Summary
Circular RNAs (circRNAs) are stable non-coding RNAs with a closed circular structure. One of the best studied circRNAs is ciRS-7 (CDR1as), which acts as a regulator of the microRNA miR-7; however, its biosynthetic pathway has remained an enigma. Here we delineate the biosynthetic pathway of ciRS-7. The back-splicing events that form circRNAs are often facilitated by flanking inverted repeats of the primate-specific Alu elements. The ciRS-7 gene lacks these elements, but, instead, we identified a set of flanking inverted elements belonging to the mammalian-wide interspersed repeat (MIR) family. Splicing reporter assays in HEK293 cells demonstrated that these inverted MIRs are required to generate ciRS-7 through back-splicing, and CRISPR/Cas9-mediated deletions confirmed the requirement of the endogenous MIR elements in SH-SY5Y cells. Using bioinformatic searches, we identified several other MIR-dependent circRNAs and confirmed them experimentally. We propose that MIR-mediated RNA circularization is used to generate a subset of mammalian circRNAs.
Subject Areas: Biological Sciences, Molecular Biology
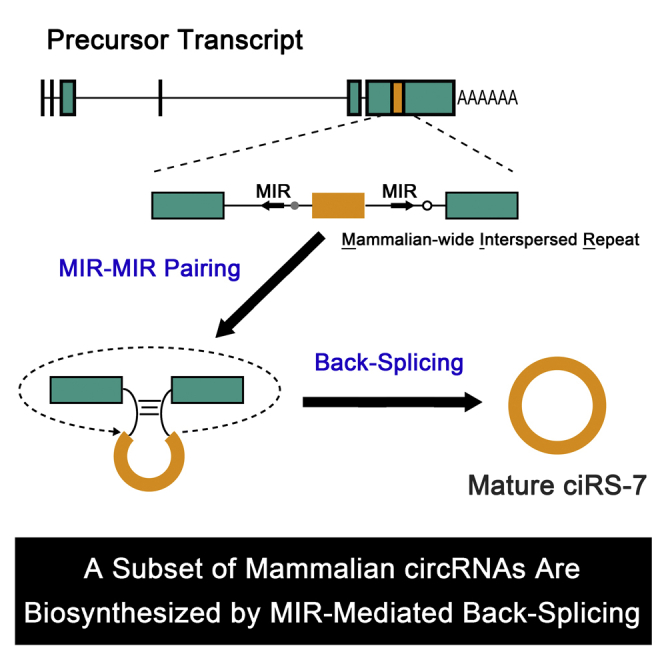
Graphical Abstract
Highlights
-
•
The circular RNA, ciRS-7 (CDR1as), functions as a regulator of miR-7
-
•
ciRS-7 is generated by back-splicing, not via intra-lariat splicing
-
•
Back-splicing of ciRS-7 is promoted by the flanking inverted MIR elements
-
•
The biosynthesis of a subset of mammalian circRNAs could be mediated by MIRs
Biological Sciences; Molecular Biology
Introduction
Endogenous circular RNAs (circRNAs) were first observed as scrambled exon transcripts (Nigro et al., 1991), and these transcripts were found to have circular structures with covalently closed ends (Capel et al., 1993; Cocquerelle et al., 1993). For two decades, however, they were disregarded as rare oddities, or regarded as poorly expressed mis-spliced products.
High-throughput sequencing of full transcriptomes (RNA sequencing; RNA-seq) has identified thousands of circRNAs in eukaryotes, and these are now considered common by-products of many protein-coding genes (Salzman et al., 2012; Jeck et al., 2013; Memczak et al., 2013). The significance of the vast majority of circRNAs remains unknown; however, some circRNAs have important biological functions. For instance, certain circRNAs control the stability and activity of micro RNAs (miRNAs), regulate transcription or alternative splicing, affect translation of host genes, or can even be translated and produce proteins themselves (reviewed in Wilusz, 2018). ciRS-7 (also known as CDR1as) was one of the first functionally annotated circular RNAs. It is conserved among mammals and is mainly expressed in the brain. ciRS-7 has many binding sites for a particular miRNA, miR-7, and a single binding site for miR-671 that triggers Argonaute2-catalyzed slicing of ciRS-7 (Hansen et al., 2011). In cellulo experiments suggested that ciRS-7 may function as a sponge, or decoy, that reduces the available free miR-7 and thus prevents repression of miR-7 targeted mRNAs (Hansen et al., 2013; Memczak et al., 2013). Knockout mice lacking the ciRS-7 genomic locus down-regulated miR-7 in the brain, suggesting that ciRS-7 has a role in stabilizing or transporting miR-7 (Piwecka et al., 2017). As a result, the ciRS-7 knockout mice had impaired sensorimotor gating, which is associated with neuropsychiatric disorders.
Recent gene editing experiments revealed a comprehensive regulatory network in mouse brain between a long non-coding RNA (lncRNA), circRNA, and two miRNAs (Kleaveland et al., 2018). This network involves the Cyrano lncRNA that promotes the degradation of miR-7, which in turn enhances the miR-671-directed degradation of ciRS-7, meaning that Cyrano causes an accumulation of ciRS-7. ciRS-7 is apparently a key component in a gene-regulatory network in the brain, but understanding the biosynthesis and transport of this particular circRNA remains an important challenge.
It is particularly important to understand the biosynthesis of endogenous ciRS-7. In an artificial ciRS-7 expression vector, the insertion of 800 nucleotides (nt) of perfectly complementary sequences into flanking introns is technically sufficient to circularize ciRS-7 (Hansen et al., 2013); however there are no such extensive stretches of complementarity found near the endogenous ciRS-7 exon. In human, the pairing of inverted Alu repeats, a primate-specific repetitive element, has been shown to promote direct back-splicing in a subgroup of circRNAs (Jeck et al., 2013; Liang and Wilusz, 2014; Zhang et al., 2014; Venø et al., 2015; Zheng et al., 2016). Here we showed that highly conserved mammalian-wide interspersed repeat (MIR) sequences, but not Alu sequences, in the flanking introns of the ciRS-7 are required for back-splicing to generate the circular RNA structure. Bioinformatics analyses followed by reporter assays furthermore identified additional distinct circRNAs generated by inverted MIR sequences. Here, we demonstrate that a subset of mammalian circRNAs are generated by mammalian-wide MIR-mediated back-splicing.
Results
Experimental Evidence Supports ciRS-7 Generation through Back-Splicing
Two major circRNA biosynthetic pathways, “Intra-lariat splicing pathway” and “Back-splicing pathway,” have been proposed (Figure S1A; reviewed in Jeck and Sharpless, 2014; Wilusz, 2018). Both pathways involve a splicing reaction between downstream 5′ and upstream 3′ splice sites of the circularized exon(s); however, this splicing event occurs either directly on the loop structure formed via the flanking intronic complementary sequences in a Back-splicing pathway, or on the lariat-structure generated by exon-skipping splicing in an Intra-lariat splicing pathway (Figure S1A).
To determine the pathway used to generate ciRS-7, we first examined the ciRS-7 precursor transcript (∼80 kb) that includes six non-coding exons and five introns (Figure S2). The last exon contains the entire ciRS-7 sequence, and we identified new flanking alternative 5′ and 3′ splice sites (Figures S1C and S2). We could detect closed circular form of ciRS-7 (C) together with several linear spliced products (M) due to multiple alternative 5′ and 3′ splice sites (Figure S1B). The precursor transcript (P) was also detected, and this could overlap either the lariat precursor (L1) from Intra-lariat splicing pathway or the looped form of precursor (P′) from Back-splicing pathway (Figures S1A and S1B).
The two possible pathways can be distinguished by blocking of specific splicing events using antisense oligoribonucleotides (ASOs). The ASOs 1 + 2 targeting the flanking 5′ and 3′ splice sites of ciRS-7 exon (Figures S1A and S1C; blue bars) can prevent circular ciRS-7 generation via both pathways and thus serve as the positive control (Figure S1A). In contrast, the ASOs 3–6 targeting alternative 5′ and 3′ splice sites outside of ciRS-7 exon (red bars) can prevent the exon-skipping and the following circular ciRS-7 generation in the Intra-lariat pathway (Figure S1A; left panel). However, in the Back-splicing pathway, these same ASOs 3–6 cannot interfere with the back-splicing event and would therefore allow circular ciRS-7 generation (Figure S1A; right panel). We found that ASOs 1 + 2 significantly repressed ciRS-7 production (given the efficiencies of transfection and annealing of ASOs), whereas none of the ASOs 3–6 inhibited ciRS-7 production (Figure S1D). These data thus imply that the Back-splicing pathway but not Intra-lariat pathway produces circular ciRS-7 (Figure S1A).
To further validate this finding, we made a genomic deletion of the same relevant splice sites by CRISPR-Cas9-mediated technology (Figures S3A and S3B). The deletion of either the alternative 5′ splice sites (between gRNA2 and gRNA3) or the 3′ splice sites (between gRNA4 and gRNA5) outside of ciRS-7 exon barely prevented the generation of ciRS-7 (Figure S3C), confirming that circular ciRS-7 is biosynthesized through the Back-splicing pathway but not through the Intra-lariat pathway.
The ciRS-7 Locus Is Flanked by Inverted MIR Sequences
Since our results demonstrated that the Back-splicing pathway but not the Intra-lariat splicing pathway is responsible for generating ciRS-7, we first searched for the inverted Alu elements thought to be required for the Back-splicing pathway. Using the UCSC RepeatMasker (in human hg19 coordinates) we identified two human Alu elements upstream of ciRS-7 within exon 5 and exon 6 (Figure S4) but found no downstream Alu element that could generate the required base pairing around the ciRS-7 exon. Therefore, flanking inverted Alu elements are unlikely to account for the generation of ciRS-7.
However, we did find two relatively conserved regions upstream and downstream of the ciRS-7 exon in an inverted orientation that are conserved both in human and mouse (Figure 1A). These regions overlapped with MIRs, an ancient short interspersed nuclear elements (SINE) family conserved among mammals and marsupials (Jurka et al., 1995; Krull et al., 2007). We found significant complementarities in these inverted pairs of MIRs (analyzed later), suggesting that ciRS-7 could be generated via a back-splicing pathway facilitated by inverted MIRs.
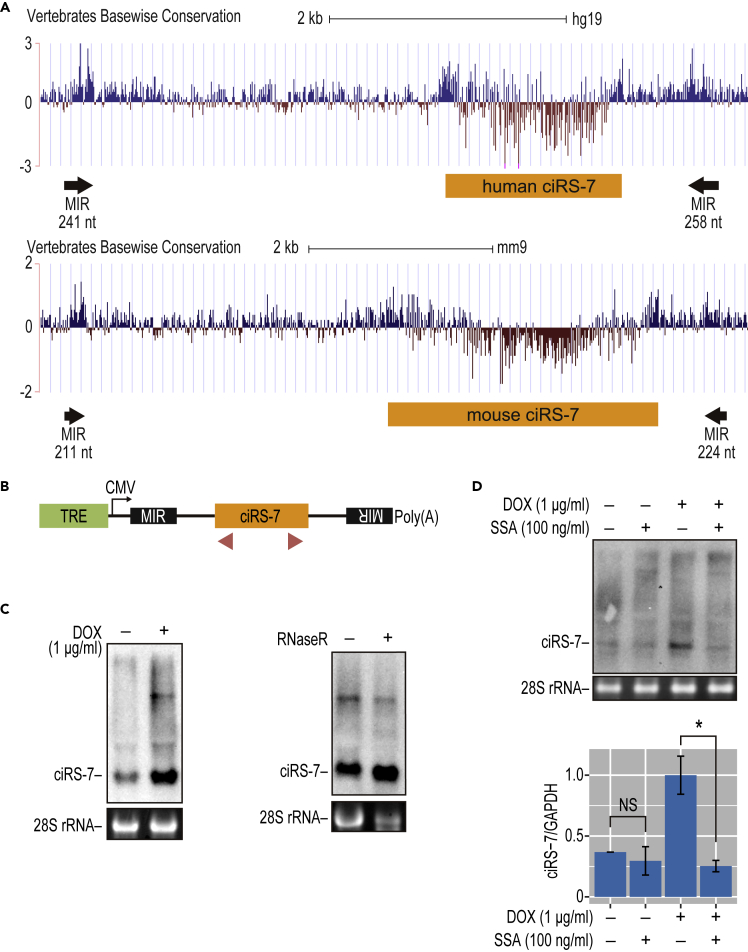
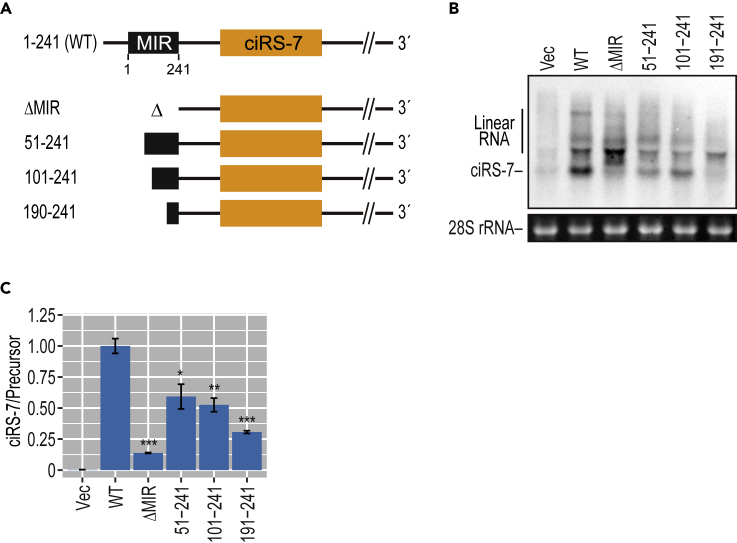
Figure 1.
The Production of ciRS-7 Depends on Inverted MIRs in the Flanking Regions
(A) The ciRS-7 locus (human/mouse ciRS-7) and the identified conserved inverted MIR elements (MIRs with the lengths are indicated). The complementarity of these inverted MIRs was modeled in Figure S8. The vertebrate base-wise conservations of human (upper) and mouse (lower) genomic sequences around the ciRS-7 precursor are displayed in the UCSC Genome Browser (GRCh37/hg19 version).
(B) Schematic structure of ciRS-7-expression plasmids. Transcription is driven by a CMV promoter, and an arrow denotes the initiation site. The tetracycline response element (TRE), inverted MIR elements, ciRS-7 coding exon (ciRS-7) are indicated. Red triangle indicates the positions of PCR primers used to detect ciRS-7 and its precursor, respectively.
(C) Detection of ciRS-7 from expression plasmid in stably transduced HEK293 cells. The cells were treated with DOX and extracted total RNA was analyzed by Northern blotting (left panel). 28S rRNA was visualized by ethidium bromide staining as a loading control. To validate the circular structure of ciRS-7, total RNA was treated with RNase R prior to the Northern blotting analysis (right panel).
(D) The effect of splicing inhibitor, SSA, on the production of ciRS-7. The same stably transduced cells described in (C) were treated with DOX (+lanes) followed by SSA addition (+lanes). After 12 h culture, extracted total RNA was analyzed by Northern blotting to detect ciRS-7 (upper panel). Ethidium bromide-stained 28S rRNA was shown as a loading control. The total RNA was also analyzed by RT-qPCR, and the graph shows quantification of the ciRS-7 generation (lower panel). ciRS-7 expression was normalized to GAPDH (ciRS-7/GAPDH) and plotted as ratios to the value of DOX-induced cells. Means ± SD are given for three independent experiments (∗p < 0.05, NS = not significant).
Inverted MIRs Promote ciRS-7 Biosynthesis
To test this hypothesis, we generated a stable HEK293 cell line that expresses a doxycycline (DOX)-inducible 5-kb transcript containing the ciRS-7 exon flanked by upstream and downstream inverted MIRs (Figure 1B).
The ciRS-7 circular product and its precursor RNA were both detected by Northern blot 24 h after induction with DOX (Figure 1C, left panel). We verified that the final ciRS-7 product was indeed circular by pre-treatment with RNase R (Figure 1C, right panel). These results demonstrate that our mini-gene, covering the ciRS-7 exon and its inverted MIRs, recapitulates the endogenous generation of circular ciRS-7. To test if ciRS-7 generation from the mini-gene system is splicing dependent, we used the general pre-mRNA splicing inhibitor Spliceostatin A (SSA) (Kaida et al., 2007; Yoshimoto et al., 2017). SSA clearly inhibited DOX-induced ectopic ciRS-7 production, suggesting that ciRS-7 is indeed generated by splicing (+DOX lanes, Figure 1D).
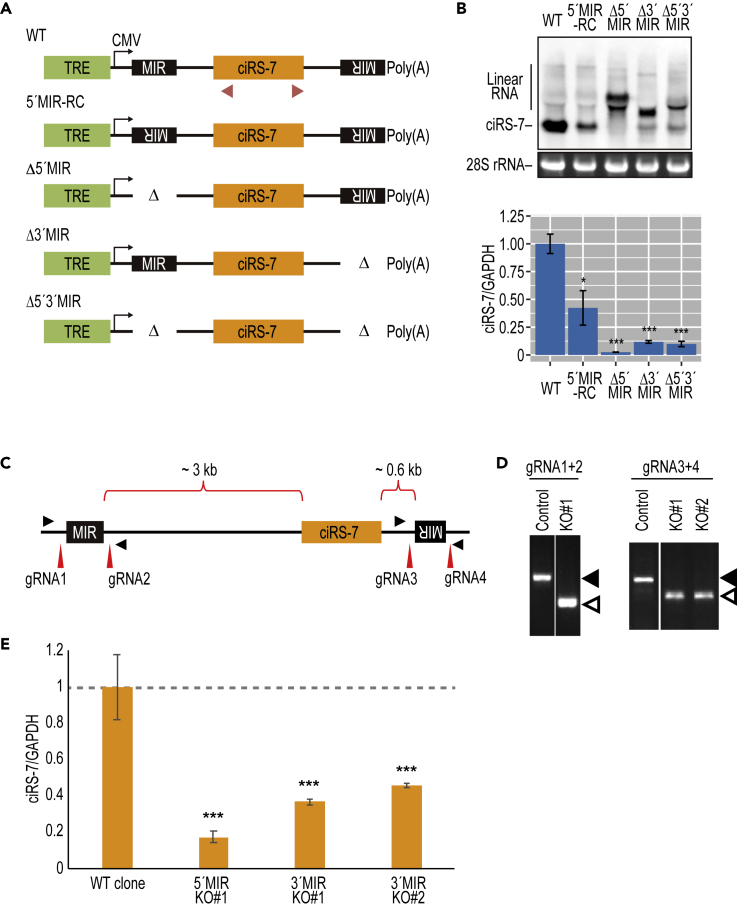
Using this stable cell line system, we next made a series of MIR-deleted mini-genes to analyze the role of MIRs in human ciRS-7 biosynthesis (Figure 2A). The wild-type mini-gene efficiently generated ciRS-7 (Figure 2B; WT), whereas the deletion of either the upstream or downstream MIR element (Δ5′MIR, Δ3′MIR, Δ5′3′MIR) and the reversion of the upstream MIR element to the same orientation as the downstream MIR element (5′MIR-RC) seriously impaired ciRS-7 production (Figures 2B and S5), demonstrating that ciRS-7 generation depends on both inverted MIRs.
Figure 2.
Deletion of Either Upstream or Downstream MIRs Diminishes the Production of ciRS-7
(A) Schematic structures of ciRS-7-expression plasmids with or without MIR sequence (see the legend of Figure 2B for CMV and TRE). Red triangles indicate the positions of PCR primers to detect ciRS-7 in (B).
(B) Generation of ciRS-7 from reporters, with or without MIR sequences, in stably transduced HEK293 cells. Total RNA extracted from the DOX-treated cells was analyzed by Northern blotting. 28S rRNA was detected by ethidium bromide staining as a control. The band corresponding to circular ciRS-7 is indicated. The upper bands observed, especially in the Δ5′/Δ3′MIR mutants, were identified as linear precursor RNAs by RNase R digestion and RT-qPCR (Figure S5). Total RNA was also quantified by RT-qPCR (lower panel). ciRS-7 expression was normalized to GAPDH (ciRS-7/GAPDH) and plotted as ratios to the value of control wild-type (WT) plasmid-expressing cells. Means ± SD are given for three independent experiments (∗∗∗p < 0.001, ∗p < 0.05).
(C) Schematic showing the genomic structure of the ciRS-7 locus with the flanking inverted MIR sequences. The positions of guide RNAs (gRNA1–gRNA4) to delete each MIR element are indicated with red vertical arrowheads. PCR primers for detecting deleted sites are indicated with filled triangles.
(D) The genomic deletions of the flanking MIR elements in SH-SY5Y cell clones were verified by genomic PCR. The indicated two pairs of gRNAs were used to delete the 5′ and 3′ MIR elements. PCR primers indicated in (A) were used to detect the MIR deletion. Open and filled triangles point to the deleted (KO#1, KO#2) and non-deleted (Control) alleles, respectively.
(E) The effect of the homozygous MIR deletions (5′MIR KO and 3′MIR KO; #1,2 denotes cell clones in D) on ciRS-7 production was quantified by RT-qPCR. ciRS-7 expression was normalized to GAPDH in the same clones (ciRS-7/GAPDH). Values are relative to the value of control wild-type clones (WT). Means ± standard deviation (SD) are given for three independent experiments (∗∗∗p < 0.001).
Finally, we used CRISPR-Cas9-mediated editing to delete the endogenous MIR sequences in the neuronal SH-SY5Y cell line. Two guide RNAs (gRNAs) sets were designed to target the boundaries of the 5′ and 3′ MIR sequences (Figure 2C) and co-transfected with the Cas9 expression vector to delete the MIRs. The efficiencies of targeted genomic deletion were determined by PCR analyses using primers flanking the targeted regions and the PCR-amplified fragments were resolved by gel electrophoresis (Figure 2D). We observed that circularization was markedly inhibited when either the upstream or downstream MIR sequence was deleted (Figure 2E). Together, we conclude that the ciRS-7 is generated via the back-splicing pathway promoted mainly by its flanking inverted MIR sequences.
Other MIR-Dependent and MIR-Independent circRNAs Were Identified
Since MIRs are ancient SINEs and ubiquitous in mammalian genomes (Jurka et al., 1995), we speculated that the utilization of MIRs in promoting back-splicing to generate “MIR-dependent” circRNAs could be a globally conserved strategy rather than a gene-specific event.
Upon bioinformatics analysis, we indeed found that MIRs neighboring ciRS-7 are conserved among Primates, Euarchontoglires, Laurasiatheria, Afrotheria, and Armadillo. Here, we show the maps of ciRS-7 with the MIRs of human and mouse (Figure 1A). We went on to search for other potential MIR-dependent circRNAs. Using the RepeatMasker track in human hg19 coordinates, we found a total of 595,094 MIRs. Comparing with the mouse mm9 coordinates, 104,074 are conserved between human and mouse. Previously, 7,771 circRNAs were identified from RNA-seq data of foreskin cells (Jeck et al., 2013). Using this dataset, we identified 846 circRNA candidates with conserved inverted intronic MIRs within 3,000 nt upstream and downstream of the circularized exons.
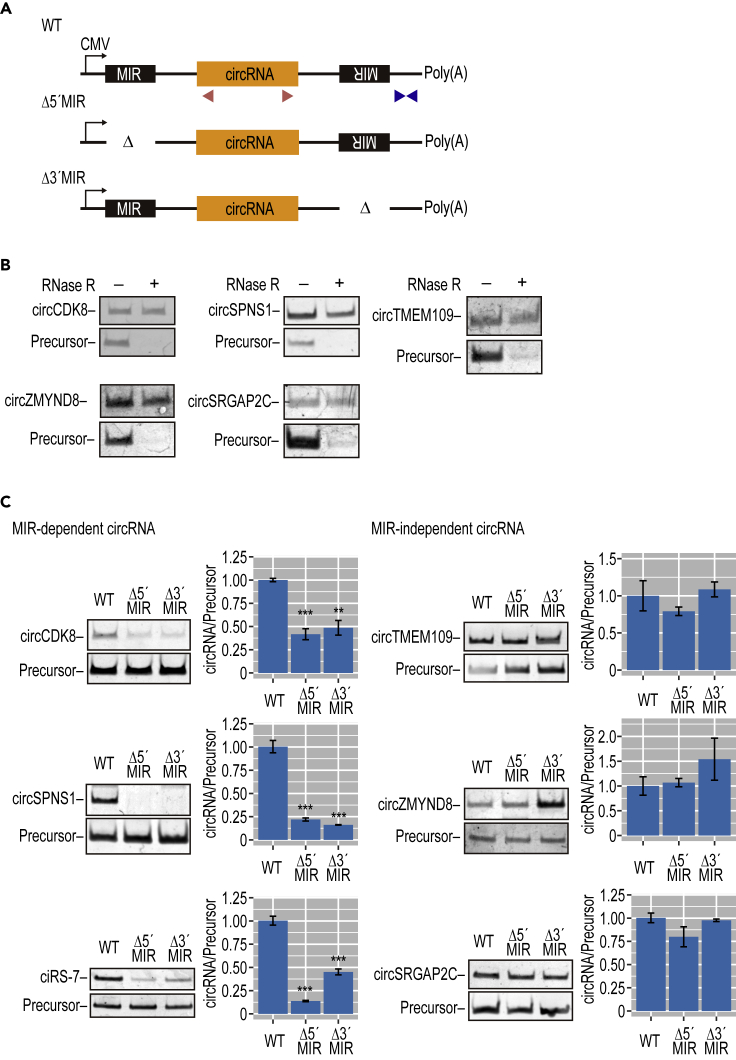
From these 846 circRNA candidates, we first eliminated circRNAs with very large precursors (>5 kb) since these are hard to validate using mini-gene reporters. Among the circRNAs showing ubiquitous expression, we arbitrarily chose five circRNAs as representatives; circCDK8, circSPNS1, circTMEM109, circZYMND8, and circSRGAP2C (Figure S6 for the maps with identified MIRs). To test whether biosynthesis of these circRNAs is MIR dependent or not, we made a series of MIR-deleted mini-genes as we had done for ciRS-7 (Figure 3A). Plasmids expressing these mini-genes were transfected into mouse N2A cells, in which ectopically expressed human circRNAs could be discriminated from the endogenously expressed mouse circRNAs. All five circRNAs were successfully detected from the wild-type mini-genes, and their circular structure was verified by RNase R digestion (Figure 3B). Deletion of either upstream or downstream MIRs prevented the production of circCDK8 and circSPNS1 (Figure 3C; MIR-dependent circRNAs) but had no effect on the production of circTMEM109, circZYMND8, and circSPGAP2C (MIR-independent circRNAs).
Figure 3.
Several Other Human MIR-Dependent circRNAs Were Identified
(A) Schematic structure of plasmid expressing wild-type (WT) or MIR deletions (Δ5′MIR, Δ3′MIR), which was constructed for five circRNAs. Red and blue triangles indicate the positions of PCR primers to detect circRNA and precursors, respectively.
(B) RT-PCR detection of five circRNAs in mouse N2A cells transfected with the plasmids depicted in (A). Total RNA from the cells was treated with (+) or without (−) RNase R to discriminate the circular from the linear structure.
(C) Identification of MIR-dependent and MIR-independent circRNAs. RT-PCR analysis of circRNAs and their precursors, expressed from the plasmid types depicted in (A). Total RNA was also quantified by RT-qPCR. The circRNA expression levels were normalized to the expression levels of precursor RNA as controls (circRNA/Precursor). Values are relative to the value of control wild-type clones (WT). Means ± SD are given for three independent experiments (∗∗∗p < 0.001, ∗∗p < 0.01).
To confirm this in an endogenous setting, CRISPR-Cas9-mediated editing was applied to delete the MIR elements flanking the circCDK8 (Figure S7A). The effective deletion was confirmed by PCR (Figure S7B). Using RT-qPCR, we observed marked inhibition of circularization when either the upstream or downstream MIR sequences were deleted (Figure S7C).
MIR-MIR Base-Pairing Stability Is Critical for circRNA Generation
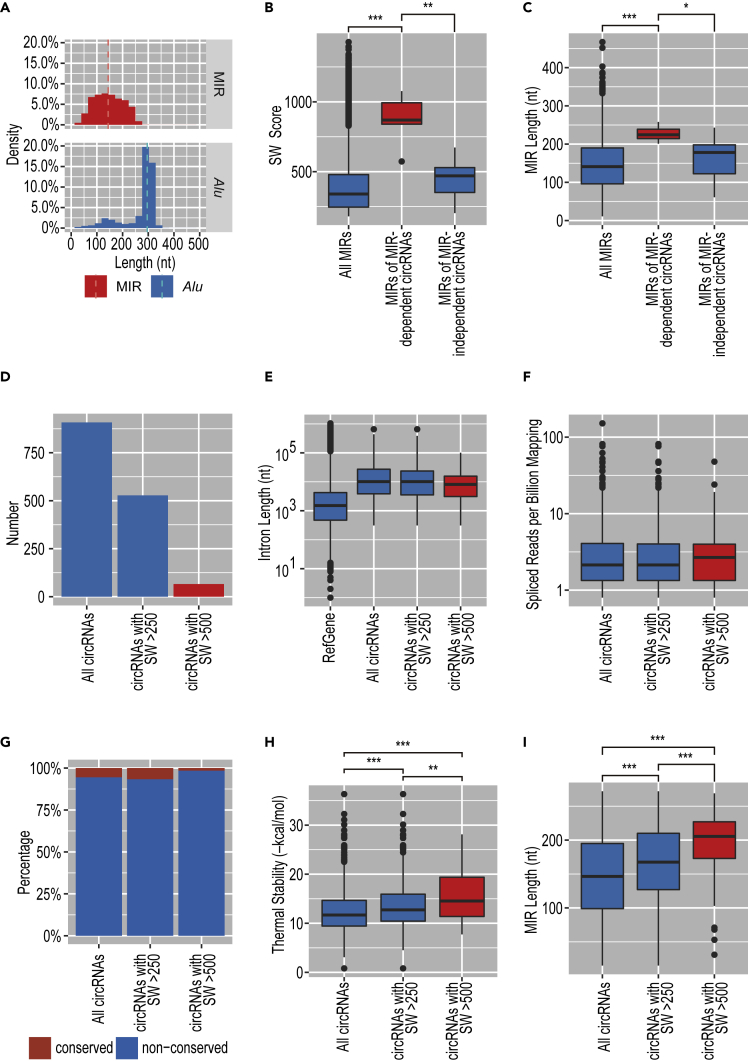
MIRs are an older SINE family than Alus (∼130 versus ∼60 million years ago), and MIR elements have often become truncated during this time (Jurka et al., 1995). In agreement with this history, the length distribution of MIRs is much broader than that for the Alus (Figure 4A). We assumed that such truncations in MIRs would cause instability of the MIR-MIR base pairing required for efficient circRNA production.
Figure 4.
Biosynthesis of MIR-Dependent circRNAs Likely Depends on Stable Pairing between Highly Conserved MIRs
(A) The length distribution for MIRs is broader than for Alus. The bin width used in the histogram is 20 nt. A broken line indicates the median length.
(B) The verified MIR-dependent circRNAs have MIRs with near-consensus features. Boxplot of SW alignment scores of all reported MIRs, three MIR-dependent circRNAs, and three MIR-independent circRNAs (∗∗∗p < 0.001; ∗∗p < 0.01). See Figure S6 for each SW score of these MIRs.
(C) The verified MIR-dependent circRNAs have longer MIRs than those of MIR-independent circRNAs. Boxplot of the MIR lengths of all reported MIRs, three MIR-dependent circRNAs, and three MIR-independent circRNAs (∗∗∗p < 0.001; ∗p < 0.05).
(D) We identified 846 circRNAs (left bar) that have inverted MIRs within 3,000 nt of the circRNA-producing exon. Among these circRNAs, 528 circRNAs had MIRs with SW score >250 (middle bar) and 66 circRNAs had MIRs with SW score >500 (right bar).
(E) MIR-dependent circRNAs are not characterized by the flanking intron length. Boxplot shows the flanking intron lengths (nt) of the circRNAs (from the groups shown in D).
(F) MIR-dependent circRNAs are not characterized by the expression levels of circRNAs. Boxplot shows the spliced reads per billion mapping of the circRNAs (from the groups shown in D).
(G) MIR-dependent circRNAs are not characterized by the conservation rate. The orthologous circRNAs found in both human and mouse were identified as “conserved” circRNAs and the number was calculated. Bar graph shows the ratio of the conserved and non-conserved circRNAs (from the groups shown in D).
(H) MIR-dependent circRNAs are characterized by the thermal stability, suggesting that inverted MIRs with higher SW scores facilitate circularization through stable MIR-MIR pairing. Boxplot shows the thermal stabilities (–kcal/mol) of inverted MIRs of the circRNAs (from the groups shown in D), which are calculated by the RNAup program (∗∗∗p < 0.001; ∗∗p < 0.01).
(I) MIR-dependent circRNAs are characterized by the relative length of the MIRs. Boxplot shows the MIR length of the circRNAs (from the groups shown in D) (∗∗∗p < 0.001).
To test this hypothesis, we evaluated the integrity of MIRs located around MIR-dependent and MIR-independent circRNAs by comparing them with the MIR consensus sequence using a Smith-Waterman (SW) alignment score (Smith and Waterman, 1981). As predicted, MIRs of MIR-dependent circRNAs had markedly higher alignment scores than those of MIR-independent circRNAs (Figures 4B and S6 for the individual score of all MIRs). We also observed that the MIRs of MIR-dependent circRNAs are significantly longer than those of MIR-independent circRNAs (Figure 4C).
Using these criteria of the SW alignment score, we estimated the number of MIR-dependent circRNAs from our identified 846 circRNAs with flanking inverted MIRs. We found that 528 and 66 circRNAs have higher SW scores than 250 and 500, respectively (Figure 4D). The high MIR-SW scores (573–1,077; Figures 3C and S6A and the legend) seen for the experimentally confirmed MIR-dependent circRNAs suggest that these 66 circRNAs (>500 MIR-SW scores) are all likely to be biosynthesized by an MIR-dependent pathway. Then we analyzed these two sets of circRNAs (SW > 250 and SW > 500) in relation to length of the flanking intron (Figure 4E), the expression level (Figure 4F), the conservation rate (Figure 4G), and the thermal stability of MIR-MIR pairing (Figure 4H). The comparisons demonstrate that these two sets of circRNAs (SW > 250 and SW > 500) can be well characterized by the thermal stability. Together, circRNAs with higher SW alignment scores can form more stable MIR-MIR pairing (Figure 4H), which might be attributed to the relatively long length of MIRs (Figure 4I). To validate this notion, the 5′ MIR of the ciRS-7 expressing plasmid was shortened by stepwise trimming from the 5′ end (Figure 5A) and the ciRS-7 generation was examined (Figures 5B and 5C). The ciRS-7 generation was gradually decreased from the deleted MIR length of 191 nt (51–241), 141 nt (101–241), and 51 nt (190–241), indicating that the efficient back-splicing requires at least 200 nt length of the inverted MIRs, which is exactly consistent with the MIR length distribution of MIR-dependent circRNAs (Figure 4C).
Figure 5.
Shortening the Flanking MIRs Reduces circRNA Biosynthesis
(A) Schematic structures of MIR-deleted ciRS-7-expression plasmids. The numbers indicate the lengths of the deleted 5′ MIR elements.
(B) Generation of ciRS-7 from reporters, with various MIR lengths, in transiently transfected ciRS-7 knockout HEK293 cells. Total RNA extracted from DOX-treated cells was analyzed by Northern blotting. 28S rRNA was detected by ethidium bromide staining as a control. The band corresponding to ciRS-7 is indicated on the left (see Figure 2B legend for the upper products).
(C) ciRS-7 formation was also quantified by RT-qPCR. Expression levels were normalized to the level of precursor RNA (ciRS-7/Precursor) and plotted as ratios to the value of control wild-type (WT) plasmid-expressing cells. Therefore, these quantified results do not exactly match up to the apparent view of Northern data in (B). Means ± SD are given for three independent experiments (∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05).
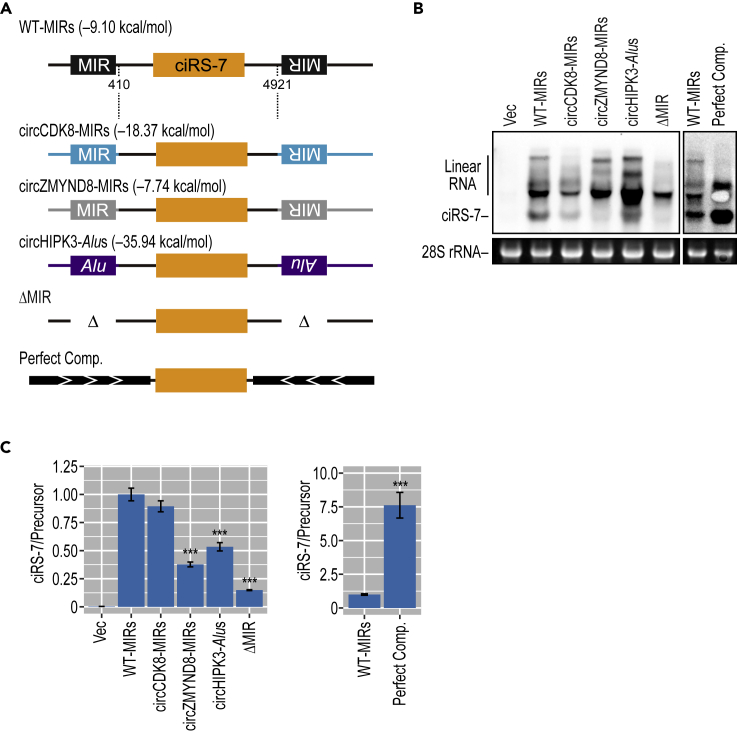
The predicted complementarity of inverted MIRs of all six selected circular RNAs together with three Alu-dependent circRNAs revealed that the MIR-MIR base pairings of MIR-dependent circRNAs are generally more stable than those of MIR-independent circRNAs, although not so much as those of Alu-dependent circRNAs (Figure S8). We experimentally tested the efficiency of the flanking MIRs and Alus elements in generating ciRS-7 using heterologous ciRS-7 expressing plasmids (Figure 6). Notably, the insertion of heterologous Alu elements (derived from circHIPK3 intron) in the ciRS-7 expression construct supported even weaker back-splicing activity compared with the original ciRS-7 construct (Figure 6), suggesting that not only the stability of base pairing but also the combination between flanking inverted sequences and the ciRS-7 exon affects in the efficiency of circularization. Together, we conclude that a subset of mammalian circRNAs contains evolutionally conserved, flanking, and inverted MIRs that mediate circularization via stable pairing.
Figure 6.
The Flanking Inverted Repeats and the ciRS-7 Exon Are Cooperative in ciRS-7 Circularization
(A) Schematic structures of chimeric ciRS-7-expressing reporters with heterologous flanking repeated elements. The indicated heterologous MIR and Alu elements were inserted in flanking introns of the ciRS-7 expressing plasmid (used in Figure 1B). The ciRS-7 expressing plasmid with ∼800-nt flanking inverted elements (Hansen et al., 2013) was used for comparison.
(B) Generation of ciRS-7 from reporters, with heterologous flanking repeated elements, in transiently transfected ciRS-7 knockout HEK293 cells. Total RNA extracted from the transfected cells was analyzed by Northern blotting. 28S rRNA was detected by ethidium bromide staining as a control. Produced ciRS-7 and linear RNA are indicated on the left (see Figure 2B legend for the linear products).
(C) ciRS-7 formation was also quantified by RT-qPCR. Expression levels were normalized to the level of precursor RNA (ciRS-7/Precursor) and plotted as ratios to the value of control WT-MIRs plasmid-expressing cells. Therefore, these quantified results do not exactly match up to the apparent view of Northern data in (B). Means ± SD are given for three independent experiments (∗∗∗p < 0.001).
Discussion
We and others first characterized the circular RNA ciRS-7 as a regulator of a specific miRNA, miR-7 (Hansen et al., 2013; Memczak et al., 2013). Here, we describe the biosynthetic pathway for this important circRNA. Circular ciRS-7 is formed via a back-splicing process that is facilitated by flanking inverted sequences derived from MIR, a conserved mammalian SINE. Furthermore, we identified MIR-derived elements that are associated with a number of circRNAs and promote back-splicing and circularization, illustrating a distinct subset of mammalian circRNAs whose formation is driven by MIRs but not by Alus.
Previous Results Are Consistent with the MIR-Dependent Back-Splicing of ciRS-7
Previous ciRS-7 back-splicing assays used reporter plasmids that include the endogenous flanking genomic sequence, 1 kb upstream and 0.2 kb downstream of the ciRS-7 exon (Hansen et al., 2013). This reporter could not produce ciRS-7, consistent with the fact that the inverted MIRs required for back-splicing are located ∼3.0 kb and ∼0.6 kb away from the 3′ and 5′ splice sites of ciRS-7 exon, respectively.
The genomic structure around the ciRS-7 precursor has been independently reported (Barrett et al., 2017). In agreement with our analysis, this study showed that the promoter region of ciRS-7 overlaps an upstream long non-coding transcript (LINC00632 locus), suggesting that the ciRS-7 precursor is a long non-coding RNA (Barrett et al., 2017). The BAC clones obtained (185 and 200 kb) cover from the promoter of this non-coding transcript (LINC00632) to far downstream of the ciRS-7 exon and they were found to produce mature ciRS-7. The rationale is that these two BAC clones include upstream and downstream MIRs that are essential for ciRS-7 generation as we showed here.
MIR-Dependent Back-Splicing Is Widely Utilized for Mammalian circRNA Biosynthesis
The direct back-splicing pathway is commonly used to generate circRNA in metazoans, and flanking inverted elements were shown to promote this event (reviewed in Wilusz, 2018). Although non-repetitive complementary sequences were reported to promote particular back-splicing events in human and fruit fly (Zhang et al., 2014; Kramer et al., 2015), Alus are major players in the biosynthesis of human circRNAs (Jeck et al., 2013; Zhang et al., 2014) because of their abundance (∼10% in human genome; Price et al., 2004). However, the Alu-SINEs are relatively young elements (∼60 million years) and exist only in primates. MIR-SINEs are much older (∼130 million years) and globally functional in mammals (Krull et al., 2007), making it likely that MIRs are commonly used in the biosynthesis of mammalian circRNAs.
Owing to their age, many MIR sequences have lost their 5′ and 3′ regions, reducing their potential to form complementary duplexes, whereas the more recently emerged Alu sequences have remained more complementary to one another. An earlier study found that short complementary sequences (30–40 nt) in the inverted Alu elements are required to support efficient back-splicing (Liang and Wilusz, 2014). According to this criterion, the MIR-independent circRNAs may not have to possess complementary sequences longer than 30 nt. Notably, the difference between MIR-dependent and MIR-independent circRNAs was reflected in the predicted secondary structures of their MIR sequences (Figure S8). We assume that other types of flanking complementary elements with the possible aid of RNA-protein interactions may explain the formation of these MIR-independent circRNAs (reviewed in Wilusz, 2018).
In human, it is possible that the MIR-dependent circRNAs are also flanked by more stable inverted Alu elements. Indeed, we found that 24 of all 66 MIR-dependent circRNAs (including circCDK8) are flanked by inverted Alu elements. Nevertheless, the CRISPR-Cas9-mediated deletion of only either the 5′ or 3′ MIR element was sufficient to prevent its circularization. We assume that inverted Alu-Alu pairing within either upstream or downstream flanking introns could prevent inverted Alu-dependent back-splicing as reported previously (Zhang et al., 2014) and we found that is exactly the case in 12 MIR-dependent circRNAs.
MIR Deletion Could Be an Effective Way to Investigate Function of Targeted circRNA
Our bioinformatics analysis identified 66 circRNAs with conserved flanking MIRs with high complementarity (SW score >500), and these are thus expected to be formed via an MIR-dependent pathway. As the representative of these, we verified that circCDK8 and circSPNS1, as well as ciRS-7, were indeed generated through flanking inverted MIR elements.
The host genes of these MIR-dependent circRNAs, circCDK8 and circSPNS1, have important biological roles. CDK8 kinase is a component of the Mediator kinase complex that regulates transcription by RNA Polymerase II (reviewed in Dannappel et al., 2019). The SPNS1 protein is a putative lysosomal H+-carbohydrate transporter that is required for autophagy (Sasaki et al., 2014; Sasaki et al., 2017; and references therein). Since exon circularization via back-splicing and linear mRNA production via authentic splicing from the same pre-mRNA are mutually exclusive events in back-splicing pathway (see Figure S1A), circRNA formation could potentially regulate the expression of the host gene (Ashwal-Fluss et al., 2014; reviewed in Wilusz, 2018).
To examine the unknown functions of these MIR-derived circRNAs, it is essential to knock out the target circRNA while retaining the host gene expression, or the normal spliced mRNA. Previously, it was shown that the CRISPR-Cas9-mediated deletion of an intronic complementary element successfully blocked circRNA formation without any effect on the linear mRNA splicing (Zhang et al., 2016). Therefore, we will take advantage of the detected MIRs to specifically attenuate circRNA expression using the CRISPR-Cas9 technique. Our investigation of the phenotype in the MIR-targeted knockdown cells will shed light on the biological and physiological functions of circCDK8 and circSPNS1.
Limitations of the Study
We demonstrate the biosynthetic pathway of the functionally important circRNA, ciRS-7 (CDR1as). The essential back-splicing process is facilitated by flanking inverted elements of MIR but not by those of the previously known primate-specific Alu repeats. Our bioinformatics analysis suggests that the MIR-dependent biosynthesis applies to a large number of circRNAs but a full experimental validation for the entire subset remains to be performed.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Akila Mayeda (mayeda@fujita-hu.ac.jp).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze datasets/code.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We are grateful to Drs. Kinji Ohno and Hitomi Tsuiji for SH-SY5Y cells and N2A cells, respectively. Spliceostatin A (SSA) was a generous gift from Dr. Minoru Yoshida. We thank Dr. Karoline Ebbesen for sharing unpublished data; Drs. Anne Nielsen, Shinichi Nakagawa, and Julian Venables for critical reading of the manuscript; Dr. Makoto K. Shimada for helpful suggestions for bioinformatics analysis; Taiwa Komatsu and Mayuko Tanahashi for providing instruction of the gene editing techniques, and members of our laboratory for constructive discussion.
R.Y. was supported by a Research Grant from the Hori Sciences and Arts Foundation, a Research Grant from the Nitto Foundation, a Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan (PMAC), and Grants-in-Aid for Scientific Research (C) (JP18K05563). A.M. was partly supported by Grants-in-Aid for Scientific Research (B) (JP16H04705) and Grants-in-Aid for Challenging Exploratory Research (JP16K14659) from the Japan Society for the Promotion of Science (JSPS). K.R. was supported by Villum Foundation.
Author Contributions
R.Y. and A.M. conceived and designed the experiments; R.Y. conducted most of the experiments, organized the data, and drafted the manuscript; K.R. performed CRISPR-Cas9-mediated genomic deletion of the MIR elements; T.B.H. and J.K. provided useful information and revised the manuscript; A.M. analyzed the data and edited the manuscript. A.M. coordinated and supervised the whole project. All authors read, corrected, and approved the final paper.
Declaration of Interests
The authors declare no competing interests.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101345.
Contributor Information
Rei Yoshimoto, Email: rei.yoshimoto@setsunan.ac.jp.
Akila Mayeda, Email: mayeda@fujita-hu.ac.jp.
Supplemental Information
References
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Barrett S.P., Parker K.R., Horn C., Mata M., Salzman J. ciRS-7 exonic sequence is embedded in a long non-coding RNA locus. PLoS Genet. 2017;13:e1007114. doi: 10.1371/journal.pgen.1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- Dannappel M.V., Sooraj D., Loh J.J., Firestein R. Molecular and in vivo functions of the CDK8 and CDK19 kinase modules. Front. Cell Dev. Biol. 2019;6:171. doi: 10.3389/fcell.2018.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Zietkiewicz E., Labuda D. Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 1995;23:170–175. doi: 10.1093/nar/23.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre–mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull M., Petrusma M., Makalowski W., Brosius J., Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Piwecka M., Glazar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- Price A.L., Eskin E., Pevzner P.A. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Lian S., Khan A., Llop J.R., Samuelson A.V., Chen W., Klionsky D.J., Kishi S. Autolysosome biogenesis and developmental senescence are regulated by both Spns1 and v-ATPase. Autophagy. 2017;13:386–403. doi: 10.1080/15548627.2016.1256934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Lian S., Qi J., Bayliss P.E., Carr C.E., Johnson J.L., Guha S., Kobler P., Catz S.D., Gill M. Aberrant autolysosomal regulation is linked to the induction of embryonic senescence: differential roles of Beclin 1 and p53 in vertebrate Spns1 deficiency. PLoS Genet. 2014;10:e1004409. doi: 10.1371/journal.pgen.1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Venø M.T., Hansen T.B., Venø S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J.E. A 360 degrees view of circular RNAs: from biogenesis to functions. WIREs RNA. 2018;9:e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto R., Kaida D., Furuno M., Burroughs A.M., Noma S., Suzuki H., Kawamura Y., Hayashizaki Y., Mayeda A., Yoshida M. Global analysis of pre-mRNA subcellular localization following splicing inhibition by spliceostatin A. RNA. 2017;23:47–57. doi: 10.1261/rna.058065.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.