Significance
Insects are dependent on olfactory cues to complete biological processes, such as foraging, oviposition, and mate choice. While extensive experimental evidence supports the importance of chemical cues in these processes, genes involved in chemosensory integration of complex behavioral responses remain largely unknown. Using a combination of differential gene expression and genome-wide signals of gene flow, we describe the chemosensory expression profiles of sensory tissues and identify candidate genes for mate and host plant recognition in a pair of Heliconius butterflies. We find that candidate chemosensory genes are physically unlinked from color-pattern genes. Our results suggest the independent evolution of loci associated with the chemosensory and visual systems of Heliconius, both potentially mediating behaviors that promote reproductive isolation and downstream speciation.
Keywords: Lepidoptera, smell, taste, butterfly, speciation
Abstract
Chemosensory communication is essential to insect biology, playing indispensable roles during mate-finding, foraging, and oviposition behaviors. These traits are particularly important during speciation, where chemical perception may serve to establish species barriers. However, identifying genes associated with such complex behavioral traits remains a significant challenge. Through a combination of transcriptomic and genomic approaches, we characterize the genetic architecture of chemoperception and the role of chemosensing during speciation for a young species pair of Heliconius butterflies, Heliconius melpomene and Heliconius cydno. We provide a detailed description of chemosensory gene-expression profiles as they relate to sensory tissue (antennae, legs, and mouthparts), sex (male and female), and life stage (unmated and mated female butterflies). Our results untangle the potential role of chemical communication in establishing barriers during speciation and identify strong candidate genes for mate and host plant choice behaviors. Of the 252 chemosensory genes, HmOBP20 (involved in volatile detection) and HmGr56 (a putative synephrine-related receptor) emerge as strong candidates for divergence in pheromone detection and host plant discrimination, respectively. These two genes are not physically linked to wing-color pattern loci or other genomic regions associated with visual mate preference. Altogether, our results provide evidence for chemosensory divergence between H. melpomene and H. cydno, two rarely hybridizing butterflies with distinct mate and host plant preferences, a finding that supports a polygenic architecture of species boundaries.
How do animals perceive the natural world? While we know that they use multisensory cues to integrate their surroundings and perform basic biological routines, our current understanding of these communication channels is heavily influenced by our own sensory biases. As such, work describing important communication strategies is dominated by descriptions of visual and auditory signals. Comparatively less understood, chemical sensing, which involves both volatile and tactile cues, plays an essential role in a variety of fundamental biological processes (1, 2). For example, chemical communication is important for mate choice (3–5), food choice (6), and host plant choice (7). Moreover, chemosensory communication plays an important role during the early stages of speciation, where it establishes chemical prezygotic barriers that may precede morphological divergence (8, 9). For example, the relatively simple enzymatic mechanisms controlling pheromone synthesis in insects (10) [e.g., Nasonia (11, 12)] allow the fast evolution of olfactory signals that drive assortative mating patterns [e.g., Drosophila (13); Lepidoptera (14)]. Furthermore, in host-specialized insect systems, chemical perception of host plant signals can serve to further establish ecological species barriers (9). In this way, chemosensory signals may play important roles in both routine behaviors and speciation processes. Despite the fundamental role of chemically mediated behaviors in speciation, few studies have identified chemosensory genes involved in reproductive isolation (9, 15, 16).
To date, most of the work on the genetic basis of chemosensory signaling has been conducted on insects, with an emphasis on Drosophila and moths (e.g., Heliothis and Bombyx). However, in the past few years, the growing accessibility of whole-genome and transcriptome sequencing has allowed us to describe chemosensory genes for a number of new butterfly species. These advances have improved our understanding of the number, diversity, and evolution of chemosensory genes. Interestingly, what has emerged from these studies is a novel view of the chemosensory molecular repertoire of butterflies that reveals an unanticipated, highly complex system, that rivals that found in moths. Heliconius butterflies, despite their diurnal and highly visual lifestyle, have more olfaction-related chemosensory genes than moths, whose nocturnal lifestyle requires navigating the landscape while relying largely on chemical cues (17). This apparent paradox likely stems from our limited understanding of how chemosensory genes shape and inform butterfly behaviors, life history, and evolutionary diversity.
Heliconius butterflies, typically recognized for their wing color and pattern mimicry, harbor an impressive diversity of chemosensory molecules, which can be broadly classified into olfactory binding proteins (OBPs), chemosensory proteins (CSPs), olfactory receptors (ORs), gustatory receptors (GRs), and ionotropic receptors (IRs) (Table 1). In Heliconius, these molecules likely play important roles in mediating various complex behaviors. For example, Heliconius males produce and administer antiaphrodisiacs to females that reduce remating and male harassment (18–20). Additionally, Heliconius butterflies feed and oviposit exclusively on Passiflora plants for which they display varying degrees of specialization; some are Passiflora host plant specialists, and others generalists (21). Furthermore, female butterflies have been documented to evaluate the suitability of host plants prior to ovipositing by “drumming” their forelegs, which contain special sensilla (7), to probe the leaf surface (22, 23). As such, sensory integration of chemical cues emitted by congeners and host plants are essential to the survival of Heliconius butterflies.
Table 1.
Reference table for chemosensory gene categories in Heliconius
| Categories | Families | Abbreviation | Review |
| Transporters | Olfactory binding proteins | OBP | Selective in terms of the chemicals they transport, these mainly include volatiles, such as pheromones [e.g., Drosophila (24); Bombyx (25)]. |
| Small, soluble carrier proteins that move mostly nonsoluble hydrophobic chemicals through the sensillar lymph and to the receptors (26). | Chemosensory proteins | CSP | Less selective than OBPs, they may bind nonvolatile compounds and semiochemicals (27). They also play a role during cuticle development (28) and in immune-related processes (29). |
| Receptors | Olfactory receptors | OR | Expressed in sensory neurons, these constitute the backbone of the sense of smell. They are responsible for the sensory integration of a wide array of odorant molecules. |
| Molecules whose function is chemical detection, catalyzing the sensory cascade. | Gustatory receptors | GR | Primarily involved in tasting sweet and bitter compounds (7), they also function as CO2 receptors (30) and are proposed to be involved in heat avoidance (31). |
| Ionotropic receptors | IR | The most primitive class of receptors and, while primarily involved in the smelling and tasting of amines and acids (32, 33), they are also essential for salt detection (34). |
For each chemosensory gene category, we provide its abbreviation and a brief review of its role in chemosensation.
While current work on Heliconius chemosensation has focused on describing the evolutionary diversity of chemosensory genes (CSPs, OBPs, ORs, GRs, and IRs) (7, 35, 36), it has yet to identify individual genes associated with chemically dependent behavioral or biological processes. To better understand the role of chemosensory genes in the establishment of species barriers, we have focused on the recently diverged [∼1 Mya (37)], sympatric species pair of Heliconius melpomene rosina and Heliconius cydno chioneus (Fig. 1) (38–40). These species differ in altitudinal range, host plant ecology, mimicry patterns, microhabitat preference, pheromone chemistry, flight pattern, and wing shape (41). Despite these differences, interspecies mating between H. melpomene and H. cydno (while rare on a per individual basis) does occur and is generally observed to occur between H. melpomene males and H. cydno females. These crosses result in sterile females, but fertile F1 males that can back-cross with either species (40, 42, 43). Perhaps because a low rate of hybridization has been ongoing for such a long period, this has resulted in extensive signals of postdivergence gene flow (37, 44, 45). As a result, these two species provide an opportunity to examine the chemosensory correlates of speciation in wild, naturally hybridizing animals.
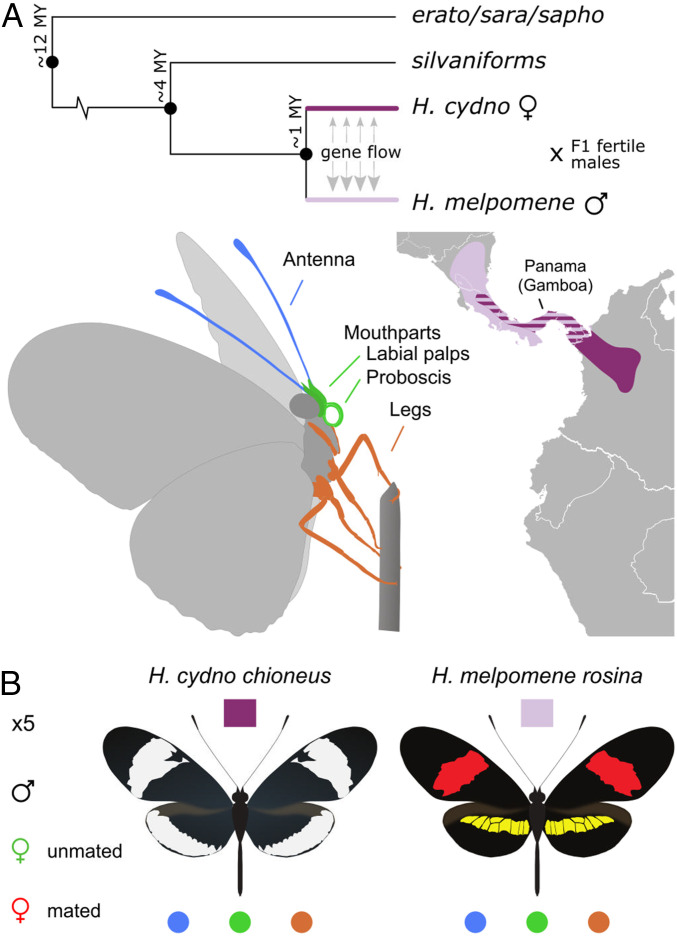
Fig. 1.
Phylogenetic relation, geographic distribution, and experimental design to study divergence of gene expression in chemosensory tissues of H. cydno and H. melpomene. (A) H. cydno and H. melpomene split ∼1 Mya, but male H. cydno still occasionally hybridize with female H. melpomene, resulting in fertile hybrid males and sterile hybrid females. Genomic patterns of admixture reflect this hybridization, with a generally stronger signal of gene flow from H. cydno into H. melpomene. We sampled the antennae, legs, and mouthparts (labial palps and proboscis) of H. cydno and H. melpomene individuals from Panama (Gamboa) in order to study divergence in chemosensory gene expression. (B) The experimental design included five replicates for each combination of species, tissues (antennae, legs, and mouthparts) and life stage (males, mated, and unmated females).
Our main goal is to identify candidate chemosensory genes underlying reproductive isolation. Previous work has shown that androconial chemicals are important for mate choice, and that there are consistent differences between these species in their androconial wing chemistry across their wide geographic ranges, supporting the hypothesis that chemical signaling is important in reproductive isolation (19, 46). Toward this goal, we first improved the annotation of chemosensory genes using a targeted resequencing approach and later analyzed RNA sequencing (RNA-seq) gene-expression data to identify candidate mate and host plant choice genes. Our experimental design includes three sensory tissue types (antenna, legs, and mouthparts) and three biological groups (males, unmated females, and mated females), representing the recently diverged species pair of H. melpomene and H. cydno (38–40). With this design we were able to describe species-, sex-, and life-stage– (mated vs. unmated) specific expression profiles that reflect important differences in mate and host plant choice for H. melpomene and H. cydno.
Combining our gene-expression data with recently published whole-genome sequences for our study species (44), we then examined patterns of differential expression as they relate to genome-wide patterns of admixture. Based on both their expression profiles and associated genomic patterns of speciation, we identified a small number of candidate genes and suggest two strong candidates for species barriers through mate and host plant choice (47, 48). Interestingly, these chemosensory candidates are not in physical proximity to known wing-color pattern genes or other genomic regions associated with visual mate preference. Our work thus suggests that chemosensory traits may have evolved to contribute to reproductive isolation independently from the color-pattern genes and visual discrimination system, providing an additional means for reinforcement in a system characterized by visual mimicry.
Results
Tissue-Specific Expression Recapitulate Sensory Function.
We found chemosensory genes belonging to all five families to be expressed in all three tissue types, underscoring the role of these tissues in chemosensation (SI Appendix, Table S2). Of the total 252 chemosensory genes, we found more to be expressed in the antennae relative to the legs or mouthparts (45% in antennae, 29.8% in legs, 25% in mouthparts). Nearly 60% were expressed equally in all of the three sensory tissue types, although often at very low levels. The remaining 40% showed variable expression and half of them showed tissue-specific expression (SI Appendix, Table S2). ORs and OBPs largely comprised antenna-specific genes, whereas CSPs, GRs, and OBPs generally comprised mouthpart- and leg-specific genes. One of the most prominent features in the antennae was the high expression of ORs, OBPs, and IRs (Fig. 2 and SI Appendix, Figs. S2, S3, and S6 and Table S2), a finding that highlights the predominantly olfactory function of antennae. In contrast, the legs and mouthparts displayed very similar chemosensory profiles and were characterized by GR expression (SI Appendix, Fig. S4). The latter is in line with results published by Briscoe et al. (7) and underscores the role of legs and mouthparts in gustation. Interestingly, we found HmGR22, a Heliconius-specific gene and putative bitter receptor, to be highly expressed in all three tissue types, suggesting that this gene might be broadly important for chemosensation. In the mouthparts, we observed that HmGR56 was overexpressed in both H. cydno unmated females and males. It is important to emphasize that while specific expression patterns characterized individual tissue types, we did not observe the exclusive expression of any chemosensory gene family in any of the tissues.
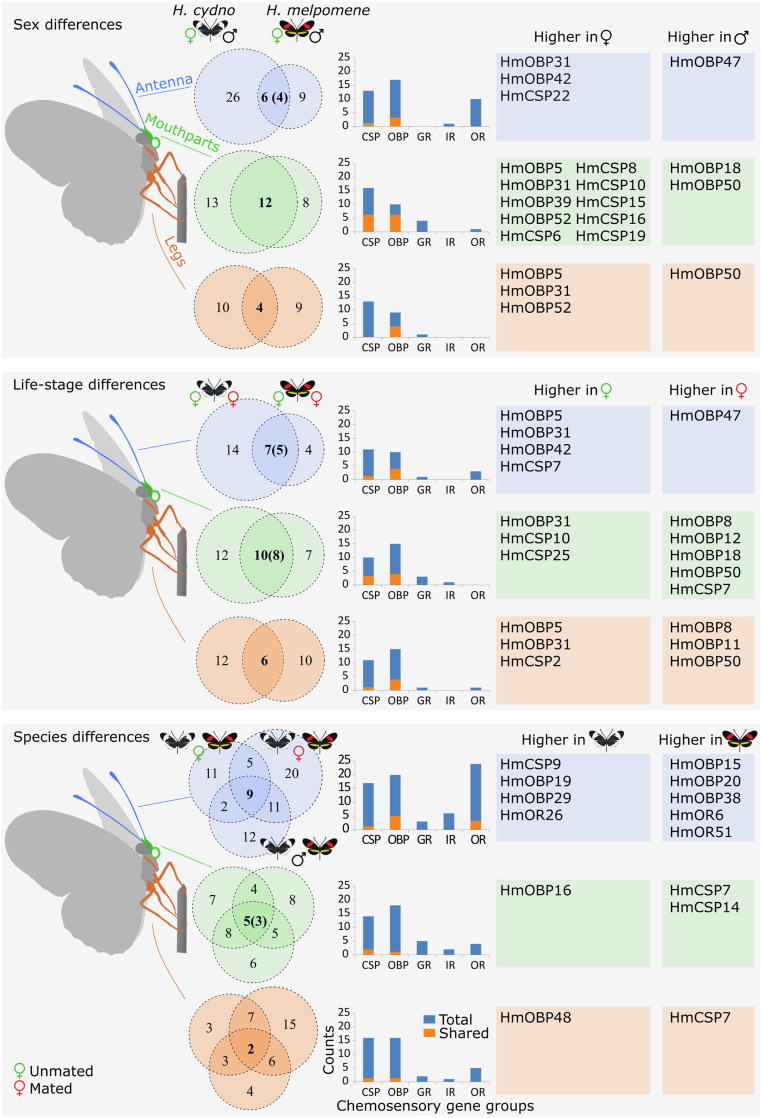
Fig. 2.
Overview of differentially expressed genes by tissue type, sex, life-stage, and species in H. cydno and H. melpomene. Venn diagrams show the number of genes differentially expressed and shared among comparisons (numbers between parentheses match the genes that are consistently differentially expressed in the same direction across sex, life-stage, or species). Bar plots indicate differentially expressed genes in each chemosensory gene family. The blue bars indicate the total gene count and the nested orange bars correspond to the genes in the overlap region of each respective Venn diagram. Tables on the right list the names of genes that are consistently differentially expressed in the same direction across sex, life-stage, or species; these match the overlap region of each respective Venn diagram.
Sex-specific Expression Patterns Suggest OBPs as Mate Recognition Candidates.
Approximately 24% of chemosensory genes showed significant expression differences as a function of sex (males vs. unmated females) (Fig. 2 and SI Appendix, Tables S3 and S4). We found some of these genes to be differentially expressed between the sexes (regardless of species), suggesting the existence of conserved sex-based chemosensation (Fig. 2). Sex-based expression differences observed between males and females may stem from their different evolutionary needs to recognize odorant volatiles during mate choice and host plant selection. Moreover, genes showing conserved sex-based differences may precede the speciation of H. melpomene and H. cydno, making them more likely to correspond to broad differences in sex-specific traits. Although we observed a few differentially expressed genes belonging to each chemosensory gene family (Fig. 2 and SI Appendix, Tables S3 and S4), OBPs and ORs are the strongest candidates for mate recognition due to their role in detecting volatile compounds, such as pheromones (Table 1). Congruently, the antenna showed the largest number of sex-based expression differences (Fig. 2 and SI Appendix, Tables S3 and S4).
Of all of the chemosensory gene families, only OBPs and CSPs showed consistent sex-based differences in expression (Fig. 2). CSPs, however, serve functions outside of chemosensation [e.g., during development (28) and immunity (29)]; this is reflected in their expression across many nonsensory tissues (26), making them less-compelling candidates for mate recognition. Generally, we found OBPs to be expressed in all three tissue types while CSPs were mostly in the mouthparts (Fig. 2). We also observed that genes with consistent sex-based expression patterns were overexpressed in females relative to males, with the mouthparts showing the largest number of differentially expressed genes (Fig. 2). Notably, HmOBP31 was the only gene to be consistently overexpressed in females of both species in all three tissue types. Three other genes—HmOBP5, HmOBP52, and HmOBP50—were consistently differentially expressed between the two sexes in at least two tissue types (Fig. 2 and SI Appendix, Table S3). Sex-based patterns of gene expression in ORs, GRs, and IRs were observed only within species; none of the genes showed sex-based expression patterns in both H. melpomene and H. cydno (SI Appendix, Tables S3 and S4). Generally, sex-specific differential expression patterns of ORs were restricted to the antennae, except for HmOR40, which was overexpressed in the mouthparts of H. melpomene males. HmOR19 and HmOR5, two genes previously reported as female-specific ORs in H. melpomene legs (7), were found to be overexpressed in H. cydno female antennae. Sex-based differential expression patterns of GRs were restricted to the legs and mouthparts. Namely, HmGR22 was found to be overexpressed in H. cydno female legs and mouthparts. The only sex-based difference observed in the IRs was for HmIR40a, which was found overexpressed in H. cydno males.
Life Stage–Specific Expression Suggests Sensory Shift in Antennae and Mated Females.
Approximately 21% of genes showed significant differences as a function of life-stage (unmated vs. mated females) (Fig. 2 and SI Appendix, Tables S3 and S4). Differences in chemosensory gene expression between unmated and mated females might reflect a change in sensory priorities from mate choice (in unmated females) to host plant choice for oviposition (in mated females). Like the sex-based expression patterns, we only observed OBPs and CSPs to be consistently differentially expressed between life-stages for both H. melpomene and H. cydno (Fig. 2 and SI Appendix, Tables S3 and S4). Four OBPs (HmOBP5, HmOBP31, HmOBP42, and HmOBP47) were consistently differentially expressed between unmated and mated female antennae across species. HmOBP5, HmOBP31, and HmOBP42 were overexpressed in unmated females, while HmOBP47 was overexpressed in mated females (Fig. 2 and SI Appendix, Table S3). HmOBP31 is particularly interesting because it is overexpressed in all three tissues for unmated females (Fig. 2). Moreover, HmOBP31 and HmOBP42 are strong candidates for female mate choice because they are overexpressed in unmated female antennae relative to both mated females and males. Several genes showed similar expression patterns in both the sex-specific and life-stage analyses (Fig. 2 and SI Appendix, Tables S3 and S4). The latter is due to similarities between the expression patterns of mated females and males; this causes life-stage patterns of gene expression to greatly overlap with the sex-specific patterns.
Similar to what we observed in the sex-specific analysis, life-stage—based patterns of gene expression in ORs, GRs, and IRs were observed only within species; none of the genes showed life-stage–based expression patterns across species (SI Appendix, Tables S3 and S4). Most life-stage expression differences in ORs were restricted to the antenna, except for HmOR37, which was overexpressed in the legs of H. melpomene-mated females. All life-stage differences for GRs show a higher expression in unmated females relative to mated females. The only life-stage based difference observed in the IRs was for HmIR25a, which was overexpressed in the mouthparts of H. cydno unmated females. More generally, we again observed significant overlap between sex-specific and life-stage–specific patterns.
Species-Specific Expression Patterns Identify Potential Species Barriers.
Approximately 40% of genes show species-specific differential expression patterns for at least one biological group (males, unmated females or mated females) (Fig. 2 and SI Appendix, Tables S3 and S4). A subset of these genes showed expression patterns strictly as a function of species (H. melpomene vs. H. cydno); these were conserved across all three biological groups (males, unmated females, and mated females). Most of these species-specific differences were observed in the antenna, particularly in ORs and OBPs. More specifically, in the antennae, HmOBP15, HmOBP20, HmOBP38, HmOR6, and HmOR51 were overexpressed in H. melpomene and HmCSP9, HmOBP19, HmOBP29, and HmOR26 were overexpressed in H. cydno. In the legs, we found HmCSP7 to be overexpressed in H. melpomene and HmOBP48 to be overexpressed in H. cydno. In the mouthparts, HmCSP7 and HmCSP14 were overexpressed H. melpomene and HmOBP16 was overexpressed in H. cydno.
More broadly, the number of overexpressed transporters (OBPs and CSPs) and receptors (ORs, GRs, IRs) in the antenna of H. cydno (n = 60) was greater than that observed in H. melpomene (n = 46). This difference was mainly driven by the male and mated female categories. We found 13 overexpressed transporters in H. cydno males compared to 5 in H. melpomene males (SI Appendix, Table S3), and 15 overexpressed receptors in H. cydno mated females compared to 5 in H. melpomene (SI Appendix, Table S4). It is possible that this increased chemosensory expression corresponds to both host plant generalist behavior and strict conspecific mate choice preferences in H. cydno (21, 40, 42, 43), behaviors that might require increasingly specialized and sensitive chemosensory discrimination genes.
Patterns of Differential Expression in the Context of Admixture.
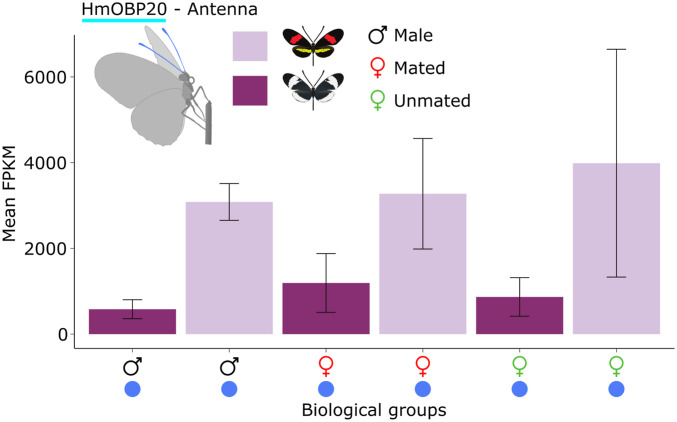
Of the 252 chemosensory genes in our dataset, we were able to confidently assign genomic admixture levels (fd) to 212 (44) (Fig. 3A). From these 212 genes, we extracted 149 genes that were significantly differentially expressed between H. melpomene and H. cydno in any of the three tissues (antennae, legs, and mouthparts) and for any of the three biological groups (males, mated females, and unmated females) (Fig. 2, species differences). These 149 genes were sorted into “genes overexpressed in H. melpomene” and “genes overexpressed in H. cydno.” Of these 149 chemosensory genes, 8.72% (n = 13) were both significantly overexpressed and had low admixture (fd ≤ 0) (Fig. 4). This subset was composed of one OBP, three GRs, and four ORs. Only one chemosensory gene, however, showed species-level expression patterns consistent across biological groups and tissue type: HmOBP20 (q-value ≤ 0.001 for all comparisons). HmOBP20 was found to be overexpressed in H. melpomene antennae relative to H. cydno, and this pattern was observed in males, mated females, and unmated females (Fig. 5). As such, HmOBP20 presents a compelling candidate for species-specific recognition in H. melpomene.
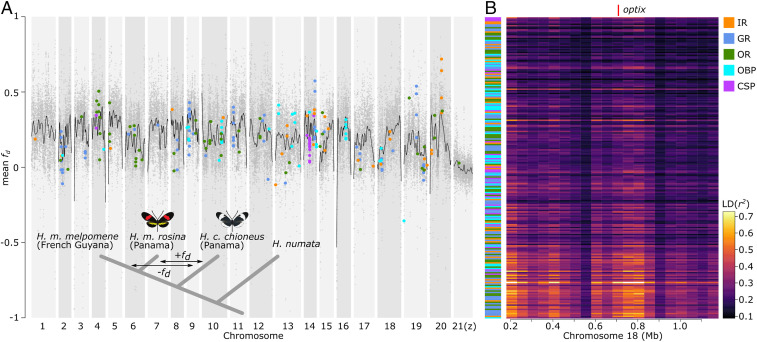
Fig. 3.
Genomic patterns of divergence between H. melpomene and H. cydno. (A) Per chromosome genomic patterns of admixture between H. cydno and H. melpomene (bottom left corner; population and relationships used in the admixture analysis). For all 21 chromosomes in the Heliconius genome (x axis), we show genome-wide admixture values (y axis; mean fd) calculated in 20-kb windows and per chromosome cubic splines (black lines). Chemosensory gene families are color-coded as indicated in the legend. (B) LD (r2) map of chemosensory genes and the color-pattern gene optix. On the y axis chemosensory gene families are color-coded as per the legend and ordered according to their fd value (highest fd at bottom). The x axis indicates a 1.2-Mb window around optix (red line) analyzed in 50-kb windows. For chemosensory gene names associated with B, see SI Appendix, Fig. S7 and Table S5.
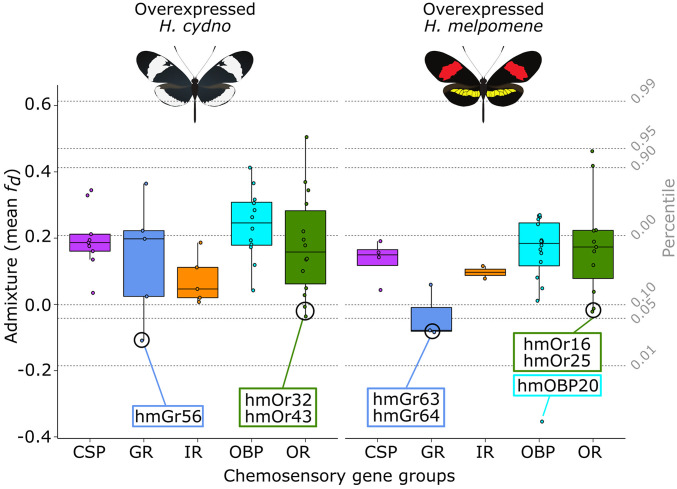
Fig. 4.
Admixture of chemosensory genes overexpressed in H. cydno and H. melpomene. Only chemosensory genes significantly overexpressed for each species are plotted. Admixture (y axis) is measured using a mean fd value was calculated from a 50-kb window around the transcription start site of each gene. Genes are considered to not show signals of admixture when fd ≥ 0; chemosensory genes with fd ≤ 0 are identified by their name. Mean genome fd and percentiles are indicated using dashed lines.
Fig. 5.
Expression levels for the odorant binding protein HmOBP20. HmOBP20 shows increased expression in the antennae for all three biological groups (males and mated and unmated females) of H. melpomene relative to H. cydno. Expression of HmOBP20 is significant (and almost exclusive) to the antennae and very low (or not present) in the legs and mouthparts. Error bars in both figures were calculated using SD values.
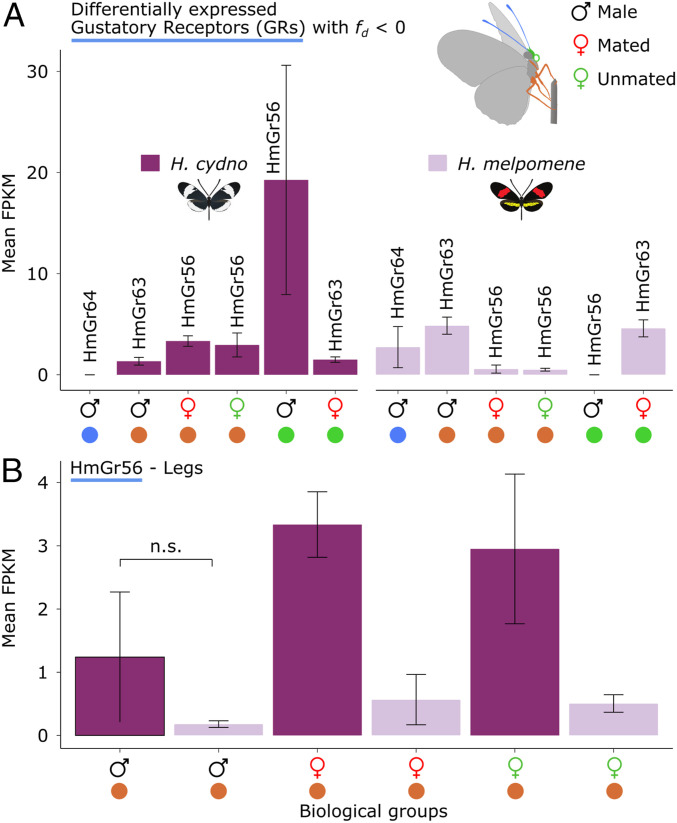
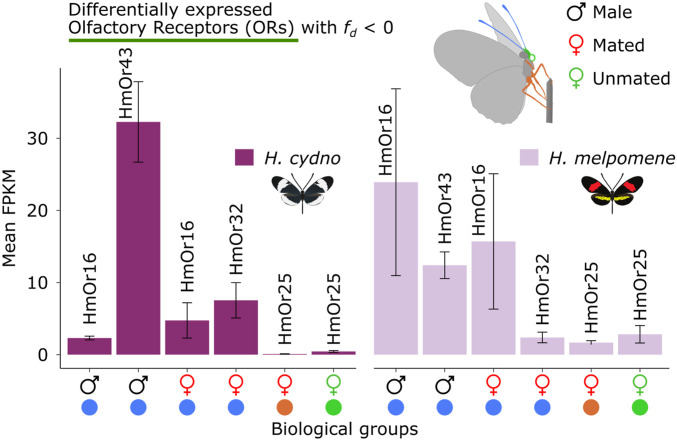
Three GRs were significantly differentially expressed with low admixture (fd ≤ 0) (HmGR63, HmGR64, and HmGR56) (Fig. 6A). Here, the most pronounced difference in expression is observed for HmGR56 in H. cydno male mouthparts. Generally, HmGR56 presents a trend toward increased expression in H. cydno legs relative to H. melpomene (Fig. 6B). The latter is noteworthy because HmGR56 is a Heliconius-specific GR putatively involved in synephrine (a common plant alkaloid) recognition (7) and may present an interesting candidate for host plant recognition. Four ORs were both significantly overexpressed and had low admixture (fd ≤ 0) (HmOr16, HmOr25, HmOr32, and HmOr43) (Fig. 7). Here, the most pronounced differences in expression are observed for ORs expressed in the antennae (HmOr16, HmOr32, and HmOr43).
Fig. 6.
Expression levels for GRs of interest. (A) Expression levels of differentially expressed GRs with fd ≤ 0 between H. cydno and H. melpomene. (B) Expression levels of HmGR56 in the legs. Note that HmGR56 was not found to be significantly differentially expressed in male legs but displays the same trend. Error bars were calculated using SE values.
Fig. 7.
Expression levels for ORs of interest. Expression levels of differentially expressed ORs with fd ≤ 0. Error bars were calculated using SE values.
Genomic Locations of Chemosensory Genes and Linkage Disequilibrium.
Generally, we found that chemosensory genes occur as small clusters, were present on every chromosome, and were spread across the genome (Fig. 3A). These clustering patterns were in line with expectations for genes originating from duplications as per the birth–death model of gene family evolution (49). On a finer scale, some clear differences between gene families emerge. CSPs, for example, showed a strong clustering pattern to specific areas of the genome. This contrasts with GRs, which were represented by many small clusters of genes across the genome. Similarly, OBPs appeared in small clusters, while ORs and IRs form no clusters but were spread out across the genome. Generally, genes clustering together were more closely related. One gene (HmOR3) mapped to the sex chromosome (Fig. 3A). HmOR3 is overexpressed in the in antenna of H. cydno-mated females relative to H. melpomene-mated females (SI Appendix, Table S4).
Genetic associations between preference and mating cue loci may promote assortative mating in diverging species (9, 43, 50, 51). In Heliconius, for example, the major color-pattern gene optix is associated with visual assortative mating behavior (51). As such, we identified chemosensory genes within 1 Mb of key wing-pattern formation genes (WntA, optix, and cortex); these genes are also involved in divergent natural selection (52, 53). Near WntA, a gene involved in variation in forewing band shape (52), we identified three chemosensory genes: HmOR23 (89 kb), HmGR62 (772 kb), and HmOR49 (952 kb). Near optix, a transcription factor controlling ommochrome development (53), we found one gene: HmOBP40 (632 kb). Near cortex, a gene involved in determining the presence of a yellow hindwing bar and white forewing (54), two chemosensory genes were identified: HmOR55 (498 kb) and HmOR12 (546 kb). While close proximity (within 1 Mb) could indicate that the chemosensory genes are physically linked to the wing-pattern genes (in linkage disequilibrium, LD), none of these chemosensory genes were identified as candidates based on expression and admixture data.
Due to the association of color pattern with mate-preference behavior in H. melpomene and H. cydno (51), we specifically explored the levels of LD between our chemosensory genes (n = 252) and the optix color pattern interval (Fig. 3B and SI Appendix, Fig. S7 and Table S5). Additionally, we tested LD levels between a group of randomly selected, nonchemosensory genes (n = 300) and the optix color pattern interval. Compared to the randomly selected group of 300 genes, the 252 chemosensory genes did not show an increase in LD among each other or with the divergently selected optix locus. The average r2 among the chemosensory genes was 0.19 ± 0.07 and closely matches the r2 observed among the randomly selected genes (0.19 ± 0.09). The average r2 of the chemosensory and random genes with the divergent cis-regulatory region of optix was 0.27 ± 0.11 and 0.29 ± 0.13, respectively. Of the chemosensory loci identified using differential expression and admixture, we found that HmGR56 (r2 = 0.45) and HmOr32 (r2 = 0.46) had the strongest association with optix and were in the top 12% highest LD with optix. HmOBP20 also showed increased LD with the optix region (r2 = 0.35) and was in the top 26% highest LD with optix. Among the chemosensory genes, the strongest association, however, was observed for HmIR75d (r2 = 0.64), a gene that has no salient expression pattern (Fig. 3B and SI Appendix, Fig. S7 and Table S5).
Discussion
Despite the essential role of chemosensory communication in insect behavior, we still know relatively little about the genetics underlying chemosensory discrimination of mate and host plant choice and their potential involvement in speciation. Heliconius butterflies exhibit chemically mediated behaviors (e.g., mate choice, mate searching, and host plant choice for oviposition) that are vital for survival and reproduction. Recently diverged species within this genus provide an excellent opportunity to study the role of chemosensation in speciation. Here, we have investigated chemosensory divergence in two recently diverged Heliconius butterfly species (H. melpomene and H. cydno) that show both strong assortative mating and use different larval host plants. By combining gene expression and genetic divergence data in this species pair, we have identified strong candidate genes for mate and host plant choice that may have important implications for the establishment of chemically mediated species barriers.
A strength of this study is the use of both RNA expression and DNA admixture data to identify chemosensory genes correlated with the early stages of speciation. Species-based expression differences may stem from a variety of life-history variables: For example, differences in adaptations for mate choice, as well as host plant preferences. These differences are particularly important for butterflies on the verge of speciation, where chemically mediated signals may be essential for the establishment of species barriers. Multiple studies support the importance of chemically mediated species boundaries (9), which indicate that species can differ in both the type and relative abundance of chemosensory genes that facilitate the detection of species-specific chemical cues, generally volatile odorants. In agreement with this, most species-specific differences in expression were observed in the antennae and were characterized by OBPs and ORs, as would be expected if these genes were involved in the recognition of volatiles (e.g., pheromones).
Despite being closely related, H. cydno and H. melpomene display key differences in reproductive behavior. They show strong premating isolation and, in laboratory crosses, the two species hybridize asymmetrically, with hybrids more often resulting from H. melpomene males and H. cydno females (40, 42, 43). Second, H. cydno is a host plant generalist and, in Central America, H. melpomene is a host plant specialist (21). These differences in mating and oviposition may cause broad patterns of differential expression between H. cydno and H. melpomene. In particular, we observed three times more genes overexpressed in the antenna of H. cydno males and mated females relative to the same categories in H. melpomene. This increased chemosensory expression in H. cydno might, for example, confer males and females of this species a greater ability to distinguish conspecific mates or recognize a wider range of host plants. Future work on populations of H. melpomene from eastern parts of the range (which are more host plant generalist) might provide a means to test whether expansions in gene expression are associated with host plant use.
In order to select the strongest candidates for mate and host plant choice, we identified genes that showed both strong differential expression and were located in regions of the genome with low admixture (fd ≤ 0). We hypothesized that chemosensory genes found in this cross-section of our data were likely to be involved in species-specific processes. Using this strategy, we identified eight candidate genes for species-specific processes: OBPs (n = 1), ORs (n = 4), and GRs (n = 3). OBPs and ORs are known to be involved in pheromone detection and GRs are likely under strong divergent selection due to specialized host-plant usage. This contrasts with CSPs and IRs, which are understood to have a more general role in chemosensation (Table 1). HmOBP20, however, stands out for being the only gene showing both significant differential expression at the species-level and low admixture (fd ≤ 0). HmOBP20 is overexpressed in H. melpomene antennae relative to H. cydno and this pattern is consistently found across biological groups (Fig. 5). The observed antennae-specific expression of HmOBP20 is in line with the hypothesized role of this gene in volatile pheromone detection. Work on the silkmoth (Bombyx mori) homolog BmOBP20 underscores the importance of this gene in chemosensation (55). More recently, work on the Drosophila homolog (OBP69a) suggests that the expression of OBP69a is regulated by cis-vaccenyl acetate (a male-specific Drosophila pheromone) and experimentally demonstrates that expression levels of OBP69a modulate male-male aggression and female receptivity (56). Moreover, HmOBP20 was previously identified to be in a region of high divergence between Heliconius elevatus and Heliconius pardalinus (57), two other recently diverged species in the genus. As such, HmOBP20 is a strong candidate for species-specific recognition of pheromones in Heliconius. The molecular mechanisms controlling divergence patterns at this gene, however, are likely very complex. Sequence-level comparisons of HmOBP20 across Heliconius species indicate that the protein structure is highly conserved (no amino acid changes), suggesting that patterns of divergence at HmOBP20 are driven by gene regulatory elements.
Due to the highly specialized relationship between Heliconius and Passiflora, chemosensory genes involved in plant volatile and nonvolatile recognition are likely under strong divergent selection and may represent important species barriers. This is particularly true for females, who select ovipositing sites in part by tasting the putative host plant surface with their forelegs, a behavior known as “drumming” (22, 23). In agreement with these behavioral observations, we find that in H. cydno (a plant generalist) HmGR22, HmGR3, HmGR56, and HmGR5 are overexpressed in the legs and mouthparts of females. It is noteworthy that no GRs were found to be overexpressed in the males, suggesting that overexpression of GRs may be exclusive to specialized female sensilla. Of these four GRs, HmGr56 emerges as a candidate for host plant discrimination in H. cydno. HmGr56 was found to have low admixture (fd ≤ 0) and to be overexpressed in H. cydno male mouthparts and mated/unmated female legs (Fig. 6A). Generally, however, HmGr56 expression was higher in H. cydno legs relative to H. melpomene (Fig. 6B). Moreover, the expression of HmGr56 was restricted to the legs and mouthparts, as would be expected if the gene were involved in recognizing contact cues associated with plant identity. Briscoe et al. (7) had previously identified HmGr56 to be highly expressed in female legs and described the gene as part of a cluster of putative paralogues of a Papilio xuthus GR known to be involved in synephrine recognition, a common plant alkaloid (58). Taken together, these data suggest that HmGr56 may be involved in host plant discrimination.
As a result of the sophisticated visual system (59) and wing-pattern diversity in Heliconius butterflies (60), speciation work in the Heliconius has focused on the three genes thought to control most wing coloration (optix [chromosome 18, chr18], WntA [chr10], and cortex [chr15]). In terms of mating behavior, a recent study identified three large-effect quantative trait loci [QTLs] associated with largely visual-based male mating preference, including one in close physical proximity to optix, the gene responsible for differences in red pattern variation between H. cydno and H. melpomene (51). While this and other studies (40, 43) support a strong role for visual cues in HeIiconius butterfly behavior, the role of chemosensory communication in establishing and maintaining species barriers remains understudied. We propose that HmOBP20 represents an independently evolved strategy for discrimination and reinforcement of species barriers in Heliconius. In addition to presenting strong species-specific genomic signals, HmOBP20 is not physically linked with any of the classic color pattern genes (optix [chr18], WntA [chr10], and cortex [chr15]), nor is it linked to any of the previously identified male preference QTLs (chr1, chr17, chr18). Instead, it is located on a different chromosome (chr19). This finding supports a multimodal system of speciation that involves both chemical and visual cues, a conclusion that is emerging from studies looking at chemical differences in wing pheromones and the role that they play in reproductive isolation (46, 61–63).
This work provides a foundation for elucidating the mechanistic basis of chemosensation in Lepidoptera, as well as understanding how chemosensory communication shapes insect behaviors and establishes species barriers. The admixture and expression-level genomic signatures observed at these candidate genes suggest that key changes underlying chemically mediated adaptations are likely found in the regulatory architecture controlling the time, level, and place of expression of these chemosensory genes. This mechanism is similar to what has been recently reported for wing-color pattern variation in Heliconius (64, 65). Our chemosensory candidates are not found in physical proximity to color pattern genes or genomic regions associated with visual mate preference but show increased LD with the optix interval. We thus suggest their likely independent evolution from the visual reinforcement system and propose that these chemosensory genes work in tandem with color pattern (62, 66) and vision genes (59, 67) to mediate reproductive isolation in Heliconius. While our data indicate that OBPs, ORs, and GRs are the most compelling candidates for mate and host plant choice, these candidates have yet to be validated. Future efforts to elucidate the molecular basis of chemosensory perception will focus on the role of individual genes in these behaviors.
Materials and Methods
Sampling, RNA Extraction, and Sequencing.
We bred 15 H. melpomene and 15 H. cydno butterflies in seminatural conditions from November 2013 to March 2014 in Gamboa, Panama. Unmated females were 2-d-old at sampling and both males and mated females were 5-d-old; females were mated within the first 2 d after eclosing. For each individual we collected tissues of three sensory tissue types: Antenna, legs (all six legs), and mouthparts (including the proboscis and labial palps). We extracted RNA separately from each of the three sensory tissue types for a total of 90 individual RNA extractions representing 3 biological groups (5 males, 5 unmated females, and 5 mated females) of 2 closely related species (15 H. melpomene and 15 H. cydno) (SI Appendix, Fig. S1). RNA was extracted using a TRIzol RNA isolation protocol followed by additional purification using a RNeasy minikit (Qiagen). Two Illumina libraries, each representing 45 individually barcoded RNA extractions for either H. melpomene or H. cydno, were generated using the Illumina TruSeq RNA sample preparation kit. Each library was sequenced three times on a HiSeq2500 Illumina system for a total of six Illumina sequencing lanes (50 bp single read) (SI Appendix, Detailed Materials and Methods).
Targeted Resequencing and Chemosensory Gene-Annotation Improvement.
We improved the chemosensory gene models by producing a target RNA resequencing dataset for the genes of interest (SI Appendix, Fig. S1). For a subset of the sampled H. melpomene butterflies (two males, two unmated females, and two mated females), we pooled equal amounts of RNA extracted from the antenna, legs, and mouthparts for each individual. The pooled samples (n = 6) were sent to Roche for targeted enrichment of sequencing libraries using the NimbleGen SeqCap protocol (68) followed by sequencing on the Illumina Hiseq2500 platform (100 bp paired end). We then used the targeted capture data to improve the annotation of the chemosensory genes, in particular the beginnings and ends of genes, and to find previously unannotated genes in the Hmel2 genome (SI Appendix, Table S1). With this improved chemosensory gene data set, we were able to accurately annotate genes and analyze our RNA-seq data (SI Appendix, Detailed Materials and Methods).
Raw Data Processing and Differential Expression Analyses.
The Illumina RNA-seq data were aligned to the Hmel2 genome using “TopHat” (q-value threshold = 0.05; for per gene q-values, see Dataset S1) and differential expression analyses were performed with Cufflinks (69). Differential expression analyses were conducted by tissue and biological group as follows. For the tissue-specific expression analyses, we combined data for all 30 butterflies and compared across tissue types. For species-, sex-, and life-stage–specific expression analyses, the data were analyzed for each tissue type independently (antennae, legs, or mouthparts). Species-specific differences were assessed by comparing males, unmated females, or mated females of H. melpomene to their biological equivalents in H. cydno. Here, differences in chemosensory gene expression between closely related species were hypothesized to reflect differences in mate or host plant choice. Sex-specific differences were assessed by comparing males and unmated females. This comparison was established to identify genes potentially central to both female- and male-choice. Life-stage–specific differences were assessed by comparing unmated and mated females. Here, differences between unmated and mated females were hypothesized to reflect a shift in sensory focus from mate searching (unmated females) to host plant detection for oviposition (mated female). Expression plots based on FPKM (fragments per kilobase of transcript per million mapped reads) values were created using the “heatmap.2” function of the “gplots” package in R (70) (SI Appendix, Detailed Materials and Methods).
Admixture and LD Analyses.
In order to further narrow down genes underlying species-level recognition, we compared results from our species-level differential expression analyses to admixture estimates for the sympatric H. melpomene–H. cydno pair from Panama (44). The admixture values (fd) are based on the ABBA-BABA test (71), which measures an excess of derived allele sharing between the sympatric nonsister taxa compared to an allopatric population of H. melpomene from French Guiana. We included H. numata as an outgroup (Fig. 3A). Negative fd values indicate low admixture or sharing of derived alleles between the sympatric Panama populations of H. melpomene and H. cydno, whereas positive fd values indicate high admixture. The fd values were calculated for 100-kb windows with a step size of 20 kb. For chemosensory genes, overlapping windows were averaged. With this approach, we were able to identify chemosensory genes that were both significantly differentially expressed (overexpressed) and presented low admixture between the two species. Chemosensory genes at the intercept of these two estimates are likely to be involved in species-specific processes. Finally, because the color-pattern locus optix has been shown to be associated with mate preference behavior in H. melpomene and H. cydno (51), and linkage between chemosensory and color-pattern genes could facilitate the forming of smell-based prezygotic barriers (43), we tested for the nonrandom association of chemosensory loci (50 kb around the transcription start site) at and around the optix gene (1,000-kb region) in 50-kb windows. Pairwise comparisons of SNPs within each 50-kb window were averaged to produce a mean r2 (SI Appendix, Detailed Materials and Methods). Finally, we evaluated the strength of LD associations for our chemosensory genes by calculating background r2 levels using 300 randomly sampled, nonchemosensory genes across the genome. We then compared r2 levels for our chemosensory genes against this null r2 distribution.
Data Availability.
The data that support the findings of this study are openly available on the National Center for Biotechnology Information Sequence Read Archive repository under the BioProject accession numbers PRJNA577441 (RNA-seq data) (47) and PRJNA577716 (targeted resequencing data) (48). Editable excel and text files associated with the data here presented are available on the Open Science Framework database (DOI: 10.17605/OSF.IO/2MB38) (72).
Supplementary Material
Acknowledgments
We thank the Smithsonian Tropical Research Institute for supporting this research; Dr. John Davey for providing additional information on the Hmel2 genome; Dr. Simon Martin for sharing the admixture data; and the University of Puerto Rico-Rio Piedras, Sequencing and Genomics Facility staff for their assistance with RNA extractions. This study was supported by an Institutional Development Award Networks of Biomedical Research Excellence (INBRE) Grant P20GM103475 from the National Institute of General Medical Sciences, a component of the NIH; the Bioinformatics Research Core of the INBRE, an NSF-Established Program to Stimulate Competitive Research RII Track-2 FEC OIA-1736026 (to R.P.); and an NSF Postdoctoral Research Fellowship in Biology (NSF‐PRFB: DBI‐1811008) (to J.M.-R.). Research reported in this publication was also supported by The Puerto Rico Science, Technology, and Research Trust under Agreement ARG 2020-00138.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject accession nos. PRJNA577441 and PRJNA577716), and in the Open Science Framework (DOI: 10.17605/OSF.IO/2MB38).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921318117/-/DCSupplemental.
References
- 1.Johansson B. G., Jones T. M., The role of chemical communication in mate choice. Biol. Rev. Camb. Philos. Soc. 82, 265–289 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Wyatt T. D., Pheromones and Animal Behaviour, (Cambridge University Press, ed. 1, 2003). [Google Scholar]
- 3.Larsdotter-Mellström H. et al., It’s all in the mix: Blend-specific behavioral response to a sexual pheromone in a butterfly. Front. Physiol. 7, 68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pliske T. E., Eisner T., Sex pheromone of the queen butterfly: Biology. Science 164, 1170–1172 (1969). [DOI] [PubMed] [Google Scholar]
- 5.Dion E., Pui L. X., Weber K., Monteiro A., Early-exposure to new sex pheromone blends alters mate preference in female butterflies and in their offspring. Nat. Commun. 11, 53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson H. M., Wanner K. W., The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briscoe A. D. et al., Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9, e1003620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horth L., Sensory genes and mate choice: Evidence that duplications, mutations, and adaptive evolution alter variation in mating cue genes and their receptors. Genomics 90, 159–175 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Smadja C., Butlin R. K., On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity 102, 77–97 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Symonds M. R. E., Elgar M. A., The evolution of pheromone diversity. Trends Ecol. Evol. 23, 220–228 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Niehuis O. et al., Behavioural and genetic analyses of Nasonia shed light on the evolution of sex pheromones. Nature 494, 345–348 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Lassance J. M., Löfstedt C., Chemical communication: A jewel sheds light on signal evolution. Curr. Biol. 23, R346–R348 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Higgie M., Chenoweth S., Blows M. W., Natural selection and the reinforcement of mate recognition. Science 290, 519–521 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Groot A. T. et al., Experimental evidence for interspecific directional selection on moth pheromone communication. Proc. Natl. Acad. Sci. U.S.A. 103, 5858–5863 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leary G. P. et al., Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coughlan J. M., Matute D. R., Speciation: On the scent of mate discrimination genes. Curr. Biol. 28, R1389–R1391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogt R. G., Große-Wilde E., Zhou J. J., The Lepidoptera odorant binding protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62, 142–153 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Darragh K., et al. , A novel terpene synthase produces an anti-aphrodisiac pheromone in the butterfly. bioRxiv: 10.1101/779678 (11 October 2019). [DOI]
- 19.Darragh K. et al., Male sex pheromone components in Heliconius butterflies released by the androconia affect female choice. PeerJ 5, e3953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz S., Estrada C., Yildizhan S., Boppré M., Gilbert L. E., An antiaphrodisiac in Heliconius melpomene butterflies. J. Chem. Ecol. 34, 82–93 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Merrill R. M., Naisbit R. E., Mallet J., Jiggins C. D., Ecological and genetic factors influencing the transition between host-use strategies in sympatric Heliconius butterflies. J. Evol. Biol. 26, 1959–1967 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Silva D. S., Barp E. A., Kucharski L. C. R., Moreira G. R. P., Sensing the plant surface prior to feeding and oviposition: Differences in external ultrastructure and function among tarsi of Heliconius erato. Neotrop. Entomol. 47, 85–95 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Thiele S. C., Rodrigues D., Moreira G. R. P., Oviposition in Heliconius erato (Lepidoptera, Nymphalidae): How essential is drumming behavior for host-plant selection? J. Insect Behav. 29, 283–300 (2016). [Google Scholar]
- 24.Xu P., Atkinson R., Jones D. N. M., Smith D. P., Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Laughlin J. D., Ha T. S., Jones D. N. M., Smith D. P., Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R., Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 93, 184–200 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Mei T., Fu W. B., Li B., He Z. B., Chen B., Comparative genomics of chemosensory protein genes (CSPs) in twenty-two mosquito species (Diptera: Culicidae): Identification, characterization, and evolution. PLoS One 13, e0190412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wanner K. W., Isman M. B., Feng Q., Plettner E., Theilmann D. A., Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Chroistoneura fumiferana. Insect Mol. Biol. 14, 289–300 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Drurey C., et al. , Chemosensory proteins in the CSP4 clade evolved as plant immunity suppressors before two suborders of plant-feeding hemipteran insects diverged. bioRxiv: 10.1101/173278 (22 April 2019). [DOI]
- 30.Freeman E. G., Wisotsky Z., Dahanukar A., Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc. Natl. Acad. Sci. U.S.A. 111, 1598–1603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni L. et al., A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh T. W. et al., The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rytz R., Croset V., Benton R., Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y. V., Ni J., Montell C., The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dasmahapatra K. K. et al.; Heliconius Genome Consortium , Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Schooten B., Jiggins C. D., Briscoe A. D., Papa R., Genome-wide analysis of ionotropic receptors provides insight into their evolution in Heliconius butterflies. BMC Genomics 17, 254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohse K., Chmelik M., Martin S. H., Barton N. H., Efficient strategies for calculating blockwise likelihoods under the coalescent. Genetics 202, 775–786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavárez J. et al., Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Mallet J., Beltrán M., Neukirchen W., Linares M., Natural hybridization in heliconiine butterflies: The species boundary as a continuum. BMC Evol. Biol. 7, 28 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiggins C. D., Naisbit R. E., Coe R. L., Mallet J., Reproductive isolation caused by colour pattern mimicry. Nature 411, 302–305 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Jiggins C. D., Lamas G., The Ecology and Evolution of Heliconius Butterflies, (Oxford University Press, 2016). [Google Scholar]
- 42.Naisbit R. E., “Ecological divergence and speciation in Heliconius cydno and H. melpomene,” PhD thesis, University College London, London, United Kingdom (2001).
- 43.Merrill R. M., Van Schooten B., Scott J. A., Jiggins C. D., Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc. Biol. Sci. 278, 511–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin S. H., Davey J. W., Salazar C., Jiggins C. D., Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17, e2006288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin S. H. et al., Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23, 1817–1828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darragh K. et al., Species specificity and intraspecific variation in the chemical profiles of Heliconius butterflies across a large geographic range. Ecol. Evol. 10, 3895–3918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Schooten B., et al. , Differential expression of Heliconius melpomene and Heliconius cydno sensory tissues. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA577441. Deposited 14 October 2019.
- 48.van Schooten B., et al. , Heliconius melpomene targeted resequencing raw reads. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA577716. Deposited 15 October 2019.
- 49.Sánchez-Gracia A., Vieira F. G., Rozas J., Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Kronforst M. R. et al., Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl. Acad. Sci. U.S.A. 103, 6575–6580 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrill R. M. et al., Genetic dissection of assortative mating behavior. PLoS Biol. 17, e2005902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin A. et al., Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. U.S.A. 109, 12632–12637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed R. D. et al., Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Nadeau N. J. et al., The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534, 106–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dani F. R. et al., Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem. Senses 36, 335–344 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Bentzur A. et al., Odorant binding protein 69a connects social interaction to modulation of social responsiveness in Drosophila. PLoS Genet. 14, e1007328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kryvokhyzha D., “Whole genome resequencing of Heliconius butterflies revolutionizes our view of the level of admixture between species,” Masters thesis, Uppsala University, Uppsala, Sweden (2014).
- 58.Ozaki K. et al., A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nat. Commun. 2, 542 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Briscoe A. D. et al., Positive selection of a duplicated UV-sensitive visual pigment coincides with wing pigment evolution in Heliconius butterflies. Proc. Natl. Acad. Sci. U.S.A. 107, 3628–3633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joron M., Jiggins C. D., Papanicolaou A., McMillan W. O., Heliconius wing patterns: An evo-devo model for understanding phenotypic diversity. Heredity 97, 157–167 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Jiggins C. D., Ecological speciation in mimetic butterflies. Bioscience 58, 541–548 (2008). [Google Scholar]
- 62.Mérot C., Frérot B., Leppik E., Joron M., Beyond magic traits: Multimodal mating cues in Heliconius butterflies. Evolution 69, 2891–2904 (2015). [DOI] [PubMed] [Google Scholar]
- 63.González-Rojas M. F. et al., Chemical signals act as the main reproductive barrier between sister and mimetic Heliconius butterflies. Proc. Biol. Sci. 387, 20200587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Belleghem S. M. et al., Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis J. J. et al., Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc. Natl. Acad. Sci. U.S.A. 116, 24174–24183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Servedio M. R., Van Doorn G. S., Kopp M., Frame A. M., Nosil P., Magic traits in speciation: “Magic” but not rare? Trends Ecol. Evol. 26, 389–397 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Zhang W. et al., Comparative transcriptomics provides insights into reticulate and adaptive evolution of a butterfly radiation. Genome Biol. Evol. 11, 2963–2975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samorodnitsky E. et al., Comparison of custom capture for targeted next-generation DNA sequencing. J. Mol. Diagn. 17, 64–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C. et al., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warnes G. R., et al. , gplots: Various R programming tools for plotting data. https://cran.r-project.org/web/packages/gplots/index.html. Accessed 24 June 2020.
- 71.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Schooten B., et al. , Supplemental files: Divergence of chemosensing during the early stages of speciation. Open Science Framework. https://osf.io/2mb38/. Deposited 14 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available on the National Center for Biotechnology Information Sequence Read Archive repository under the BioProject accession numbers PRJNA577441 (RNA-seq data) (47) and PRJNA577716 (targeted resequencing data) (48). Editable excel and text files associated with the data here presented are available on the Open Science Framework database (DOI: 10.17605/OSF.IO/2MB38) (72).