Abstract
With documented global declines in insects, including wild bees, there has been increasing interest in developing and expanding insect monitoring programs. Our objective here was to organize, validate, and share an analysis-ready version of one of the few existing long-term monitoring datasets for wild bees in the United States. Since 1999, the Native Bee Inventory and Monitoring Lab (BIML) of the United States Geological Survey has sampled wild-bee communities in the Mid-Atlantic U.S., but samples were collected in multiple studies and the datasets are not fully integrated. Furthermore, critical information about sampling methodology was often lacking, though these factors can significantly influence collection outcomes and must be considered in analyses. We cleaned and verified BIML data from Maryland, Delaware, and Washington DC, USA, and generated sampling methodology for over 84% of the 99,053 pan-trapped occurrences in this region. We enthusiastically invite creative analyses of this rich dataset to advance understanding of the biology and ecology of wild bees, inform conservation efforts, and perhaps help design a nationwide bee monitoring program.
Subject terms: Conservation biology, Biodiversity, Entomology
| Measurement(s) | Abundance |
| Technology Type(s) | digital curation |
| Factor Type(s) | site • year |
| Sample Characteristic - Organism | Apoidea |
| Sample Characteristic - Location | United States of America |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12473270
Background & Summary
Wild bees are crucial pollinators of many crop and wild-plant species. Globally, 87 out of 115 major food crops require insect pollination and 78% of temperate plant species and 94% of tropical plant species require insect pollination for reproductive success and persistence1,2. Wild-bee conservation is crucial for global biodiversity as pollinators represent several extremely diverse taxa and have a vital co-evolved role in supporting and selecting for diverse plant communities3.
In Europe and the United States, there has been a surge of interest in developing programs to monitor wild-bee populations4–7. There have been documented declines in several North American bumble bees populations and species8, but we largely lack baseline data to assess declines in many other taxa. In response to heavy colony losses since 2006, monitoring of honey bee (Apis mellifera L.) populations has greatly increased9. These data have greatly contributed to our understanding of honey bee population dynamics, ecological interactions, colony stressors, and effective management.
We expect coordinated, wild-bee monitoring will enable similar advances in understanding wild-bee diversity, natural history, and conservation strategies, but there are still several challenges in collecting and analysing monitoring data. Ideally, a wild-bee monitoring program, or any monitoring program for that matter, would utilize consistent methods over time to detect long-term population trends. In practice, monitoring protocols often shift over time, and many existing datasets were compiled from studies using multiple sampling methods10.
Our goal here was to organize, validate, and share an analysis-ready version of one of the few existing long-term, wild-bee monitoring datasets in the United States. Since 1999, the Native Bee Inventory and Monitoring Lab (BIML) of the United States Geological Survey has collected, curated, and identified over 99,000 specimens of more than 314 wild bee species from over 1400 sites in Maryland, Delaware, and Washington DC. The BIML collection is among the most geographically dense, recent sampling of wild bees in the country. Also, all specimens were lethally collected and expert-identified to species, which provides a crucial comparison for observation-based datasets with coarser taxonomic information, including rapidly growing citizen science data11,12.
BIML data contain considerable diversity in wild-bee-sampling methods and critical sampling information is often missing or not easily accessible to end-users. One goal of BIML research was to test a variety of passive sampling methods to establish a standard protocol for wild-bee monitoring. They studied sampling efficiency of passive pan traps (bee bowls) with varying trap color, volume, and liquid. Pan traps are a reliable method for monitoring wild bees13,14, particularly in combination with net collections or transect walks, but the specific pan-trapping method, especially trap color and visibility, influences the abundance and composition of wild bees collected15–17. To fully utilize the BIML dataset collected with multiple methods, trap color should be included as a variable in statistical analyses. Currently, this is not possible with the publicly available BIML data18 as 90% of the pan-trapped occurrences in Maryland, Delaware, and DC have no trap color data. Similarly, 36% of occurrences are missing trap volume and 21% lack sampling effort (the number of traps used during each sampling period). Fortunately, much of the missing information was recorded in BIML field notes, but not in a consistent, accessible format.
To fill these gaps, we cleaned and verified BIML data and extracted trap color, volume, and sampling effort for more than 84% of the pan-trapped occurrences. We focused on occurrences collected in pan traps because they represent more than 82% of the larger BIML dataset. We also created, for all pan-trapped occurrences, identifier variables and verified species binomials, date, and locality. We enthusiastically invite analyses of this rich dataset to advance understanding of bee biology and ecology, inform current wild bee conservation efforts, and perhaps even help design a national wild bee monitoring program.
Methods
Identifier variables
To facilitate aggregation and analysis of the BIML data, we added ‘site’, ‘site-year’, ‘sampling event’, and ‘transect’ identifier variables. We defined ‘sites’ as unique combinations of latitude and longitude, and ‘site-years’ as unique combinations of site and year of sampling. Within site-years, we defined ‘sampling events’ according to the date of sampling and ‘transects’ as unique combinations of sampling event and text field notes. For some specimens, field notes included a transect ID, indicating that the BIML used multiple sets of pan traps at the same site. In other cases, field notes recorded differing sampling methods, or different information on the number of missing traps (traps that were cracked, tipped over, or otherwise compromised). If field notes recorded different methods or number of lost traps, we assumed that the BIML deployed multiple sets of traps (transects). We reviewed the field notes for all sampling events with multiple transects and reassigned these occurrences to a single transect if there was no evidence of multiple transects in the field notes.
Locality and taxonomic identification
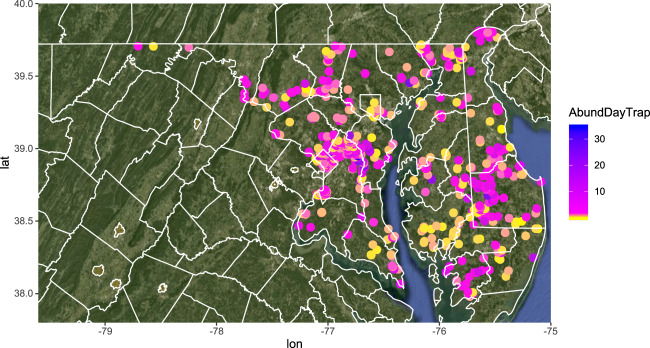
Next, we reviewed and excluded occurrences lacking critical date and locality information. We removed all occurrences lacking sampling date or latitude and longitude of sampling location and occurrences with duplicated specimen identifiers. We filtered occurrences to a limited geographic area (Maryland, Delaware, and Washington DC, Fig. 1) that represents the densest region of BIML sampling (39.6% of dataset). This filtering removed wild-bee communities collected in desert or tropical biomes, which are likely governed by very different floral resource and climate dynamics19,20, and within the Mid-Atlantic USA, limited sampling locations to a region with a consistent dominant forest type21. Bee occurrences in 1999 and 2001 represented fewer than three sites per year, so we removed these years, retaining sites sampled from 2002–2016.
Fig. 1.
Abundance per day per trap of wild bees at locations surveyed between 2002 and 2016 by the United States Geological Survey Native Bee Inventory and Monitoring Lab (USGS BIML).
We also filtered data to our taxa of interest. We removed non-bee occurrences (species outside superfamily Apoidea, clade Anthophila) and records lacking species-level identity, discarding occurrences identified to family or genus (Online-only Table 1). Almost all non-bee occurrences we removed were wasps in the Vespidae, Crabronidae, and Sphecidae families, which are primarily predators, rather than pollen-collectors like most wild bees. For the transect-level dataset22 (see ‘Data Records’ below), we calculated the abundance of Apis mellifera L., then removed A. mellifera from the dataset before calculating total bee abundance per transect, since often A. mellifera specimens likely originated from managed colonies and are not considered to be wild bees.
Online-only Table 1.
List of all specimen names that were modified or removed during data cleaning with justification for each change.
| Original species name | Cleaned name | Grouped name | Genus | species | Family | Reason for modification/removal |
|---|---|---|---|---|---|---|
| Modified species names | ||||||
| Andrena arabis/algida | Andrena algida/arabis | Andrena algida/arabis | Andrena | algida/arabis | Andrenidae | Reordered multiple species names |
| Lasioglossum carlini | Andrena carlini | Andrena carlini | Andrena | carlini | Andrenidae | Incorrect genus |
| Andrena tridens/erythronii | Andrena erythronii/tridens | Andrena erythronii/tridens | Andrena | erythronii/tridens | Andrenidae | Reordered multiple species names |
| Amdrema imitatrix/morrisonella | Andrena imitatrix/morrisonella | Andrena imitatrix/morrisonella | Andrena | imitatrix/morrisonella | Andrenidae | Corrected spelling |
| Andrena morisonella | Andrena morrisonella | Andrena imitatrix/morrisonella | Andrena | morrisonella | Andrenidae | Corrected spelling |
| Melissodes nivalis | Andrena nivalis | Andrena nivalis | Andrena | nivalis | Andrenidae | Incorrect genus |
| Lasioglossum perplexa | Andrena perplexa | Andrena perplexa | Andrena | perplexa | Andrenidae | Incorrect genus |
| Nomada personata | Andrena personata | Andrena personata | Andrena | personata | Andrenidae | Incorrect genus |
| Andrena puni | Andrena pruni | Andrena pruni | Andrena | pruni | Andrenidae | Corrected spelling |
| Pseudopanurgus rudbeckiae | Pseudopanurgus near rudbeckiae | Pseudopanurgus near rudbeckiae | Pseudopanurgus | rudbeckiae | Andrenidae | Recommended by data source (S. Droege) |
| apis mellifera | Apis mellifera | Apis mellifera | Apis | mellifera | Apidae | Standardized format |
| bombus bimaculatus | Bombus bimaculatus | Bombus bimaculatus | Bombus | bimaculatus | Apidae | Standardized format |
| ceratina calcarata | Ceratina calcarata | Ceratina calcarata/dupla/mikmaqi | Ceratina | calcarata | Apidae | Standardized format |
| Ceratina mikmaqi/calcarata | Ceratina calcarata/mikmaqi | Ceratina calcarata/dupla/mikmaqi | Ceratina | calcarata/mikmaqi | Apidae | Reordered multiple species names |
| ceratina dupla | Ceratina dupla | Ceratina calcarata/dupla/mikmaqi | Ceratina | dupla | Apidae | Standardized format |
| Ceratina miqmaki | Ceratina mikmaqi | Ceratina calcarata/dupla/mikmaqi | Ceratina | mikmaqi | Apidae | Corrected spelling |
| Ceratina Strenua | Ceratina strenua | Ceratina strenua | Ceratina | strenua | Apidae | Standardized format |
| ceratina strenua | Ceratina strenua | Ceratina strenua | Ceratina | strenua | Apidae | Standardized format |
| Melissodes tineta | Melissodes tinctus | Melissodes tinctus | Melissodes | tinctus | Apidae | Corrected spelling |
| Nomada sayi/illinoensis | Nomada illinoensis/sayi | Nomada illinoensis/sayi | Nomada | illinoensis/sayi | Apidae | Reordered multiple species names |
| Nomada sayi/illinoense | Nomada illinoensis/sayi | Nomada illinoensis/sayi | Nomada | illinoensis/sayi | Apidae | Corrected spelling |
| Nomada sulphurata/luteola | Nomada luteola/sulphurata | Nomada luteola/sulphurata | Nomada | luteola/sulphurata | Apidae | Reordered multiple species names |
| Nomada luteolodies | Nomada luteoloides | Nomada luteoloides | Nomada | luteoloides | Apidae | Corrected spelling |
| hylaeus affinis/modestus | Hylaeus affinis/modestus | Hylaeus affinis/modestus | Hylaeus | affinis/modestus | Colletidae | Standardized format |
| hylaeus ornatus | Hylaeus ornatus | Hylaeus ornatus | Hylaeus | ornatus | Colletidae | Standardized format |
| Agapostemon angelicus/texanus | Agapostemon texanus | Agapostemon texanus | Agapostemon | angelicus/texanus | Halictidae | One species of set not found in Mid-Atlantic USA |
| augochlorella aurata | Augochlorella aurata | Augochlorella aurata | Augochlorella | aurata | Halictidae | Standardized format |
| Augochlora aurata | Augochlorella aurata | Augochlorella aurata | Augochlorella | aurata | Halictidae | Incorrect genus |
| Lasioglossum aurata | Augochlorella aurata | Augochlorella aurata | Augochlorella | aurata | Halictidae | Incorrect genus |
| Augochlorellagratiosa | Augochlorella gratiosa | Augochlorella gratiosa | Augochlorella | gratiosa | Halictidae | Standardized format |
| Augochloropsis Metallica | Augochloropsis metallica | Augochloropsis metallica | Augochloropsis | metallica | Halictidae | Standardized format |
| Halictus poeyi/ligatus | Halictus ligatus/poeyi | Halictus ligatus/poeyi | Halictus | ligatus/poeyi | Halictidae | Reordered multiple species names |
| Halitcus poeyi/ligatus | Halictus ligatus/poeyi | Halictus ligatus/poeyi | Halictus | ligatus/poeyi | Halictidae | Corrected spelling |
| Haclictus tectus | Halictus tectus | Halictus tectus | Halictus | tectus | Halictidae | Corrected spelling |
| Andrena bruneri | Lasioglossum bruneri | Lasioglossum bruneri | Lasioglossum | bruneri | Halictidae | Incorrect genus |
| Ceratina callidum | Lasioglossum callidum | Lasioglossum callidum | Lasioglossum | callidum | Halictidae | Incorrect genus |
| lasioglossum coreopsis | Lasioglossum coreopsis | Lasioglossum coreopsis | Lasioglossum | coreopsis | Halictidae | Standardized format |
| Lasioglossum coeropsis | Lasioglossum coreopsis | Lasioglossum coreopsis | Lasioglossum | coreopsis | Halictidae | Corrected spelling |
| Lasioglossum geminum | Lasioglossum geminatum | Lasioglossum geminatum | Lasioglossum | geminatum | Halictidae | Corrected spelling |
| Lasioglossum Gotham | Lasioglossum gotham | Lasioglossum gotham | Lasioglossum | gotham | Halictidae | Standardized format |
| Andrena hitchensi | Lasioglossum hitchensi | Lasioglossum hitchensi/weemsi | Lasioglossum | hitchensi | Halictidae | Incorrect genus |
| Nomada lustrans | Lasioglossum lustrans | Lasioglossum lustrans | Lasioglossum | lustrans | Halictidae | Incorrect genus |
| lasioglossum pilosum | Lasioglossum pilosum | Lasioglossum pilosum | Lasioglossum | pilosum | Halictidae | Standardized format |
| Sphecodes cressonii/atlantis | Sphecodes atlantis/cressonii | Sphecodes atlantis/banksii/cressonii | Sphecodes | atlantis/cressonii | Halictidae | Reordered multiple species names |
| Hoplitis producta/pilosifrons | Hoplitis pilosifrons/producta | Hoplitis pilosifrons/producta | Hoplitis | pilosifrons/producta | Megachilidae | Reordered multiple species names |
| hoplitis spoliata | Hoplitis spoliata | Hoplitis spoliata | Hoplitis | spoliata | Megachilidae | Standardized format |
| Megachile apicata | Megachile apicalis | Megachile apicalis | Megachile | apicalis | Megachilidae | Corrected spelling |
| megachile brevis | Megachile brevis | Megachile brevis/mendica | Megachile | brevis | Megachilidae | Standardized format |
| Megachile mendica/brevis | Megachile brevis/mendica | Megachile brevis/mendica | Megachile | brevis/mendica | Megachilidae | Reordered multiple species names |
| Osmia atriventis | Osmia atriventris | Osmia atriventris/pumila | Osmia | atriventris | Megachilidae | Corrected spelling |
| Osmia pumila/atriventris | Osmia atriventris/pumila | Osmia atriventris/pumila | Osmia | atriventris/pumila | Megachilidae | Reordered multiple species names |
| Osmia callinsia | Osmia collinsiae | Osmia collinsiae | Osmia | collinsiae | Megachilidae | Corrected spelling |
| Lasioglossum conjuncta | Osmia conjuncta | Osmia conjuncta | Osmia | conjuncta | Megachilidae | Incorrect genus |
| Osmia conjucta | Osmia conjuncta | Osmia conjuncta | Osmia | conjuncta | Megachilidae | Corrected spelling |
| osmia sandhouseae | Osmia sandhouseae | Osmia sandhouseae | Osmia | sandhouseae | Megachilidae | Standardized format |
| Removed species names | ||||||
| Lasioglossum melanopus | Not found in Mid-Atlantic USA | |||||
| Lasioglossum incompletum | Not found in Mid-Atlantic USA | |||||
| Hylaeus cressonii | Species epithet could not be verified | |||||
| Lasioglossum kevensi | Species epithet could not be verified | |||||
| Osmia composita | Species epithet could not be verified | |||||
| Removed, Genus-only identifications | ||||||
| Aculeata | Specimen identified to genus or subgenus | |||||
| Agapostemon | Specimen identified to genus or subgenus | |||||
| Ancistrocerus | Specimen identified to genus or subgenus | |||||
| Andrena | Specimen identified to genus or subgenus | |||||
| Andrena (Melandrena) | Specimen identified to genus or subgenus | |||||
| Andrena (Scrapteropsis) | Specimen identified to genus or subgenus | |||||
| Andrena (Trachandrena) | Specimen identified to genus or subgenus | |||||
| Andrena melandrena | Specimen identified to genus or subgenus | |||||
| Anthophila | Specimen identified to genus or subgenus | |||||
| Apidae | Specimen identified to genus or subgenus | |||||
| Apoidea | Specimen identified to genus or subgenus | |||||
| Arachnida | Specimen identified to genus or subgenus | |||||
| Augochlorella | Specimen identified to genus or subgenus | |||||
| Bombus | Specimen identified to genus or subgenus | |||||
| Carabidae | Specimen identified to genus or subgenus | |||||
| Ceratina | Specimen identified to genus or subgenus | |||||
| Chalcid | Specimen identified to genus or subgenus | |||||
| Chrysidid | Specimen identified to genus or subgenus | |||||
| Chrysididae | Specimen identified to genus or subgenus | |||||
| Coelioxys | Specimen identified to genus or subgenus | |||||
| Colletes | Specimen identified to genus or subgenus | |||||
| Figitidae/Eucoilinae | Specimen identified to genus or subgenus | |||||
| Formicidae | Specimen identified to genus or subgenus | |||||
| Halictus | Specimen identified to genus or subgenus | |||||
| Hedychridium | Specimen identified to genus or subgenus | |||||
| Heriades | Specimen identified to genus or subgenus | |||||
| Hoplitis | Specimen identified to genus or subgenus | |||||
| Hylaeus | Specimen identified to genus or subgenus | |||||
| Ichneumonidae | Specimen identified to genus or subgenus | |||||
| Ichneumonidae/Ichneunominae | Specimen identified to genus or subgenus | |||||
| Ichneumonidae/Phygadeuontinae | Specimen identified to genus or subgenus | |||||
| Ichneumonidae/Tryphoninae | Specimen identified to genus or subgenus | |||||
| Larrinae | Specimen identified to genus or subgenus | |||||
| lasioglossum | Specimen identified to genus or subgenus | |||||
| Lasioglossum | Specimen identified to genus or subgenus | |||||
| Lasioglossum male | Specimen identified to genus or subgenus | |||||
| Megachile | Specimen identified to genus or subgenus | |||||
| Melissodes | Specimen identified to genus or subgenus | |||||
| Nomada | Specimen identified to genus or subgenus | |||||
| Osmia | Specimen identified to genus or subgenus | |||||
| Panurginus | Specimen identified to genus or subgenus | |||||
| Parancistrocerus | Specimen identified to genus or subgenus | |||||
| Perdita | Specimen identified to genus or subgenus | |||||
| Pompilidae | Specimen identified to genus or subgenus | |||||
| Pseudopanurgus | Specimen identified to genus or subgenus | |||||
| Pteromalidae | Specimen identified to genus or subgenus | |||||
| Scoliidae | Specimen identified to genus or subgenus | |||||
| Sphecidae | Specimen identified to genus or subgenus | |||||
| sphecodes | Specimen identified to genus or subgenus | |||||
| Sphecodes | Specimen identified to genus or subgenus | |||||
| Sphecodogastra | Specimen identified to genus or subgenus | |||||
| Svastra | Specimen identified to genus or subgenus | |||||
| Tenthredinidae | Specimen identified to genus or subgenus | |||||
| Tenthredo | Specimen identified to genus or subgenus | |||||
| Tiphia | Specimen identified to genus or subgenus | |||||
| Tiphiidae | Specimen identified to genus or subgenus | |||||
| Tiphiidae/Tiphiinae | Specimen identified to genus or subgenus | |||||
| Tiphiinae | Specimen identified to genus or subgenus | |||||
| Triepeolus | Specimen identified to genus or subgenus | |||||
| Vespidae | Specimen identified to genus or subgenus | |||||
| Zanysson | Specimen identified to genus or subgenus | |||||
| Removed, species that are not bees | ||||||
| Acmaeodera ornata | Species outside superfamily Apoidea, clade Anthophila | |||||
| Acmaeodera pulchella | Species outside superfamily Apoidea, clade Anthophila | |||||
| Acmaeodera tubulus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ammophila fernaldi | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ammophila nigricans | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ammophila pictipennis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ammophila procera | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ammophila urnaria | Species outside superfamily Apoidea, clade Anthophila | |||||
| Anacrabro ocellatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ancistrocerus adiabatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ancistrocerus albophaleratus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ancistrocerus antilope | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ancistrocerus campestris | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ancistrocerus cressonii | Species outside superfamily Apoidea, clade Anthophila | |||||
| Argogorytes nigrifrons | Species outside superfamily Apoidea, clade Anthophila | |||||
| Astata unicolor | Species outside superfamily Apoidea, clade Anthophila | |||||
| Bicyrtes ventralis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Caenochrysis doriae | Species outside superfamily Apoidea, clade Anthophila | |||||
| Caenochrysis doriae/Cleptes alienus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ceratochrysis declinis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ceratochrysis enhuycki | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ceratochrysis kansensis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chalybion californicum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis angolensis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis antennalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis cembricola | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis conica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis derivata | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis inaequidens | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis montana | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis nisseri | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis nitidula/coerulans | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis pattoni | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis pellucidula | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis propria | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis remissa | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis scitula | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis smaragdala | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysis tripartita | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysura cobaltina | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysura cobaltina/pacifica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysura kyrae/pacifica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Chrysura pacifica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Cicindella sexguttata | Species outside superfamily Apoidea, clade Anthophila | |||||
| Cinthophora abrupta | Species outside superfamily Apoidea, clade Anthophila | |||||
| Compsomeris plumipes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Dolichovespula arenaria | Species outside superfamily Apoidea, clade Anthophila | |||||
| Dolichovespula maculata | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ectemnius lapidarius | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ectemnius rufipes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Ectemnius scaber | Species outside superfamily Apoidea, clade Anthophila | |||||
| Epinysson mellipes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Eremnophila aureonotata | Species outside superfamily Apoidea, clade Anthophila | |||||
| Eumenes fraternus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Eumenes reticalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus annulatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus boscii | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus castigatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus crypticus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus foraminatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus hidalgo | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus hidalgo boreoorientalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus leucomelas | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus megaera | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodynerus schwarzi | Species outside superfamily Apoidea, clade Anthophila | |||||
| Euodyrnus megaera | Species outside superfamily Apoidea, clade Anthophila | |||||
| Hedychridium dimidiatum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Hedychrum confusum/violaceum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Hedychrum parvum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Hedychrum violaceum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Isodontia apicalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Isodontia auripes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Isodontia mexicana | Species outside superfamily Apoidea, clade Anthophila | |||||
| Isodontia philadelphica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Leptochilus acolhuus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Leptochilus republicans | Species outside superfamily Apoidea, clade Anthophila | |||||
| Leptochilus republicanus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Lestica confluenta | Species outside superfamily Apoidea, clade Anthophila | |||||
| Lestica producticollis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Liris argentatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Lyroda subita | Species outside superfamily Apoidea, clade Anthophila | |||||
| Monobia quadridens | Species outside superfamily Apoidea, clade Anthophila | |||||
| Oxybelus emarginatum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Oxybelus emarginatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Oxybelus subulatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Palmodes collinsiae | Species outside superfamily Apoidea, clade Anthophila | |||||
| Palmodes dimidiatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Palmodes pumila | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus fulvipes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus histrio | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus leionotus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus pedestris | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus pedestris or pensylvanicus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus pedestris pedestris | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus pensylvanicus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus perennis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus salcularis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parancistrocerus vagus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parazumia perennis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Parazumia symmorpha | Species outside superfamily Apoidea, clade Anthophila | |||||
| Philanthus gibbosus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes bellicosus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes dominula | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes dorsalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes exclamans | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes fuscatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes hirsuticornis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes metricus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Polistes parametricus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Pompilidae Ctenocerinae | Species outside superfamily Apoidea, clade Anthophila | |||||
| Prionyx parkeri | Species outside superfamily Apoidea, clade Anthophila | |||||
| Pseduoanthidium nanum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Pseudodynerus quadrisectus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Scolia dubia | Species outside superfamily Apoidea, clade Anthophila | |||||
| Sphex nudus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Sphex pensylvanicus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus ammonia | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus ammonia paraensis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus anormis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus blepharus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus fundatiformis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus fundatiformis gonosceles | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus histrionalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus krombeini | Species outside superfamily Apoidea, clade Anthophila | |||||
| Stenodynerus oculeus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Symmorphus canadensis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Synnevrus plagiatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex acutus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex antennatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex belfragei | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex mundus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex similis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex tarsatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachysphex terminatus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachytes abdominalis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachytes chrysopyga obscurus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachytes distinctus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachytes guatemalensis | Species outside superfamily Apoidea, clade Anthophila | |||||
| Tachytes validus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Trypoxylon clavicerum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Trypoxylon frigidum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Trypoxylon pennsylvanicum | Species outside superfamily Apoidea, clade Anthophila | |||||
| Typocerus velutinus | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespa crabro | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula consobrina | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula flavopilosa | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula germanica | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula maculifrons | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula squamosa | Species outside superfamily Apoidea, clade Anthophila | |||||
| Vespula vidua | Species outside superfamily Apoidea, clade Anthophila | |||||
| Zethus spinipes | Species outside superfamily Apoidea, clade Anthophila | |||||
| Zethus spinipes variegatus | Species outside superfamily Apoidea, clade Anthophila | |||||
We verified species names by cross-referencing all species binomials with the Discover Life database23. We corrected genus and species names that were clear spelling errors (Online-only Table 1) and consulted the original data source (S. Droege) for remaining species binomials that did not exist on Discover Life. We also referenced Discover Life occurrence maps to confirm that all species in the BIML dataset occur in the Mid-Atlantic US. After these data cleaning steps, we removed six occurrences of the remaining five unknown or out-of-region species (Online-only Table 1). Some species in the BIML data were identified singularly and as part of a species set. To avoid double counting these species, we created a new variable with cleaned, mutually exclusive species names (termed ‘grouped name’). In ‘grouped name’, we combined singular species names with their associated species sets (Online-only Table 1). For example, we reclassified occurrences identified as Halictus ligatus/poeyi, Halictus ligatus, or Halictus poeyi to Halictus ligatus/poeyi to avoid inflating future species richness estimates when occurrences might be the same species. In the final occurrence datasets, we included the cleaned, singular names (‘name’) and cleaned, grouped names (‘grouped_name’), so future analysts can select the appropriate taxonomic aggregation for their research objectives. Voucher specimens for most species in the BIML dataset are housed in the Smithsonian collection, but some are not yet permanently archived. We suggest interested parties contact Sam Droege (current email: sdroege@usgs.gov) to access voucher specimens. We also included, to the best of our knowledge, current affiliations for individuals who identified BIML specimens (Supplementary Information, Table S1), and standardized names of identifiers (‘identifiedBy’) in the final datasets.
Sampling method and effort
To describe sampling method and effort, we used regular expressions to extract these data from field notes. We sought to compare bee communities sampled with a standard methodology, so we discarded bee occurrences collected with vane traps or nets, only retaining occurrences sampled with pan traps (i.e., bee bowls). Using the stringr package in R24,25, we searched the text of field notes to document trap volume, trap color, total number of traps, and the number of traps missing or disturbed. The most common BIML pan-trapping method involved setting out traps of multiple colors and combining the bees in all traps into one sample. Consequently, BIML recorded trap color in field notes as the number of traps of each color used for a specific sampling event. We designed regular expressions to extract the number of traps for the eight most common colors (white, blue, yellow, pale blue, fluorescent yellow, fluorescent blue, and florescent pale blue). For some occurrences, our regular expressions yielded no sampling information, so we manually reviewed these field notes and recorded any data missed by the automated search.
Next, we simplified the trap color and volume classification to facilitate future statistical analyses. To reduce the number of trap volume categories, we rounded trap volume to the nearest 0.5 ounces, and removed trap volumes greater than 40 ounces, assuming these were errors in data entry or extraction. When the trap color or volume used at a specific site changed within a year, we manually reviewed the field notes and corrected color or volume classifications when necessary. After correcting these discrepancies, we found the BIML very rarely changed sampling methods within a year, so we filled in most missing trap color or volume information by assuming a constant sampling method for all transects within a site-year. Finally, we combined rarely used color/volumes (fewer than 1% of transects) into an ‘Other’ category. In the archived datasets with sampling information, we included original and simplified variables for trap color and volume.
Lastly, we summarized sampling effort and calculated effort-adjusted abundance of wild bees. We calculated the total number of traps for each sampling event, and, when available, we also described the number of traps missing or disturbed. If there was no documentation of missing or disturbed traps, we assumed all traps were recovered successfully. When the total number of traps differed between the field notes and ‘number of traps’ column, we selected the lower value. We defined the final number of traps in each transect as the original number minus the number missing or disturbed. We calculated the duration of sampling as the difference between the date traps were collected and date traps were set. If traps were set and collected on the same day, we set the duration of sampling to one day. For each occurrence in the BIML dataset, we converted bee abundance to abundance day−1 trap−1. We conducted all data manipulation and aggregation with the R statistical and computing language 3.6.025,26
Data Records
We produced two primary datasets and a third analytical dataset22 (details below), which are all archived at figshare.com. The first dataset (‘Mid-Atlantic USA wild bee occurrences, all records’) contains all 99,053 wild bee occurrences collected in pan traps by BIML from 2002 to 2016 in Maryland, Delaware, and Washington DC, USA. We describe all variables included in dataset one in Table 1. The second dataset (‘Mid-Atlantic USA wild bee occurrences, records with sampling info’) is all pan-trapped occurrences with sampling effort information. We retained 83,583 wild bee occurrences in dataset two representing 300 species from 1225 sites sampled over 1322 site-years. Dataset two documents the trap color and volume when these data were available. See Online-only Table 2 for full meta-data for dataset two.
Table 1.
Description of all fields in dataset 1, all wild bee specimens in Maryland, Delaware, and Washington DC. Variables marked as ‘Original’ are unmodified from Droege and Sellers18.
| Field Name | Unit | Description |
|---|---|---|
| Identifiers | ||
| identifier | Original, specimen ID | |
| id | Original, specimen URL | |
| TransectID | transect ID | |
| SamplEvent | sampling event ID | |
| SiteID | site ID | |
| SiteID_Year | site ID and year of bee sampling | |
| year | year of bee sampling | |
| Taxonomy | ||
| name | taxonomic classification | |
| Genus | taxonomic classification | |
| species | taxonomic classification | |
| grouped_name | grouped taxonomic classification | |
| orig_name | Original, taxonomic classification | |
| sex | Original, specimen sex | |
| identifiedBy | Individual who determined specimen identity | |
| Locality and Date | ||
| latitude | decimal degrees | Original |
| longitude | decimal degrees | Original |
| coordinateUncertaintyInMeters | meters | Original |
| time1 | Original, start date and time | |
| time2 | Original, end date and time | |
| startdate | month/day/year | sampling start date |
| enddate | month/day/year | sampling end date |
| country | Original | |
| countryCode | Original | |
| state | Original | |
| county | Original | |
| municipality | Original | |
| Field notes | ||
| habitat | Original, habitat in location of sampling | |
| field_note | Original | |
| note | Original | |
| modif_fieldnote | modified field note used for automated string search | |
| modif_note | modified note used for automated string search | |
| Sampling method and effort | ||
| SampleType | type of sampling method | |
| TrapLiquid | Original, liquid used in traps | |
| TrapColor | Original, color of trap | |
| TrapVolume | Original, volume of trap | |
| Ntraps | Original, number of traps | |
| Wild bee abundance | ||
| Abundance | wild bee abundance | |
Online-only Table 2.
Description of all fields in archived dataset 2, wild bee specimens in Maryland, Delaware, and Washington DC with recorded sampling effort.
| Field Name | Unit | Description |
|---|---|---|
| Identifiers | ||
| identifier | Original, specimen ID | |
| id | Original, specimen URL | |
| TransectID | transect ID | |
| SamplEvent | sampling event ID | |
| SiteID | site ID | |
| SiteID_Year | site ID and year of bee sampling | |
| year | year of bee sampling | |
| Taxonomy | ||
| name | taxonomic classification | |
| Genus | taxonomic classification | |
| species | taxonomic classification | |
| grouped_name | grouped taxonomic classification | |
| orig_name | Original, taxonomic classification | |
| sex | Original | |
| identifiedBy | Individual who determined specimen identity | |
| Locality and Date | ||
| latitude | decimal degrees | Original |
| longitude | decimal degrees | Original |
| coordinateUncertaintyInMeters | meters | Original |
| time1 | Original, start date and time | |
| time2 | Original, end date and time | |
| startdate | month/day/year | sampling start date |
| enddate | month/day/year | sampling end date |
| trapdays | days | length of sampling period |
| country | Original | |
| countryCode | Original | |
| state | Original | |
| county | Original | |
| municipality | Original | |
| Field notes | ||
| habitat | Original, habitat in location of sampling | |
| field_note | Original | |
| note | Original | |
| modif_fieldnote | modified field note used for automated string search | |
| modif_note | modified note used for automated string search | |
| Sampling method and effort | ||
| SampleType | type of sampling method | |
| TrapLiquid | Original, liquid used in traps | |
| TrapColor | Original, color of trap | |
| TrapVolume | Original, volume of trap | |
| Ntraps | Original, number of traps | |
| nwhite | number of white pan traps | |
| nblue | number of blue pan traps | |
| nyellow | number of yellow pan traps | |
| nFLblue | number of fluorescent blue pan traps | |
| nFLyellow | number of fluorescent yellow pan traps | |
| nFLwhite | number of fluorescent white pan traps | |
| isblue | T/F | sampling method = blue pan traps? |
| isyellow | T/F | sampling method = yellow pan traps? |
| iswhite | T/F | sampling method = white pan traps? |
| isFLblue | T/F | sampling method = fluorescent blue pan traps? |
| isFLyellow | T/F | sampling method = fluorescent yellow pan traps? |
| isFLwhite | T/F | sampling method = fluorescent white pan traps? |
| ispaleblue | T/F | sampling method = pale blue pan traps? |
| isFLpaleblue | T/F | sampling method = fluorescent pale blue pan traps? |
| TrapVolumeParsed | trap volume (from regex string search) | |
| TrapVolumeFinal | trap volume (combined original and regex)* | |
| Nparse | number of traps set (from regex string search) | |
| Nmissing | number of traps missing (from regex string search) | |
| NTrapsFinal | number of successful traps (set-missing)* | |
| Wild bee abundance | ||
| Abundance | specimens | wild bee abundance |
| AbundDay | specimens day−1 | wild bee abundance day-1 |
| AbundTrap | specimens trap−1 | wild bee abundance trap-1 |
| AbundDayTrap | specimens day−1 trap−1 | wild bee abundance day-1 trap-1* |
Variables marked as ‘Original’ are unmodified from Droege and Sellers18. *Variable recommended for analysis (within set of similar variables).
We also generated an analytical dataset22 (‘Mid-Atlantic USA wild bee abundance per transect, records with sampling info’) derived from dataset two. Dataset three documents wild-bee abundance, trapping method, and sampling effort per transect (see ‘Identifier Variables’ methods above). At the transect level, we defined trap color and volume as the combination of all trap colors and volumes associated with specimens captured in a transect. Table 2 describes all meta-data for dataset three.
Table 2.
Description of all fields in archived dataset 3, wild bee abundance per transect.
| Field Name | Unit | Description |
|---|---|---|
| Identifiers | ||
| TransectID | transect ID | |
| SamplEvent | sampling event ID | |
| SiteID_Year | site ID and year of bee sampling | |
| Apis mellifera abundance | ||
| apis_abund | Apis mellifera abundance | |
| prop_apis | Apis mellifera proportional abundance | |
| Sampling method and effort | ||
| TstVolume | trap volumes used in this transect | |
| TstColor | trap colors used in this transect | |
| ColorVolume | color and volume of traps in transect | |
| VolumeSimple | simplified classification of ‘TstVolume' | |
| ColorSimple | simplified classification of ‘TstColor' | |
| trapdays | days | length of sampling period |
| NTrapsFinal | number of successful traps (set-missing) | |
| Wild bee abundance | ||
| Abundance | specimens | wild bee abundance |
| AbundDayTrap | specimens day−1 trap−1 | wild bee abundance day−1 trap−1 |
Technical Validation
We verified our sampling method and effort data in two ways. First, as described above (see ‘Sampling method and effort’), we manually reviewed field notes for all occurrences lacking sampling effort information. This allowed us to correct instances where sampling data were missed by our automated processing. Second, for occurrences that had sampling effort information, we quantified the accuracy of our processing procedure. We selected a random sample of 1000 occurrences and compared field-note text and sampling-method variables from the original dataset with our sampling effort and method data. We noted whether trap color, trap volume, and the total number of traps were assigned correctly and quantified how much information we created compared with the original BIML dataset.
Overall, we were able to generate or verify pan-trap sampling effort and methodology for 84% of BIML specimens in the Mid-Atlantic U.S. We improved data coverage for trap color, trap volume, and sampling effort by 71%, 4.7%, and 6.1%, respectively. The percentage of occurrences with present, accurate sampling data was good to excellent for all three variables (trap color: 82.2%, trap volume: 88.6%, sampling effort: 95.9%,). Specifically, 888 observations in our validation set were missing trap color data. Using our automated text search, we accurately described trap color for 714 occurrences. Of the remaining 174 occurrences, we generated partial or incorrect data for 27 and 2 occurrences, respectively, and 145 had no color data in the text field notes. For trap volume and sampling effort (the total number of traps used in each sampling event), the original BIML dataset was much more complete. Out of 1000 specimens in our validation dataset, sampling effort and trap volume were missing for 50 and 161 occurrences, respectively. We generated accurate sampling effort for 41 occurrences and corrected BIML data for an additional 20. Of the 161 occurrences lacking trap volume, we filled in 47.
Usage Notes
The BIML data is a rich source of information on wild-bee biodiversity but it has some notable limitations. First, the BIML sampled few locations more than three years. The main goal of BIML collections was to establish baseline estimates of wild-bee species richness in the Mid-Atlantic region. They selected sites representing a wide variety of habitats, focusing on sampling many sites rather than repeated sampling at the same sites over time. Consequently, the BIML dataset has excellent geographic coverage in Maryland, Delaware, and Washington DC (Fig. 1), but lacks long time-series of bee abundance or community composition in consistent locations. Similarly, the BIML did not sample an equal number of sites for all habitat types represented in the dataset. For example, there are relatively few sites in highly agricultural landscapes compared with forested areas, so an analysis of the whole dataset may fail to reveal processes specific to agricultural land. For analyses focused on specific habitat types, we recommend sub-sampling or otherwise adjusting for uneven representation among habitat types.
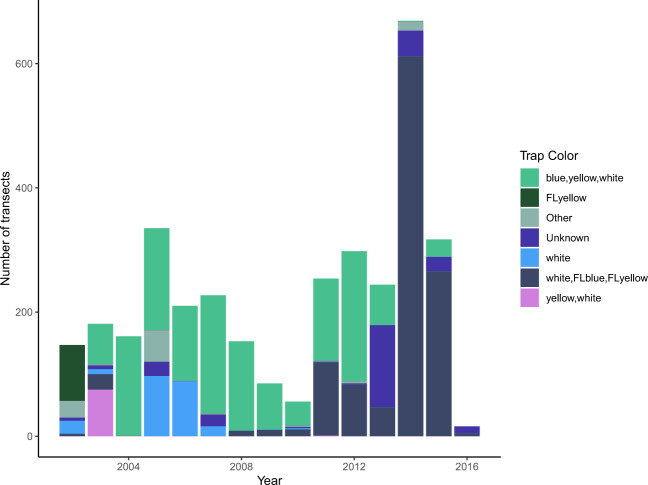
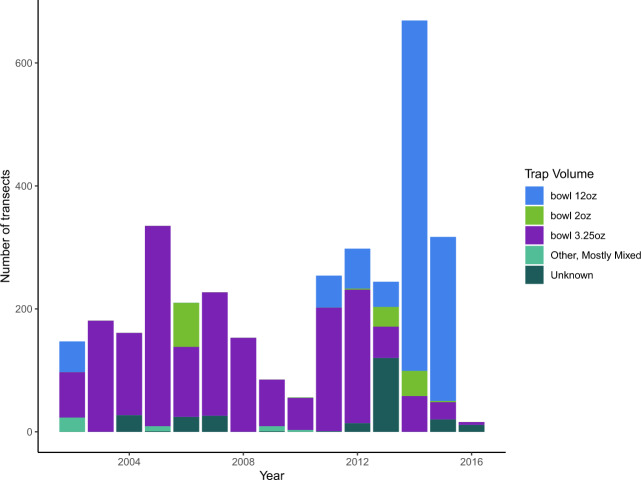
Second, the dominant sampling method changed over the study period. We found that the BIML utilized different colors and volumes of pan traps over time (Figs. 2 and 3). Before 2003, the BIML used a variety of pan-trap colors and volumes. From 2003–2013, the most common sampling method was 3.25 ounce white, blue, and yellow pans, and in 2014, the BIML transitioned almost all their sampling to 12-ounce white, fluorescent blue, and fluorescent yellow bowls.
Fig. 2.
Color of wild bee traps used by USGS BIML from 2002–2016.
Fig. 3.
Volume of wild bee traps used by USGS BIML from 2002–2016.
These features limit, or at best complicate, interpretation of temporal patterns of bee abundance or community composition in the BIML data. For example, a pattern of declining bee abundance over time could be explained by a less effective sampling method or sampling poorer sites or less attractive habitats in later years. In our view, the BIML data are not well suited for assessing temporal trends, as researchers must account for sampling and location changes over time or use a small subset of the data sampled with consistent methodology.
The BIML data we archive here were sampled with pan traps, which impose some inherent biases and limitations. Pan traps significantly under sample large-bodied bees15,27, probably because larger-bodied individuals are able to break surface tension in a container of liquid and fly away. Consequently, Bombus sp., Xylocopa virginica, and other large-bodied bees are less common in pan-trapped data than net-collected or observational datasets. Additionally, some evidence suggests pan traps perform poorly in areas with highly abundant flowering plants28–30, such as field edges adjacent to mass-flowering crops. In proximity to flowering plants, pan traps are less attractive to wild bees than real flowers, leading to an artificially depauperate wild-bee community captured by the pan traps.
Consequently, the BIML datasets we produced are not well suited for studies on large-bodied taxa or in areas with highly abundant flower resources, however these limitations could likely be addressed by combining this dataset with net collections or direct observations. Net collecting has its own limitations of more active time spent per specimen and observer biases, but there are few limitations in common between the two methods. In addition to pan-trapped specimens, the BIML net-collected approximately 16,000 specimens from 2002–2016 in Maryland, Delaware, and Washington DC18, but these collections did not necessarily occur at pan-trapping sites. Also, we did not include net-collected occurrences in our data processing pipeline, so species names should be verified before analysis, and sampling effort information may not exist.
We also advise future analysts that some BIML sampling locations are very close to each other. We created ‘site’ designations as all unique combinations of latitude and longitude, no matter the distance from other sites. In the format we provide, the BIML data violates the assumption of independent observations imposed by a frequentist statistical framework. Not all analytical frameworks assume independent observations, so we did not combine or thin sites, but refer interested parties to statistical methods or software designed for this task (k-means clustering or similar, spThin31 for spatial thinning).
Supplementary information
Acknowledgements
We are indebted to Sam Droege and the USGS BIML team for generously sharing data from their pioneering monitoring effort. We also thank Sarah Goslee, Maggie Douglas, DJ McNeil and Grozinger lab members for helpful discussions on project framing and an earlier version of this manuscript. We thank Shelby Kilpatrick, David Biddinger, and Sam Droege for assistance verifying species names and two anonymous reviewers for feedback that substantially improved the manuscript. Funding was provided by the United State Department of Agriculture National Institute for Food and Agriculture (USDA NIFA) pre-doctoral fellowship PENW-2017-07007 to MK, Foundation for Food and Agriculture Research grant #549032 to CMG, and the Pennsylvania State University Intercollege Graduate Degree Program in Ecology.
Online-only Tables
Author contributions
M.K., J.F.T. and C.M.G. originated concepts. M.K. accessed, cleaned, and processed data and prepared archival version of data and accompanying code. M.K., J.F.T. and C.M.G. wrote the manuscript.
Code availability
All R code used to generate the archived data is available at Zenodo26.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-020-00577-0.
References
- 1.Klein A-M, et al. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Biol. Sci. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ollerton J, Erenler H, Edwards M, Crockett R. Extinctions of aculeate pollinators in Britain and the role of large-scale agricultural changes. Science. 2014;346:1360–1362. doi: 10.1126/science.1257259. [DOI] [PubMed] [Google Scholar]
- 3.Fontaine C, Dajoz I, Meriguet J, Loreau M. Functional diversity of plant–pollinator interaction webs enhances the persistence of plant communities. PLoS Biol. 2005;4:e1. doi: 10.1371/journal.pbio.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvell, C. et al. Design and Testing of a National Pollinator and Pollination Monitoring Framework. Final summary report to the Department for Environment, Food and Rural Affairs (Defra), Scottish Government and Welsh Government: Project WC1101, https://www.ceh.ac.uk/our-science/projects/pollinator-monitoring (2016).
- 5.Droege, S., Engler, J. D., Sellers, E. A. & O’Brien, L. National protocol framework for the inventory and monitoring of bees, https://pubs.er.usgs.gov/publication/70176107 (2016).
- 6.Scherber C, Beduschi T, Tscharntke T. Novel approaches to sampling pollinators in whole landscapes: a lesson for landscape-wide biodiversity monitoring. Landsc. Ecol. 2019;34:1057–1067. doi: 10.1007/s10980-018-0757-2. [DOI] [Google Scholar]
- 7.Lebuhn, G. et al. A standardized method for monitoring Bee Populations – The Bee Inventory (BI) Plot, http://online.sfsu.edu/beeplot/pdfs/Bee%20Plot%202003.pdf (2003).
- 8.Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odoux J-F, et al. ECOBEE: a tool for long-term honey bee colony monitoring at the landscape scale in West European intensive agroecosystems. J. Apic. Res. 2014;53:57–66. doi: 10.3896/IBRA.1.53.1.05. [DOI] [Google Scholar]
- 10.Magurran AE, et al. Long-term datasets in biodiversity research and monitoring: assessing change in ecological communities through time. Trends Ecol. Evol. 2010;25:574–582. doi: 10.1016/j.tree.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Theobald EJ, et al. Global change and local solutions: Tapping the unrealized potential of citizen science for biodiversity research. Biol. Conserv. 2015;181:236–244. doi: 10.1016/j.biocon.2014.10.021. [DOI] [Google Scholar]
- 12.Chandler M, et al. Contribution of citizen science towards international biodiversity monitoring. Biol. Conserv. 2017;213:280–294. doi: 10.1016/j.biocon.2016.09.004. [DOI] [Google Scholar]
- 13.Westphal C, et al. Measuring bee diversity in different European habitats and biogeographical regions. Ecol. Monogr. 2008;78:653–671. doi: 10.1890/07-1292.1. [DOI] [Google Scholar]
- 14.Nielsen A, et al. Assessing bee species richness in two Mediterranean communities: importance of habitat type and sampling techniques. Ecol. Res. 2011;26:969–983. doi: 10.1007/s11284-011-0852-1. [DOI] [Google Scholar]
- 15.Joshi NK, et al. Comparative Trapping Efficiency to Characterize Bee Abundance, Diversity, and Community Composition in Apple Orchards. Ann. Entomol. Soc. Am. 2015;108:785–799. doi: 10.1093/aesa/sav057. [DOI] [Google Scholar]
- 16.Leong JM, Thorp R. The pan trap colour preferences of oligolectic and nonoligolectic bees associated with a vernal pool plant. Ecol. Entomol. 1999;24:329–335. doi: 10.1046/j.1365-2311.1999.00196.x. [DOI] [Google Scholar]
- 17.Tuell JK, Isaacs R. Elevated pan traps to monitor bees in flowering crop canopies. Entomol. Exp. Appl. 2009;131:93–98. doi: 10.1111/j.1570-7458.2009.00826.x. [DOI] [Google Scholar]
- 18.Droege, S. & Sellers, E. USGS PWRC - Native Bee Inventory and Monitoring Lab (BIML). Version 1.5. United States Geological Survey. Occurrence Dataset, 10.15468/6autvb (2017).
- 19.Minckley, R. L., Roulston, T. H. & Williams, N. M. Resource assurance predicts specialist and generalist bee activity in drought. Proc. R. Soc. B Biol. Sci. 280 (2013). [DOI] [PMC free article] [PubMed]
- 20.Stone GN, et al. Windows of opportunity and the temporal structuring of foraging activity in a desert solitary bee. Ecol. Entomol. 1999;24:208–221. doi: 10.1046/j.1365-2311.1999.00181.x. [DOI] [Google Scholar]
- 21.Dyer JM. Revisiting the Deciduous Forests of Eastern North America. BioScience. 2006;56:341. doi: 10.1641/0006-3568(2006)56[341:RTDFOE]2.0.CO;2. [DOI] [Google Scholar]
- 22.Kammerer M, Tooker JF, Grozinger CM. 2020. A long-term dataset on wild bee abundance in Mid-Atlantic United States. figshare. [DOI] [PMC free article] [PubMed]
- 23.Ascher, J. S. & Pickering, J. Discover Life bee species guide and world checklist (Hymenoptera:Apoidea:Anthophila), http://www.discoverlife.org/mp/20q?guide=Apoidea_species (2013).
- 24.Wickham, H. stringr: Consistent, Wrappers for Common String Operations. (2018).
- 25.R Core Team. R: A Language and Environment for Statistical Computing. (2019).
- 26.Kammerer, M. melaniekamm/CleanParseDroegeBees: Source code for: A long-term dataset on wild bee abundance in Mid-Atlantic United States. Zenodo, 10.5281/zenodo.3528323 (2020). [DOI] [PMC free article] [PubMed]
- 27.Roulston TH, Smith SA, Brewster AL. A Comparison of Pan Trap and Intensive Net Sampling Techniques for Documenting a Bee (Hymenoptera: Apiformes) Fauna. J. Kans. Entomol. Soc. 2007;80:179–181. doi: 10.2317/0022-8567(2007)80[179:ACOPTA]2.0.CO;2. [DOI] [Google Scholar]
- 28.Baum KA, Wallen KE. Potential Bias in Pan Trapping as a Function of Floral Abundance. J. Kans. Entomol. Soc. 2011;84:155–159. doi: 10.2317/JKES100629.1. [DOI] [Google Scholar]
- 29.Rhoades P, et al. Sampling technique affects detection of habitat factors influencing wild bee communities. J. Insect Conserv. 2017;21:703–714. doi: 10.1007/s10841-017-0013-0. [DOI] [Google Scholar]
- 30.Popic TJ, Davila YC, Wardle GM. Evaluation of Common Methods for Sampling Invertebrate Pollinator Assemblages: Net Sampling Out-Perform Pan Traps. PLoS ONE. 2013;8:e66665. doi: 10.1371/journal.pone.0066665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B. & Anderson, R. P. spThin: Functions for Spatial Thinning of Species Occurrence Records for Use in Ecological Models. (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kammerer M, Tooker JF, Grozinger CM. 2020. A long-term dataset on wild bee abundance in Mid-Atlantic United States. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All R code used to generate the archived data is available at Zenodo26.