigation, as mass controls increase, provides useful information for the trend of global immunity, and allows us to do a systematic check of disease relapse in immunized people [20],as well as to evaluate viral spread.
Although coronavirus literature reports possible advantages for the female sex [21], the current 2019-nCoV (novel coronavirus) does not seem to show significant frequency in one sex compared to the other; however, in other aspects, there are statistically distinctive data for lethality [22]. The latter aspect has not been analyzed here, despite that this study confirms infection incidence as being comparable between males and females (Table S1 Supplementary).
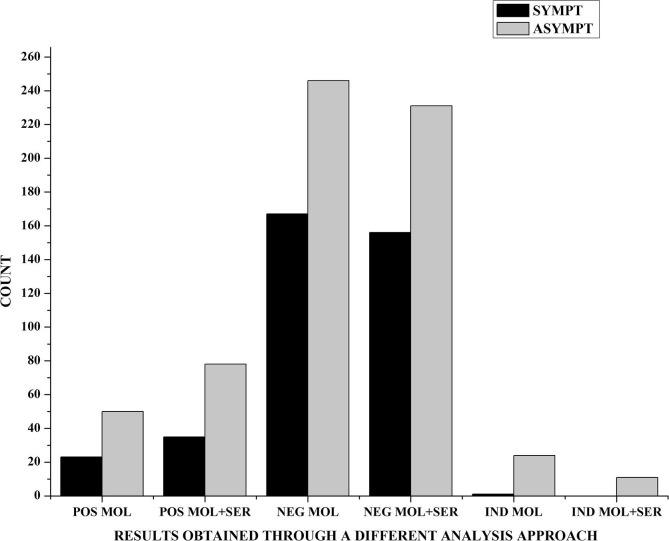
Analyzing the two S/P and As sets(Table 3), it was observed that there were no statistically significant differences between exclusive molecular investigation and that for immunoglobulins (p greater than 0.05). This confirms how seeking viral RNA in the swab sample is considered the gold standard for a diagnosis, but the total number of positive cases (POS MOL + SER) is higher than that identified by analysis of molecular status alone (POS MOL). This is noted in a viral spreading scenario to detect the greatest number of potentially infectious people (Graph 2 ).
Graph 2.
Different results obtained through molecular (MOL) and serological (SER) analysis in S/P (SYMPT) and As sets (ASYMPT). Absolute count of positive molecular results obtained by the swab test (POS MOL) is less than that by swabsandserological test (POS MOL + SER). We observe the same trend for negative results with the swab test (NEG MOL) vs. the duplex approach (NEG MOL + SER),or for indeterminate ones (IND MOL vs. IND MOL + SER).However, there are not statistically significant differences among groups (p greater than 0.05).
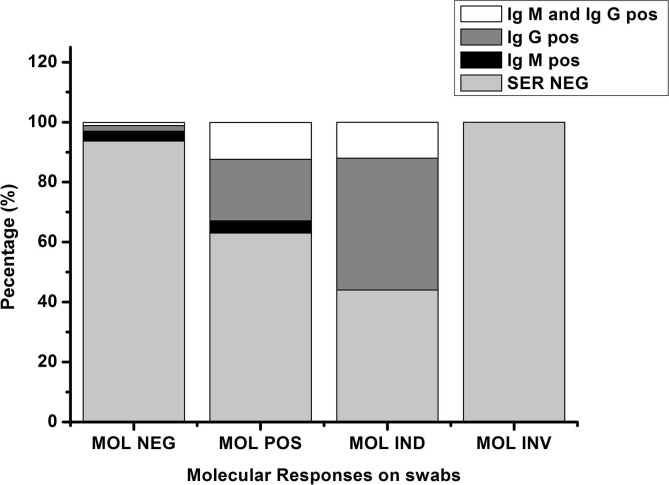
The cumulative positivity of molecular and serological tests must be considered in population screening to reduce false positives and to increase diagnostic strength. Serological positivity incidence in the MOL NEG and the MOL IND indicate that a combination of dual methodological approaches would provide an efficient instrument for laboratory investigations - while at the same time, the data show low predictivity for single use serological tests (Graph 3 ).
Graph 3.
Serological response distribution (%) among molecular results.
The dual analysis (molecular and serological) should be considered as a prudent approach to reduce false negatives. In this period, we performed a number of swabs, with various mass control managed by local health administrations, while mass screening is debated in its ability to solve the actual crisis [23], [24]. The need to increase oropharyngeal swabs and the consequent need to rapidly train staff, lacking practical experience,emphasizes an unsuitable sample. This aspect is particularly problematic, considering that the most widespread molecular analysis kits do not allow verification of the integrity of the collected material, so there is a concrete possibility of providing false negatives [25].
In our findings, the serological testcould be important to confirm negative and to clarify indeterminate molecular results,so health systems will have to consider the role of serology in their evaluation algorithms. In other hand, considering large part of serological negative results among molecular positive cases, it's undoubtedly necessary to conduct further research into serology approach. Novel antibodies, more specific than used, and use of CLIA (Chemiluminescent immunoassay) kits should be tested to raise predictivity value. In this study the molecular test has been confirmed gold standard method to define and to evaluate the disease status and the potential single patient infection power.
The immunopositivity for older patients among MOL POS of the S/P set emerged in this study (Graph 1), as this data must be understood. First, a possible confounding effect linked to this case study must be considered. In a pandemic scenario, older patients (over 75) are considered the population most at risk for lethality, related to SARS-CoV-2 infection. Given this logic, it is also possible that there is distrust in visiting the hospital, which is a potentially risky environment. This approach was carried out by patients themselves, family members, home assistants,while family doctors may have selected a group of patients who should have turned to health facilities in an advanced disease state;this may have been determined by immunopositivity data as being higher in older subjects.
If we exclude this confounding effect, the biological interpretation of the data is complex and needs more studies, but we suggest some hypotheses: early literature data report that binding and neutralizing antibodies are higher in older adults [26]; these patients might have been exposed to coronavirus contacts for a longer time than younger ones. While examining the Gorse et al. study (2020), this condition reveals a more frequent and specific immunoglobulin presence in older patients “due to the stimulation of antibodies against conserved cross‐reactive antigens expressed by the current infecting strain of HCoV and, due to recall of immune memory, to other strains that previously infected the person earlier in life.” If corroborated by other studies, this data could enhance the validity of serological tests in older patients. Molecular and serological analyses help the virologist to determine disease status and to support clinical decisions.
The use of more accurate and sensitive serological systems, derived from recent studies on isotypes [27]would allow us to perform a more efficient dual approach for the patient’s diagnosis, prognosis, and management.
5. Conclusion
These data confirm that serological and molecular approaches should not be considered as two alternative systems for the monitoring of virus spreading, rather they seem to be two necessary applications. The molecular test allows to identify the most part of infected patients and the simultaneous serological investigation helps to clarify diagnostic response of the indeterminate molecular cases.This fact confirms that dual approach of molecular and serological tests must be recommended for a complete screening of population in pandemic scenario.
Moreover, considering improving specificity of serological test in the future, it is probable that it will be confirmed a high incidence of coronavirus seropositivity in older people among symptomatic/paucisympthomatic patients. This data could clarify epidemiology of the COVID and it could be useful to define correct diagnostic approach especially in emergency situation.
CRediT authorship contribution statement
Alessandro Pancrazzi: Writing - review & editing, Data curation, Conceptualization, Methodology, Formal analysis. Pasqualino Magliocca: Data curation, Investigation. Maria Lorubbio: Methodology, Data curation. Guendalina Vaggelli: Methodology, Data curation. Angelo Galano: Methodology, Data curation. Manuela Mafucci: Data curation, Investigation. Diletta Duranti: Methodology, Data curation. Monica Cortesi: Methodology. Erica Mazzeschi: Methodology. Sara Fabbroni: Resources. Gianluca Viti: Resources. Alessandro Tartaglia Polcini: Methodology, Data curation. Emanuela Tripodo: Methodology, Data curation. Paola Sanchini: Data curation. Silvana Gervino: Data curation. Danilo Tacconi: Data curation, Investigation. Simona Dei: Resources, Data curation. Monica Mazzierli: Methodology. Antonio D'Urso: Resources, Data curation. Agostino Ognibene: Resources, Data curation, Supervision, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clinbiochem.2020.07.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hui D.S., Azhar E.I., Kim Y.J., Memish Z.A., Oh M.D., Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis. 2018;18:e217–e227. doi: 10.1016/S1473-3099(18)30127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfaraj S.H., Al-Tawfiq J.A., Altuwaijri T.A., Alanazi M., Alzahrani N., Memish Z.A. Middle East respiratory syndrome coronavirus transmission among health care workers: Implication for infection control. Am J Infect Control. 2018;46:165–168. doi: 10.1016/j.ajic.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus in the last two years: Health care workers still at risk. Am J Infect Control. 2019;47:1167–1170. doi: 10.1016/j.ajic.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://www.gazzettaufficiale.it/eli/id/2020/02/25/20A01272/sg.
- 5.https://github.com/pcm-dpc/COVID-19/blob/master/schede-riepilogative/regioni/dpc-covid19-ita-scheda-regioni-20200324.pdf.
- 6.Z. Li Y. Yi X. Luo N. Xiong Y. Liu S. Li R. Sun Y. Wang B. Hu W. Chen Y. Zhang J. Wang B. Huang Y.e. Lin J. Yang W. Cai X. Wang J. Cheng Z. Chen K. Sun W. Pan Z. Zhan L. Chen F. Ye Development and Clinical Application of A Rapid IgM‐IgG Combined Antibody Test for SARS‐CoV‐2 Infection Diagnosis J Med Virol 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed]
- 7.Rubens Costa Lima J., Rouquayrol M.Z., Monteiro Callado M.R., Florindo Guedes M.I., Pessoa C. Interpretation of the presence of IgM and IgG antibodies in a rapid test for dengue: Analysis of dengue antibody prevalence in Fortaleza City in the 20th year of the epidemic. Rev Soc Bras Med Trop. 2012;45:163–167. doi: 10.1590/s0037-86822012000200005. [DOI] [PubMed] [Google Scholar]
- 8.Warkad S.D., Song K.S., Pal D., Nimse S.B. Developments in the HCV Screening Technologies Based on the Detection of Antigens and Antibodies. Sensors (Basel). 2019;19:E4257. doi: 10.3390/s19194257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections: state of the art. Emerg Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.http://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2020&codLeg=73714&parte=1%20&serie=null.
- 12.Y.W. Tang J.E. Schmitz D.H. Persing C.W. Stratton Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges J Clin Microbiol. 2020:JCM.00512-20. 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed]
- 13.Y.Y. Wang Y.H. Jin X.Q. Ren Y.R. Li X.C. Zhang X.T. Zeng X.H. Wang Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team. Updating the diagnostic criteria of COVID-19 “suspected case” and “confirmed case” isnecessary Mil Med Res. 2020 Apr 4;7(1):17. 10.1186/s40779-020-00245-9. [DOI] [PMC free article] [PubMed]
- 14..http://www.centerforhealthsecurity.org/resources/COVID-19/serology/Serology-based-tests-for-COVID-19.html.
- 15.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S., Kim H.M., Han M.G., Kim S.Y., Chin B.S. Viral Load Kinetics of SARS-CoV-2 Infection in the First Two Patients in Korea. JKorean Med Sci. 2020;35 doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395:1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D., Xu W., Lei Z., Huang Z., Liu J., Gao Z., Peng L. Recurrence of positive SARS-CoV-2 RNA in COVID-19: A case report. Int J Infect Dis. 2020;93:297–299. doi: 10.1016/j.ijid.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020 Feb 15;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peto J. Covid-19 mass testing facilities could end the epidemic rapidly. Brit Med J. 2020;368 doi: 10.1136/bmj.m1163. [DOI] [PubMed] [Google Scholar]
- 24.Kwon K.T., Ko J.H., Shin H., Sung M., Kim J.Y. Drive-Through Screening Center for COVID-19: A Safe and Efficient Screening System against Massive Community Outbreak. J Korean Med Sci. 2020;35 doi: 10.3346/jkms.2020.35.e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancrazzi A., Perticucci R., Vecchietti S., Magrini G., Vaggelli G., Magliocca P., Galano A., Mafucci M., Galanti I.A., Mol O.A.J., Biol Potential False Negatives in the Molecular Diagnostics of COVID-19 Infection: Experience of an Italian Laboratory during the COVID-19 Epidemic. Methods. 2020;3(1) doi: 10.37532/jmbm.2020.3(1).106. [DOI] [Google Scholar]
- 26.Gorse G.J., Donovan M.M., Patel G.B. Antibodies to coronaviruses are higher in older adults compared with younger adults, and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus: Associated illnesses. J Med Virol. 2020;92:512–517. doi: 10.1002/jmv.25715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.https://www.centerforhealthsecurity.org/resources/COVID-19/serology/Serology-based-tests-for-COVID-19.html#sec2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.