Abstract
The increasing prevalence of type 2 diabetes poses a major challenge to societies worldwide. Blood-based factors like serum proteins are in contact with every organ in the body to mediate global homeostasis and may thus directly regulate complex processes such as aging and the development of common chronic diseases. We applied a data-driven proteomics approach, measuring serum levels of 4,137 proteins in 5,438 elderly Icelanders, and identified 536 proteins associated with prevalent and/or incident type 2 diabetes. We validated a subset of the observed associations in an independent case-control study of type 2 diabetes. These protein associations provide novel biological insights into the molecular mechanisms that are dysregulated prior to and following the onset of type 2 diabetes and can be detected in serum. A bidirectional two-sample Mendelian randomization analysis indicated that serum changes of at least 23 proteins are downstream of the disease or its genetic liability, while 15 proteins were supported as having a causal role in type 2 diabetes.
Introduction
Type 2 diabetes is a progressive disease characterized by decreasing sensitivity of peripheral tissues to plasma insulin, accompanied by compensatory hyperinsulinemia, and a gradual failure of the pancreatic islet β-cells to maintain glucose homeostasis. In the past decade, the use of data-driven omics technologies has led to a significant advancement in the discovery of new biomarker candidates and biological insights for complex diseases. More than 240 genetic loci have been associated with type 2 diabetes in genome-wide association studies (GWAS) (1–5), and blood-based biomarker candidates for type 2 diabetes have begun to emerge, perhaps most notably the branched-chain amino acids (BCAAs) (6,7), the catabolism of which has recently been proposed as a novel treatment target for obesity-associated insulin resistance (8). However, only fragmentary data are available for serum protein links to type 2 diabetes (9). While few biomarker candidates provide much improvement in type 2 diabetes prediction over conventional measures of glycemia and adiposity (9), they may provide insight into biological processes that are important in the disease pathogenesis.
Proteins are the key functional units of biology and disease; however, high-throughput detection and quantification of serum proteins in a large human population have been hampered by the limitations of available proteomic profiling technologies. The Slow-Off rate Modified Aptamer (SOMAmer)-based technology has emerged as a powerful proteomic profiling platform in terms of sensitivity, dynamic range of detection, and multiplex capacity (10–12). A custom-designed SOMAscan platform was recently developed to measure 5,034 protein analytes in a single serum sample, of which 4,782 SOMAmers bind specifically to 4,137 distinct human proteins (13). We applied this platform to 5,457 subjects of the Age, Gene/Environment Susceptibility (AGES)-Reykjavik study (13,14) and demonstrate novel serum protein associations with prevalent and incident type 2 diabetes. A subset of these associations replicated in the Qatar Metabolomics Study on Diabetes (QMDiab) with use of a different version of the SomaLogic platform. By applying a bidirectional Mendelian randomization (MR) analysis, we identify a subset of proteins that may be causally related to type 2 diabetes and another set of proteins that may be affected by the disease itself or its genetic liability.
Research Design and Methods
Study Populations
An overview of the cohort and study workflow is shown in Supplementary Fig. 1. Cohort participants aged 66–96 years were included from the AGES-Reykjavik Study (14), a prospective study of deeply phenotyped subjects (Northern Europeans). After exclusion of individuals without a fasting glucose measurement or with established type 1 diabetes, 5,438 individuals remained for analysis in the current study (mean age 76.6 ± 5.6 years). Type 2 diabetes was defined from self-reported diabetes, diabetes medication use, or fasting plasma glucose ≥7 mmol/L (15). Of 4,784 AGES-Reykjavik participants free of diabetes at baseline, 2,940 attended a 5-year follow-up visit (AGESII). Manifest type 2 diabetes at the follow-up visit was classified as an incident case, using the same criteria as for the baseline visit. Blood samples were collected at the baseline visit after overnight fasting and serum lipids, glucose, HbA1c, and insulin measured using standard protocols. Serum creatinine was measured with the Roche Hitachi 912 instrument and estimated glomerular filtration rate (eGFR) derived with the four-variable MDRD study equation (16).
External validation analysis was performed in the QMDiab study, which is a cross-sectional case-control study for type 2 diabetes that was carried out in 2012 at the Dermatology Department in Hamad Medical Corporation (HMC) (Doha, Qatar). This study has previously been described and comprises 388 study participants from Arab and Asian ethnicities (17). A subset of 356 participants with proteomics data were used in this study (ncases = 179).
Protein Profiling
Each protein has its own detection reagent selected from chemically modified DNA libraries, referred to as SOMAmers (18). The custom version of the SOMApanel platform included proteins known or predicted to be found in the extracellular milieu (18). Serum levels of 4,137 human proteins, targeted by 4,782 SOMAmers, were determined at SomaLogic (Boulder, CO) in samples from 5,457 AGES-Reykjavik participants as previously described (13). Sample collection and processing for protein measurements were randomized and all samples run as a single set. The SOMAmers that passed quality control had median intra-assay and interassay coefficients of variation (CV) <5%, similar to that reported on variability in the SOMAscan assays (11,13). In addition to multiple types of inferential support for SOMAmer specificity toward target proteins including cross-platform validation and detection of the cis-acting genetic effects (13), direct measures of the SOMAmer specificity for 779 of the SOMAmers in complex biological samples were performed using tandem mass spectrometry (13). Hybridization controls were used to correct for systematic variability in detection, and calibrator samples of three dilution sets (40%, 1%, and 0.005%) were included so that the degree of fluorescence was a quantitative reflection of protein concentration. In the QMDiab cohort, the kit-based SOMAscan platform run by the Weill Cornell Medicine - Qatar (WCM-Q) proteomics core was used to quantify a total of 1,129 protein measurements in 356 plasma samples from QMDiab (19), with median intra- and interassay CVs <7%. Here, 1,079 SOMAmers targeting 1,050 proteins were shared with the platform used in AGES-Reykjavik. Protocols and instrumentation were provided and certified using reference samples by SomaLogic. Experiments were conducted under supervision of SomaLogic personnel. No samples or probes were excluded.
Genotyping and Imputation
Genetic data were available for 3,219 AGES-Reykjavik participants. Genotyping was performed using the Illumina 370CNV BeadChip array, and genotype calling was performed using the Illumina BeadStudio. Samples were excluded based on sample failure, genotype mismatch with reference panel, and sex mismatch on genotypes (20). Imputation (1000 Genomes Phase 1 v3 reference panel) was performed using MaCH (version 1.0.16), and the following quality control filtering was applied at the variant level: call rate (<95%), Hardy Weinberg equilibrium (P < 1 × 10−6), PLINK mishap haplotype-based test for nonrandom missing genotype data (P < 1 × 10−9), and mismatched positions between Illumina, dbSNP, and/or HapMap.
Statistical Analysis
Box-Cox transformation was applied on the protein data (21). Extreme outlier values were excluded, defined as values above the 99.5th percentile of the distribution of 99th percentile cutoffs across all proteins after scaling, resulting in the removal of an average 11 samples per SOMAmer. Associations between serum protein levels and prevalent or incident type 2 diabetes were determined using a logistic regression adjusted for age and sex (base model), where Bonferroni-corrected P < 0.05/4,782 SOMAmers = 1.1 × 10−5 was considered statistically significant. In subsequent models, the following covariates were included: fasting glucose (only for incident type 2 diabetes), BMI, fasting insulin, HDL, triglycerides (TG), eGFR, systolic blood pressure (SBP), abdominal circumference, and parental history of diabetes. These covariates include the components of the Framingham Offspring Risk Score (FORS) clinical prediction model (22) in addition to fasting insulin and eGFR. Fasting insulin and TG were log transformed due to a skewed distribution. Most of these clinical risk factors were significantly different between individuals with type 2 diabetes and those without (Supplementary Table 1). When more than one SOMAmer was available for the same protein, the one with the lowest P value in the age- and sex-adjusted model was retained in downstream analyses. Similar analysis was performed in the QMDiab cohort, except a P value threshold of P < 0.05/1,129 SOMAmers was considered statistically significant and the following study-specific covariates were included: the first three principal components (PCs) of genotyping data and the first three PCs of the proteomics data. These PCs are considered as standard covariates of the QMDiab study (19). The genetic PCs account for the ethnic variability of the QMDiab cohort, and the proteomics PCs account for a moderate level of observed cell lysis. Sex-specific protein associations were defined as follows: P < 0.05/4,782 in one sex but P > 0.05 for the other sex, and sex × protein interaction term P < 0.05 in the combined sample. Functional enrichment analysis was performed with g:Profiler (23), using the full set of proteins targeted by the SOMApanel as background and a significance threshold of Benjamini-Hochberg false discovery rate (FDR) <0.05. Tissue-specific gene expression enrichment analysis was performed using TissueEnrich (24) with data from the Human Protein Atlas (25). K-means clustering was used to group AGES-Reykjavik participants with prevalent type 2 diabetes into subgroups based on clinical variables (age of diagnosis, BMI, HbA1c, HOMA of insulin resistance and HOMA of β-cell function), as proposed by Ahlqvist et al. (26) (Supplementary Material).
For the two-sample bidirectional MR analysis, we identified genetic instruments as follows. For each protein, single nucleotide polymorphisms (SNPs) within a cis window of 100 kb up- or downstream of the respective protein-encoding gene were tested for an association with protein levels in a linear regression model adjusted for age and sex and assuming an additive genetic model. SNPs were included as genetic instruments if the association was window-wide significant (P < 0.05/n SNPs in window, as previously described [13]) and F statistic ≥10. The genetic instruments per protein were filtered to only include independent signals (r2 > 0.1, >500 kb), using the clump_data command in the TwoSampleMR R package (27). Genetic instruments for type 2 diabetes were selected from the DIAMANTE (DIAbetes Meta-ANalysis of Trans-Ethnic association studies) GWAS in European individuals (5). Of 403 independent variants, 319 with minor allele frequency >5% and passing quality filters in AGES-Reykjavik were used as instruments for type 2 diabetes. The strength of the instruments was evaluated by the association of a polygenic risk score (constructed from the 319 SNPs and weighted by the β-values available from DIAMANTE summary statistics) with type 2 diabetes in AGES-Reykjavik using a logistic regression adjusted for age and sex. We investigated cell type–specific enhancer enrichment of the genetic instruments for proteins compared with established GWAS loci through HaploReg v4.1 (28) using the SNP with the lowest association P value per protein.
The bidirectional two-sample MR analysis was performed using the TwoSampleMR R package (27). To test a causal effect of proteins on type 2 diabetes, we used the DIAMANTE GWAS (5) for type 2 diabetes without adjustment for BMI in European individuals as a primary outcome (effective sample size [Neff] = 231,436) and type 2 diabetes adjusted for BMI as a secondary outcome (Neff = 157,401). To test the reverse causal effect of type 2 diabetes on proteins, we used SNP-protein associations in AGES-Reykjavik as outcome. The inverse variance–weighted (IVW) method was used for the main MR analysis unless only one genetic instrument was available, in which case the Wald ratio was used, and a Benjamini-Hochberg FDR <0.05 was considered statistically significant. For sensitivity analyses we used MR-Egger and IVW with penalization (29), which minimizes the effect of genetic variants with heterogeneous causal estimates. Cochran Q statistic was used to evaluate heterogeneity of instruments and MR-Egger regression intercept for indication of horizontal pleiotropy.
Ethics Approval and Consent
The study was conducted in concordance with the Declaration of Helsinki of ethical principles for medical research involving human subjects. AGES-Reykjavik was approved by the National Bioethics Committee in Iceland (approval number VSN-00-063), the National Institute on Aging Intramural Institutional Review Board (U.S.), and the Data Protection Authority in Iceland. Informed consent was obtained from all study participants. QMDiab was approved by the institutional review boards of HMC and WCM-Q under research protocol number 11131/11. All study participants provided written informed consent.
Data and Resource Availability
The custom-design Novartis SOMAscan is available through a collaboration agreement with the Novartis Institutes for BioMedical Research (lori.jennings@novartis.com). Data from the AGES-Reykjavik study are available through collaboration (AGES_data_request@hjarta.is) under a data usage agreement with the Icelandic Heart Association. All data supporting the conclusions of the article are presented in the main text and Supplementary Material.
Results
The full AGES-Reykjavik cohort included 654 individuals with prevalent type 2 diabetes and 4,784 individuals free of diabetes at baseline (Supplementary Table 1). Out of 2,940 individuals without diabetes at baseline who participated in the 5-year AGESII follow-up visit, 112 developed type 2 diabetes within the period. As expected, both individuals with prevalent and individuals with incident type 2 diabetes differed markedly from individuals free of diabetes in terms of metabolic phenotypes at baseline and many already had prediabetes at the baseline visit (Supplementary Table 1). Characteristics for the QMDiab cohort are shown in Supplementary Table 2.
Serum Protein Profile of Prevalent Type 2 Diabetes
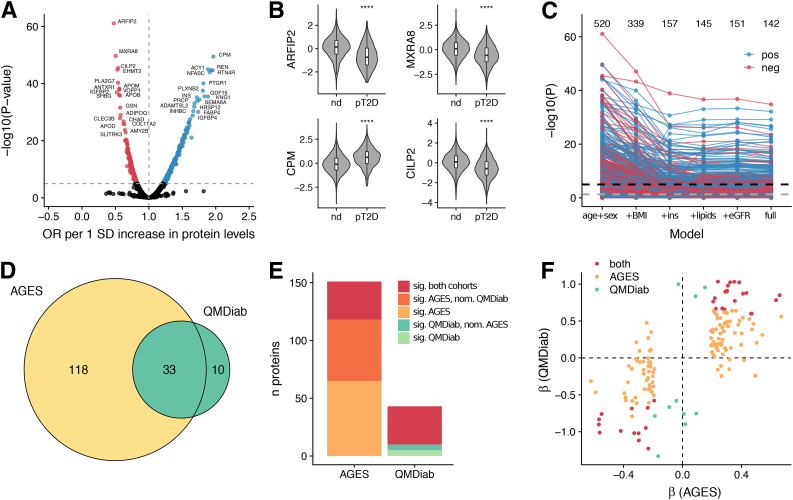
We first compared the serum protein profile of individuals with prevalent type 2 diabetes with that of individuals without. (See study workflow in Supplementary Fig. 1.) We identified 520 unique proteins that were significantly (Padjusted < 0.05) associated with prevalent type 2 diabetes (Supplementary Table 3), with odds ratios ranging from 0.47 (for ARFIP2) to 1.96 (for CPM) per SD increase of protein levels (Fig. 1A) and the most significant associations observed for ARFIP2, MXRA8, CPM, and CILP2 (Fig. 1B). In a second model including adjustment for BMI, 339 proteins were statistically significant and 157 in the model additionally adjusted for fasting insulin, indicating a large effect of these two variables on the overall protein profile of prevalent type 2 diabetes (Fig. 1C and Supplementary Table 3). In the fully adjusted model, 142 proteins remained robustly associated with prevalent type 2 diabetes (Fig. 1C), of which 30 had not reached statistical significance in the base model. Many of the 520 proteins associated with prevalent type 2 diabetes (base model) were intercorrelated, with pairwise Pearson r ranging from −0.60 to 0.97 (Supplementary Fig. 2A), and were enriched for proteins involved in extracellular matrix (ECM)-receptor interaction, complement and coagulation cascades, metabolic processes, and extracellular region (Supplementary Fig. 3A and Supplementary Table 4). The genes encoding the 520 proteins were furthermore enriched for liver-specific gene expression, followed by other tissues such as kidney, gastrointestinal tract, and pancreas (Supplementary Fig. 4A).
Figure 1.
Serum protein associations for prevalent type 2 diabetes. A: Volcano plot demonstrating positive (blue) and negative (red) serum protein associations with prevalent type 2 diabetes; points are colored where P < 0.05/4,782. B: Violin and box plots showing serum protein levels for the AGES cohort (n = 5,438) stratified by prevalent type 2 diabetes status for the top four most significant proteins. Box plots indicate median value, 25th and 75th percentile. Whiskers extend to smallest/largest value no further than 1.5× interquartile range. Outliers not shown. ****P < 0.0001. C: Spaghetti plot providing an overview of the P values (−log10 [y-axis]) for SOMAmer associations with prevalent type 2 diabetes, using six different models (x-axis). The full model includes age, sex, BMI, fasting insulin, TG, HDL, SBP, eGFR, abdominal circumference, and parental history of diabetes. The black and gray dashed lines denote the Bonferroni-corrected (P < 0.05/4,782) and nominal (P < 0.05) significance thresholds, respectively. The numbers above denote how many unique proteins are significant at a Bonferroni-corrected threshold in each model. The color of the points/lines indicates positive (blue) or negative (red) associations. D: Venn diagram illustrating the overlap between 161 proteins that were significantly (Padjusted < 0.05) associated with type 2 diabetes in either the AGES or the QMDiab cohort and measured in both. E: For each cohort, the proportion of significant proteins that were also significant (Padjusted < 0.05) in the other cohort are shown, as well as the proportion of proteins that were nominally significant (P < 0.05) and directionally consistent in the other cohort. F: Comparison of β-coefficients for prevalent type 2 diabetes between the two cohorts. The colors indicate significant (Padjusted < 0.05) associations in either or both cohorts. ins, fasting insulin; nd, nondiabetic; neg, negative; nom, nominally significant; OR, odds ratio; pos, positive; pT2D, prevalent type 2 diabetes; sig, significant.
We next sought to externally validate the observed associations in an independent population. In the QMDiab study (n = 356), measurements for 1,050 of the proteins measured in AGES-Reykjavik were available. In the base model, 43 proteins were associated with type 2 diabetes at a Bonferroni-corrected P < 0.05/1,129 in the QMDiab study (Supplementary Table 5), of which 33 were also significantly associated with prevalent type 2 diabetes in AGES-Reykjavik (Fisher exact test P = 3.5 × 10−21), amounting to 22% and 77% of the significantly associated proteins included in the comparison between AGES-Reykjavik and QMDiab, respectively (Fig. 1D). With consideration of a second validation tier as being nominally significant and directionally consistent in the other cohort, these proportions amounted to 57% and 88% for AGES-Reykjavik and QMDiab, respectively (Fig. 1E). Notably, of the 161 proteins that were significantly associated with type 2 diabetes in either cohort and measured in both, 143 (89%) were directionally consistent between the two cohorts (binomial test P = 2.4 × 10−25) (Fig. 1F).
Type 2 diabetes is a heterogeneous disease, and further subgrouping of patients has been proposed that may better represent the primary biological defects driving disease onset (26). Such subgroups are likely to differ in terms of their serum proteomic profile. We therefore used a similar approach as described by Ahlqvist et al. (26) to cluster the type 2 diabetes patients in AGES-Reykjavik into five subgroups based on their clinical features (Supplementary Material, Supplementary Fig. 5, and Supplementary Fig. 6A). A PC analysis (PCA) of the 520 proteins associated with prevalent type 2 diabetes distanced the two subgroups distinguished by high BMI or high insulin resistance (subgroups 3 and 4) from those with seemingly milder disease (groups 1 and 5) (Supplementary Fig. 6B), in line with the large effect of adjustments for these covariates on the protein associations described above.
Serum Protein Associations With Incident Type 2 Diabetes
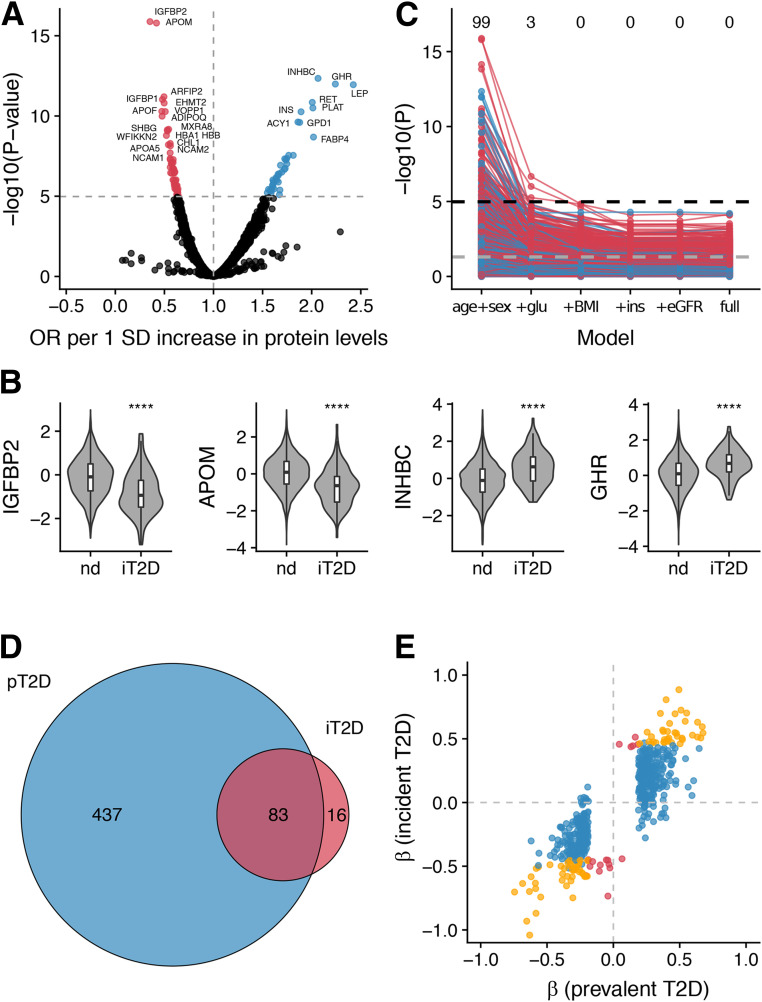
The serum protein profiles of patients with type 2 diabetes observed in a cross-sectional analysis may represent shifts that occurred either before or after the onset of the disease. To identify serum protein signatures that precede the onset of type 2 diabetes, we next focused our analysis on the 2,940 AGES-Reykjavik participants without diabetes at baseline who participated in the AGESII follow-up visit. We identified 99 unique proteins significantly (P adjusted < 0.05) associated with incident type 2 diabetes, with odds ratios ranging from 0.35 (IGFBP2) to 2.42 (LEP) per SD increase of protein levels (Fig. 2A and Supplementary Table 6) and the most significant associations observed for IGFBP2, APOM, INHBC, and GHR (Fig. 2B). The majority of protein associations with incident type 2 diabetes were attenuated after adjustment for fasting glucose (Fig. 2C and Supplementary Table 6), which is not surprising, as fasting glucose is a defining feature of diabetes. No single protein remained significant at a Bonferroni-corrected threshold after further adjustment for BMI (Fig. 2C and Supplementary Table 6). Again, we observed extensive correlations between many of the serum proteins, with pairwise Pearson r ranging from −0.55 to 0.97 (Supplementary Fig. 2B). Many of the proteins associated with incident type 2 diabetes were also associated with prevalent type 2 diabetes (84 of 99 proteins or 85%) (Fig. 2D), and the direction of effect was generally consistent (Spearman correlation coefficient = 0.82 [Fig. 1E]). The 99 proteins associated with incident type 2 diabetes were enriched for numerous gene ontology terms related to metabolism, lipid transport, and response to insulin, while enriched pathways included leptin signaling and adipogenesis (Supplementary Fig. 3B and Supplementary Table 4). Tissue expression enrichment analysis revealed enrichment for genes expressed in liver, followed by adipose tissue (Supplementary Fig. 4B). The 99 proteins associated with incident type 2 diabetes yielded a similar separation of type 2 diabetes patient subgroups as the proteins associated with prevalent type 2 diabetes (Supplementary Fig. 6B). Thus, the functional annotation of the serum proteins associated with incident type 2 diabetes was characterized by tissue-specific signatures and pathways that reflect dyslipidemia and insulin resistance. We compared our findings with previously described protein biomarker candidates for incident type 2 diabetes (11). Of 58 previously suggested candidates that were targeted in our study, we found 26 to be at least nominally associated (P < 0.05) with incident type 2 diabetes in our data and an additional 15 with prevalent type 2 diabetes (Supplementary Table 7).
Figure 2.
Serum protein associations for incident type 2 diabetes. A: Volcano plot demonstrating positive (blue) and negative (red) serum protein associations with incident type 2 diabetes; points are colored where P < 0.05/4,782. B: Violin and box plots showing serum protein levels for the AGES cohort with follow-up data (n = 2,940) stratified by incident type 2 diabetes status for the top four most significant proteins. Box plots indicate median value, 25th and 75th percentile. Whiskers extend to smallest/largest value no further than 1.5× interquartile range. Outliers not shown. ****P < 0.0001. C: Spaghetti plot providing an overview of the P values (−log10 [y-axis]) for SOMAmer associations with incident type 2 diabetes, using six different models (x-axis). The full model includes age, sex, fasting glucose, BMI, fasting insulin, TG, HDL, SBP, eGFR, abdominal circumference, and parental history of diabetes. The black and gray dashed lines denote the Bonferroni-corrected (P < 0.05/4,782) and nominal (P < 0.05) significance thresholds, respectively. The numbers above denote how many unique proteins are significant at a Bonferroni-corrected threshold in each model. The color of the points/lines indicates positive (blue) or negative (red) associations. D: Venn diagram showing the overlap between unique proteins associated with prevalent (blue) and incident (red) type 2 diabetes. E: β-Coefficients for associations between proteins and prevalent or incident type 2 diabetes. The colors denote significant associations with prevalent type 2 diabetes (blue), incident type 2 diabetes (red), or both (yellow). glu, fasting glucose; ins, fasting insulin; iT2D, incident type 2 diabetes; nd, nondiabetic; OR, odds ratio; pT2D, prevalent type 2 diabetes; T2D, type 2 diabetes.
Sex-Specific Serum Protein Associations for Type 2 Diabetes
Sex differences in cardiometabolic disorders have previously been described (30), and we therefore investigated whether any proteins exhibited sex-specific associations with incident or prevalent diabetes in the AGES-Reykjavik cohort. The β-coefficients from a sex-stratified analysis (age-adjusted model) were strongly correlated between males and females (Spearman correlation coefficient 0.85 and 0.73 for prevalent and incident type 2 diabetes, respectively). A sex-stratified analysis yielded 15 female-specific and 6 male-specific protein associations for prevalent type 2 diabetes and 4 female-specific protein associations for incident type 2 diabetes (Supplementary Fig. 7 and Supplementary Table 8). Of the 25 proteins with sex-specific associations for type 2 diabetes, 11 were not Bonferroni significant in the original combined analysis. The proteins with sex-specific associations included numerous hormones, growth factors, and related proteins, such as growth hormone 2 (GH2), follicle-stimulating hormone (CGA FSHB), the thyroid hormone carrier protein thyroxine-binding globulin (SERPINA7), a component of the progesterone-binding protein complex (PGRMC1), the epidermal growth factor betacellulin (BTC), and the hepatocyte growth factor receptor (MET). Of seven proteins measured in the QMDiab cohort, the protein * sex interaction was validated (P < 0.05) for four proteins (CGA FSHB, MET, CHRDL1, and MATN2), all of which had stronger inverse association with type 2 diabetes in females than males (Supplementary Table 8).
Potentially Causal Associations Between Serum Proteins and Type 2 Diabetes
While it is not a requirement for clinically useful biomarkers to be causally related to disease, identifying causal disease pathways can provide important insights for the development of new therapeutic strategies. We therefore performed a bidirectional two-sample MR analysis (31) to identify proteins with a potentially causal role in the development of type 2 diabetes and proteins with changes that may be downstream of the disease (Supplementary Fig. 8). Of 536 proteins significantly associated with prevalent or incident type 2 diabetes, we identified suitable genetic instruments for 246 proteins (Supplementary Table 9). Of those, 164 proteins had GWAS summary statistics available from the independent INTERVAL study (32), where 162 (99%) had a directionally consistent estimate for its lead SNP as identified in AGES-Reykjavik and 138 (84%) were nominally significant (P < 0.05) (Supplementary Table 9). On average, we identified 5 (range 1–20) genetic instruments per protein, where the lead variant per protein explained on average 10% (range 0.4–48) of the variance in their respective protein levels and with a mean F statistic of 229 (range 13–3,014). Of note, the genetic variants regulating the levels of the type 2 diabetes–associated proteins were enriched within enhancer regions mapped in liver and hepatocytes from the ENCyclopedia Of DNA Elements (ENCODE) and the Roadmap Epigenomics Project (Supplementary Fig. 4C and D), supporting the previously observed enrichment for liver expression of the genes encoding the same proteins.
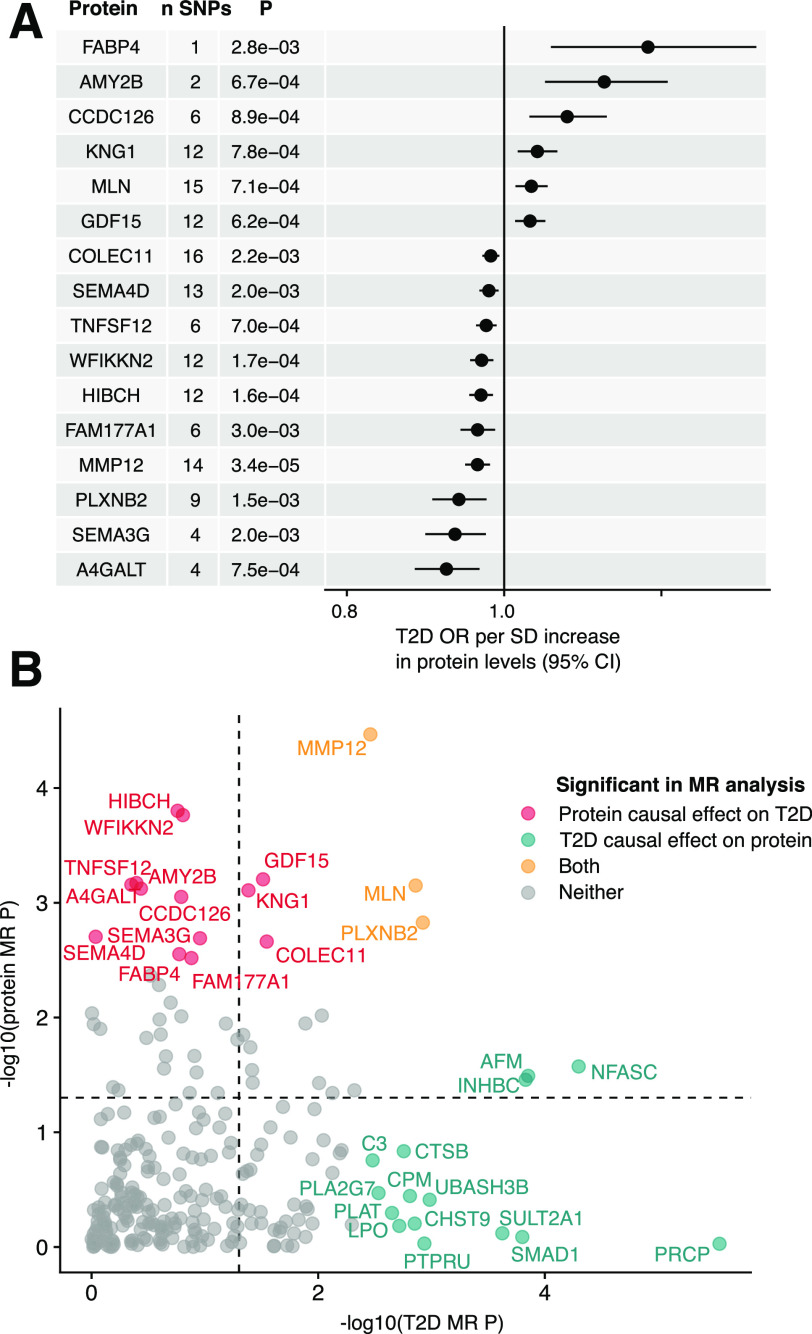
In the two-sample MR analysis, 16 proteins were supported (FDR <0.05) as potentially having a causal effect on the development of type 2 diabetes (Fig. 3A), of which 15 showed no significant signs of heterogeneity or horizontal pleiotropy (Supplementary Fig. 9 and Supplementary Table 10). Three of those (WFIKKN2, TNFSF12, and PLXNB2) remained significant (FDR <0.05) in a secondary MR analysis with a smaller sample size using type 2 diabetes adjusted for BMI as outcome, and one additional protein (CRTAC1) reached statistical significance (Supplementary Table 10). We next investigated the reverse direction, i.e., whether a genetic predisposition to type 2 diabetes has an effect on serum protein levels. A polygenic risk score of 319 type 2 diabetes SNPs selected from the DIAMANTE GWAS (5) (research design and methods) was associated with type 2 diabetes in AGES-Reykjavik (β = 0.82, SE = 0.09, P = 2.3 × 10−19, likelihood ratio test statistic = 85.6), indicating a suitable instrument. The two-sample MR analysis indicated a significant (FDR <0.05) effect of type 2 diabetes on serum levels of 40 proteins (Supplementary Table 11), of which three (MMP12, MLN, and PLXNB2) also had a significant causal estimate for type 2 diabetes (Fig. 3B). Sensitivity analyses indicated that 17 of the 40 proteins showed significant evidence of heterogeneity or horizontal pleiotropy (Supplementary Table 11), leaving 23 proteins with support for being affected by genetic predisposition to type 2 diabetes. However, it should be noted that the MR-Egger method is more sensitive to outlier variants (33), such as here: the TCF7L2 variant rs7903146 that by far has the largest effect for type 2 diabetes (β = 0.31, SE = 0.007) of all the SNPs included as type 2 diabetes instruments, which could contribute to some of the observed support for pleiotropic effects on protein levels.
Figure 3.
Bidirectional Mendelian randomization analysis supports causal associations for proteins on type 2 diabetes and vice versa. A: Forest plot for the 16 proteins supported as causal (FDR <0.05) for type 2 diabetes in the two-sample MR analysis, together with the number of SNPs used as instruments and the MR P value. MR estimates were obtained using the inverse variance–weighted method when more than one SNP was available for a given protein but otherwise with the Wald ratio. B: Comparison of bidirectional two-sample MR P values (−log10) for the 246 proteins that could be investigated in both directions, where the x-axis indicates the P value for a causal effect of type 2 diabetes on protein levels and the y-axis indicates the P value for a causal effect of protein levels on type 2 diabetes. Proteins with FDR <0.05 are colored as indicated in the legend, while the dashed lines indicate P = 0.05. OR, odds ratio.
We compared the IVW MR and observational estimates for all proteins that were indicated as significant in either direction in the MR analysis. We found that all 40 type 2 diabetes–to–protein causal estimates were directionally consistent with observational estimates for prevalent type 2 diabetes, supporting that their levels may be changed downstream of the disease or its genetic liability (Supplementary Fig. 10A). By contrast, the same was true for only 9 out of 16 (56%) protein–to–type 2 diabetes causal estimates in comparison with the observational estimates for incident type 2 diabetes (Supplementary Fig. 10B), to exclude any possible effect of prevalent type 2 diabetes on protein levels, where differing direction of effect was observed even for proteins with very strong instruments such as COLEC11 and HIBCH (lead variant F statistics = 917 and 775, respectively). However, the observational estimates for the seven proteins that were not consistent with the protein–type 2 diabetes causal estimate were instead all directionally consistent with their type 2 diabetes–to–protein causal estimates (Supplementary Fig. 10C), which were statistically significant (FDR <0.05) for two (PLXNB2 and MMP12, Supplementary Table 11). Similar results were obtained using observational estimates for prevalent type 2 diabetes (Supplementary Fig. 10D and E), with the exception of one protein (SEMA4D) that had divergent directions of effect for incident and prevalent type 2 diabetes (but only statistically significant for the latter).
Finally, we took advantage of the combined genetic and protein data in the AGES-Reykjavik study to investigate cis-acting associations in established type 2 diabetes GWAS loci (5). Of 319 established risk variants for type 2 diabetes included here, 127 were within 100 kb of a gene encoding a protein targeted by the SOMApanel. Of those, 10 were associated with one or more proteins acting in cis at a genome-wide significant threshold (P < 5 × 10−8) (Supplementary Table 12). For eight of the observed associations, the protein-coding gene was not the same as the nearest gene, thus implicating potentially novel causal candidates at those loci.
Discussion
To our knowledge, the primary data used in the current study comprise the largest protein data set described to date in terms of number of proteins measured and human samples screened. In the literature there are few descriptions of plasma protein-based biomarkers and drug targets for type 2 diabetes, and those available have been limited to relatively few protein measurements (34–38). In this study of a population-based sample of 5,438 elderly Icelanders, we describe hundreds of proteins significantly associated with prevalent and incident type 2 diabetes. Both obesity and insulin resistance contribute considerably to the serum protein changes associated with type 2 diabetes, but one-third of the protein associations for prevalent type 2 diabetes were robust to adjustment for clinical factors and reflect a major systemic shift in the serum proteome in the diabetic state. Most protein associations for incident type 2 diabetes were explained by fasting glucose at baseline and may thus be directly related to the pathophysiological pathways leading to type 2 diabetes.
Importantly, when considering proteins measured in both cohorts, we replicated 33 of 151 (22%) significant protein associations for prevalent type 2 diabetes in AGES-Reykjavik in the smaller QMDiab cohort and 33 of 43 (77%) QMDiab associations were replicated in AGES-Reykjavik. The remarkably high directional consistency between the two cohorts indicates robust patterns across populations, but the difference in proportions replicated between the two cohorts indicates that the statistical power in QMDiab is a limiting factor for finding the true overlap of associations. Given the enriched pathways among these proteins, the proteomic shift in the diabetic state to some extent reflects inflammatory processes and ECM alterations. By contrast, those pathways were not enriched among proteins associated with incident type 2 diabetes, suggesting they may be secondary to the onset of the disease. Further studies are required to understand whether and how these proteomic changes may affect downstream complications of type 2 diabetes, as diabetes-induced changes of the ECM may for example contribute to cardiovascular disease (39). In addition, several sex-specific protein associations were observed in our data that can be further explored to understand sex differences in relation to type 2 diabetes onset and outcomes.
The proteins associated with 5-year incident type 2 diabetes represent changes in the serum proteome that take place already in individuals free of diabetes. Most of these associations were attenuated after adjustment for fasting glucose at baseline, which is not surprising given that fasting glucose is the strongest predictor of incident type 2 diabetes and is essentially a part of the progression toward the disease rather than comorbidity. Therefore, these proteins may still hold important biological information relevant to the disease. The proteins associated with incident type 2 diabetes were mainly involved in lipid transport, metabolism, and insulin response, supporting the involvement of these pathways during the preclinical stage of type 2 diabetes. Both sets of proteins associated with prevalent and incident type 2 diabetes were enriched for liver-specific gene expression compared with the full set of measured proteins, consistent with the genetic variants regulating their levels being enriched in enhancers mapped in liver tissue and hepatocyte cell lines. These results underscore that the diabetic serum proteomic signatures identified here may mainly reflect processes ongoing in the liver, although other tissues also contribute as demonstrated by the enrichment of adipose tissue expression among proteins associated with incident type 2 diabetes. Currently, similar proteomic data in cohorts with information on incident type 2 diabetes are lacking, and future efforts will have to be made for replication of our findings in independent populations.
We used a bidirectional MR analysis to prioritize causal relationships between proteins and type 2 diabetes. One limitation of this analysis was that approximately one-half of the proteins did not have a suitable instrument and could thus not be tested for causality. The majority of genetic instruments for protein levels could be validated in external data (32) and, as cis-SNPs in the vicinity of the respective protein-coding genes, are likely to be directly involved in the regulation of the protein levels. We did not observe much evidence for heterogeneity or pleiotropy when considering the effects of proteins on type 2 diabetes, whereas the opposite was the true when we investigated the effect of genetic predisposition to type 2 diabetes on protein levels, likely due to the more complex instrument used for type 2 diabetes consisting of 319 variants that may affect the disease through a myriad of biological pathways. Thus, many of the type 2 diabetes–to–protein effects require further study, although notably there was complete agreement between the directionality of those causal estimates and the observational estimates for prevalent type 2 diabetes in our data. By contrast, when considering the causal effects of protein levels on type 2 diabetes, we often found causal and observational estimates to disagree, even for proteins with very strong instruments. As an example, we found serum levels of MMP12 to be increased in patients with type 2 diabetes, consistent with previous reports (40), whereas our MR estimate suggested a protective effect of MMP12 on type 2 diabetes risk. Similarly, a protective MR estimate for MMP12 and risk of coronary heart disease has been reported (32) whereas clinical and experimental studies have consistently shown higher levels of MMP12 in cardiovascular disease (40,41). In all such cases in our data, we found the observational estimates instead to be directionally consistent with the reverse causal effect of predisposition to type 2 diabetes on the proteins. As this was the case even when we considered observational estimates for incident type 2 diabetes (thus, protein changes occurring before the onset of disease), these results may suggest that the genetic liability to type 2 diabetes, and the related physiological changes that may develop before overt disease, already have an effect on these proteins and that those effects may be greater than the effects of the proteins themselves on the disease. Others have furthermore suggested that an effect of a disease polygenic risk score on gene or protein levels may represent convergent genetic effects on important disease pathways (42,43). Further work is needed to establish the complex causal chain from individual proteins to convergent pathways, intermediate phenotypes, and overt type 2 diabetes, which may then in turn affect serum protein levels.
The two-sample MR analysis revealed 15 proteins that may be causally related to type 2 diabetes and did not exhibit significance evidence of pleiotropy. Many of these associations were attenuated in a secondary MR analysis using type 2 diabetes adjusted for BMI as outcome, which may partly be because of the smaller sample size and thus reduced statistical power for this outcome but could also indicate some causal effects being mediated through BMI. Interestingly, the causal candidates included the BCAA catabolic enzyme HIBCH, for which the causal estimate suggested a protective effect on risk of type 2 diabetes. Circulating BCAA levels have consistently been shown to predict type 2 diabetes (44), although the underlying mechanisms are complex and remain to be fully understood (45). Our findings support a model where higher protein expression of the BCAA catabolic pathway reduces risk of type 2 diabetes. Another interesting causal candidate is WFIKKN2, also supported by a recent MR study using the same outcome data as here but different instruments (43). WFIKKN2 is a follistatin domain–containing protein that binds GDF8/GDF11 proteins with high affinity (46)—both of which have been implicated in diabetes (47,48). Genetic variants in the WFIKKN2 region regulate serum GDF8/11 levels in trans via WFIKKN2 protein levels (13,32), although in the current study we did not find a significant association between GDF8/11 and type 2 diabetes, so additional studies are required to understand the mechanisms by which WFIKKN2 may affect risk of type 2 diabetes. Other notable causal candidates from the MR analysis included FABP4, a member of the PPAR signaling pathway and a suggested inhibitor target for novel therapeutic strategies for obesity and type 2 diabetes (49), and GDF15, consistently implicated in cardiometabolic diseases (50), but previous MR studies have failed to observe support for a causal effect on type 2 diabetes (51,52). Here, using a different set of genetic instruments than in the previous MR studies, we find suggestive evidence for a causal effect of higher circulating GDF15 levels on type 2 diabetes risk.
To conclude, our results demonstrate a major shift in the serum proteome before and during the diabetes stage. Furthermore, proteins supported as potentially causal for type 2 diabetes in our data could be of particular interest as novel therapeutic targets, although in some cases their effect may be masked by the downstream effects of type 2 diabetes or its genetic liability on the serum proteome.
Article Information
Acknowledgments. The authors thank the staff of the Icelandic Heart Association for their contribution to AGES-Reykjavik and the staff of the HMC dermatology department and of WCM-Q for their contribution to QMDiab. Finally, the authors are grateful to all study participants of AGES-Reykjavik and QMDiab for their invaluable contributions to this study.
Funding. The study was funded by Icelandic Heart Association contract HHSN271201200022C, National Institute on Aging contract N01-AG-12100, and Althingi (the Icelandic Parliament). Va.G. is supported by the Icelandic Centre for Research (grant no. 184845-051). Work on the QMDiab cohort was supported by the Biomedical Research Program at WCM-Q, a program funded by the Qatar Foundation. K.S. is also supported by Qatar National Research Fund grant NPRP11C-0115-180010.
Duality of Interest. The study was supported by the Novartis Institute for Biomedical Research, and protein measurements for the AGES-Reykjavik cohort were performed at SomaLogic. J.R.L. and L.L.J. are employees and stockholders of Novartis. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. Va.G., V.E., and Vi.G. designed the study. Va.G., M.I., T.A., E.F.G., S.M.J., and N.R.Z. performed data analysis within AGES. K.S. and S.B.Z. contributed QMDiab data and performed validation analysis. J.R.L. and L.L.J. provided expertise on proteomics data and contributed to discussion. Va.G. and V.E. wrote the first draft of the manuscript, with all coauthors contributing to revisions. Vi.G. and V.E. supervised the project. Vi.G. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 55th Annual Meeting of the European Association for the Study of Diabetes, 16–20 September 2019, Barcelona, Spain.
Footnotes
V.G., S.B.Z., and V.E. contributed equally as joint first authors.
K.S., L.L.J., and V.G. contributed equally as joint senior authors.
This article contains supplementary material online at https://doi.org/10.2337/figshare.12249884.
References
- 1.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan A, Go MJ, Zhang W, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium . Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott RA, Scott LJ, Mägi R, et al. ; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017;66:2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol 2014;2:65–75 [DOI] [PubMed] [Google Scholar]
- 7.Merino J, Leong A, Liu CT, et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia 2018;61:1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M, Shao J, Wu C-Y, et al. Targeting BCAA catabolism to treat obesity-associated insulin resistance. Diabetes 2019;68:1730–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbasi A, Sahlqvist AS, Lotta L, et al. A systematic review of biomarkers and risk of incident type 2 diabetes: an overview of epidemiological, prediction and aetiological research literature. PLoS One 2016;11:e0163721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies DR, Gelinas AD, Zhang C, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A 2012;109:19971–19976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hathout Y, Brody E, Clemens PR, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2015;112:7153–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, Gene/Environment Susceptibility-Reykjavik study: multidisciplinary applied phenomics. Am J Epidemiol 2007;165:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care 2014;37(Suppl. 1):S81–S90 [DOI] [PubMed] [Google Scholar]
- 16.Levey AS, Greene T, Kusek J, Beck G. A simplified equation to predict glomerular filtration rate from serum creatinine. J Am Soc Nephrol 2000;11:A0828 [Google Scholar]
- 17.Mook-Kanamori DO, Selim MME-D, Takiddin AH, et al. 1,5-anhydroglucitol in saliva is a noninvasive marker of short-term glycemic control. J Clin Endocrinol Metab 2014;99:E479–E483 [DOI] [PubMed] [Google Scholar]
- 18.Candia J, Cheung F, Kotliarov Y, et al. Assessment of variability in the SOMAscan assay. Sci Rep 2017;7:14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psaty BM, O’Donnell CJ, Gudnason V, et al.; CHARGE Consortium . Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M, Johnson K. Applied Predictive Modeling. New York, Springer-Verlag New York, 2013, Chapter 3, p. 1–600 [Google Scholar]
- 22.Wilson PWF, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 23.Reimand J, Arak T, Adler P, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 2016;44(W1):W83–W89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain A, Tuteja G. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics 2019;35:1966–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 26.Ahlqvist E, Storm P, Käräjämäki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 2018;6:361–369 [DOI] [PubMed] [Google Scholar]
- 27.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016;44:D877–D881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rees JMB, Wood AM, Dudbridge F, Burgess S. Robust methods in Mendelian randomization via penalization of heterogeneous causal estimates. PLoS One 2019;14:e0222362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerdts E, Regitz-Zagrosek V. Sex differences in cardiometabolic disorders. Nat Med 2019;25:1657–1666 [DOI] [PubMed] [Google Scholar]
- 31.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature 2018;558:73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowak C, Sundström J, Gustafsson S, et al. Protein biomarkers for insulin resistance and type 2 diabetes risk in two large community cohorts. Diabetes 2016;65:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raynor LA, Pankow JS, Duncan BB, et al. Novel risk factors and the prediction of type 2 diabetes in the Atherosclerosis Risk in Communities (ARIC) study. Diabetes Care 2013;36:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herder C, Baumert J, Zierer A, et al. Immunological and cardiometabolic risk factors in the prediction of type 2 diabetes and coronary events: MONICA/KORA Augsburg case-cohort study. PLoS One 2011;6:e19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao C, Song Y, Cook N, et al. The lack of utility of circulating biomarkers of inflammation and endothelial dysfunction for type 2 diabetes risk prediction among postmenopausal women: the Women’s Health Initiative Observational Study. Arch Intern Med 2010;170:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salomaa V, Havulinna A, Saarela O, et al. Thirty-one novel biomarkers as predictors for clinically incident diabetes. PLoS One 2010;5:e10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law B, Fowlkes V, Goldsmith JG, Carver W, Goldsmith EC. Diabetes-induced alterations in the extracellular matrix and their impact on myocardial function. Microsc Microanal 2012;18:22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goncalves I, Bengtsson E, Colhoun HM, et al.; SUMMIT Consortium . Elevated plasma levels of MMP-12 are associated with atherosclerotic burden and symptomatic cardiovascular disease in subjects with type 2 diabetes. Arterioscler Thromb Vasc Biol 2015;35:1723–1731 [DOI] [PubMed] [Google Scholar]
- 41.Mahdessian H, Perisic Matic L, Lengquist M, et al.; IMPROVE study group . Integrative studies implicate matrix metalloproteinase-12 as a culprit gene for large-artery atherosclerotic stroke. J Intern Med 2017;282:429–444 [DOI] [PubMed] [Google Scholar]
- 42.Võsa U, Claringbould A, Westra H-J, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. 19 October 2018 [preprint]. bioRxiv:447367.
- 43.Ritchie SC, Liu Y, Lambert SA, et al. Integrative analysis of the plasma proteome and polygenic risk of cardiometabolic diseases. 19 December 2019 [preprint].bioRxiv:2019.12.14.876474. [DOI] [PMC free article] [PubMed]
- 44.Guasch-Ferré M, Hruby A, Toledo E, et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care 2016;39:833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White PJ, Newgard CB. Branched-chain amino acids in disease. Science 2019;363:582–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondás K, Szláma G, Trexler M, Patthy L. Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J Biol Chem 2008;283:23677–23684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Li Y, Xiang L, et al. GDF11 attenuates development of type 2 diabetes via improvement of islet β-cell function and survival. Diabetes 2017;66;1914–1927 [DOI] [PubMed] [Google Scholar]
- 48.Guo T, Bond ND, Jou W, Gavrilova O, Portas J, McPherron AC. Myostatin inhibition prevents diabetes and hyperphagia in a mouse model of lipodystrophy. Diabetes 2012;61:2414–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 2008;7:489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adela R, Banerjee SK. GDF-15 as a target and biomarker for diabetes and cardiovascular diseases: a translational prospective. J Diabetes Res 2015;2015:490842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Au Yeung SL, Luo S, Schooling CM. The impact of GDF-15, a biomarker for metformin, on the risk of coronary artery disease, breast and colorectal cancer, and type 2 diabetes and metabolic traits: a Mendelian randomisation study. Diabetologia 2019;62:1638–1646 [DOI] [PubMed] [Google Scholar]
- 52.Cheung C-L, Tan KCB, Au PCM, Li GHY, Cheung BMY. Evaluation of GDF15 as a therapeutic target of cardiometabolic diseases in human: a Mendelian randomization study. EBioMedicine 2019;41:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]