Abstract
A key aspect of rice breeding programs is the optimization of days to heading (DTH) for maximizing grain productivity in cultivation areas. Here, the effects of genotypes for heading date on yield-related traits in rice (culm and panicle length (CL and PL), panicle number (PN), and total number of seeds) were investigated. Heading date 1 (Hd1) and Days to heading 8 (DTH8) are the main controllers of the variation in heading date in the rice population of Hokkaido, Japan. Thus, an F2 population (n = 192) derived from a cross between Kitaibuki (Hd1dth8) and Akage (hd1DTH8) was developed. Significant differences in DTH were found among all combinations. Each genotype for heading date showed variations in the yield-related traits without a significant difference. However, DTH exhibited high positive coefficient values (more than 0.709) with the yield-related traits except for PN, which had a negative coefficient value of –0.431. A later heading date resulted in a longer growth duration and a higher yield with a combination of longer PL and CL and lower PN. These results suggest that DTH limits the yield-related traits rather than the genotype for heading date.
Keywords: rice, heading date, yield, days to heading (DTH), Hd1, DTH8
Introduction
Heading date in crop species is a limiting factor of species range and cropping season. Variations in the heading date are determined by the response to seasonal cues from changing environmental factors, including day length and temperature (Blümel et al. 2015, Fujino et al. 2019a, Hill and Li 2016, Hu et al. 2019). Expanding the species range is a major goal for crop breeding programs in improving the food supply for the growing worldwide population and under the conditions of global warming. Many genes for the control of heading date in rice have been identified (Gramene, http://www.gramene.org), and a genetic network for heading date has been proposed (Hori et al. 2016, Tsuji et al. 2011). However, it is unclear whether the genotype for heading date effects the yield in agriculture.
Asian cultivated rice, Oryza sativa L., originated in the tropics (Choi et al. 2017, Fuller 2011). Extensive efforts in rice breeding programs to optimize the heading date have made the stable production of rice possible under various climatic conditions at latitudes ranging from 53°N to 40°S (Fujino et al. 2015, 2017, 2019a, 2019b, Lu and Chang 1980, Shinada et al. 2014). An earlier heading date, by a decrease in the photoperiod sensitivity, may have played an important role in expanding the range of rice (Fujino et al. 2019a, 2019b, 2019c, Izawa 2007, Shrestha et al. 2014, Zheng et al. 2016).
Based on a series of genetic analyses for the heading date in rice, QTLs/genes for an extremely early heading date were previously identified. Only varieties with an extremely early heading date were adapted to Hokkaido (41–45°N latitude), Japan, which has a long natural day length of more than 15 hours in summer (Fujino and Sekiguchi 2005a, 2005b, 2008, Fujino et al. 2013, 2019a, 2019b, 2019c, Nonoue et al. 2008). The loss-of-function alleles at Oryza sativa Pseudo-Response Regulator37 (OsPRR37) and Grain number, plant height and heading date 7 (Ghd7) accounted for the extremely early heading date in EARLY-DUO (Fujino et al. 2019a, Gao et al. 2014, Koo et al. 2013, Xue et al. 2008).
In addition to the adaptability to local environmental conditions, genes shaping the adaptability have been selected for stable rice production in modern agriculture. A loss-of-function allele at Heading date 1 (Hd1), hd1, was selected for in the early phase of expanding the rice range to northern Japan (Yokoo et al. 1980). Rice breeding programs in Hokkaido introduced functional alleles to the Hd1 locus, Hd1, in the Hokkaido rice population. Under long-day conditions, a functional Hd1 delays the heading date (Hayama et al. 2003, Yano et al. 2000), whereas a functional Hd1 promotes an earlier heading date in EARLY-DUO (Fujino et al. 2019b).
Furthermore, the gene controlling the cropping season in Hokkaido has been identified. A loss-of-function allele at Heading date 5 (Hd5), hd5, accounted for the earlier heading date in the Hokkaido rice population in an early-maturing type (Fujino 2003, Fujino et al. 2013). Hd5 is allelic to Days to heading 8 (DTH8), encoding a putative HAP3 subunit of a CCAAT-box-binding transcription factor (Fujino et al. 2013, Wei et al. 2010).
Rice breeding programs in Hokkaido had great success in the development of new varieties expressing good eating quality with a unique adaptability to Hokkaido’s climate (Fujino et al. 2019a, 2019c). The heading date and yield are major determinants for the development of commercial rice. The yield-related traits (panicle length (PL), culm length (CL), panicle number (PN), and total number of seeds (TS)) are indicators of yield in rice. Due to the significance of rice in agriculture, many mappings and molecular cloning of QTLs for heading date and yield have been reported, revealing the natural variations of the traits. Some have shown common associations between heading date and yield; delaying the heading date causes an increase in yield.
The genes and genotype for the heading date established by rice breeding programs in Hokkaido have been investigated (Fujino et al. 2019a). However, the genetic base for rice yield in agriculture was still unclear. Pleiotropic effects of the heading date genes on the developmental phase in panicles have been reported (Endo-Higashi and Izawa 2011, Fujino and Ikegaya 2020, Fujino et al. 2019a, Wei et al. 2010, Weng et al. 2014, Xue et al. 2008). Furthermore, it was unclear whether the genotype for heading date regulates the yield-related traits as a genetic background. Here, we examined the effects of Hd1 and DTH8 on yield in the Hokkaido rice population.
Materials and Methods
Plant material
Two japonica rice varieties from Hokkaido, Kitaibuki (KT) and Akage (AG), were used as the parental varieties. KT was registered in 1990 in rice breeding programs in Hokkaido. AG is a landrace variety in Hokkaido. To identify the effects of Hd1 and DTH8 on the yield-related traits, an F2 population (n = 192) derived from a cross between KT (Hd1dth8) and AG (hd1DTH8) was developed.
Cultivation methods
Seeds of these rice varieties were provided by the Local Independent Administrative Agency Hokkaido Research Organization Hokkaido Central Agricultural Experiment Station (Takikawa, Japan). All plant materials were cultivated in an experimental paddy field at Hokkaido Agricultural Research Center (Sapporo, Hokkaido, Japan, 43°00ʹ N latitude) in 2015 with 15.0 cm spacing between the plants within each row and 30.0 cm spacing between rows. Cultivation management followed the standard procedures used at Hokkaido Agricultural Research Center. Sowing and transplanting were performed on April 27 and May 21, 2015, respectively.
Trait evaluation
The heading date was measured for all F2 plants as days to heading (DTH). DTH is the number of days from sowing to heading of the earliest heading panicle on an individual. As yield-related traits, CL, PL, PN, and TS were evaluated in accordance with Fujino et al. (2017). These yield-related traits were measured in plants with the four genotypes homozygous in both Hd1 and DTH8. In addition, the internode length and panicle components were measured according to Fujino and Ikegaya (2020). The phenotypes were averaged for each line/variety and were compared using Tukey–Kramer HSD test among genotypes. Genetic interactions between Hd1 and DTH8 on the yield-related traits were calculated by two-way ANOVA.
DNA analysis
Total DNA was isolated from young leaves using the CTAB method (Murray and Thompson 1980). The genotypes of Hd1 and DTH8 were determined using the primers for functional nucleotide polymorphisms in these genes (Fujino et al. 2013, 2019a). PCR and electrophoresis were performed as described previously (Fujino et al. 2004, 2005).
Results
Variation in heading dates by Hd1 and DTH8
There was a significant difference (p < 0.001, t-test) in DTH between the parental varieties (Fig. 1, Table 1). The DTH of KT was 87.7 ± 0.7 days, whereas that of AG was 91.5 ± 0.9 days. A wide and continuous variation was observed in the frequency distribution of DTH among the F2 populations, ranging from 80 to 100 days (Fig. 1). According to the genotypes of Hd1 and DTH8, plants in this population were classified into nine genotypes with significantly different DTH (Table 1).
Fig. 1.

Frequency distribution of days to heading among the F2 populations (n = 192) derived from a cross between Kitaibuki (KT) and Akage (AG). Vertical and horizontal bars show the mean and range of days to heading, respectively, in the parents.
Table 1.
Average days to heading (DTH) among nine genotypes for heading date
| Genotype | No. of plants |
DTH | |||
|---|---|---|---|---|---|
| DTH8 | Hd1 | Range | |||
| Minimum | Maximum | ||||
| A | A | 14 | 96.4 ± 2.1d | 92 | 100 |
| A | H | 26 | 94.4 ± 2.7cd | 90 | 100 |
| A | B | 19 | 91.3 ± 3.0bc | 86 | 96 |
| H | A | 21 | 96.0 ± 1.9d | 92 | 100 |
| H | H | 44 | 92.1 ± 3.3bc | 82 | 98 |
| H | B | 25 | 89.2 ± 3.7b | 80 | 94 |
| B | A | 13 | 90.5 ± 2.7bc | 86 | 96 |
| B | H | 17 | 84.7 ± 3.1a | 80 | 92 |
| B | B | 13 | 80.9 ± 1.0a | 80 | 82 |
A, B, and H in genotype indicate the Akage (DTH8hd1), Kitaibuki (dth8Hd1) and heterozygous alleles, respectively.
DTH is expressed as mean ± standard deviation.
Different letters indicate significant differences at p < 0.05.
Fifty-five plants in the four classes homozygous in both the Hd1 and DTH8 alleles were selected and used for further analysis of the associations between DTH and yield-related traits. First, the effects of Hd1 and DTH8 on the heading date were examined (Table 1). The genotype Hd1DTH8 was defined as a wild type (WT) with DTH of 91.3. hd1 delayed DTH by 5.4 days in comparison to WT, whereas dth8 significantly decreased DTH by 10.2 days in comparison to WT. hd1dth8 decreased DTH by 0.6 days in comparison to WT, suggesting that hd1 and dth8 epistatically controlled DTH.
Comparison of the yield-related traits among the genotypes for heading date
To elucidate the role of genotypes for heading date on the yield-related traits, means of the four traits, PL, CL, PN, and TS, were compared among the four genotypes of Hd1 and DTH8 (Table 3). No abnormality in the yield-related traits evaluated in this experiment was observed in genotypes with the recessive alleles of genes for heading date, hd1 and dth8 with EARLY-DUO, ghd7, and osprr37 (Fujino et al. 2019a).
Table 3.
Coefficient in the yield-related traits among the four genotypes for heading date in the F2 population
| DTH | PL | CL | PN | TS | |
|---|---|---|---|---|---|
| DTH | 1 | ||||
| PL | 0.763212 | 1 | |||
| CL | 0.825262 | 0.770695 | 1 | ||
| PN | –0.42837 | –0.33047 | –0.42183 | 1 | |
| TS | 0.90472 | 0.740094 | 0.77523 | –0.36027 | 1 |
DTH: days to heading, CL: culm length, PL: panicle length, PN: panicle number, TS: total number of seeds.
All values indicate significant differences at p < 0.05.
The variation in PL in the four genotypes for heading date was 12.2–20.5 cm (Table 2). PL increased by 0.6 cm and dth8 decreased by 3.8 cm in hd1 in comparison to WT (17.4). PL epistatically exhibited a 0.2-cm increase in hd1dth8 (Supplemental Table 1). The variation in CL was 51.5–92.5 cm. CL increased by 9.1 cm in hd1 and decreased by 16.4 cm in dth8 in comparison to WT (75.5). hd1dth8 epistatically exhibited a 0.4-cm decrease in CL (Supplemental Table 1). The variation in PN was 10–32. PN decreased by 0.9 in hd1and increased by 6.2 in dth8 in comparison to WT (16.7). hd1dth8 additively exhibited a 2.5 increase in PN (Supplemental Table 1). In TS, WT increased by 8.1 in hd1 and decreased by 46 in dth8 in comparison to WT (87.4). hd1dth8 epistatically exhibited a 5.4 decrease in TS.
Table 2.
Differences in yield-related traits among genotypes for heading date
| Genotype | No. of plants |
Trait | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DTH | PL | CL | PN | TS | ||||||||||||||||
| Avarage | Minimum | Maximum | Avarage | Minimum | Maximum | Avarage | Minimum | Maximum | Avarage | Minimum | Maximum | Avarage | Minimum | Maximum | ||||||
| DTH8hd1 | 13 | 96.5 ± 2.2a | 92.0 | 100.0 | 18.0 ± 1.2b | 16.5 | 20.5 | 84.6 ± 5.6c | 74.0 | 92.5 | 15.8 ± 3.1b | 12 | 22 | 95.5 ± 10.7b | 79 | 112 | ||||
| Dth8Hd1 | 18 | 91.1 ± 3.0b | 86.0 | 96.0 | 17.4 ± 1.1b | 15.0 | 19.4 | 75.5 ± 6.8b | 65.0 | 85.0 | 16.7 ± 3.1b | 11 | 21 | 87.4 ± 14.0b | 60 | 125 | ||||
| dth8hd1 | 11 | 90.5 ± 2.4b | 86.0 | 96.0 | 17.6 ± 1.7b | 14.1 | 19.9 | 75.1 ± 5.1b | 67.0 | 85.0 | 19.2 ± 6.5ab | 10 | 32 | 82.0 ± 15.1b | 56 | 108 | ||||
| dth8Hd1 | 13 | 80.9 ± 1.0c | 80.0 | 82.0 | 13.6 ± 0.8a | 12.2 | 14.9 | 59.1 ± 5.1a | 51.5 | 67.5 | 22.9 ± 3.1a | 19 | 30 | 41.4 ± 9.4a | 28 | 60 | ||||
DTH: days to heading, PL: panicle length, CL: culm length, PN: panicle number, TS: total number of seeds.
Values in the traits are expressed as mean ± standard deviation.
Different letters indicate significant differences at p < 0.05.
Hd1dth8 exhibited the earliest heading date with the smallest standard deviation, 80.9 ± 1.0 in DTH. PL, CL, TS, and PN in the Hd1dth8 genotype were significantly smaller than those in the other genotypes (Table 2). Although there were no significant differences in yield-related traits among the three genotypes for heading date, Hd1DTH8, hd1DTH8, and hd1dth8, DTH had high positive coefficient values for PL, CL, and TS of more than 0.709 (Fig. 2, Table 3), whereas PN had a negative coefficient for DTH of –0.431.
Fig. 2.
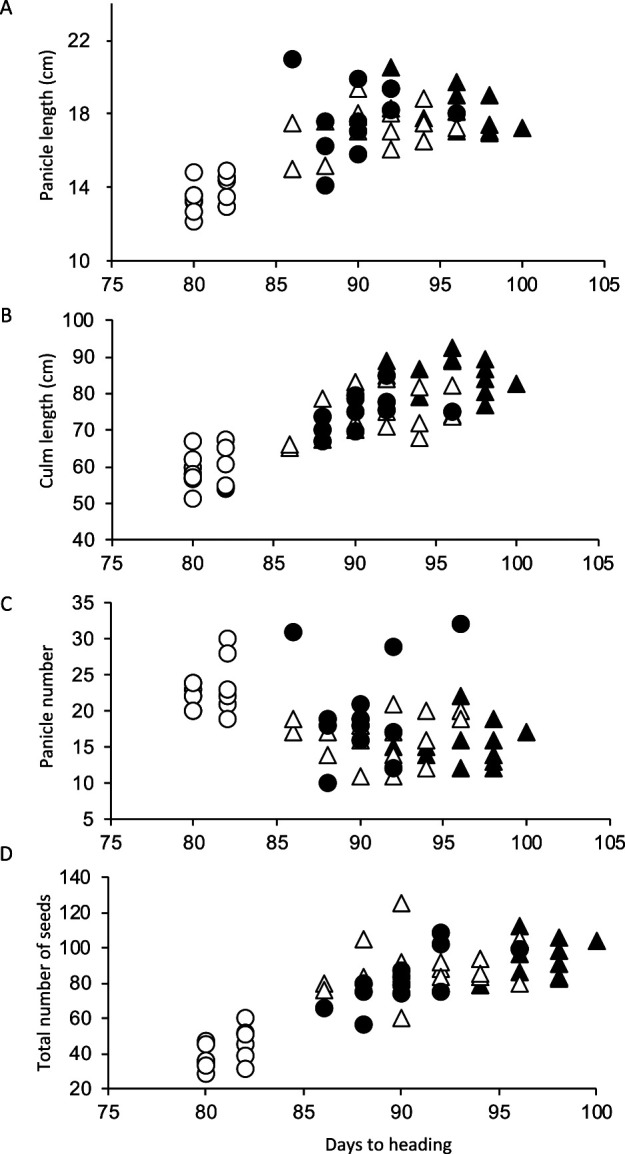
Associations of days to heading with the yield-related traits of the four genotypes for heading date, dth8Hd1 (open circle), dth8hd1 (closed triangle), DTH8Hd1 (open triangle), and DTH8hd1 (closed circle). (A) panicle length, (B) culm length, (C) panicle number, and (D) total number of seeds.
Variation of the component traits in CL and TS
CL and TS were compared among the four genotypes for heading date. In CL, variations in the internode length were in the ranges of 28.0–42.5 in I, 17.5–27.0 in II, 4.0–20.5 in III, and 0–10.5 in IV (Supplemental Table 2). The lengths of the II, III, and IV internodes were regulated additively by Hd1 and DTH8, whereas that of I was regulated epistatically (Supplemental Table 1). dth8 significantly decreased the internode lengths of I–III, whereas hd1 significantly increased the internode length of specifically IV. High positive coefficient values were observed between DTH and the internode length of more than 0.646 (Supplemental Fig. 1, Supplemental Table 3).
TS was expressed as the sum of seeds on the primary branches (Pb) and secondary branches (Sb). dth8 had significantly smaller numbers of both Pb and Sb, decreasing TS. The variations in Pb and Sb were 4–12 and 3–22, respectively (Supplemental Table 4). Pb increased by 1.4 in hd1 and decreased by 3.1 in dth8 in comparison to WT (8.3). Pb epistatically decreased by 0.2 in hd1dth8. For Sb, hd1 and dth8 decreased by 0.2 and 8.3, respectively, in comparison to WT (13.9). Sb epistatically decreased by 1.2 in hd1dth8.
Similar to the numbers of Pb and Sb, the numbers of seeds on the Pb (SPb) and Sb (SSb) depended on the genotype for heading date. The variations in SPb and SSb were 18–68 and 8–64, respectively (Supplemental Table 3). SPb increased by 8.4 in hd1 and decreased by 21.6 in dth8 in comparison to WT (48.1). SPb epistatically decreased by 2.5 in hd1dth8. For SSb, hd1 and dth8 decreased by 0.4 and 24.6, respectively, in comparison to WT (39.4). SSb epistatically decreased by 3.6 in hd1dth8.
Pb and SPb had higher coefficients for DTH than Sb and SSb (Supplemental Fig. 2, Supplemental Table 5). TS also had a high positive coefficient value for DTH (0.826).
Discussion
Optimization of the heading date to maximize yield in agriculture has been shown in differentiated rice populations from different environments. Rice breeding programs generate novel genotypes other than those in natural populations for exploring the adaptability beyond native species ranges and produce stable rice production (Fujino et al. 2019a, Hua et al. 2019, Huang et al. 2018). Hd1 and DTH8 generated a variation in the heading date within the Hokkaido rice population (Fujino and Sekiguchi 2005a, Fujino et al. 2013, 2019a, 2019c). Here, the effects of Hd1 and DTH8 on yield were examined.
Hd1 and DTH8 generated a significant variation in heading date within the growth period in Hokkaido (Tables 1, 2). There were no significant differences in the yield-related traits among the genotypes. However, higher coefficient values were found between DTH and yield-related traits (Supplemental Tables 3, 5). DTH positively increased PL, CL, and PL, and negatively increased PN (Table 3). Hd1 and DTH8 may have roles additively and epistatically on the regulation of the yield-related traits and exhibit opposite but different degrees of effects on these traits (Table 2, Supplemental Table 1). Variations in DTH in each genotype were similar. DTH showed high positive coefficients with yield-related traits, suggesting that it limits yield rather than the genotype for heading date.
Genes for yield-related traits have epistatic effects on each other (Ando et al. 2008, Takai et al. 2014, Xu et al. 2015). In addition, they may be involved in the pleiotropy of DTH. The genetic role of yield makes their control difficult and complex in rice breeding programs. Previously, associations of DTH with yield-related traits have been demonstrated using series of NILs (Fujino and Ikegaya 2020, Fujino et al. 2019a). The higher coefficient values in these studies indicate that the significance of genes/QTLs for the heading date plays a role not only in the control of heading date but also in the regulation of yield.
Fig. 3 shows the model for the selection of desirable lines in rice breeding programs in Hokkaido is proposed. Desirable lines may exhibit a heading date within the optimum range to produce rice grain with good eating quality. A later heading date produces a maximum growth duration and a higher yield with a combination of longer PL and CL and lower PN. The total seed number is determined as panicle architecture depending on the DTH.
Fig. 3.

Model of the selection during rice breeding programs in Hokkaido. Days to heading (DTH) of the breeding line is within the optimum range (black and bold arrow) of DTH in Hokkaido. The breeding line whose yield is limited by DTH is selected, which is more favorable than those of the current commercial varieties.
During the process of rice breeding programs in Hokkaido, exotic germplasms may generate novel phenotypes (Fujino et al. 2015, 2017, Shinada et al. 2014). Genes controlling the heading date were under selection for their unique adaptability to environmental conditions and stable rice production (Fujino et al. 2019a). In addition, this study confirmed that the genes for heading date may regulate not only an optimized heading date but also maximize the fitness to agriculture. The pleiotropy of DTH may clarify the yield complex in modern rice breeding programs to develop new rice varieties. Current genomic tools may facilitate rice breeding programs for future demands.
Author Contribution Statement
Conceived and designed the experiments: KF. Performed the experiments, analyzed the data, and wrote the manuscript: KF.
Supplementary Material
Acknowledgments
I thank M. Obara for help with the DNA experiments and T. Ikegaya and K. Komatsu for the statistical analysis.
Literature Cited
- Ando T., Yamamoto T., Shimizu T., Ma X.F., Shomura A., Takeuchi Y., Lin S.Y. and Yano M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116: 881–890. [DOI] [PubMed] [Google Scholar]
- Blümel M., Dally N. and Jung C. (2015) Flowering time regulation in crops—what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32: 121–129. [DOI] [PubMed] [Google Scholar]
- Choi J.Y., Platts A.E., Fuller D.Q., Hsing Y.-I., Wing R.A. and Purugganan M.D. (2017) The rice paradox: multiple origins but single domestication in Asian rice. Mol. Biol. Evol. 34: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo-Higashi N. and Izawa T. (2011) Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 52: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K. (2003) Photoperiod sensitivity gene controlling heading date in rice cultivars in the northernmost region of Japan. Euphytica 131: 97–103. [Google Scholar]
- Fujino K., Sekiguchi H., Sato T., Kiuchi H., Nonoue Y., Takeuchi Y., Ando T., Lin S.Y. and Yano M. (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 108: 794–799. [DOI] [PubMed] [Google Scholar]
- Fujino K. and Sekiguchi H. (2005a) Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.). Theor. Appl. Genet. 111: 393–398. [DOI] [PubMed] [Google Scholar]
- Fujino K. and Sekiguchi H. (2005b) Identification of QTLs conferring genetic variation for heading date among rice varieties at the northern-limit of rice cultivation. Breed. Sci. 55: 141–146. [Google Scholar]
- Fujino K., Sekiguchi H. and Kiguchi T. (2005) Identification of an active transposon in intact rice plants. Mol. Genet. Genomics 273: 150–157. [DOI] [PubMed] [Google Scholar]
- Fujino K. and Sekiguchi H. (2008) Mapping of quantitative trait loci controlling heading date among rice cultivars in the northernmost region of Japan. Breed. Sci. 58: 367–373. [Google Scholar]
- Fujino K., Yamanouchi U. and Yano M. (2013) Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 126: 611–618. [DOI] [PubMed] [Google Scholar]
- Fujino K., Obara M., Ikegaya T. and Tamura K. (2015) Genetic shift in local rice populations during rice breeding programs in the northern limit of rice cultivation in the world. Theor. Appl. Genet. 128: 1739–1746. [DOI] [PubMed] [Google Scholar]
- Fujino K., Nishimura T., Kiuchi H., Hirayama Y. and Sato T. (2017) Phenotypic changes during 100-year rice breeding programs in Hokkaido. Breed. Sci. 67: 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K., Obara M. and Ikegaya T. (2019a) Establishment of adaptability to the northern-limit of rice production. Mol. Genet. Genomics 294: 729–737. [DOI] [PubMed] [Google Scholar]
- Fujino K., Yamanouchi U., Nonoue Y., Obara M. and Yano M. (2019b) Switching genetic effects of the flowering time gene Hd1 in LD conditions by Ghd7 and OsPRR37 in rice. Breed. Sci. 69: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K., Hirayama Y. and Kaji R. (2019c) Marker-assisted selection in rice breeding programs in Hokkaido. Breed. Sci. 69: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K. and Ikegaya T. (2020) A novel genotype DATTO5 developed using the five genes exhibits the fastest heading date designed in rice. Breed. Sci. 70: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller D.Q. (2011) Pathways to Asian civilizations: tracing the origins and spread of rice and rice cultures. Rice 4: 78–92. [Google Scholar]
- Gao H., Jin M., Zheng X.M., Chen J., Yuan D., Xin Y., Wang M., Huang D., Zhang Z., Zhou K. et al. (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 111: 16337–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M. and Shimamoto K. (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Hill C.B. and Li C. (2016) Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Front. Plant Sci. 7: 1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Matsubara K. and Yano M. (2016) Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129: 2241–2252. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li S. and Xing Y. (2019) Lessons from natural variations: artificially induced heading date variations for improvement of regional adaptation in rice. Theor. Appl. Genet. 132: 383–394. [DOI] [PubMed] [Google Scholar]
- Hua Z., Olatoyea M.O., Marlaa S. and Morris G.P. (2019) An integrated genotyping-by-sequencing polymorphism map for over 10,000 sorghum genotypes. Plant Genome 12: 1–15. [DOI] [PubMed] [Google Scholar]
- Huang C., Sun H., Xu D., Chen Q., Liang Y., Wang X., Xu G., Tian J., Wang C., Li D. et al. (2018) ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 115: E334–E341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. (2007) Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58: 3091–3097. [DOI] [PubMed] [Google Scholar]
- Koo B.H., Yoo S.C., Park J.W., Kwon C.T., Lee B.D., An G., Zhang Z., Li J., Li Z. and Paek N.C. (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6: 1877–1888. [DOI] [PubMed] [Google Scholar]
- Lu, J.J. and T.T. Chang (1980) Rice in its temporal and spatial perspectives. In: Luh, B.S. (ed.) Rice: Production and Utilization, AVI Publishing Co., Inc., Westport, CT, pp. 1–74. [Google Scholar]
- Murray M.G. and Thompson W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoue Y., Fujino K., Hirayama Y., Yamanouchi U., Lin S.Y. and Yano M. (2008) Detection of quantitative trait loci controlling extremely early heading in rice. Theor. Appl. Genet. 116: 715–722. [DOI] [PubMed] [Google Scholar]
- Shinada H., Yamamoto T., Yamamoto E., Hori K., Yonemaru J., Matsuba S. and Fujino K. (2014) Historical changes in population structure during rice breeding programs in the northern limits of rice cultivation. Theor. Appl. Genet. 127: 995–1004. [DOI] [PubMed] [Google Scholar]
- Shrestha R., Gómez-Ariza J., Brambilla V. and Fornara F. (2014) Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114: 1445–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T., Ikka T., Kondo K., Nonoue Y., Ono N., Arai-Sanoh Y., Yoshinaga S., Nakano H., Yano M., Kondo M. et al. (2014) Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines. BMC Plant Biol. 14: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Taoka K. and Shimamoto K. (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14: 45–52. [DOI] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H. and Wan J. (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X., Wang L., Wang J., Hu Y., Du H., Xu C., Xing Y., Li X., Xiao J. and Zhang Q. (2014) Grain Number, Plant Height, and Heading Date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164: 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Chen W. and Xu Z. (2015) Relationship between grain yield and quality in rice germplasms grown across different growing areas. Breed. Sci. 65: 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X. et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y. et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo M., Kikuchi F., Nakane A. and Fujimaki H. (1980) Genetical analysis of heading time by aid of close linkage with blast, Pyricularia oryzae, resistance in rice. Bull. Natl. Inst. Agric. Sci. Ser. D 31: 95–126. [Google Scholar]
- Zheng X.M., Feng L., Wang J., Qiao W., Zhang L., Cheng Y. and Yang Q. (2016) Nonfunctional alleles of long-day suppressor genes independently regulate flowering time. J. Integr. Plant Biol. 58: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


